Significance
The regeneration of entire plants from explants is an important step in plant production and plant transformation protocols. Despite recent advances in the knowledge on the molecular basis of regeneration, many aspects of the process and the causes of regeneration recalcitrance are still poorly understood. We combined linkage with association mapping to find genes underlying the natural variation of shoot regeneration in Arabidopsis. With this approach, we identified and confirmed the involvement of RECEPTOR-LIKE PROTEIN KINASE1 as a previously unknown determinant of shoot regeneration. Because this gene is implicated in abscisic acid signaling, it seems that this hormone might be an important player in this developmental process.
Keywords: regeneration recalcitrance, SNP, ABA, natural variation, QTG
Abstract
De novo shoot organogenesis (i.e., the regeneration of shoots on nonmeristematic tissue) is widely applied in plant biotechnology. However, the capacity to regenerate shoots varies highly among plant species and cultivars, and the factors underlying it are still poorly understood. Here, we evaluated the shoot regeneration capacity of 88 Arabidopsis thaliana accessions and found that the process is blocked at different stages in different accessions. We show that the variation in regeneration capacity between the Arabidopsis accessions Nok-3 and Ga-0 is determined by five quantitative trait loci (QTL): REG-1 to REG-5. Fine mapping by local association analysis identified RECEPTOR-LIKE PROTEIN KINASE1 (RPK1), an abscisic acid-related receptor, as the most likely gene underlying REG-1, which was confirmed by quantitative failure of an RPK1 mutation to complement the high and low REG-1 QTL alleles. The importance of RPK1 in regeneration was further corroborated by mutant and expression analysis. Altogether, our results show that association mapping combined with linkage mapping is a powerful method to discover important genes implicated in a biological process as complex as shoot regeneration.
The capacity to regenerate in vitro adventitious shoots is of major importance for biotechnological breeding and commercial in vitro initiation and propagation of plants. Unfortunately, shoot regeneration is not always easy to achieve: among plant species, varieties, and cultivars, it is highly variable and currently unpredictable. The impact of shoot regeneration for horticulture and agriculture is illustrated by the numerous studies that assess the natural allelic variation and map quantitative trait loci (QTL) for the regeneration capacity in diverse crops, such as tomato (Solanum lycopersicum), wheat (Triticum aestivum), rice (Oryza sativa), barley (Hordeum vulgare), sunflower (Helianthus annuus), cabbage (Brassica oleracea), and potato (Solanum tuberosum) (1–12). However, it is difficult to draw general conclusions from these studies because of the low-resolution linkage maps and little detailed knowledge about gene functions in these crops.
Therefore, the use of the model plant Arabidopsis thaliana is more appropriate. In a widely applied two-step regeneration procedure, root explants are first incubated on an auxin-rich callus-inducing medium (CIM) and subsequently transferred to a cytokinin-rich shoot-inducing medium (SIM) (13). Genome-wide analyses of the gene expression profiles accompanying the successive steps in the regeneration process revealed multiple key regulators and genes implicated in phytohormonal signaling during shoot regeneration (14–18). Reporter gene fusions with marker genes allowed visualization of their spatiotemporal expression patterns during regeneration, contributing to the elucidation of the function of important shoot-related genes, such as CUP SHAPED COTYLEDON1, CUP SHAPED COTYLEDON2, SHOOT MERISTEMLESS, WUSCHEL (WUS), and CLAVATA3 (14, 19–22). By means of classical forward and reverse genetics approaches, additional genes involved in shoot regeneration have been identified (23).
Shoot regeneration in Arabidopsis has also been studied by QTL mapping with recombinant inbred lines (RILs) of Ler × Col (24, 25) or Ler × Cvi (26). These studies revealed multiple QTL, but thus far, no quantitative trait gene (QTG) or quantitative trait nucleotide (QTN) responsible for any of these QTL has been reported. Indeed, linkage mapping studies often fail to identify the causal gene because of their limiting mapping resolution (27).
Recently, genome-wide association studies with an increased mapping resolution have received much attention for the identification of QTL in plants, particularly in Arabidopsis, as an alternative to or combined with linkage mapping approaches (28–31). Here, we aimed at identifying QTGs underlying the natural shoot regeneration variation in Arabidopsis by using linkage mapping complemented with association mapping. Furthermore, early parameters, such as callus and root formation, explant greenness, and shoot primordia development, were examined in a set of 88 Arabidopsis accessions. We calculated pairwise correlations between the different parameters and shoot formation to assess whether the early observations could predict regeneration. We phenotyped 86 RILs derived from a cross of Nok-3 and Ga-0, accessions with high- and low-regeneration abilities, respectively, and mapped five regeneration QTL. A local association mapping revealed that RECEPTOR-LIKE PROTEIN KINASE1 (RPK1) is the most likely gene underlying the major QTL REG-1, which was supported by mutant analysis and a quantitative complementation test.
Results
Shoot Regeneration Has a Low Correlation with Related Traits.
