Significance
Microbial pathogens cause food and water-borne disease through infection of the intestine. Many of these pathogens invade intestinal cells to replicate, but they ultimately need to exit out of these cells to spread to new hosts. We use the transparent nematode Caenorhabditis elegans to show that a natural intracellular pathogen called Nematocida parisii escapes from intestinal cells by using host trafficking pathways and a host small GTPase protein called RAB-11 for directional exocytosis into the lumen. N. parisii belongs to a large phylum of pathogens called microsporidia that can infect all animals, including humans, so our findings may be broadly applicable to understanding how agriculturally and medically relevant diseases are spread.
Abstract
Pathogen exit is a key stage in the spread and propagation of infectious disease, with the fecal-oral route being a common mode of disease transmission. However, it is poorly understood which molecular pathways provide the major modes for intracellular pathogen exit and fecal-oral transmission in vivo. Here, we use the transparent nematode Caenorhabditis elegans to investigate intestinal cell exit and fecal-oral transmission by the natural intracellular pathogen Nematocida parisii, which is a recently identified species of microsporidia. We show that N. parisii exits from polarized host intestinal cells by co-opting the host vesicle trafficking system and escaping into the lumen. Using a genetic screen, we identified components of the host endocytic recycling pathway that are required for N. parisii spore exit via exocytosis. In particular, we show that the small GTPase RAB-11 localizes to apical spores, is required for spore-containing compartments to fuse with the apical plasma membrane, and is required for spore exit. In addition, we find that RAB-11–deficient animals exhibit impaired contagiousness, supporting an in vivo role for this host trafficking factor in microsporidia disease transmission. Altogether, these findings provide an in vivo example of the major mode of exit used by a natural pathogen for disease spread via fecal-oral transmission.
After invasion and replication inside of host cells, intracellular pathogens must escape back into the environment to find new hosts and propagate disease. Although pathogen exit is not as well understood as pathogen entry, there are a variety of exit strategies that have been described recently, including both lytic and nonlytic egress from host cells (1, 2). These studies in tissue culture cells have identified a diversity of host pathways and processes in pathogen exit. In some cases, multiple modes of exit have been implicated for the same pathogen, although it is still poorly understood which modes of exit are crucial for disease transmission of any microbial pathogen in a whole animal host.
One of the major sites for pathogen infection in animals is the intestine, which can be invaded by intracellular pathogens that cause food and water-borne disease (3). These pathogens must exit from intestinal cells back into the lumen to be released by defecation for disease transmission. Intestinal pathogens are often studied in tissue culture models, but their life cycles can proceed differently in intact intestinal cells. For example, the bacterial pathogen Listeria monocytogenes is initially entrapped in a membrane-bound compartment after entering host cells. When infecting tissue culture cells, Listeria escapes from this compartment into the host cytosol, and then spreads between cells by a protrusion mechanism (4). In contrast, it uses a different strategy in vivo. When infecting mice, Listeria does not escape from the membrane-bound compartment and instead remains membrane-bound, ultimately exiting basolaterally from intestinal cells by exocytosis to spread systemically into the host (5). However, it is not known how Listeria escapes back into the intestinal lumen for transmission to new hosts. These differing results of Listeria trafficking in vitro and in vivo highlight the importance of investigating pathogen exit in a whole-animal host.
The nematode C. elegans provides an accessible whole-animal host in which to dissect the life cycle and transmission of intestinal pathogens (6, 7). The C. elegans intestine consists of 20 nonrenewable epithelial cells that share many morphological similarities with mammalian intestinal epithelial cells (Fig. 1A). These cells are polarized, with microvilli on the apical side facing the lumen of the intestinal tract. Like in humans, these microvilli are anchored into a cytoskeletal structure called the terminal web that is composed of actin and intermediate filaments (8). C. elegans is transparent, which facilitates analysis of infection in these cells within intact animals. We recently described a natural intracellular pathogen that infects the C. elegans intestine and showed that this pathogen defines a new genus and species of microsporidia, which are obligate, fungal-related intracellular pathogens (9, 10). The microsporidia phylum comprises more than 1,400 species of pathogens that can infect a wide variety of animals including humans, where they commonly infect the intestine and can cause lethal diarrhea in immunocompromised hosts (11, 12). We named the C. elegans-infecting species of microsporidia Nematocida parisii, or nematode killer from Paris, because it was isolated from wild-caught C. elegans in a compost pit near Paris and it eventually kills its host. Wild-caught Caenorhabditis nematodes infected with microsporidia have been isolated from environmental regions around the globe, indicating that microsporidia are a common cause of infection for C. elegans in the wild (9, 13).
