Significance
This report reveals that the more than 200 genes controlled by Bacillus subtilis global regulatory protein CodY are controlled in a hierarchical manner that reflects the extent of activation of CodY by its ligands. The results suggest that B. subtilis uses hierarchical regulation by CodY to achieve strategic goals.
Keywords: BCAA, ILV, RNA-seq, metabolite analysis
Abstract
Global regulators that bind strategic metabolites allow bacteria to adapt rapidly to dynamic environments by coordinating the expression of many genes. We report an approach for determining gene regulation hierarchy using the regulon of the Bacillus subtilis global regulatory protein CodY as proof of principle. In theory, this approach can be used to measure the dynamics of any bacterial transcriptional regulatory network that is affected by interaction with a ligand. In B. subtilis, CodY controls dozens of genes, but the threshold activities of CodY required to regulate each gene are unknown. We hypothesized that targets of CodY are differentially regulated based on varying affinity for the protein’s many binding sites. We used RNA sequencing to determine the transcription profiles of B. subtilis strains expressing mutant CodY proteins with different levels of residual activity. In parallel, we quantified intracellular metabolites connected to central metabolism. Strains producing CodY variants F71Y, R61K, and R61H retained varying degrees of partial activity relative to the WT protein, leading to gene-specific, differential alterations in transcript abundance for the 223 identified members of the CodY regulon. Using liquid chromatography coupled to MS, we detected significant increases in branched-chain amino acids and intermediates of arginine, proline, and glutamate metabolism, as well as decreases in pyruvate and glycerate as CodY activity decreased. We conclude that a spectrum of CodY activities leads to programmed regulation of gene expression and an apparent rerouting of carbon and nitrogen metabolism, suggesting that during changes in nutrient availability, CodY prioritizes the expression of specific pathways.
Adaptation of bacteria to fluctuating environmental conditions is a fundamental biological process that is mediated at the level of gene expression in most cases. Genetic switches, controlled, in part, by global regulatory factors that bind key metabolites, allow bacteria to integrate diverse signals to alter their gene expression programs in response to changes in nutrient availability, exposure to reactive oxygen species, osmotic stress, and other environmental factors (1–3). Sensing the quality and availability of nutrients is not only important for survival and biomass production but also for successful competition against other cell types.
CodY is a global regulatory protein that controls dozens of genes in a variety of low guanine + cytosine (G + C) Gram-positive bacterial species (4–10). First identified in Bacillus subtilis as a factor that represses stationary phase genes during exponential growth in rich medium (11), CodY is now known to provide a critical regulatory link between metabolism and pathogenesis in bacteria important for human health (5, 6, 12–16). CodY is activated as a DNA-binding protein in vitro by at least two classes of metabolites: the branched-chain amino acids [BCAAs; i.e., isoleucine, leucine, and valine (ILV)] and GTP (17–21). Changes in the intracellular pools of these effectors correlate with altered CodY activity (13, 17, 20, 22–24). CodY typically behaves as a repressor of gene expression by binding to a specific sequence located in or near the promoter region of target genes (4, 25), by competing with a positive regulator (26), or by causing premature termination of transcription via a roadblock mechanism (27). CodY also activates transcription of some target genes (8, 24, 28).
Extensive characterization of targets in B. subtilis has revealed that medium-dependent changes in steady-state CodY activity lead to varying extents of gene expression (17, 21, 22, 25), likely due to varying intracellular pools of CodY effectors and variable affinity of CodY for target sites. Indeed, Belitsky and Sonenshein (29) recently performed a genome-wide in vitro binding experiment that not only identified all CodY-binding sites in the B. subtilis chromosome but also determined relative binding strengths for these sites. Although the targets of CodY are generally known, the threshold concentrations of activated CodY that regulate each gene are not. Theoretically, one can determine such thresholds by monitoring the global transcriptional response at multiple stages of nutrient exhaustion or by titration of a regulatory protein (30). However, such experimental design strategies can be problematic. First, changes in medium composition and growth rate may cause transcriptional perturbations due to factors other than the regulator being analyzed. Second, altering the cellular abundance of a transcription factor by ectopic expression creates a nonphysiological state and may lead to heterogeneity in the population if the factor is expressed from a plasmid (31). We report an alternative approach in which we conducted genome-wide profiling of transcript abundances in strains producing mutant CodY proteins with varying residual activities during steady-state growth under conditions that maximize WT CodY activity. We thereby identified genes that are turned on or off at different levels of CodY activity and coupled this analysis with MS-based metabolite quantification, providing a framework for determining the bacterium’s strategy for altering the activities of multiple metabolic pathways when faced with changing levels of nutrient availability.
Results and Discussion
Strand-Specific RNA Sequencing Reveals Additional CodY Targets Not Previously Observed.
The B. subtilis CodY regulon has been previously defined by microarray analysis (8) and theoretical genomic reconstruction (32). To quantify the dynamic ranges of CodY target genes, we extracted RNA from WT and codY null strains during steady-state exponential phase growth in defined glucose-ammonium medium supplemented with a 16-amino acid mixture that fully activates CodY (33) and subjected cDNA versions of the RNAs to RNA-sequencing (RNA-seq) analysis (34).
We generated a dataset of sequence reads associated with each ORF and used DESeq (35) to calculate the expression for all CodY targets corrected for effective library size (Dataset S1). After calculating significance levels (P < 0.05) adjusted for multiple hypothesis testing, we identified 196 genes that were overexpressed and 27 genes that were underexpressed at least threefold in the codY null strain relative to its parent strain (Datasets S2 and S3). These genes include both direct and indirect targets of CodY; the vast majority were identified previously (8, 29). Negatively regulated targets (i.e., those overexpressed in the codY null strain) exhibited the highest dynamic ranges (e.g., frlO was regulated 894-fold) (Dataset S2). The target of strongest positive regulation (underexpressed in the codY null strain) was nhaC, an indirect target (29) that was regulated 34-fold (Dataset S3). The RNA-seq approach revealed several CodY target genes that were missed in the microarray analysis. These genes include the argCJBD-carAB-argF operon (four- to 40-fold derepression in the codY mutant strain) and the mtnAKU operon, whose products play a role in methionine salvage (36).
RNA-Seq Reveals a Graded Transcriptional Response Mediated by CodY.
Previous results indicated that CodY binds to different target sites with varying affinity (29, 37). Furthermore, significant variation has been observed in the extent to which mutations in CodY that affect its DNA-binding activity alter the regulation of a limited number of its direct targets (17, 37, 38). These findings suggest that not all genes regulated by CodY respond equally to a given extent of nutritional limitation (27, 37). That is, if one assumes that the intracellular concentration of active (ligand-bound) CodY molecules is the limiting factor for determining whether CodY binds to and regulates a given gene, regulation of genes with low-affinity (i.e., weak) binding sites would be lost when the concentration of active CodY molecules decreases even moderately. On the other hand, high-affinity (i.e., strong) binding sites would remain occupied by CodY even when the intracellular concentration of active CodY is relatively low.