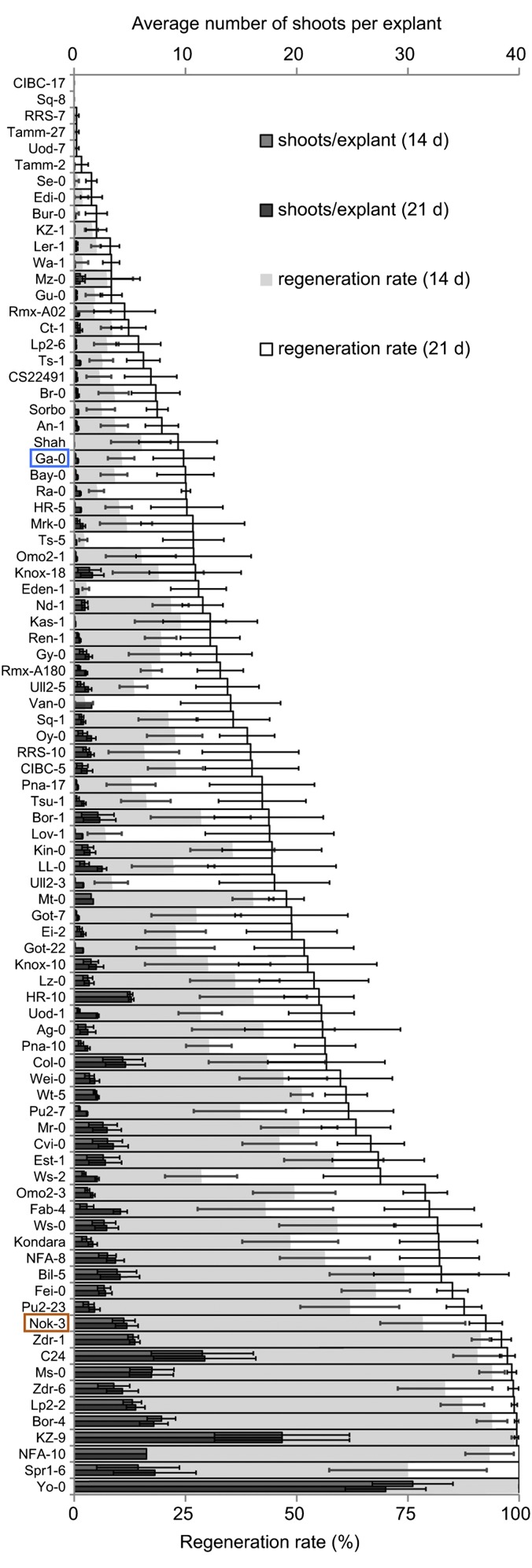
A set of 88 Arabidopsis accessions was evaluated for variations in the number of shoots, shoot primordia, and roots as well as the extent of callus formation and greenness of root explants after different periods of SIM incubation (Figs. 1 and 2 A–D, SI Materials and Methods, and Table S1). The accessions varied considerably in the regeneration rate that was calculated as the proportion of regeneration-responsive explants, ranging from 0% (accessions CIBC-17 and Sq-8) to 100% (accessions NFA-10, Spr1-6, and Yo-0) (Fig. 1), and the other evaluated parameters (Fig. 2 C and D and Table S1).
Fig. 1.
Shoot regeneration capacity of 88 Arabidopsis accessions. Means ± SEMs for the number of shoots per root explant (top axis) and the regeneration rate [i.e., the average number of explants producing at least one shoot (bottom axis)] after 14 and 21 d of SIM incubation. Each experiment was replicated three (number of shoots) or six (regeneration rate) times, with 30 explants per accession. The more recalcitrant Ga-0 (blue) and the regenerative Nok-3 (orange) accession were selected for QTL analysis.
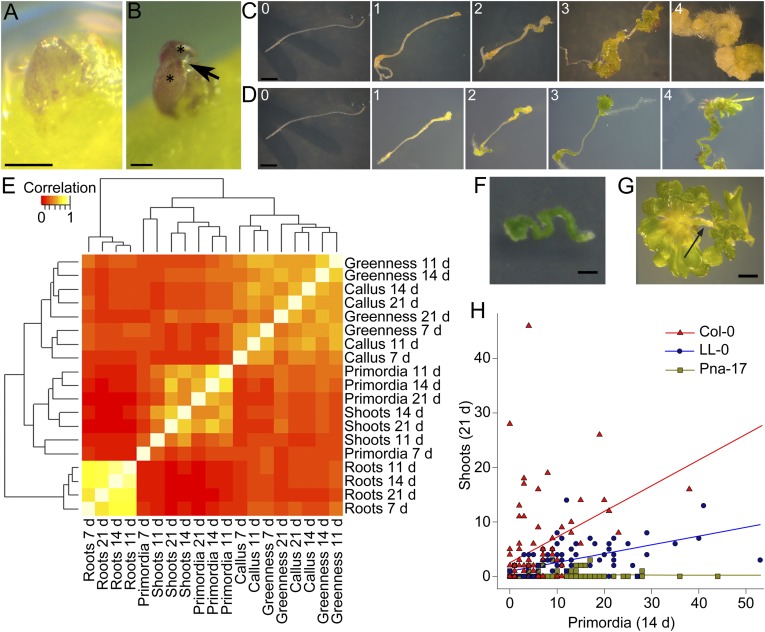
Fig. 2.
Shoot, primordium, root, and callus formation and greening during the regeneration process and correlation between these different responses in 88 Arabidopsis accessions. (A) Representative primordium: green or purple dome-shaped or spherical structure with a smooth surface and a cellular organization. (Scale bar: 100 µm.) (B) Representative shoot: structure originating from a primordium with two or more leaves (asterisks) emerging from one meristem; arrow indicates trichomes. (Scale bar: 100 µm.) (C) Classification of callus formation; class 0, no callus; class 1, callus present at explant ends; class 2, callus present and covering maximum one-third of the explant; class 3, abundant callus but not covering the whole explant; class 4, excessive callus covering the whole explant. (Scale bar: 1 mm.) (D) Evaluation of different classes of explant greenness excluding shoots: class 0, white explant; class 1, yellow explant; class 2, (partially) pale green explant; class 3, completely green explant; class 4, completely dark green explant. (Scale bar: 1 mm.) (E) Correlation of different responses accompanying shoot regeneration (number of shoots, number of primordia, number of lateral roots, callus, and greenness) at different time points (7, 11, 14, and 21 d) of SIM incubation. The heat map represents the Spearman’s rank correlation matrix for the various responses; the dendrograms are distance trees. The experiment was done in triplicate with 30 explants per accession per repeat. Because almost no shoots were observed after 7 d of SIM incubation, this information was not included. (F) The more recalcitrant accession Eden-1 forming dark green callus without shoots. The picture was taken after 21 d of SIM incubation. (Scale bar: 1 mm.) (G) The regeneration-competent Wei-0 developing little callus, with a pale remaining explant and the formation of a lot of shoots. The picture was taken after 14 d of SIM incubation. The arrow marks the root explant. (Scale bar: 1 mm.) (H) Scatter plot of the number of primordia on an explant after 14 d plotted against the number of shoots on the same explant after 21 d of SIM incubation measured in 90 explants of the accessions Col-0, LL-0, and Pna-17. A linear regression was fitted to each accession: the more recalcitrant accession Pna-17 forms a lot of primordia but few shoots compared with Col-0, which has a high primordia-to-shoot development rate, and LL-0, which has an intermediate relation.