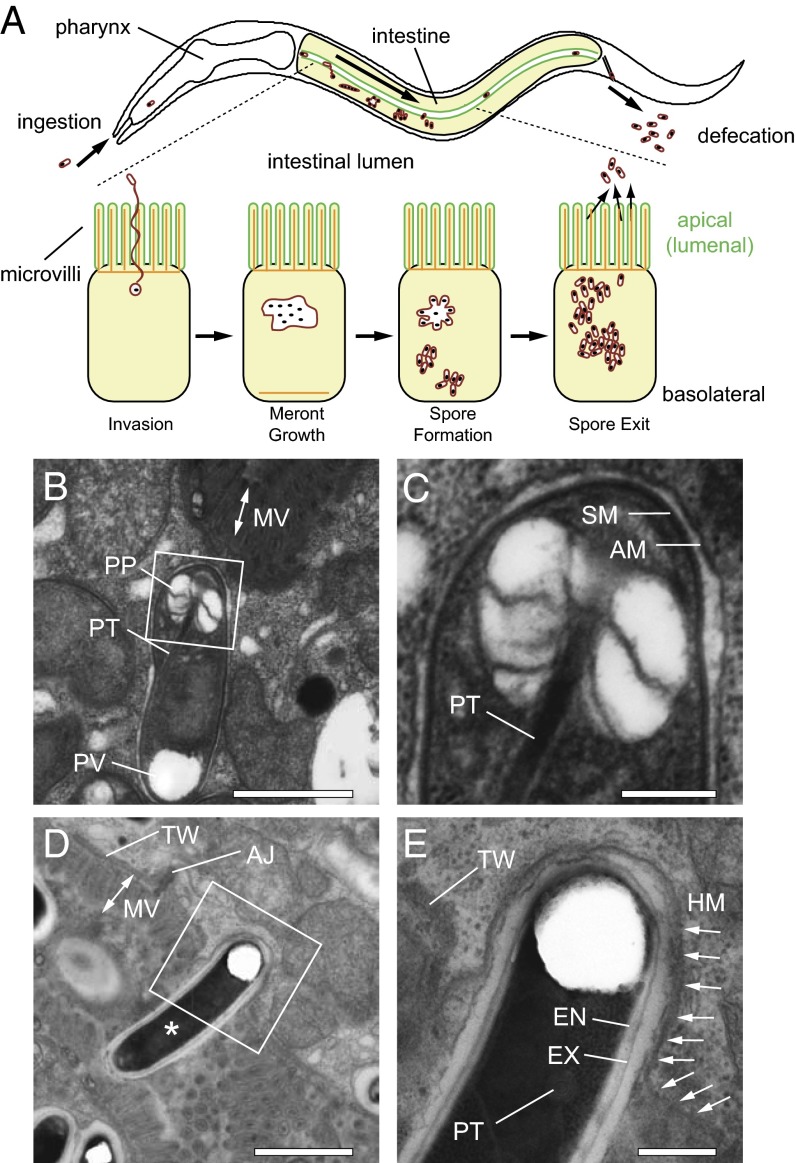
Fig. 1.
N. parisii spores are contained in an additional membrane-bound compartment inside C. elegans intestinal cells. (A) The C. elegans intestine (in yellow) is comprised of polarized epithelial cells. The apical side (green) faces the lumen and contains actin-rich microvilli anchored into a terminal web (actin labeled orange). Ingested N. parisii spores invade intestinal cells by using a polar tube that delivers spore contents directly in host cytosol, where it replicates in the multinucleate meront stage. The pathogen eventually develops back into the spore form, which exits back into the lumen for defecation from the animal. Arrow within intestine indicates the progression of infection over time. (B) TEM of an immature N. parisii spore. (C) Magnification of box from B shows the additional membrane (AM) that surrounds the spore membrane (SM). (D) A mature spore (asterisk) spanning the luminal membrane. (E) Magnification of box from D. Arrows indicate continuous host apical membrane (HM) that surrounds the spore. (B–E) AJ, apical junction; EN, endospore; EX, exospore; MV, microvilli; PP, polaroplast; PT, polar tube; PV, polar vacuole; TW, terminal web. (Scale bars: B and D, 1 µm; C and E, 500 nm.)
Microsporidia are obligate intracellular pathogens, meaning they must be inside of a host cell to replicate. Microsporidia survive outside the host as transmissible spores, which deploy a dramatic invasion mechanism to enter host cells. These spores contain a specialized structure called a polar tube, which “fires” to pierce a host cell, and then the parasite nucleus and sporoplasm are injected through this tube directly into the host cytosol (Fig. 1A). In the case of N. parisii and in most microsporidia species, the pathogen then grows inside the host cell in a replicative meront form that appears to be in direct contact with the host cytosol (9, 14). N. parisii meronts become large and multinucleate as they develop and eventually differentiate back into mononucleate cells that become spores, which are shed to transmit the infection (9).
How do N. parisii spores exit from host intestinal cells? Previously we showed that extensive cytoskeletal rearrangements occur during the meront stage, with relocalization of C. elegans actin from the apical to the basolateral side of intestinal cells (15). This relocalization appears to trigger gap formation in the terminal web as a mechanism to remove this barrier to pathogen exit. We found that N. parisii spore exit is nonlytic and that spores do not bud out of the host cell, because spores are released into the lumen free of host membrane. However, the precise exit mechanism was not clear. Our results were consistent with either an exocytic mechanism, which would involve membrane-bound spores fusing with the host plasma membrane, or an ejectosome mechanism, which would involve an actin-rich structure at the plasma membrane that delivers microbes directly from the cytosol into the extracellular environment (16). Here, we show that N. parisii spores are in a separate membrane-bound compartment, supporting an exocytic mode of exit. We performed a screen for C. elegans small GTPases required for N. parisii spore exit and found that components of the endocytic recycling pathway are critical for exit, with the RAB-11 protein acting as a key player in this process. RAB-11 localizes to apically polarized spores, is required for spore-containing compartments to fuse with the apical plasma membrane, and is critical for pathogen exit and for contagiousness. Altogether these studies define endocytic recycling and exocytosis to be key processes for microsporidia exit, and the major mechanism of exit for fecal-oral transmission of an intestinal pathogen in vivo.
Results
N. parisii Spores Are Contained in Separate Compartments Inside of Host Intestinal Cells.
As noted above, our previous findings implied that N. parisii exit involves either an ejectosome-like mechanism, or an exocytic event in which an intracellular compartment containing a spore fuses with the host apical membrane. To distinguish between these possibilities, we used transmission electron microscopy (TEM) to examine whether there is membrane localization around newly replicated N. parisii spores while inside of C. elegans intestinal cells. Indeed, we found that intracellular spores were always contained in an additional membrane (n = 42 spores examined; immature intracellular spore shown in Fig. 1 B and C and more mature spores shown in Fig. S1 A–D). In addition, we observed plasma membrane surrounding mature spores that appear to be in the process of exiting through host microvilli into the lumen. Occasionally we were able to see that the membrane around these exiting spores is continuous with the host plasma membrane, indicating that the spore-containing compartment (SCC) is fusing with the apical membrane of the host intestine (Fig. 1 D and E).