To modify the activity of CodY without changing the medium composition or the codY gene copy number, we used a set of codY mutant strains that have alterations in the GAF domain (18) that is responsible for ILV binding (37, 38, and this study). The mutant strains were expected to behave as though ILV levels were limiting even though excess ILV was provided in the medium. A similar set of mutant strains was shown previously to provide highly variable regulation of a target gene (37). To verify that the variable activities of the mutant set used for the experiments reported here was related to the presence of ILV, we measured transcript abundance for bcaP (a direct CodY target) in strains producing WT CodY and the mutant forms F71Y, R61K, and R61H (ref. 38 and this study), all of which have single amino acid substitutions in the ILV-binding pocket (Fig. S1). Transcript abundance was monitored during growth in a minimal glucose-ammonium medium (known as TSS) and TSS supplemented with l-isoleucine. The bcaP gene was derepressed to varying degrees in the variants in TSS in which the only source of ILV is endogenous synthesis. Moreover, all three mutants responded to the addition of isoleucine, but none of them to the same extent as the WT. The strain producing the variant F71Y behaved most like the WT, and the strain producing the variant R61H was most similar to the null mutant. We verified that the amino acid substitutions did not affect the cellular abundance of CodY, its mobility under denaturing conditions, or its oligomeric state (Fig. S2). CodYR61H ran slightly faster than the other forms of CodY under nondenaturing conditions, as expected from the change in net charge.
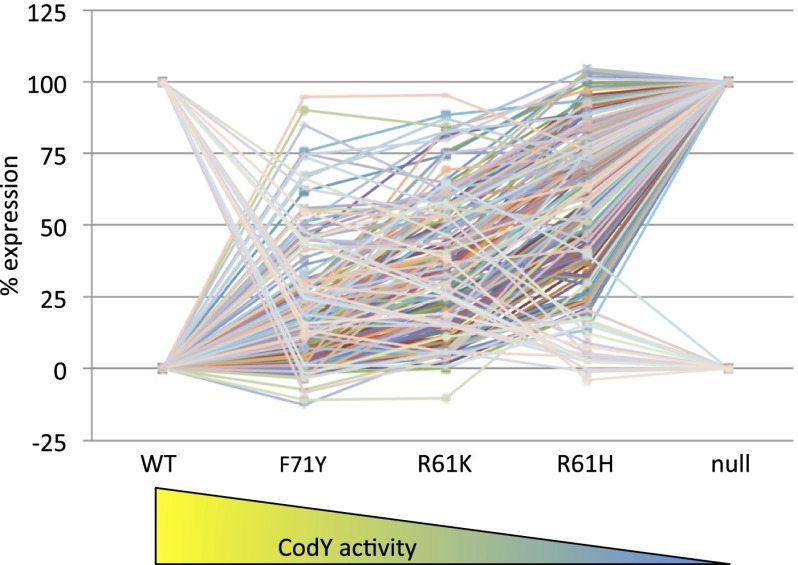
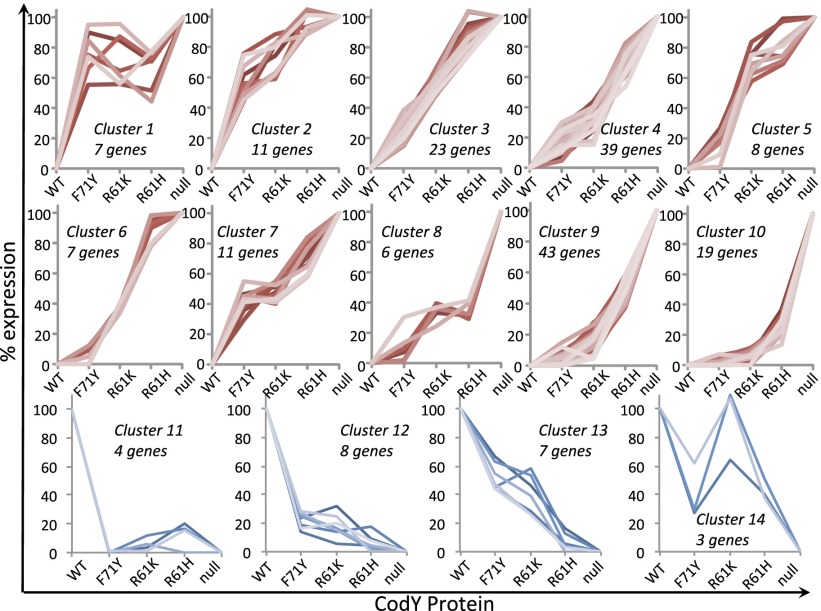
We then used RNA-seq to measure the variability in expression of the entire CodY regulon in the set of mutant strains. We calculated reads per kilobase per million ORF read (RPKMO) values; values in the variant strains were converted to a percentage of the RPKMO value obtained when expression was maximal (i.e., in the null strain for negatively regulated targets or in the WT strain for positively regulated targets) (Dataset S4). The results (Fig. 1) demonstrate that CodY target genes exhibit a broad range of sensitivities to changes in CodY activity. Direct targets of CodY [determined by genome-wide analysis of CodY-binding sites (29)] could be aggregated into 36 negatively regulated transcription units and seven positively regulated transcription units, whereas indirect targets of CodY could be aggregated into 16 negatively regulated transcription units and 10 positively regulated transcription units. Products of some genes that are direct (rapA, phrA, rapF, phrF, yobQ, kinB) and indirect (hpr, ywaE) targets of CodY regulate other genes. The dataset was subjected to k-means clustering (39–41), set to 10 clusters for negatively regulated genes and four clusters for positively regulated genes, to identify distinct expression patterns (Fig. 2 and Dataset S5). Some negatively regulated genes were fully derepressed in the strain producing F71Y (cluster 1), whereas others were only fully derepressed in the codY null strain (cluster 10). Similarly, some positively regulated genes were not expressed above background levels in the F71Y strain (cluster 11), whereas others were partially expressed in all strains except the null strain (cluster 14). All positively regulated genes were affected by the F71Y mutation, which seemed to have the least effect on CodY activity.
Fig. 1.
Reductions in transcription factor activity generate a diverse set of responses throughout the CodY regulon. RPKMO values for 197 differentially expressed targets with values >10 in at least one strain were plotted as a percentage of maximal expression (i.e., expression in the codY null mutant for negatively regulated genes and expression in the WT for positively regulated genes). Some CodY target genes, including adcA, frlR, rpmE2, rpsN, trpD, yczL, yrpE, ybfAB, yciABC, yjbA, and yrhP, were omitted because the data were not reproducible or the patterns were complex. Note that the positions of the point mutant strains on the x axis are arbitrary and are not meant to imply a linear relationship between position and residual CodY activity. Data points are the means of two independent experiments.