A correlation analysis between the different parameters and subsequent clustering revealed three distinct groups: a root, a primordia/shoot, and a callus/greenness cluster (Fig. 2E and Table S2). Unexpectedly, no high correlations were found either between callus and shoots (r = 0.049–0.422) or between greenness and shoots (r = 0.05–0.432) (Fig. 2E and Table S2). Apparently, considering the accessions analyzed, the capacity to form callus on SIM or develop chloroplasts does not correlate with efficient regeneration. Indeed, for example, accession Eden-0, which is regeneration recalcitrant, forms dark green callus but few shoots (Fig. 2F and Table S1), whereas accession Wei-0, which regenerates well, forms little pale green callus but a lot of shoots (Fig. 2G and Table S1). Within the callus/greenness cluster, only moderate correlations were found (r = 0.192–0.633) (Fig. 2E), indicating that callus formation and chloroplast development do not necessarily occur simultaneously (Fig. 2 C and D). The highest correlation coefficients were found in the root cluster (r = 0.679–0.863) (Fig. 2E): all accessions developed roots mainly during the first 7 d of SIM incubation, and their number did not change at later time points (Fig. S1 and Table S1). In the primordia/shoot cluster, the correlation coefficients for each developmental phase over time were lower than the coefficients in the root cluster (Fig. 2E), illustrating that the timing of shoot formation differed between accessions (Fig. 1). Indeed, when the number of shoots produced per explant after 14 and 21 d of SIM incubation was compared in the different accessions, very clear differences were observed, reflecting their relative degree of regeneration capacity (Fig. 1). Intriguingly, the correlations between primordia and shoots were quite low as well (r = 0.089–0.737) (Fig. 2E), suggesting that the potential to form primordia can be uncoupled from the subsequent development into shoots under our experimental conditions. Additional evidence was provided by scatter plots for three accessions, in which the number of primordia present on a specific explant after 14 d of SIM incubation was plotted against the number of shoots present on the same explant 7 d later. Although accession Pna-17 produced many primordia, it had the lowest number of shoots per explant, whereas accession LL-0 produced many primordia as well but many developed into shoots. Accession Columbia-0 (Col-0) had fewer primordia but a higher number of shoots, showing that the development of primordia into shoots occurred rapidly and efficiently (Figs. 1 and 2H and Table S1).
Linkage Mapping of the Regeneration Rate Reveals Five QTL.
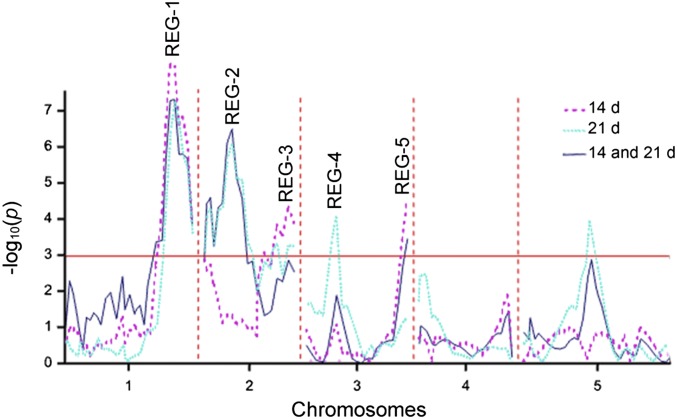
To identify QTL underlying the regeneration rate variation, we accessed the Nok-3 × Ga-0 RIL population (32). The choice for this population was motivated by the large difference in regeneration capacity measured between Ga-0 (recalcitrant) and Nok-3 (regenerative) (Fig. 1). Linkage mapping revealed QTL REG-1, REG-3, and REG-5 at 14 d and REG-1, REG-2, and REG-4 at 21 d of SIM incubation (Fig. 3 and Table S3). Multitrait linkage analysis across both time points resulted in QTL REG-1, REG-2, and REG-5 (Fig. 3 and Table S3). The apparent QTL on chromosome 5 was not significant after the final backward selection (SI Materials and Methods).
Fig. 3.
QTL significance map for the regeneration rate in an Nok-3 × Ga-0 RIL population. Composite interval mapping was used to identify QTL for the single trait (dotted lines) and multitrait (solid line) analyses of the regeneration rate after 14, 21, or 14 and 21 d of SIM incubation, respectively. The red line marks the genome-wide significance threshold (α = 0.05).
Complementing Linkage Mapping with a Local Association Mapping Reveals RPK1 as a Candidate QTG for REG-1.