N. parisii Spore-Containing Compartments Fuse with Host Membrane To Access the Intestinal Lumen.
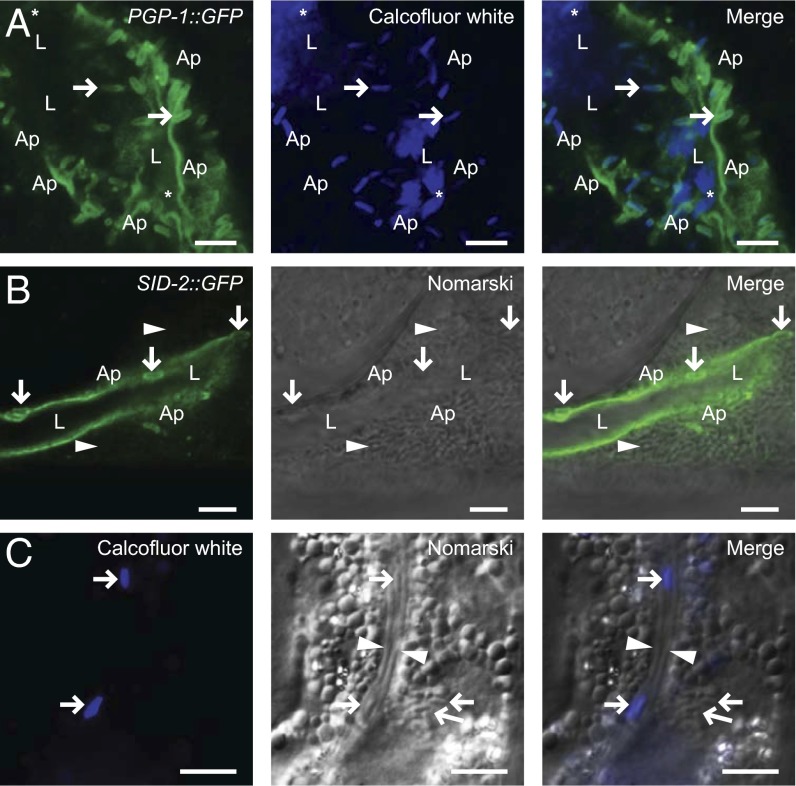
To confirm that the additional membrane around apically fused N. parisii spores was continuous with host-derived membrane, we examined the localization of GFP-tagged C. elegans apical membrane markers in infected animals. We infected animals expressing the apical membrane transporter protein PGP-1::GFP with N. parisii spores, let the infection progress to generate new spores, and then looked at these spores to determine whether they colocalized with the host GFP-tagged membrane (17). Indeed, we found that PGP-1::GFP colocalizes with SCCs fused along the apical cell surface that appear to be exiting into the lumen (Fig. 2A). Similarly, the apical plasma membrane marker SID-2::GFP (18) also localizes to spores along the apical cell surface (Fig. 2B). However, we did not observe colocalization of the apical membrane transporter PEPT-1::DsRed (19) with N. parisii spores (Fig. S1E), suggesting that some membrane markers may not mix as easily with the N. parisii-containing compartments, or that there is heterogeneity of protein components in the plasma membrane regions where N. parisii exits. These experiments suggest that N. parisii spores arrive at the apical membrane in compartments that fuse with the apical plasma membrane of C. elegans intestinal cells.
Fig. 2.
N. parisii spores are surrounded by host apical membrane and have access to the intestinal lumen. (A) PGP-1::GFP localizes to SCCs fused with host apical membrane. Calcofluor white stains spores in contact with the lumen. Asterisk marks spores free in the lumen, rightward facing arrows mark putative exiting spores that stain with Calcofluor white and PGP-1::GFP. (B) SID-2::GFP surrounds SCCs fused to apical membrane (downward arrows). Arrowheads indicate intracellular spores not stained by Calcofluor white. Ap, apical; L, lumen. (C) Calcofluor white does not stain intracellular spores (leftward facing arrows), but does stain spores in lumen (rightward facing arrows). Lumen is indicated by arrowheads. (Scale bars: A and B, 5 µm; C, 10 µm.)
After fusion with the plasma membrane, the cargo inside an exocytic vesicle should have access to the luminal environment via the fusion pore. We therefore tested whether spores marked with apical membrane marker were in contact with the intestinal lumen by feeding infected animals the cell-impermeable dye Calcofluor white, which binds chitin found in the spore walls of microsporidia. When fed to C. elegans, Calcofluor white only stains spores in contact with the lumen, and not spores inside of intestinal cells (Fig. 2C). We found that spores cloaked in the apical membrane marker PGP-1::GFP also were marked with Calcofluor white (Fig. 2A). Taken together, these results show that intracellular spores are contained in compartments that are able to fuse with the host apical membrane and gain access to the intestinal lumen for exit.
The results described above, together with our previous results (9), indicate that N. parisii replicates in direct contact with the cytosol during its meront stage, and then is contained in exocytic compartments at the differentiated spore stage. One likely candidate pathway to mediate this transition from membrane-free to membrane-bound is the autophagy pathway (20). To determine whether the autophagy pathway is important for N. parisii exit from the host, we knocked down expression of key host autophagy components using feeding RNAi, infected these animals, and then quantified spore exit. Previously, we demonstrated that an individual infected C. elegans animal can shed thousands of N. parisii spores per hour, and that spores exit only from the apical side of host cells (15). To measure this exit, we use spore-shedding assays starting at 40 h postinoculation (hpi), when N. parisii has differentiated from meronts into spores that are exiting from cells into the lumen and are being defecated out by the host (Fig. S2A). Using this assay, we found that RNAi against the autophagy components atg-18, bec-1, and lgg-1 had no significant effect on spore exit (Fig. S2B). The RNAi clones appeared to knock down their targets, as assessed by a loss of GFP::LGG-1 protein in the intestine upon lgg-1 RNAi treatment (Fig. S2C), and a reduction of GFP::LGG-1 puncta (a standard measure of autophagy) upon atg-18 and bec-1 RNAi (Fig. S2D). Thus, these results suggest that autophagy is not important for N. parisii spore maturation and exit.