Fig. 2.
Graded response to changes in CodY activity is revealed by k-means clustering. Genes repressed by CodY were distributed to 10 clusters (clusters 1–10, red), and genes activated by CodY were distributed to four clusters (clusters 11–14, blue). Reading from cluster 1–10 and from cluster 11–14, gene clusters that responded to modest changes in CodY activity precede those that only responded to more substantial changes in activity. The relative positioning of clusters 6 and 7 was arbitrary.
In general, genes located in the same operon or with related functions fell into the same cluster (Dataset S5), lending credence to the number of clusters used in calculations. For example, the majority of the polyketide synthase genes (i.e., pksFGHIJLM, acpK) fell into cluster 9; the distal pksN gene was assigned to cluster 4. All five genes of the dipeptide permease (dpp) operon, as well as the ykfABCD genes that are transcribed by read-through from the dpp operon (8), were assigned to cluster 9; all seven genes of the ilv-leu operon were assigned to cluster 4. The unlinked BCAA biosynthesis genes ilvA and ilvD, however, were assigned to clusters 7 and 3, respectively.
Whereas the vast majority of the gene expression patterns correlate with relative CodY activity, some do not (e.g., cluster 1, cluster 14). We offer three possible explanations. First, many CodY-regulated genes are subject to multiple forms of regulation. Second, because binding of BCAAs induces a major conformational change in CodY, the population of WT protein is expected to be almost entirely in the liganded conformation, whereas different fractions of the mutant protein populations will be in the unliganded conformation. If unliganded CodY is able to bind to some CodY-binding sites but not to most, a mutant protein that has a higher unliganded fraction will bind better to those sites than a protein that has a higher liganded population. Third, if a gene has two independent CodY-binding sites with different affinities and CodY acts as a negative regulator at one site and a positive regulator at the other site, there will be intermediate levels of CodY activity at which positive regulation prevails, another level at which negative regulation prevails, and a third level at which the two modes of regulation will compete. As a result, the overall expression pattern will be unpredictable.
Validation of RNA-Seq Data.
We chose three direct targets of CodY [yoyD, ilvB, and amhX (8, 29, 42)] to validate the RNA-seq experiment by quantitative RT-PCR (qRT-PCR) analysis. The target genes fell into clusters 2, 4, and 10, respectively. Transcript abundance was normalized to that of sigA, whose expression was stable (Table 1). Fold-regulation (i.e., the ratio of transcript abundance in a codY mutant strain to that in the WT strain) compared favorably between the qRT-PCR and RNA-seq experiments (Table 1).
Table 1.
Validation of RNA-seq data by qRT-PCR
| Target | CodY protein | Copies of transcript relative to sigA* | Expression by qRT-PCR,† % | Expression by RNA-seq,† % |
| amhX | WT | 1.3 | 0.0 | 0.0 |
| F71Y | 3.0 | 1.4 | 0.9 | |
| R61K | 7.2 | 4.9 | 5.0 | |
| R61H | 45.2 | 36.2 | 36.4 | |
| Null | 122.6 | 100.0 | 100.0 | |
| ilvB | WT | 14.0 | 0.0 | 0.0 |
| F71Y | 124.9 | 23.3 | 25.5 | |
| R61K | 191.5 | 37.3 | 44.0 | |
| R61H | 342.7 | 69.1 | 74.1 | |
| Null | 489.7 | 100.0 | 100.0 | |
| yoyD | WT | 43.8 | 0.0 | 0.0 |
| F71Y | 239.0 | 46.2 | 61.6 | |
| R61K | 382.8 | 80.2 | 74.2 | |
| R61H | 489.7 | 105.5 | 101.5 | |
| Null | 466.5 | 100.0 | 100.0 |
Transcript abundances were measured by qRT-PCR using RNAs purified from cells grown in chemically defined glucose-ammonium medium with ILV and 13 other amino acids. Transcript abundances are relative to sigA. SEMs were ≤30%.
After subtracting the transcript abundances in the WT strain from those determined for the mutant strains, the increased abundance in each point mutant strain was related to the abundance in the codY null mutant strain, which was normalized to 100%.
Genes for Glutamate-Producing Pathways Are More Sensitive to CodY Activity than Genes of Glutamate Biosynthesis.
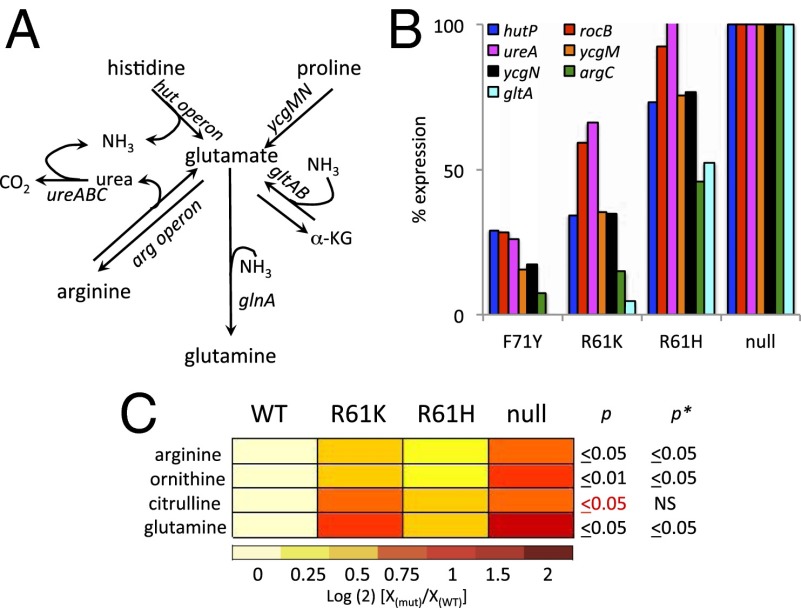
Glutamate connects carbon and nitrogen metabolism by providing the carbon skeleton for α-ketoglutarate synthesis, the entry point into carbon metabolism for certain amino acid degradation pathways, and by serving as the substrate for ammonium assimilation via glutamine synthesis (43) (Fig. 3A). A complex interplay involving several regulatory proteins, including CodY, controls the expression of nitrogen metabolism genes. We observed coregulation of genes coding for enzymes thought to be involved in uptake and utilization of urea (ureABC), histidine (hutGHIMPU), and proline (putBC/ycgMN) (Fig. 3B). rocB, postulated to convert citrulline to carbamate and ornithine (44), followed a similar pattern of expression. These pathways all lead to production of glutamate, ammonia, or both. These degradation genes were more responsive to modest reductions in CodY activity than were the operons for glutamate and arginine biosynthesis (i.e., argC, gltAB; Fig. 3B), suggesting that the cell’s strategy is to use these externally supplied amino acids both for protein synthesis and as sources of glutamate and ammonium ion before up-regulating de novo biosynthesis of glutamate or arginine.