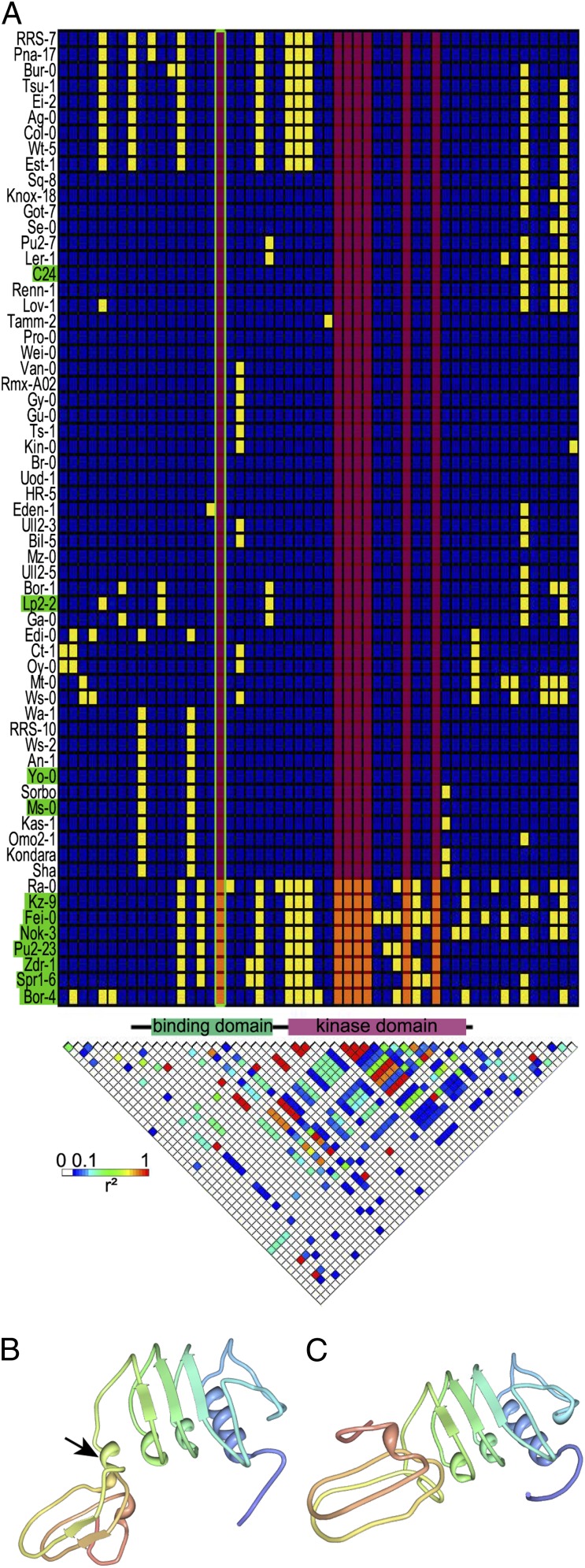
In both QTL analyses, REG-1 was the major QTL and accounted for ∼30% of the regeneration rate variation in the Nok-3 × Ga-0 population (Table S3). A local association study was conducted to identify a putative QTG for REG-1. For this purpose, we filtered the 250,000 SNP set (33) for polymorphisms between Nok-3 and Ga-0 in a 1-Mb region flanking the f5a1859436 marker that was strongly linked to the REG-1 QTL (26.65 Mb; http://www.jic.ac.uk/staff/ian-bancroft/arabidopsis.htm). Statistical analysis of the associations between the selected SNPs and the regeneration rate in the set of 88 Arabidopsis accessions (SI Materials and Methods) revealed a strong association with the SNP at 26,041,261 bp (P = 5.81E-4), located in RPK1. An additional local association study in 62 completely sequenced Arabidopsis accessions revealed seven polymorphisms in RPK1 that were strongly associated with the regeneration rate (P = 8.83E-5) (Fig. 4A). The Nok-3 allele of RPK1 conferred an increased regeneration capacity and was mainly present in accessions with a regeneration rate of more than 85% after 21 d of SIM incubation (Figs. 1 and 4A). RPK1 encodes a leucine-rich repeat receptor-like kinase consisting of an extracellular ligand-binding domain and a cytosolic kinase domain (34). A single SNP in the binding domain resulted in an amino acid change (V > L on position 162). Prediction of the structure of this extracellular domain of both alleles using PHYRE2 (35) revealed a polymorphism-dependent modification (Fig. 4 B and C) that might affect the binding efficiency of the receptor to its ligand and hence, the RPK1 activity.
Fig. 4.
Polymorphisms in RPK1. (A) Haplotype structure map and linkage disequilibrium (LD) plot for RPK1 and the surrounding region in 62 fully sequenced accessions. In the haplotype structure map, each column represents a polymorphic site with minor and major alleles (yellow and blue, respectively) and clustering of accessions by haplotype. Highly regenerative accessions (regeneration rate at 21 d > 85%) are indicated in green. Regeneration-associated polymorphisms (P = 8.83E-05) are marked by a red overlay; the only associated polymorphism resulting in an amino acid change is framed in green. The LD plot reflects r2 for each pair of polymorphisms, with the strongest LD in red. The protein model between the plots gives the coding sequence (black line) and specific structural domains (34). Positions are not in proportion, because only polymorphic positions are included in the matrix. (B and C) 3D model of the extracellular binding domain of the (B) negative and (C) positive RPK1 variants predicted by PHYRE2 (35). Of the residues, (B) 95% and (C) 94% were modeled at a >90% confidence. The arrow marks an additional helix, resulting in a different conformation.
RPK1 Is Required for Shoot Regeneration.
The relevance of RPK1 during shoot regeneration was established by subjecting root explants of the rpk1-1 and rpk1-5 Col-0 mutants (36) (SI Materials and Methods) to the two-step regeneration protocol. Indeed, rpk1-5 was almost completely recalcitrant (Fig. 5A), and rpk1-1 had a significantly reduced regeneration rate (Fig. 5A), showing that RPK1 is essential in the regeneration process. Because the potential dominance of poor regeneration alleles in Ga-0 might interfere with a transgenic complementation approach, we opted for quantitative complementation to validate RPK1 as the most likely gene underlying REG-1. This method is well-established in natural variation studies in animal systems (37–39) and has successfully been applied in plant research as well (40, 41). We crossed the parental accessions Nok-3 and Ga-0 to the WT Col-0 and the rpk1-5 mutant and analyzed the regeneration rate of the four resulting F1 genotypes. When crossed to the mutant, the natural rpk1 alleles at the mutant locus are functionally hemizygous, but when crossed to the WT, any difference between the natural alleles is reduced by the presence of the WT allele. When the natural alleles differentially affect the regeneration phenotype, a significant interaction term in a two-way ANOVA is obtained, and then, the mutation is said to fail to complement quantitatively the natural alleles (37). As shown in Fig. 5B, the difference in regeneration rate between the Nok-3 and Ga-0 QTL alleles was larger in the rpk1-5 mutant than in the WT Col-0 allele background and characterized by a significant interaction term (P < 0.001). This result supports an allelic interaction between the mutant allele at the RPK1 locus and the homologous QTL alleles and confirms that RPK1 is the most probable REG-1–underlying QTG. Finally, the RPK1 expression was monitored during the regeneration process with a pRPK1::RPK1-GFP line (36). Fluorescence was particularly visible in dividing presumptive pericycle cells during CIM incubation (Fig. 5C and Fig. S2), which precedes shoot initiation (16), and presumptive shoot primordia and shoots during subsequent SIM incubation (Fig. 5 D and E and Figs. S3 and S4), further implying a role for RPK1 in the de novo shoot formation process.