Small GTPases Involved in Apical Endocytic Recycling Are Required for Spore Exit.
To identify which host factors are required for directional exit of N. parisii from intestinal cells, we conducted a feeding RNAi screen of 41 predicted small GTPases (smGTPases) in C. elegans, because these factors are involved in many intracellular trafficking events (21). Again we used spore-shedding assays to measure N. parisii exit (Fig. S2A). Knockdown of genes that caused developmental defects in animals were excluded from analysis (ran-1, sar-1) or diluted 10-fold with L4440 empty-vector bacteria (rab-5, rab-11.1) to achieve normal development of animals assayed for pathogen exit.
We found that of the 41 genes tested, RNAi knockdown of the smGTPases rab-2, rab-5, rab-10, rab-11.1, rab-18, arf-3, arl-5, and ral-1 caused significant defects in spore shedding (Fig. S3). These hits from our screen were reanalyzed in independent experiments to confirm the screen results (Fig. 3A). Notably, most of these screen hits (rab-2, rab-5, rab-10, rab-11.1, and rab-18) are implicated in apical recycling pathways, which can direct intracellular cargo up to the apical membrane (22, 23). To ensure that RNAi clones that block spore exit do not simply block N. parisii spore development, we examined whether RNAi treatment impaired spore production by using two different methods. First, we examined individual animals semiquantitatively for the amount of intracellular spores produced, and we found no change after any RNAi treatment (Fig. 3B) (Friedman’s test; ANOVA P = 0.6081). In addition, we lysed infected animals to quantitatively assess the number of intracellular spores produced on a population level and again found no difference (Fig. 3C). Furthermore, because a defect in spore shedding could also be due to a global disruption of cell polarity, which then could cause a disruption in intracellular trafficking pathways, we also examined whether knocking down these smGTPases affected cell polarity. We analyzed the polarized distribution of the apically localized protein PGP-1::GFP and found its localization to be unaffected by any of the RNAi treatments that cause spore shedding defects (Fig. S4). Altogether, these results indicate that blocking expression of these smGTPase proteins does not disrupt spore formation or cell polarity and, instead, disrupts spore exit from the animal.
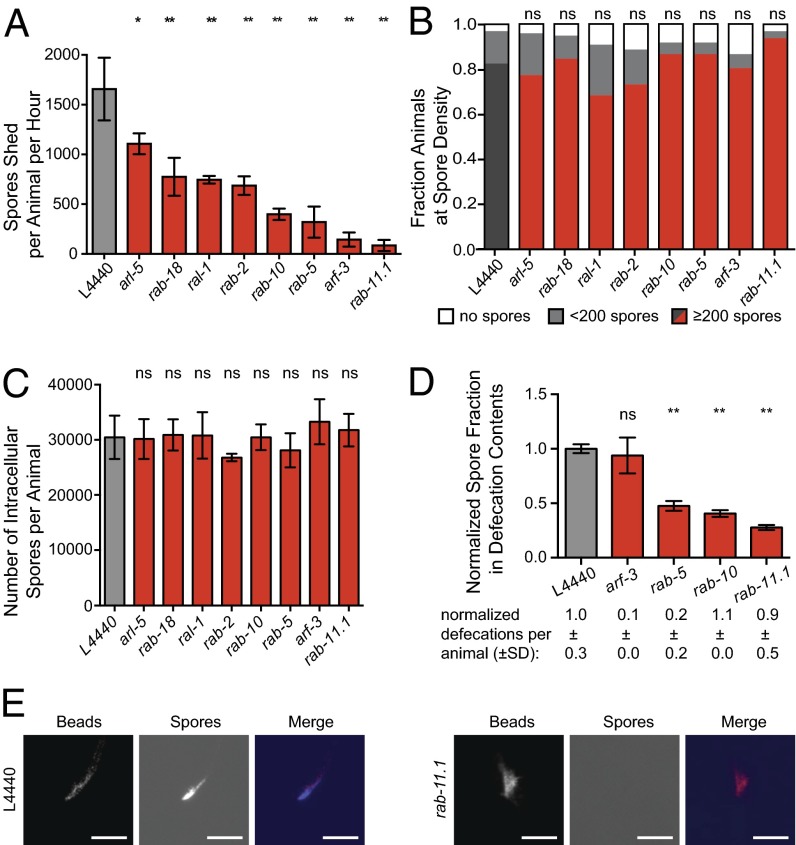
Fig. 3.
Host small GTPases in the apical endocytic recycling pathway are required for spore exit. (A) Quantification of spore shedding defects in RNAi hits. Average is from three biological replicates; error bars are SD. Experiments were repeated three independent times for rab-5, rab-10, and rab-11 and two independent times for other genes. (B) Spore production in individual assay. Dark gray or red is the fraction of animals with ≥200 spores per animal; light gray with <200 spores per animal; and white with no spores. n = 40 animals per treatment group. (C) Spore production in population assay. y axis indicates the number of intracellular spores per animal after lysis at 44 hpi. Error bars are SD. (D) Defecation contents assay. y axis indicates the normalized contribution of fluorescent signal from spores in defecated material. Average is from three independent experiments (except for arf-3, which was tested twice), error bars are SEM. See Materials and Methods for more detail. Number of defecation events is the average from 48 to 96 animals, ± SD. (E) Representative images of defecated material quantified in D showing fluorescent beads and Calcofluor white spores shed by animals. (Scale bars: 50 µm.)