Fig. 3.
CodY represses certain amino acid utilization genes less stringently than glutamate and arginine biosynthesis genes. All data here and in subsequent figures are presented as the means of at least two independent experiments. (A) Histidine, proline, and arginine are degraded to glutamate or to glutamate plus ammonium ion. (Urea is released as the first step in arginine degradation and is then degraded by urease to ammonia and CO2.) α-KG, α-ketoglutarate. (B) CodY-repressed targets coding for enzymes for degradation of histidine (hutP), citrulline (rocB), urea (ureA), and proline (ycgMN, also known as putBC), as well as CodY-repressed targets coding for biosynthesis of arginine (argC) and glutamate (gltA), are shown. (C) Heat map displays the relationship between relative levels of CodY activity and key metabolites of amino acid metabolism (Dataset S6). Changes in the abundance of each metabolite are color-coded, with red indicating an increase in intracellular abundance relative to the baseline (abundance in the WT) on a log(2) scale. p, level of statistical significance by Cuzick’s nonparametric test for trend of the abundance of each metabolite in WT (SRB109), R61K (SRB361), R61H (SRB465), and null (SRB268) strains; p*, the adjusted P value after correction for multiple hypothesis testing (65). P values in red typeface indicate the level of significance in the WT vs. codY null mutant determined by a Mann–Whitney U test but do not show a dose-dependent relationship by Cuzick’s test of trend across all isolates. NS, not significant.
Based on the observations above, one might expect that intracellular pools of histidine, proline, ornithine, and other metabolites would change as CodY becomes deactivated. To gain further insight into the physiological consequences of CodY deactivation, we used tandem liquid chromatography-TOF MS (LC-MS) in parallel with our RNA-seq experiment to measure the intracellular pools of metabolites of central and intermediary metabolism. We identified 62 named compounds by comparison against an accurate mass retention time database (Dataset S6). We observed increases (≤twofold) in intracellular arginine, ornithine, citrulline, and glutamine pools (Fig. 3C; P < 0.05 for all except citrulline) in the codY null strain relative to its WT parent. Glutamate abundance also increased, but the measured difference was not statistically significant. In the codY null mutant, uptake, degradation, and biosynthesis are all fully derepressed. At intermediate levels of CodY activity (i.e., in the CodYR61K- and CodYR61H-producing strains), we saw less of an increase in abundance of arginine, ornithine, citrulline, and glutamine. Based on transcript analyses, the levels of the amino acids in these strains are likely to reflect derepression of uptake and degradation concomitant with repression of biosynthesis. Calculating relative fluxes using labeled nutrients would help to resolve the activities of these competing pathways.
CodY Regulates BCAA Aminotransferases Differentially.
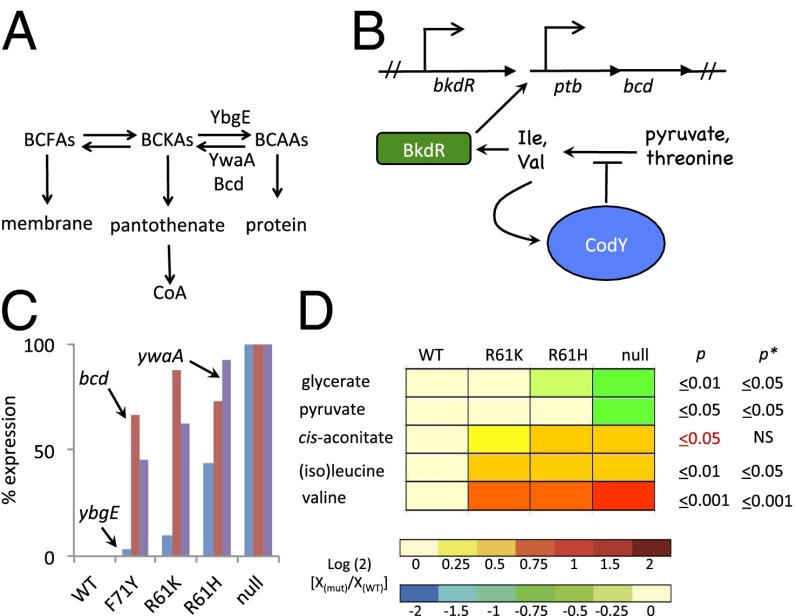
B. subtilis encodes three enzymes (YbgE, YwaA, and Bcd) that can interconvert the branched-chain keto acids (BCKAs) and BCAAs (17, 45–48) (Fig. 4A). YbgE and YwaA are aminotransferases; Bcd (also known as leucine dehydrogenase) is primarily a deaminating enzyme, but it can also aminate BCKAs. CodY represses all of these genes; ybgE and ywaA are direct targets, and bcd is an indirect target (Dataset S2). The physiological basis for the functional redundancy of these enzymes remains an open question. The Bcd enzyme is primarily used for conversion of BCAAs to BCKAs as part of the pathway for branched-chain fatty acid synthesis. The bcd gene is positively regulated by BkdR in response to isoleucine and valine availability, and thus is indirectly regulated by CodY via control of BCAA biosynthesis (46) (Fig. 4B). YbgE and YwaA both prefer ILV over nearly all other amino acids tested as an amino donor; the aminotransferase activity of YbgE is much greater than that of YwaA (45). Interestingly, the ywaA (cluster 2) and bcd (cluster 1) genes responded to relatively small changes in CodY activity, whereas ybgE (cluster 9) responded only when CodY activity was almost completely ablated (Fig. 4C and Dataset S4). Coexpression of ilv-leu (cluster 4), ctrA (bcaP, encoding the major BCAA transporter; cluster 4), ywaA, and bcd would be consistent with the hypothesis that when those genes are expressed, the cells direct a substantial portion of imported BCAAs and the newly synthesized BCKAs toward pantothenate and branched-chain fatty acids. When YbgE is added to the mix (i.e., when CodY is nearly inactive), the balance may tilt toward the BCAAs. B. subtilis likely keeps ybgE gene expression low in the presence of exogenous ILV to prevent futile cycling between the amino acid and the keto acid. When CodY is absent, the pool of pyruvate decreases as the pools of BCAAs increase, consistent with the idea that BCAA biosynthesis is a major consumer of pyruvate (Fig. 4D).
Fig. 4.