Fig. 5.
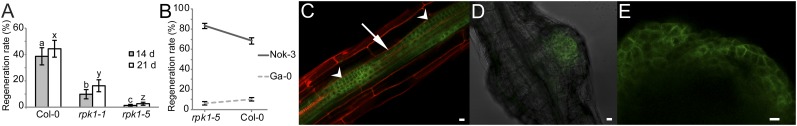
Importance and expression pattern of RPK1 during shoot regeneration. (A) Mean values ± SEMs for the regeneration rate of Col-0 (n = 3) and mutants rpk1-1 (n = 2) and rpk1-5 (n = 3) measured after 14 (gray) and 21 d (white) of SIM incubation. Different letters indicate significant differences at the α-level = 0.05. (B) Quantitative complementation test: mean values ± effective SEs (n = 12) for the regeneration rate after 14 d of four genotypes: Nok-3/rpk1-5, Ga-0/rpk1-5, Nok-3/Col-0, and Ga-0/Col-0. The difference between the Nok-3 and Ga-0 regeneration rate QTL allele effect is significantly larger (P < 0.001) in the rpk1-5 mutation background than in the WT Col-0 allele background, indicative of an interaction between the rpk1-5 allele and the Nok-3 and Ga-0 QTL alleles. (C–E) Expression of pRPK1::RPK1-GFP in Col-0 background during the shoot regeneration protocol. After 4 d of CIM incubation, expression is particularly visible in dividing (arrowheads) but not in nondividing (arrow) cells in the vasculature. Cells are counterstained with propidium iodide (red). (C) Expression in presumptive shoot primordia after (D) 7 and (E) 11 d of SIM incubation. (Scale bars: 10 µm.)
Discussion
Screening of Arabidopsis accessions for explant greening, callus development, and formation of roots, primordia, and shoots during the regeneration procedure brought to light a wide natural variation in these parameters. The recording of these phenotypes and the ranking of the accessions according to their regeneration rate provide valuable information for future studies. For instance, accessions NFA-10, Spr1-6, and Yo-0 have a 100% regeneration rate and are useful alternatives for the commonly used accessions, such as Col-0 and Ler-1, that exhibit only an intermediate- or low-regeneration rate to discover novel gene functions implicated in regeneration. Similarly, accessions CIBC-17 and Sq-8 are completely recalcitrant under our experimental conditions and can be used in mutagenesis screens for (partial) reversion of their regeneration defect. Although pairwise correlations between the parameters were generally significant and revealed three distinct correlation clusters, no high correlations were identified between traits, such as callus and shoot formation, explant greening and shoot formation, and primordium and shoot formation. Indeed, neither callus formation nor greening was predictive for subsequent shoot regeneration; hence, these responses should merely be seen as effects induced by 2,4-dichlorophenoxyacetic acid and cytokinin (42, 43). Thus, whereas for particular individual accessions, green protuberance and callus formation on SIM are related (14) and even necessary for the regeneration process (16), for other accessions, they are clearly neither related nor required. The weak correlation between primordium and shoot formation was also intriguing and implies that regeneration can be blocked very late in the developmental process. Accessions, such as Pna-17, that form primordia but almost no shoots should be the starting material of choice to study this type of recalcitrance. Thus, the natural variation in regeneration capacity across the different Arabidopsis accessions is a powerful resource to unravel the diverse developmental processes that are implicated in the formation of adventitious shoots and to examine the genetic basis of regeneration recalcitrance.
The QTL study showed the complexity and broad polygenic basis of shoot regeneration. As such, none of the QTL obtained from assessing the regeneration rate of an Nok-3 × Ga-0 RIL population corresponded to previously identified QTL in Ler × Col (24, 25) or Ler × Cvi (26) populations. Nevertheless, by combining linkage mapping with a subsequent local association mapping, we succeeded in identifying a candidate QTN in RPK1 for REG-1, which was confirmed by quantitative failure of the rpk1-5 mutation to complement the parental high- and low-regeneration QTL alleles.
RPK1 encodes a leucine-rich repeat receptor-like kinase, and this type of receptor is critical in the signal transduction pathways triggered by developmental and environmental signals (34). RPK1 has been reported to function upstream of abscisic acid (ABA) signaling (44) and be involved in diverse processes, such as stress tolerance, senescence, embryonic patterning, and formation of cotyledon primordia (34, 36, 45–48). Assessment of the regeneration rate of two rpk1 mutants in the Col-0 background clearly showed the importance of this gene in regeneration, assigning an additional function to RPK1. Whereas rpk1-5 is a nonsense mutation predicted to result in a premature stop codon, rpk1-1 is supposed to be a null mutation (36) (SI Materials and Methods). Nevertheless, in the regeneration assay, the rpk1-5 allele had the strongest phenotype, suggesting that the rpk1-1 allele is not completely null. Previous observations of RPK1 expression in the shoot apical meristem (44) and the shoot primordia (this work) further support the significance of RPK1 in shoot regeneration. All of the reported characteristics of the rpk1 mutants are related to ABA: delay in age-dependent leaf senescence, increased water loss rate of detached leaves, increased salt stress resistance, and decreased survival rate after drought stress (45, 47, 49). Other phenotypes, such as differences in germination rate, shoot and root growth rates, or stomatal aperture, occur only after ABA treatment (44, 47). Interestingly, in whole plants, RPK1 expression is induced upon treatment with ABA but not 2,4-dichlorophenoxyacetic acid and cytokinins (34, 44), the hormones present in CIM and SIM, respectively. Although ABA levels have been shown to affect shoot regeneration in rape (Brassica rapa), rice, and canola (Brassica napus) (50–52), thus far, ABA signaling has not been identified as a determining factor in the molecular mechanism underlying shoot regeneration.