An additional explanation for reduced spore shedding is that animals are simply defecating less overall volume, which then leads to fewer spores being shed. Accurately measuring the volume defecated by a tiny animal such as C. elegans is difficult, and quantifying the number of spores present in the lumen before defecation is challenging and is also confounded by defecation defects. Therefore, to investigate spore exit into the lumen, we developed an assay to measure the density of spores within the contents defecated out of the lumen. We pulse-fed infected animals fluorescent beads and then measured the ratio of spores to fluorescent beads in the defecated material. With this assay, we found a significantly reduced ratio of spores to beads in the contents defecated from rab-5, rab-10, and rab-11.1-defective animals (Fig. 3 D and E). Therefore, knockdown of these smGTPases causes a spore shedding defect because of a defect in N. parisii spore egress into the lumen, and not simply because of a reduction in defecation volume. In contrast, the spore-shedding defect of arf-3–defective animals appears to be solely due to a defecation defect, because the spore density in these defecation contents was similar to that of control animals and the number of defecations per animal was much smaller (Fig. 3D). Taken together, these experiments confirm the importance of the apical recycling components RAB-5, RAB-10, and RAB-11 for spore egress from intestinal cells into the intestinal lumen.
RAB-11 Localizes to Spores near the Apical Side of Intestinal Cells.
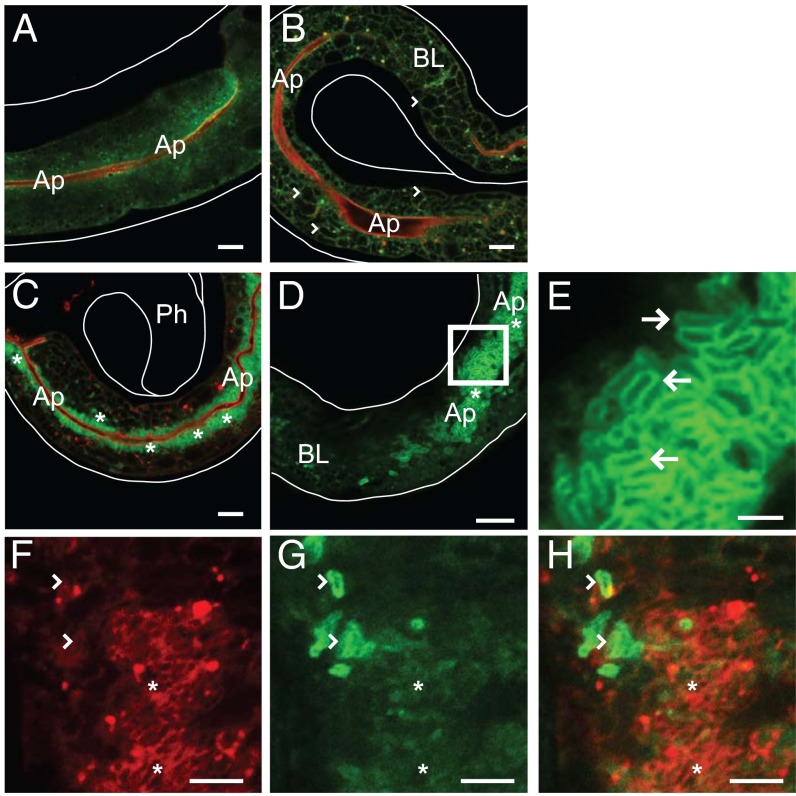
Because endocytic recycling appears to be a pathway required for spore exit, we next examined whether apical endocytic recycling factors localize to N. parisii spores. The Rab11 subfamily comprises Rab11a, Rab11b, and Rab25 in mammals, and they are well-known markers of recycling endosomes, with Rab11a in particular shown to play a key role in targeting endosomes to the apical plasma membrane (24–28). C. elegans has two RAB-11 subfamily members called RAB-11.1 and RAB-11.2, which both are most similar to mammalian Rab11a. C. elegans RAB-11.1 has been studied in intestinal cell trafficking, where it is apically localized (19, 22) (Fig. 4A). Because the rab-11.1 RNAi clone caused a stronger block in spore shedding than did rab-11.2, and because the rab-11.2 knockdown also reduces RAB-11.1 expression levels (Fig. S5) likely due to the high sequence similarity between these genes, we chose to focus on RAB-11.1. We examined RAB-11.1 localization (hereafter referred to as RAB-11) by infecting transgenic animals expressing GFP::RAB-11 (19) with N. parisii. In the intestinal cells of infected meront-stage animals, we found that GFP::RAB-11 is still apically enriched, is primarily cytosolic, and is excluded from pathogen tissue (Fig. 4B). Strikingly, however, when spores are being shed at approximately 41 hpi, we find that vast numbers of apically localized spores are coated in GFP::RAB-11 (Fig. 4 C–E). These spores are crowded around the lumen in all dimensions, always densest along the apical face of the intestinal cells. To ensure that the localization of RAB-11 to spores was not an artifact of this transgene, we confirmed that a separate RFP::RAB-11 transgene also localizes to N. parisii spores (22) (Fig. 4F). In addition, we performed immunohistochemistry by using an antibody directed against endogenous RAB-11 (29) in N2 wild-type animals, and in animals expressing GFP::RAB-11. We observed localization of RAB-11 antibody to spores in infected animals, which colocalized with the GFP::RAB-11 transgene marker (Fig. S6 A–C). Interestingly, the vast majority of RAB-11–coated SCCs do not appear to have PGP-1::GFP on them, but it appears that some PGP-1::GFP SCCs also exhibit RAB-11 staining, suggesting that SCCs may undergo a transition from RAB-11 coats to PGP-1 coats as they fuse with the apical plasma membrane (Fig. 4 F–H).
Fig. 4.