CodY prioritizes BCAA aminotransferase gene expression. (A) YbgE, YwaA, and Bcd mediate interconversion of α-keto acids (BCKAs) and their cognate amino acids (BCAAs). (B) CodY regulates the expression of bcd indirectly. Inactivation of CodY leads to increased synthesis of Ile and Val, both of which can activate BkdR, the positive regulator of bcd. (C) ywaA and bcd were substantially derepressed when CodY activity was mildly reduced, whereas ybgE was derepressed more than 50% only in the strain lacking CodY activity. (D) Heat map displays the relationship between strains with varying levels of CodY activity and the fate of pyruvate. Changes in the abundance of each metabolite (Dataset S6) are color-coded, with red and blue indicating an increase or decrease in intracellular abundance, respectively, relative to the baseline (abundance in the WT) on a log(2) scale. Strains and statistical testing are presented as described in the legend for Fig. 3.
CodY Inversely Regulates Carbon Overflow Metabolism and Reutilization of Overflow Metabolites.
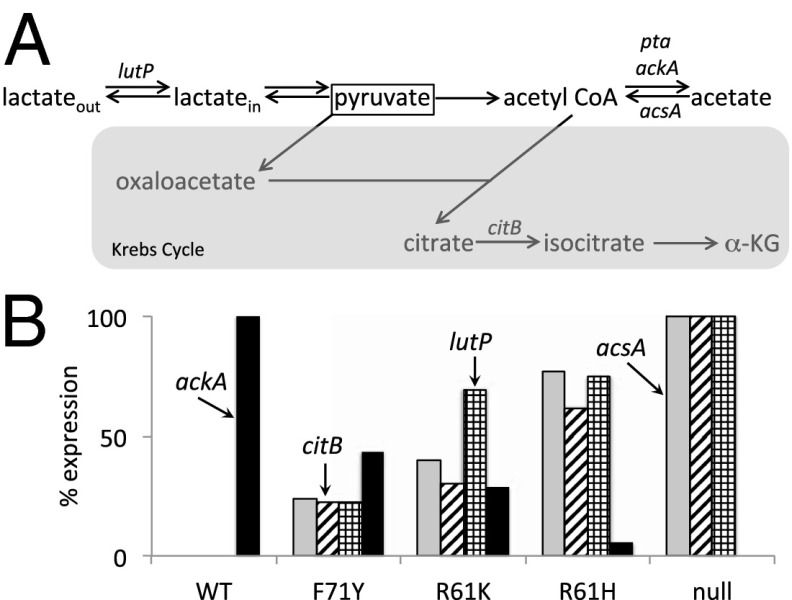
When B. subtilis is offered a mixture of glucose and other potential carbon sources, as it was in these experiments, it preferentially metabolizes the glucose. Multiple mechanisms, including inducer exclusion, catabolite repression by the global regulator CcpA, and repression by CodY of various carbon source uptake systems, contribute to the preferential use of glucose (1, 30, 49–52). Glucose is degraded via glycolysis to pyruvate, a critical intracellular metabolite that has many fates (43). For example, excess pyruvate can be diverted to overflow pathways, which results in the synthesis and excretion of lactate, acetate, and acetoin via the lct-, pta-ackA–, and acu-encoded pathways (53, 54); to the Krebs cycle; or to the synthesis of alanine, BCAAs, and other cellular building blocks. Once the supply of glucose is exhausted, excreted acetate and lactate can be reassimilated via the Ac-CoA synthetase encoded by acsA (54) and the lut gene products, respectively (54, 55) (Fig. 5A). CodY appears to be a direct positive regulator of ackA (24, 29) and a direct negative regulator of ilv-leu, acsA, the Krebs cycle gene citB, and the lactate transporter gene lutP (formerly yvfH) (29), potentially controlling the flow of carbon in and out of this important metabolic intersection (24, 29, 42, 56). Consistent with these results, we found that expression of ackA decreased as CodY activity decreased (Fig. 5B), concomitant with increases in the expression of acsA, lutP, citB, and the ilv-leu operon. The extent of expression of the latter transcription units was similar at varying levels of residual CodY activity (Fig. 5B and Dataset S4). This hierarchical coregulation reveals a role for CodY in prioritizing carbon source utilization by stimulating acetate synthesis when nutrients are in excess and in importing and metabolizing acetate and lactate when nutrients are limited.
Fig. 5.
CodY regulates carbon overflow metabolism and reutilization of overflow metabolites. (A) Metabolic paths to and from pyruvate. The terms “out” and “in” denote extracellular and intracellular lactate, respectively. Reactions of the Krebs cycle are shaded in gray. (B) Extent of expression of four genes that represent carbon overflow metabolism (ackA) and overflow metabolite reutilization (acsA, lutP, and citB) is depicted as a percentage of maximal expression. A reduction in CodY activity reduced expression of ackA, but increased expression of acsA, lutP, and citB.
Genes for utilization of other potential carbon sources are also within the CodY regulon and have different expression patterns. For instance, genes for transport of citrate/malate, BCAAs, oligopeptides, arginine/ornithine, and γ-aminobutyrate are turned on when CodY activity is slightly reduced, whereas genes for the transport and metabolism of lactate, proline, and histidine are derepressed at an intermediate level of CodY activity and genes for the transport of choline/glycine-betaine, dipeptides, and fructosamines are only activated when CodY activity is almost completely ablated (Dataset S4). Highly active CodY is a positive regulator of iron and ribose uptake.
Is the CodY Regulon Hierarchy Determined by Binding Strength?
Based on previous studies, we hypothesized that CodY affinity for DNA at a given site would dictate its relative position in the CodY expression hierarchy. We did, in fact, find a substantial but imperfect correlation between the expression hierarchy and the binding strength rankings determined by Belitsky and Sonenshein (29). Fourteen of the 22 transcription units in clusters 9 and 10 that are direct targets of CodY are among the 25 highest strength binding sites [of a total of 353; dataset S1A sheet 1(2) in ref. 29] associated with CodY-regulated genes expressed under the conditions used here. Also, of the 13 direct CodY target operons in clusters 5–8, nine are within the top 100 binding sites, and of the 42 direct targets of CodY in clusters 1–4, 39 are in the top 200 binding sites. However, there were outliers in almost every cluster. For instance, the gmuF-gmuG locus of cluster 3 ranks as the sixth highest strength binding site (29) and the argC operon of cluster 10 has a ranking of 169th. These discrepancies may be attributable to multiple factors. First, the exact positioning of a CodY-binding site with respect to the promoter may influence the efficiency with which CodY regulates transcription. Second, CodY binding to some targets, such as argC, is cooperative. At 8 nM CodY, the argC site has a ranking of 169th, but at 40 nM CodY, the argC site has a ranking of 61st. Third, if a gene has a second regulator whose role is to interfere with CodY binding, a relatively strong CodY-binding site might not have the same relative effectiveness in vivo. For instance, Hillen and coworkers (57) recently showed that CodY and CcpA, the global regulators of carbon metabolism, can be cross-linked to each other in vivo, consistent with the known coregulation or antagonistic regulation by the two proteins at the ackA, ilvB, and other promoters (8, 24, 58).