The developmental role in patterning and cotyledon formation of RPK1 seems to be functionally redundant with that of RPK2/TOADSTOOL2 (36, 46). RPK2, but not RPK1, turned out to be a key regulator of shoot apical meristem maintenance and a regulator of WUS expression (53). RPK1 and RPK2 share extensive sequence similarity in their kinase but not their ligand-binding domain (54), suggesting that they respond to different extracellular signals. Binding of the ligand seems to be imperative for the RPK1 function in shoot regeneration, because the identified QTN is predicted to affect the ligand-binding domain. Importantly, single rpk1 mutants exhibit a recalcitrant shoot regeneration phenotype, and hence, in contrast to its role in embryogenesis, RPK1 plays an autonomous role in shoot organogenesis. Finally, several aspects described for RPK1 during embryo formation (36, 46, 48) support its involvement in regeneration: (i) RPK1 expression is important for PINFORMED1 localization, which is required for the establishment of auxin maxima, (ii) an RPK1-dependent pathway is thought to initiate WUS-RELATED HOMEOBOX5 expression, and (iii) RPK1 is assumed to receive intercellular signals and mediate intracellular responses required for pattern formation. Indeed, correct PINFORMED1 localization and auxin maxima are necessary for the initiation of shoot regeneration (21, 55, 56), WUS-RELATED HOMEOBOX5 expression is associated with shoot primordium formation (18), and patterning is clearly an essential phase in the establishment of shoot primordia in root tissues (21, 56).
Conclusion
The analysis of diverse regeneration-related traits in 88 Arabidopsis accessions generated interesting datasets and valuable information for future investigations in the field of shoot regeneration. We showed that linkage mapping combined with association mapping is a powerful approach to identify QTGs, even for a highly complex trait such as shoot regeneration. By combining mutant analysis with quantitative complementation tests, we obtained strong evidence on the identity of the QTG. These combinatorial approaches revealed that RPK1 and possibly, ABA signaling are unanticipated mediators of shoot regeneration.
Materials and Methods
For shoot regeneration, 7-mm-long apical root segments were taken from 7-d-old seedlings and placed on CIM. After 4 d, explants were transferred to SIM. Shoots, primordia, roots, callus, and greenness were monitored after 7, 11, 14, and 21 d of SIM incubation. Pairwise correlations between the different parameters were calculated with the Spearman’s rank correlation coefficient. QTL analysis was carried out as implemented in GenStat, version 14 (57), and the association analysis was by means of a linear mixed model controlling for the population structure (31) as implemented in GenStat (57). Detailed experimental procedures are described in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Frans Tax for providing the rpk1 mutants and the pRPK1::RPK1-GFP line, Annick De Keyser for technical assistance, and Martine De Cock for help in preparing the manuscript. This work was supported by the University College Ghent Research Fund (H.M. and D.V.), the Agency for Innovation by Science and Technology in Flanders Project 70575 (to S.L.), and the Research Foundation–Flanders (S.D.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1404978111/-/DCSupplemental.
References
- 1.Flores Berrios E, Gentzbittel L, Kayyal H, Alibert G, Sarrafi A. AFLP mapping of QTLs for in vitro organogenesis traits using recombinant inbred lines in sunflower (Helianthus annuus L.) Theor Appl Genet. 2000;101(8):1299–1306. [Google Scholar]
- 2.Van Sint Jan V, Laublin G, Birhman RK, Cappadocia M. Genetic analysis of leaf explant regenerability in Solanum chacoense. Plant Cell Tissue Organ Cult. 1996;47(1):9–13. [Google Scholar]
- 3.Koornneef M, et al. Characterization and mapping of a gene controlling shoot regeneration in tomato. Plant J. 1993;3(1):131–141. [Google Scholar]
- 4.Mano Y, Komatsuda T. Identification of QTLs controlling tissue-culture traits in barley (Hordeum vulgare L.) Theor Appl Genet. 2002;105(5):708–715. doi: 10.1007/s00122-002-0992-3. [DOI] [PubMed] [Google Scholar]
- 5.Kwon Y-S, Kim K-M, Eun M-Y, Sohn J-K. Quantitative trait loci mapping associated with plant regeneration ability from seed derived calli in rice (Oryza sativa L.) Mol Cells. 2001;11(1):64–67. [PubMed] [Google Scholar]
- 6.Taguchi-Shiobara F, et al. Mapping quantitative trait loci associated with regeneration ability of seed callus in rice, Oryza sativa L. Theor Appl Genet. 1997;95(5-6):828–833. [Google Scholar]
- 7.Takeuchi Y, Abe T, Sasahara T. RFLP mapping of QTLs influencing shoot regeneration from mature seed-derived calli in rice. Crop Sci. 2000;40(1):245–247. [Google Scholar]
- 8.Trujillo-Moya C, Gisbert C, Vilanova S, Nuez F. Localization of QTLs for in vitro plant regeneration in tomato. BMC Plant Biol. 2011;11:140. doi: 10.1186/1471-2229-11-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tyagi N, Dahleen LS, Bregitzer P. Candidate genes within tissue culture regeneration QTL revisited with a linkage map based on transcript-derived markers. Crop Sci. 2010;50(5):1697–1707. [Google Scholar]
- 10.Ben Amer IM, Korzun V, Worland AJ, Börner A. Genetic mapping of QTL controlling tissue-culture response on chromosome 2B of wheat (Triticum aestivum L.) in relation to major genes and RFLP markers. Theor Appl Genet. 1997;94(8):1047–1052. [Google Scholar]