RAB-11 protein forms coats on apically localized intracellular spores. Uninfected (A), meront-infected (B), and spore-infected (C–E) transgenic animals expressing GFP::RAB-11 in green and apical marker mCherry::ACT-5 in red. (B) Arrowheads mark examples of meronts, which are dark because of the exclusion of GFP::RAB-11. (C) GFP::RAB-11 localizes to spores near the apical cell surface (asterisks). Ph, pharynx. (D) Spore-infected animal. Region enclosed by box is expanded in E. Ap, apical; BL, basolateral, and white lines outline the animals. (E) Arrows point to rod-shaped spores coated in GFP::RAB-11. (F) Arrowheads mark RFP::RAB-11 coated spores that are also marked by PGP-1::GFP (G). Asterisks denote spores with RFP::RAB-11 but not PGP-1::GFP. (H) Merged image. (Scale bars: A–D, 10 µm; E, 2.5 µm; F–H, 5 µm.)
We also investigated the localization of RAB-5 (early endosomes) and RAB-10 (Golgi and recycling endosomes) to N. parisii spores by using fluorescently tagged versions of these proteins (22). As in other organisms, RAB-5 and RAB-10 are known to play active roles in intracellular transport in C. elegans, including apical recycling in the intestine and neuropeptide secretion in neurons (22, 30). Although both of these proteins are required for N. parisii spore egress, we did not find these proteins localized to spores in the intestine (Fig. S7).
RAB-11 Is Required for Spore-Containing Compartment Fusion with the Apical Membrane, and Contagiousness of Infected Animals.
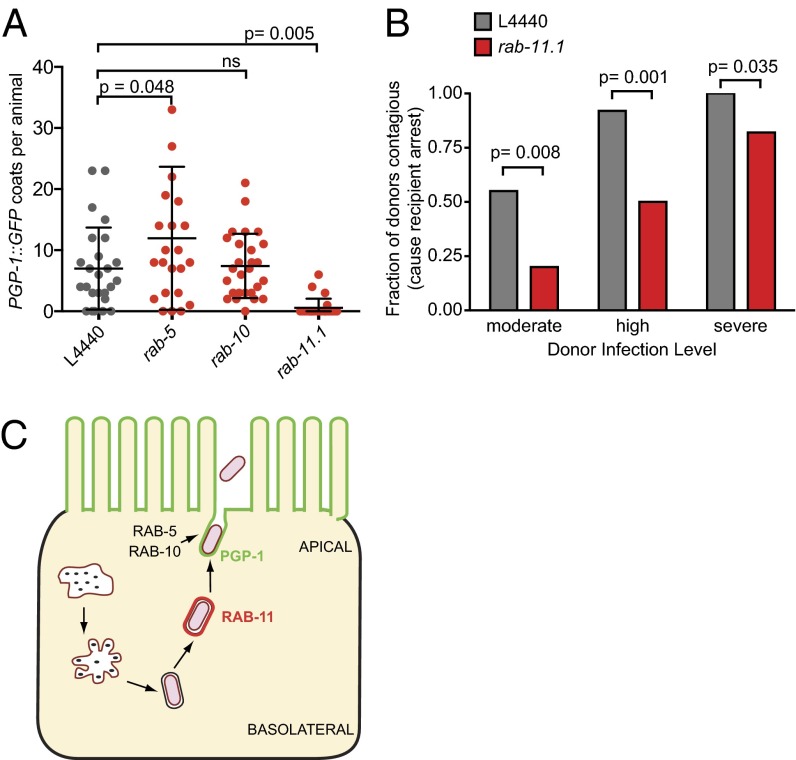
The above localization results indicate that RAB-11 acts directly on the N. parisii exit process, whereas RAB-5 and RAB-10 act more indirectly. If N. parisii spores use RAB-11 membrane compartments to traffic to the apical cell surface, we would expect that blocking RAB-11 function would decrease the number of SCCs fusing with the host apical membrane. To test this hypothesis, we compared the number of apically fused SCCs that are marked with PGP-1::GFP (as shown in Fig. 2A) in L4440 empty vector control and rab-11.1 RNAi-treated animals. Indeed, we found that depleting RAB-11 caused a near complete block in the number of SCCs that fused with the apical membrane (Fig. 5A). We also quantified the apically fused SCCs as assessed with PGP-1::GFP coats in rab-5 and rab-10 RNAi-treated animals and found there was not a block in fusion with the apical membrane. Similarly, there was not a substantial block in the formation of GFP::RAB-11 coats in rab-5 or rab-10 RNAi-treated animals (Fig. S6D). These results indicate that whereas N. parisii relies directly on RAB-11 to achieve apical fusion, RAB-5 and RAB-10 appear to either act in parallel to or downstream of fusion.
Fig. 5.
RAB-11 is required for spore fusion and contagiousness. (A) rab-11.1 RNAi blocks fusion of SCCs with apical plasma membrane, as assessed by PGP-1::GFP membrane-coated spores. rab-5 and rab-10 RNAi do not significantly impact the number of PGP-1::GFP-coated spores. Each point represents the number of PGP-1::GFP coats in a single animal. n = 30 animals per treatment group. Average is shown with error bars as SD. (B) rab-11.1 RNAi-treated animals are less contagious than control animals. Fraction of infected donor animals causing symptoms of severe infection in recipients is shown for each treatment group. n = 24–41 donor animals per infection level per RNAi treatment group (n = 184 animals total). (C) Model for RAB-11-mediated directional exocytosis of N. parisii spores from C. elegans intestinal cells.
We next determined the functional role of RAB-11 on microsporidia disease transmission by conducting contagiousness assays with rab-11.1 RNAi-treated animals (Fig. S8). These experiments demonstrated that RAB-11–defective animals are less contagious than control animals (Fig. 5B). One trivial explanation for this reduced transmission could be that rab-11.1 RNAi-treated animals die more rapidly than control. However, there was only a slight decrease in survival of rab-11.1 RNAi-treated animals compared with controls, and this decrease was not caused by infection (Fig. S9). Altogether, these results demonstrate that host RAB-11 is required for fecal-oral transmission of N. parisii to new hosts.