Conclusion
We have presented here an approach to determine in a rigorous and empirical manner the expression patterns of all genes controlled by a regulator. This approach, which is based on altering the responsiveness of the regulator to its activating ligands, is robust and generates physiologically meaningful data with minimal experimental artifacts because the cells are cultivated without nutritional limitation and are at biological steady state. It is important to note that the differential regulation observed here is independent of the dynamic range of regulation of individual CodY targets (ratio of expression in a codY null mutant to that in the parental strain). Thus, whether a gene is regulated threefold or 300-fold by CodY, its place in the expression hierarchy is determined by the fraction of the population of CodY molecules that is active. Coupling LC-MS–based metabolite profiling to the expression analysis provides further insight into the metabolic changes associated with changes in regulator activity. Taken together, the transcriptional profiling and intracellular metabolite measurements indicate that CodY is responsible, at least in part, for executing several strategies for adapting to fluctuating nutritional conditions. The approach can be applied to any biological model with a manipulatable genetic system. Because CodY is produced by low G + C Gram-positive bacterial species of both industrial and medical relevance, the approach has the potential to reveal the cell’s strategy for integrating central metabolic pathways with virulence gene expression and production of useful byproducts. These questions are the subject of current investigations in our laboratories.
Materials and Methods
Bacterial Strains and Growth Conditions.
All Escherichia coli and B. subtilis (SMY lineage) strains used in this study are listed in Table S1; growth media are described in detail in SI Materials and Methods. All B. subtilis RNA samples were isolated from cells in midexponential phase as described (22) in a defined medium (TSS) containing glucose as a carbon source and a mixture of 16 amino acids (TSS + 16 AA) (33).
DNA Manipulations and Transformations.
All molecular biology techniques were performed as described (59). Oligonucleotides were obtained from Integrated DNA Technologies (Table S2). Construction of strains was performed as previously described (17, 60) or as described in SI Materials and Methods.
RNA Sampling and Preparation.
Samples were collected and nucleic acid was extracted as previously described (22) during steady-state exponential phase growth within approximately one generation of one another, when CodY activity is maximal.
RNA-Seq Library Construction, Sequencing, and Analysis.
Libraries for Illumina sequencing were constructed using the strand-specific method described previously (61) and in SI Materials and Methods. Reads were aligned to the B. subtilis genome (RefSeq NC_000964) using Burrows–Wheeler Aligner version 5.9 (62); gene annotations were obtained from RefSeq (www.ncbi.nlm.nih.gov/refseq/). The overall read coverage of genomic regions corresponding to features such as ORFs and rRNAs was conducted as described (63). Differential expression analysis was conducted using DESeq (35). Clustering analysis was performed in MATLAB (MathWorks) using standard k-means, with the Pearson correlation as a distance measure.
qRT-PCR Analysis.
Real-time qRT-PCR was performed as described (17) with either a Roche LightCycler 480 instrument and associated SYBR green chemistry or a Bio-Rad CFX96 instrument and SensiFAST SYBR No ROX reagent (Bioline). Samples (3 μg) of nucleic acid were treated using the TURBO DNA-free kit (Invitrogen) to reduce contaminating DNA to undetectable levels. Standard curves constructed from purified PCR products or chromosomal DNA allowed for absolute quantification of transcripts over five to six orders of magnitude. Assay efficiencies were 85–110%. Specificity of amplification was assessed using a combination of melting curve analysis and agarose gel electrophoresis with ethidium bromide staining (59). Statistical tests were performed in Prism version 4.0a (GraphPad).
MS-Based Metabolomics Sample Preparation and Analysis.
Sample preparation and metabolite analyses were performed as described in SI Materials and Methods using methods similar to those of Weisenberg et al. (64).
Supplementary Material
Acknowledgments
We thank Boris Belitsky for sharing experimental results, important advice, and helpful comments; David Lazinski for sharing oligonucleotides and advice about library construction; and Deepak Sharma for technical support. This work was supported, in part, by Pathway to Independence Award K99 GM099893 (to S.R.B.) and National Institutes of Health (NIH) Research Grant R01 GM042219 (to A.L.S.). Analysis of nucleic acids for RNA-seq library construction was performed using an Agilent Technologies Bioanalyzer in the Tufts Neuroscience Core Facility (supported by NIH Grant P30 NS047243).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The RNA-sequencing transcriptional profiling data reported in this paper have been deposited in the Sequence Read Archive of the National Center for Biotechnology (accession no. SRP041677).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1321308111/-/DCSupplemental.
References
- 1.Henkin TM. The role of CcpA transcriptional regulator in carbon metabolism in Bacillus subtilis. FEMS Microbiol Lett. 1996;135(1):9–15. doi: 10.1111/j.1574-6968.1996.tb07959.x. [DOI] [PubMed] [Google Scholar]
- 2.Pratt LA, Hsing W, Gibson KE, Silhavy TJ. From acids to osmZ: Multiple factors influence synthesis of the OmpF and OmpC porins in Escherichia coli. Mol Microbiol. 1996;20(5):911–917. doi: 10.1111/j.1365-2958.1996.tb02532.x. [DOI] [PubMed] [Google Scholar]
- 3.Zheng M, Storz G. Redox sensing by prokaryotic transcription factors. Biochem Pharmacol. 2000;59(1):1–6. doi: 10.1016/s0006-2952(99)00289-0. [DOI] [PubMed] [Google Scholar]
- 4.den Hengst CD, et al. The Lactococcus lactis CodY regulon: Identification of a conserved cis-regulatory element. J Biol Chem. 2005;280(40):34332–34342. doi: 10.1074/jbc.M502349200. [DOI] [PubMed] [Google Scholar]