- 11.Holme IB, Torp AM, Hansen LN, Andersen SB. Quantitative trait loci affecting plant regeneration from protoplasts of Brassica oleracea. Theor Appl Genet. 2004;108(8):1513–1520. doi: 10.1007/s00122-003-1570-z. [DOI] [PubMed] [Google Scholar]
- 12.Nishimura A, et al. Isolation of a rice regeneration quantitative trait loci gene and its application to transformation systems. Proc Natl Acad Sci USA. 2005;102(33):11940–11944. doi: 10.1073/pnas.0504220102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Valvekens D, Van Montagu M, Van Lijsebettens M. Agrobacterium tumefaciens-mediated transformation of Arabidopsis thaliana root explants by using kanamycin selection. Proc Natl Acad Sci USA. 1988;85(15):5536–5540. doi: 10.1073/pnas.85.15.5536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cary AJ, Che P, Howell SH. Developmental events and shoot apical meristem gene expression patterns during shoot development in Arabidopsis thaliana. Plant J. 2002;32(6):867–877. doi: 10.1046/j.1365-313x.2002.01479.x. [DOI] [PubMed] [Google Scholar]
- 15.Che P, Gingerich DJ, Lall S, Howell SH. Global and hormone-induced gene expression changes during shoot development in Arabidopsis. Plant Cell. 2002;14(11):2771–2785. doi: 10.1105/tpc.006668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Che P, Lall S, Howell SH. Developmental steps in acquiring competence for shoot development in Arabidopsis tissue culture. Planta. 2007;226(5):1183–1194. doi: 10.1007/s00425-007-0565-4. [DOI] [PubMed] [Google Scholar]
- 17.Che P, Lall S, Nettleton D, Howell SH. Gene expression programs during shoot, root, and callus development in Arabidopsis tissue culture. Plant Physiol. 2006;141(2):620–637. doi: 10.1104/pp.106.081240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sugimoto K, Jiao Y, Meyerowitz EM. Arabidopsis regeneration from multiple tissues occurs via a root development pathway. Dev Cell. 2010;18(3):463–471. doi: 10.1016/j.devcel.2010.02.004. [DOI] [PubMed] [Google Scholar]
- 19.Motte H, Verstraeten I, Werbrouck S, Geelen D. CUC2 as an early marker for regeneration competence in Arabidopsis root explants. J Plant Physiol. 2011;168(13):1598–1601. doi: 10.1016/j.jplph.2011.02.014. [DOI] [PubMed] [Google Scholar]
- 20.Gordon SP, Chickarmane VS, Ohno C, Meyerowitz EM. Multiple feedback loops through cytokinin signaling control stem cell number within the Arabidopsis shoot meristem. Proc Natl Acad Sci USA. 2009;106(38):16529–16534. doi: 10.1073/pnas.0908122106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gordon SP, et al. Pattern formation during de novo assembly of the Arabidopsis shoot meristem. Development. 2007;134(19):3539–3548. doi: 10.1242/dev.010298. [DOI] [PubMed] [Google Scholar]
- 22.Heisler MG, et al. Patterns of auxin transport and gene expression during primordium development revealed by live imaging of the Arabidopsis inflorescence meristem. Curr Biol. 2005;15(21):1899–1911. doi: 10.1016/j.cub.2005.09.052. [DOI] [PubMed] [Google Scholar]
- 23.Motte H, Vereecke D, Geelen D, Werbrouck S. The molecular path to in vitro shoot regeneration. Biotechnol Adv. 2014;32(1):107–121. doi: 10.1016/j.biotechadv.2013.12.002. [DOI] [PubMed] [Google Scholar]
- 24.Schiantarelli E, De la Peña A, Candela M. Use of recombinant inbred lines (RILs) to identify, locate and map major genes and quantitative trait loci involved with in vitro regeneration ability in Arabidopsis thaliana. Theor Appl Genet. 2001;102(2-3):335–341. [Google Scholar]
- 25.Lall S, Nettleton D, DeCook R, Che P, Howell SH. Quantitative trait loci associated with adventitious shoot formation in tissue culture and the program of shoot development in Arabidopsis. Genetics. 2004;167(4):1883–1892. doi: 10.1534/genetics.103.025213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Velázquez I, Valencia S, López-Lera A, de la Peña A, Candela M. Analysis of natural allelic variation in in vitro organogenesis of Arabidopsis thaliana. Euphytica. 2004;137(1):73–79. [Google Scholar]
- 27.Mackay TFC, Stone EA, Ayroles JF. The genetics of quantitative traits: Challenges and prospects. Nat Rev Genet. 2009;10(8):565–577. doi: 10.1038/nrg2612. [DOI] [PubMed] [Google Scholar]
- 28.Sterken R, et al. Combined linkage and association mapping reveals CYCD5;1 as a quantitative trait gene for endoreduplication in Arabidopsis. Proc Natl Acad Sci USA. 2012;109(12):4678–4683. doi: 10.1073/pnas.1120811109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brachi B, et al. Linkage and association mapping of Arabidopsis thaliana flowering time in nature. PLoS Genet. 2010;6(5):e1000940. doi: 10.1371/journal.pgen.1000940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhao K, et al. An Arabidopsis example of association mapping in structured samples. PLoS Genet. 2007;3(1):e4. doi: 10.1371/journal.pgen.0030004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ehrenreich IM, et al. Candidate gene association mapping of Arabidopsis flowering time. Genetics. 2009;183(1):325–335. doi: 10.1534/genetics.109.105189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.O’Neill CM, et al. Six new recombinant inbred populations for the study of quantitative traits in Arabidopsis thaliana. Theor Appl Genet. 2008;116(5):623–634. doi: 10.1007/s00122-007-0696-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim S, et al. Recombination and linkage disequilibrium in Arabidopsis thaliana. Nat Genet. 2007;39(9):1151–1155. doi: 10.1038/ng2115. [DOI] [PubMed] [Google Scholar]