Discussion
Our studies indicate that a major mode of N. parisii spore exit from intestinal cells is to traffic through RAB-11–marked endocytic recycling compartments, followed by polarized exocytosis into the lumen for fecal-oral transmission (Fig. 5C). In particular, we found that RAB-11 is required for SCC fusion with the apical membrane of intestinal cells, whereas RAB-5 and RAB-10 act in parallel to or downstream of fusion. Although RAB-5 and RAB-10 can act upstream of RAB-11 and vesicle fusion in apical recycling events in other systems, they have also been shown to act downstream of vesicle fusion, such as in the C. elegans nervous system where they are required for neuropeptide release from dense core vesicles, but are not required for fusion (30). Thus, RAB-5 and RAB-10 may similarly act downstream of fusion for N. parisii release, whereas RAB-11 acts upstream of fusion to promote directional exit for fecal-oral transmission of infection.
Rab11 has been well characterized in many systems as the master regulator of endocytic recycling, responsible for directing intracellular cargo to the apical cell surface by acting as an upstream mediator of exocytic trafficking pathways and vesicle fusion (24, 31). Rab11 is apically localized in many polarized epithelial cells, which could aid intracellular pathogens that need to egress from such cells in a directional manner to exit from hosts. Not only intestinal pathogens, but also respiratory and urogenital pathogens, need to egress out the apical side of epithelial cells for transmission to new hosts. Interestingly, a variety of respiratory viruses such as influenza virus have been shown to use host Rab11 for trafficking of viral particles to the plasma membrane (32). Although these viruses ultimately bud from the membrane instead of exiting through exocytosis, trafficking through the host cell by Rab11 could provide apical directionality to viral egress from polarized lung epithelial cells in vivo.
How general is the N. parisii polarized exocytosis strategy likely to be for other microbial pathogens? An exocytosis-type process termed “vomocytosis” has been implicated in the exit of fungal pathogens from macrophages (33, 34), although this process is likely distinct from the polarized exocytosis used to escape from epithelial cells. Whereas exocytosis has been shown to contribute to bacterial pathogen exit in mammalian tissue culture systems of epithelial cells (35), multiple modes of exit have been proposed for the same pathogen, and the in vivo significance and contribution of exocytosis to disease transmission has not been clear. In contrast to the C. elegans intestine, the mammalian intestine regularly renews and sheds epithelial cells. Exiting through these extruded cells appears to be an exit strategy for Salmonella in vivo (36), so it is possible that mammalian pathogens do not need to exit from intact intestinal cells via exocytosis. However, doing so would greatly accelerate their transmission, because intestinal cell shedding occurs on the timescale of days (37), whereas doubling time of many bacterial pathogens occurs on the timescale of minutes. Therefore, pathogens that egress via exocytosis could have a large advantage in terms of doubling time over pathogens that wait for extrusion. Thus, the apical recycling exit process that we have found as critical for exit of N. parisii may also be important for exit, and dissemination, of pathogens that use a fecal-oral route of transmission. In particular, N. parisii belongs to the microsporidia phylum, which includes a large number of pathogen species that commonly infect and replicate in the intestine. For example, Enterocytozoon bieneusi, which has been responsible for lethal diarrhea in AIDS patients, appears to replicate only within human intestinal epithelial cells (38). The mode of exit is unknown for any species of microsporidia, but it is interesting to speculate that other microsporidia species may use exocytosis for egress from host cells.
N. parisii and C. elegans are a naturally occurring host–pathogen pair and, by using RAB-11–based exocytosis for productive exit, they appear to have coevolved a compromise between host survival and pathogen propagation. It is likely that facilitating a minimally damaging exit strategy such as exocytosis is beneficial to both the host and the intracellular pathogen: By prolonging the lifespan of the host, the replication environment is preserved for future pathogen generations. Minimizing host damage is likely to be particularly important for N. parisii, given that it cannot replicate on its own, and its replication environment is the nonrenewable epithelial cells of the C. elegans intestine. Damage to any of these 20 cells is likely to be detrimental to the host. It will be interesting to investigate how C. elegans copes with the demand from this prodigious N. parisii spore production, with thousands of spores exiting the host every hour. Presumably this process places a huge increase in demand on C. elegans intestinal trafficking pathways.
N. parisii cells appear to initially be in direct contact with the host cytoplasm of intestinal cells (9). How does this pathogen then enter the host vesicle trafficking system to ultimately exit via apical exocytosis? Recently, the autophagy pathway, which can be used to control intracellular pathogen growth, has been implicated as a “recapture” mechanism to return bacterial pathogens that have escaped into the cytoplasm back into a host endosome (39, 40). However, our data indicate that the autophagy pathway is not important for N. parisii maturation and exit (Fig. S2). Thus, this pathogen may coopt a distinct pathway to enter the host vesicle trafficking system. Further analyses of host pathways, together with studies of the recently sequenced N. parisii genome (10), should help determine how this natural pathogen so efficiently coopts the host intestinal cell for prodigious proliferation and directional exit for fecal/oral transmission.
Materials and Methods
Infection Assays and Microscopy.
In general, synchronized C. elegans L1 larvae were grown for 24 h on 6-cm NGM plates seeded with OP50 at 20 °C until approximately L3 stage, when they were infected with 2 × 106 N. parisii spores and then incubated at 25 °C. Images used for quantifying spores shed and pathogen load were captured with fluorescence on a Zeiss AxioImager at 400× or 630× magnification. Confocal images were acquired on a Zeiss LSM700 at 630× magnification by using ZEN2010 software. Transmission electron microscopy was performed as described (9).
Contagiousness and Survival Assays.