- 5.Dineen SS, McBride SM, Sonenshein AL. Integration of metabolism and virulence by Clostridium difficile CodY. J Bacteriol. 2010;192(20):5350–5362. doi: 10.1128/JB.00341-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hendriksen WT, et al. CodY of Streptococcus pneumoniae: Link between nutritional gene regulation and colonization. J Bacteriol. 2008;190(2):590–601. doi: 10.1128/JB.00917-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Majerczyk CD, et al. Direct targets of CodY in Staphylococcus aureus. J Bacteriol. 2010;192(11):2861–2877. doi: 10.1128/JB.00220-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Molle V, et al. Additional targets of the Bacillus subtilis global regulator CodY identified by chromatin immunoprecipitation and genome-wide transcript analysis. J Bacteriol. 2003;185(6):1911–1922. doi: 10.1128/JB.185.6.1911-1922.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sonenshein AL. CodY, a global regulator of stationary phase and virulence in Gram-positive bacteria. Curr Opin Microbiol. 2005;8(2):203–207. doi: 10.1016/j.mib.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 10.van Schaik W, et al. The global regulator CodY regulates toxin gene expression in Bacillus anthracis and is required for full virulence. Infect Immun. 2009;77(10):4437–4445. doi: 10.1128/IAI.00716-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Slack FJ, Serror P, Joyce E, Sonenshein AL. A gene required for nutritional repression of the Bacillus subtilis dipeptide permease operon. Mol Microbiol. 1995;15(4):689–702. doi: 10.1111/j.1365-2958.1995.tb02378.x. [DOI] [PubMed] [Google Scholar]
- 12.Bennett HJ, et al. Characterization of relA and codY mutants of Listeria monocytogenes: Identification of the CodY regulon and its role in virulence. Mol Microbiol. 2007;63(5):1453–1467. doi: 10.1111/j.1365-2958.2007.05597.x. [DOI] [PubMed] [Google Scholar]
- 13.Lobel L, Sigal N, Borovok I, Ruppin E, Herskovits AA. Integrative genomic analysis identifies isoleucine and CodY as regulators of Listeria monocytogenes virulence. PLoS Genet. 2012;8(9):e1002887. doi: 10.1371/journal.pgen.1002887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Majerczyk CD, et al. Staphylococcus aureus CodY negatively regulates virulence gene expression. J Bacteriol. 2008;190(7):2257–2265. doi: 10.1128/JB.01545-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Malke H, Steiner K, McShan WM, Ferretti JJ. Linking the nutritional status of Streptococcus pyogenes to alteration of transcriptional gene expression: The action of CodY and RelA. Int J Med Microbiol. 2006;296(4-5):259–275. doi: 10.1016/j.ijmm.2005.11.008. [DOI] [PubMed] [Google Scholar]
- 16.Pohl K, et al. CodY in Staphylococcus aureus: A regulatory link between metabolism and virulence gene expression. J Bacteriol. 2009;191(9):2953–2963. doi: 10.1128/JB.01492-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brinsmade SR, Sonenshein AL. Dissecting complex metabolic integration provides direct genetic evidence for CodY activation by guanine nucleotides. J Bacteriol. 2011;193(20):5637–5648. doi: 10.1128/JB.05510-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Levdikov VM, et al. Structural rearrangement accompanying ligand binding in the GAF domain of CodY from Bacillus subtilis. J Mol Biol. 2009;390(5):1007–1018. doi: 10.1016/j.jmb.2009.05.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Levdikov VM, Blagova E, Joseph P, Sonenshein AL, Wilkinson AJ. The structure of CodY, a GTP- and isoleucine-responsive regulator of stationary phase and virulence in gram-positive bacteria. J Biol Chem. 2006;281(16):11366–11373. doi: 10.1074/jbc.M513015200. [DOI] [PubMed] [Google Scholar]
- 20.Ratnayake-Lecamwasam M, Serror P, Wong K-W, Sonenshein AL. Bacillus subtilis CodY represses early-stationary-phase genes by sensing GTP levels. Genes Dev. 2001;15(9):1093–1103. doi: 10.1101/gad.874201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shivers RP, Sonenshein AL. Activation of the Bacillus subtilis global regulator CodY by direct interaction with branched-chain amino acids. Mol Microbiol. 2004;53(2):599–611. doi: 10.1111/j.1365-2958.2004.04135.x. [DOI] [PubMed] [Google Scholar]
- 22.Brinsmade SR, Kleijn RJ, Sauer U, Sonenshein AL. Regulation of CodY activity through modulation of intracellular branched-chain amino acid pools. J Bacteriol. 2010;192(24):6357–6368. doi: 10.1128/JB.00937-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Petranovic D, et al. Intracellular effectors regulating the activity of the Lactococcus lactis CodY pleiotropic transcription regulator. Mol Microbiol. 2004;53(2):613–621. doi: 10.1111/j.1365-2958.2004.04136.x. [DOI] [PubMed] [Google Scholar]
- 24.Shivers RP, Dineen SS, Sonenshein AL. Positive regulation of Bacillus subtilis ackA by CodY and CcpA: Establishing a potential hierarchy in carbon flow. Mol Microbiol. 2006;62(3):811–822. doi: 10.1111/j.1365-2958.2006.05410.x. [DOI] [PubMed] [Google Scholar]
- 25.Belitsky BR, Sonenshein AL. Genetic and biochemical analysis of CodY-binding sites in Bacillus subtilis. J Bacteriol. 2008;190(4):1224–1236. doi: 10.1128/JB.01780-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Belitsky BR. Indirect repression by Bacillus subtilis CodY via displacement of the activator of the proline utilization operon. J Mol Biol. 2011;413(2):321–336. doi: 10.1016/j.jmb.2011.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Belitsky BR, Sonenshein AL. Roadblock repression of transcription by Bacillus subtilis CodY. J Mol Biol. 2011;411(4):729–743. doi: 10.1016/j.jmb.2011.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Preis H, Eckart RA, Gudipati RK, Heidrich N, Brantl S. CodY activates transcription of a small RNA in Bacillus subtilis. J Bacteriol. 2009;191(17):5446–5457. doi: 10.1128/JB.00602-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Belitsky BR, Sonenshein AL. Genome-wide identification of Bacillus subtilis CodY-binding sites at single-nucleotide resolution. Proc Natl Acad Sci USA. 2013;110(17):7026–7031. doi: 10.1073/pnas.1300428110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marciniak BC, et al. High- and low-affinity cre boxes for CcpA binding in Bacillus subtilis revealed by genome-wide analysis. BMC Genomics. 2012;13:401. doi: 10.1186/1471-2164-13-401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Novick A, Weiner M. Enzyme induction as an all-or-none phenomenon. Proc Natl Acad Sci USA. 1957;43(7):553–566. doi: 10.1073/pnas.43.7.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Leyn SA, et al. Genomic reconstruction of the transcriptional regulatory network in Bacillus subtilis. J Bacteriol. 2013;195(11):2463–2473. doi: 10.1128/JB.00140-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Atkinson MR, Wray LV, Jr, Fisher SH. Regulation of histidine and proline degradation enzymes by amino acid availability in Bacillus subtilis. J Bacteriol. 1990;172(9):4758–4765. doi: 10.1128/jb.172.9.4758-4765.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sorek R, Cossart P. Prokaryotic transcriptomics: A new view on regulation, physiology and pathogenicity. Nat Rev Genet. 2010;11(1):9–16. doi: 10.1038/nrg2695. [DOI] [PubMed] [Google Scholar]