- 34.Hong SW, Jon JH, Kwak JM, Nam HG. Identification of a receptor-like protein kinase gene rapidly induced by abscisic acid, dehydration, high salt, and cold treatments in Arabidopsis thaliana. Plant Physiol. 1997;113(4):1203–1212. doi: 10.1104/pp.113.4.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kelley LA, Sternberg MJE. Protein structure prediction on the Web: A case study using the Phyre server. Nat Protoc. 2009;4(3):363–371. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
- 36.Nodine MD, Yadegari R, Tax FE. RPK1 and TOAD2 are two receptor-like kinases redundantly required for Arabidopsis embryonic pattern formation. Dev Cell. 2007;12(6):943–956. doi: 10.1016/j.devcel.2007.04.003. [DOI] [PubMed] [Google Scholar]
- 37.Mackay TFC. Complementing complexity. Nat Genet. 2004;36(11):1145–1147. doi: 10.1038/ng1104-1145. [DOI] [PubMed] [Google Scholar]
- 38.Turner TL, Miller PM, Cochrane VA. Combining genome-wide methods to investigate the genetic complexity of courtship song variation in Drosophila melanogaster. Mol Biol Evol. 2013;30(9):2113–2120. doi: 10.1093/molbev/mst111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Turner TL. Fine-mapping natural alleles: Quantitative complementation to the rescue. Mol Ecol. 2014 doi: 10.1111/mec.12719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kasulin L, Agrofoglio Y, Botto JF. The receptor-like kinase ERECTA contributes to the shade-avoidance syndrome in a background-dependent manner. Ann Bot (Lond) 2013;111(5):811–819. doi: 10.1093/aob/mct038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schwartz C, et al. Cis-regulatory changes at FLOWERING LOCUS T mediate natural variation in flowering responses of Arabidopsis thaliana. Genetics. 2009;183(2):723–732. doi: 10.1534/genetics.109.104984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schmülling T, Schäfer S, Romanov G. Cytokinins as regulators of gene expression. Physiol Plant. 1997;100(3):505–519. [Google Scholar]
- 43.Skoog F, Miller CO. Chemical regulation of growth and organ formation in plant tissues cultured in vitro. Symp Soc Exp Biol. 1957;11:118–130. [PubMed] [Google Scholar]
- 44.Osakabe Y, et al. Leucine-rich repeat receptor-like kinase1 is a key membrane-bound regulator of abscisic acid early signaling in Arabidopsis. Plant Cell. 2005;17(4):1105–1119. doi: 10.1105/tpc.104.027474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee IC, et al. Age-dependent action of an ABA-inducible receptor kinase, RPK1, as a positive regulator of senescence in Arabidopsis leaves. Plant Cell Physiol. 2011;52(4):651–662. doi: 10.1093/pcp/pcr026. [DOI] [PubMed] [Google Scholar]
- 46.Nodine MD, Tax FE. Two receptor-like kinases required together for the establishment of Arabidopsis cotyledon primordia. Dev Biol. 2008;314(1):161–170. doi: 10.1016/j.ydbio.2007.11.021. [DOI] [PubMed] [Google Scholar]
- 47.Osakabe Y, et al. Overproduction of the membrane-bound receptor-like protein kinase 1, RPK1, enhances abiotic stress tolerance in Arabidopsis. J Biol Chem. 2010;285(12):9190–9201. doi: 10.1074/jbc.M109.051938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Luichtl M, et al. Mutations in the Arabidopsis RPK1 gene uncouple cotyledon anlagen and primordia by modulating epidermal cell shape and polarity. Biol Open. 2013;2(11):1093–1102. doi: 10.1242/bio.20135991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shi C-C, et al. Overexpression of the receptor-like protein kinase genes AtRPK1 and OsRPK1 reduces the salt tolerance of Arabidopsis thaliana. Plant Sci. 2014;217-218(3):63–70. doi: 10.1016/j.plantsci.2013.12.002. [DOI] [PubMed] [Google Scholar]
- 50.Ghasemi Bezdi K, Karlov GI, Ahmadikhah A. Effects of genotype, explant type and nutrient medium components on canola (Brassica napus L.) shoot in vitro organogenesis. Afr J Biotechnol. 2007;6(7):861–867. [Google Scholar]
- 51.Hoang TG, Raldugina GN. Regeneration of transgenic plants expressing the GFP gene from rape cotyledonary and leaf explants: Effects of the genotype and ABA. Russ J Plant Physiol. 2012;59(3):406–412. [Google Scholar]
- 52.Huang W-L, Lee C-H, Chen Y-R. Levels of endogenous abscisic acid and indole-3-acetic acid influence shoot organogenesis in callus cultures of rice subjected to osmotic stress. Plant Cell Tissue Organ Cult. 2012;108(2):257–263. and erratum 108(2):265. [Google Scholar]
- 53.Kinoshita A, et al. RPK2 is an essential receptor-like kinase that transmits the CLV3 signal in Arabidopsis. Development. 2010;137(22):3911–3920. doi: 10.1242/dev.048199. and erratum 137(24):4327. [DOI] [PubMed] [Google Scholar]
- 54.Shiu S-H, Bleecker AB. Receptor-like kinases from Arabidopsis form a monophyletic gene family related to animal receptor kinases. Proc Natl Acad Sci USA. 2001;98(19):10763–10768. doi: 10.1073/pnas.181141598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Atta R, et al. Pluripotency of Arabidopsis xylem pericycle underlies shoot regeneration from root and hypocotyl explants grown in vitro. Plant J. 2009;57(4):626–644. doi: 10.1111/j.1365-313X.2008.03715.x. [DOI] [PubMed] [Google Scholar]
- 56.Cheng ZJ, et al. Pattern of auxin and cytokinin responses for shoot meristem induction results from the regulation of cytokinin biosynthesis by AUXIN RESPONSE FACTOR3. Plant Physiol. 2013;161(1):240–251. doi: 10.1104/pp.112.203166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Payne RW. GenStat Release 14 Reference Manual, Part 3 Procedure Library PL20. Hemel Hempstead, United Kingdom: VSN International; 2011. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.