- 35.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11(10):R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sekowska A, Danchin A. The methionine salvage pathway in Bacillus subtilis. BMC Microbiol. 2002;2(8):8. doi: 10.1186/1471-2180-2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Belitsky BR, Sonenshein AL. Contributions of multiple binding sites and effector-independent binding to CodY-mediated regulation in Bacillus subtilis. J Bacteriol. 2011;193(2):473–484. doi: 10.1128/JB.01151-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Villapakkam AC, et al. Genetic and biochemical analysis of the interaction of Bacillus subtilis CodY with branched-chain amino acids. J Bacteriol. 2009;191(22):6865–6876. doi: 10.1128/JB.00818-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bushel PR, Wolfinger RD, Gibson G. Simultaneous clustering of gene expression data with clinical chemistry and pathological evaluations reveals phenotypic prototypes. BMC Syst Biol. 2007;1(15):15. doi: 10.1186/1752-0509-1-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Datta S, Datta S. Comparisons and validation of statistical clustering techniques for microarray gene expression data. Bioinformatics. 2003;19(4):459–466. doi: 10.1093/bioinformatics/btg025. [DOI] [PubMed] [Google Scholar]
- 41.Tavazoie S, Hughes JD, Campbell MJ, Cho RJ, Church GM. Systematic determination of genetic network architecture. Nat Genet. 1999;22(3):281–285. doi: 10.1038/10343. [DOI] [PubMed] [Google Scholar]
- 42.Shivers RP, Sonenshein AL. Bacillus subtilis ilvB operon: An intersection of global regulons. Mol Microbiol. 2005;56(6):1549–1559. doi: 10.1111/j.1365-2958.2005.04634.x. [DOI] [PubMed] [Google Scholar]
- 43.Sonenshein AL. Control of key metabolic intersections in Bacillus subtilis. Nat Rev Microbiol. 2007;5(12):917–927. doi: 10.1038/nrmicro1772. [DOI] [PubMed] [Google Scholar]
- 44.Belitsky BR, Sonenshein AL. Modulation of activity of Bacillus subtilis regulatory proteins GltC and TnrA by glutamate dehydrogenase. J Bacteriol. 2004;186(11):3399–3407. doi: 10.1128/JB.186.11.3399-3407.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Berger BJ, English S, Chan G, Knodel MH. Methionine regeneration and aminotransferases in Bacillus subtilis, Bacillus cereus, and Bacillus anthracis. J Bacteriol. 2003;185(8):2418–2431. doi: 10.1128/JB.185.8.2418-2431.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Débarbouillé M, Gardan R, Arnaud M, Rapoport G. Role of BkdR, a transcriptional activator of the SigL-dependent isoleucine and valine degradation pathway in Bacillus subtilis. J Bacteriol. 1999;181(7):2059–2066. doi: 10.1128/jb.181.7.2059-2066.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thomaides HB, et al. Essential bacterial functions encoded by gene pairs. J Bacteriol. 2007;189(2):591–602. doi: 10.1128/JB.01381-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yoshida K, et al. Systematic study of gene expression and transcription organization in the gntZ-ywaA region of the Bacillus subtilis genome. Microbiology. 2000;146(Pt 3):573–579. doi: 10.1099/00221287-146-3-573. [DOI] [PubMed] [Google Scholar]
- 49.Brückner R, Titgemeyer F. Carbon catabolite repression in bacteria: Choice of the carbon source and autoregulatory limitation of sugar utilization. FEMS Microbiol Lett. 2002;209(2):141–148. doi: 10.1111/j.1574-6968.2002.tb11123.x. [DOI] [PubMed] [Google Scholar]
- 50.Henkin TM, Grundy FJ, Nicholson WL, Chambliss GH. Catabolite repression of alpha-amylase gene expression in Bacillus subtilis involves a trans-acting gene product homologous to the Escherichia coli lacI and galR repressors. Mol Microbiol. 1991;5(3):575–584. doi: 10.1111/j.1365-2958.1991.tb00728.x. [DOI] [PubMed] [Google Scholar]
- 51.Kleijn RJ, et al. Metabolic fluxes during strong carbon catabolite repression by malate in Bacillus subtilis. J Biol Chem. 2010;285(3):1587–1596. doi: 10.1074/jbc.M109.061747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nelson SO, Wright JK, Postma PW. The mechanism of inducer exclusion. Direct interaction between purified III of the phosphoenolpyruvate:sugar phosphotransferase system and the lactose carrier of Escherichia coli. EMBO J. 1983;2(5):715–720. doi: 10.1002/j.1460-2075.1983.tb01490.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cruz Ramos H, et al. Fermentative metabolism of Bacillus subtilis: Physiology and regulation of gene expression. J Bacteriol. 2000;182(11):3072–3080. doi: 10.1128/jb.182.11.3072-3080.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Grundy FJ, Waters DA, Takova TY, Henkin TM. Identification of genes involved in utilization of acetate and acetoin in Bacillus subtilis. Mol Microbiol. 1993;10(2):259–271. doi: 10.1111/j.1365-2958.1993.tb01952.x. [DOI] [PubMed] [Google Scholar]
- 55.Chai Y, Kolter R, Losick R. A widely conserved gene cluster required for lactate utilization in Bacillus subtilis and its involvement in biofilm formation. J Bacteriol. 2009;191(8):2423–2430. doi: 10.1128/JB.01464-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kim H-J, et al. Complex regulation of the Bacillus subtilis aconitase gene. J Bacteriol. 2003;185(5):1672–1680. doi: 10.1128/JB.185.5.1672-1680.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wünsche A, et al. CcpA forms complexes with CodY and RpoA in Bacillus subtilis. FEBS J. 2012;279(12):2201–2214. doi: 10.1111/j.1742-4658.2012.08604.x. [DOI] [PubMed] [Google Scholar]
- 58.Tojo S, et al. Elaborate transcription regulation of the Bacillus subtilis ilv-leu operon involved in the biosynthesis of branched-chain amino acids through global regulators of CcpA, CodY and TnrA. Mol Microbiol. 2005;56(6):1560–1573. doi: 10.1111/j.1365-2958.2005.04635.x. [DOI] [PubMed] [Google Scholar]
- 59.Sambrook J, Russell DW. Molecular Cloning: A Laboratory Manual. 3rd Ed. Plainview, NY: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- 60.Belitsky BR, Sonenshein AL. Role and regulation of Bacillus subtilis glutamate dehydrogenase genes. J Bacteriol. 1998;180(23):6298–6305. doi: 10.1128/jb.180.23.6298-6305.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Giannoukos G, et al. Efficient and robust RNA-seq process for cultured bacteria and complex community transcriptomes. Genome Biol. 2012;13(3):R23. doi: 10.1186/gb-2012-13-3-r23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mandlik A, et al. RNA-Seq-based monitoring of infection-linked changes in Vibrio cholerae gene expression. Cell Host Microbe. 2011;10(2):165–174. doi: 10.1016/j.chom.2011.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Weisenberg SA, Butterfield TR, Fischer SM, Rhee KY. Suitability of silica hydride stationary phase, aqueous normal phase chromatography for untargeted metabolomic profiling of Enterococcus faecium and Staphylococcus aureus. J Sep Sci. 2009;32(13):2262–2265. doi: 10.1002/jssc.200900256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Stat Soc Series B Stat Methodol. 1995;57(1):289–300. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.