Standard infection conditions were used on RNAi-treated donor animals. Contagiousness and survival assays were conducted as in described (9, 15). See Fig. S8 for details. For survival assays, the survival of 40 animals was assessed for each RNAi treatment group, each on three plates for a total of 120 animals per treatment. For each experimental replicate, results from triplicate plates were combined into one dataset. Animals that died because of desiccation on the wall of the plate rather than because of infection were excluded from analysis.
GFP::RAB-11 and PGP-1::GFP Coat Quantification.
To determine whether genes are upstream or downstream of RAB-11 and whether genes are required for SCC fusion with the apical membrane, the number of GFP::RAB-11.1 or PGP-1::GFP-coated SCCs was recorded after RNAi treatment. Animals were fixed in 4% (vol/vol) PFA at 46 hpi, and the anterior-most ring of intestinal cells was imaged in each of 30 animals per treatment group, taking 18 0.8-µm sectional confocal images per animal.
See SI Materials and Methods for additional materials and methods.
Supplementary Material
Acknowledgments
We thank K. Estes for early characterization of Calcofluor white staining of N. parisii; M. Zerial, B. Grant, K. Sato, and D. McEwan for C. elegans strains; A. Spang for RAB-11 antibodies; R. Luallen, L. Cohen, and T. Dror for feedback on the manuscript; and M. Wood and N. Olson for TEM assistance. Some strains were provided by the Caenorhabditis Genetics Center, which is funded by National Institutes of Health (NIH) Office of Research Infrastructure Programs Grant P40 OD010440. This work was supported by NIH Predoctoral Training Grant T32 GM008666 and a National Science Foundation Predoctoral Fellowship (to S.C.S.) and National Institute of Allergy and Infectious Diseases Grant R01 AI087528, the Searle Scholars Program, David and Lucile Packard Foundation, and Burroughs Wellcome Fund Fellowship (to E.R.T.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1400696111/-/DCSupplemental.
References
- 1.Friedrich N, Hagedorn M, Soldati-Favre D, Soldati T. Prison break: Pathogens’ strategies to egress from host cells. Microbiol Mol Biol Rev. 2012;76(4):707–720. doi: 10.1128/MMBR.00024-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hybiske K, Stephens RS. Exit strategies of intracellular pathogens. Nat Rev Microbiol. 2008;6(2):99–110. doi: 10.1038/nrmicro1821. [DOI] [PubMed] [Google Scholar]
- 3.Gallo RL, Hooper LV. Epithelial antimicrobial defence of the skin and intestine. Nat Rev Immunol. 2012;12(7):503–516. doi: 10.1038/nri3228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hamon M, Bierne H, Cossart P. Listeria monocytogenes: A multifaceted model. Nat Rev Microbiol. 2006;4(6):423–434. doi: 10.1038/nrmicro1413. [DOI] [PubMed] [Google Scholar]
- 5.Nikitas G, et al. Transcytosis of Listeria monocytogenes across the intestinal barrier upon specific targeting of goblet cell accessible E-cadherin. J Exp Med. 2011;208(11):2263–2277. doi: 10.1084/jem.20110560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pukkila-Worley R, Ausubel FM. Immune defense mechanisms in the Caenorhabditis elegans intestinal epithelium. Curr Opin Immunol. 2012;24(1):3–9. doi: 10.1016/j.coi.2011.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Szumowski SC, Estes KA, Troemel ER. Preparing a discreet escape: Microsporidia reorganize host cytoskeleton prior to non-lytic exit from C. elegans intestinal cells. Worm. 2012;1(4):1–5. doi: 10.4161/worm.20501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McGhee JD. The C. elegans intestine. WormBook. 2007:1–36. doi: 10.1895/wormbook.1.133.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Troemel ER, Félix MA, Whiteman NK, Barrière A, Ausubel FM. Microsporidia are natural intracellular parasites of the nematode Caenorhabditis elegans. PLoS Biol. 2008;6(12):2736–2752. doi: 10.1371/journal.pbio.0060309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cuomo CA, et al. Microsporidian genome analysis reveals evolutionary strategies for obligate intracellular growth. Genome Res. 2012;22(12):2478–2488. doi: 10.1101/gr.142802.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Didier ES, Weiss LM. Microsporidiosis: Not just in AIDS patients. Curr Opin Infect Dis. 2011;24(5):490–495. doi: 10.1097/QCO.0b013e32834aa152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Keeling PJ, Fast NM. Microsporidia: Biology and evolution of highly reduced intracellular parasites. Annu Rev Microbiol. 2002;56:93–116. doi: 10.1146/annurev.micro.56.012302.160854. [DOI] [PubMed] [Google Scholar]
- 13.Félix MA, Duveau F. Population dynamics and habitat sharing of natural populations of Caenorhabditis elegans and C. briggsae. BMC Biol. 2012;10:59. doi: 10.1186/1741-7007-10-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vávra J, Lukeš J. Microsporidia and ‘the art of living together’. Adv Parasitol. 2013;82:253–319. doi: 10.1016/B978-0-12-407706-5.00004-6. [DOI] [PubMed] [Google Scholar]
- 15.Estes KA, Szumowski SC, Troemel ER. Non-lytic, actin-based exit of intracellular parasites from C. elegans intestinal cells. PLoS Pathog. 2011;7(9):e1002227. doi: 10.1371/journal.ppat.1002227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hagedorn M, Rohde KH, Russell DG, Soldati T. Infection by tubercular mycobacteria is spread by nonlytic ejection from their amoeba hosts. Science. 2009;323(5922):1729–1733. doi: 10.1126/science.1169381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sato T, et al. The Rab8 GTPase regulates apical protein localization in intestinal cells. Nature. 2007;448(7151):366–369. doi: 10.1038/nature05929. [DOI] [PubMed] [Google Scholar]
- 18.Winston WM, Sutherlin M, Wright AJ, Feinberg EH, Hunter CP. Caenorhabditis elegans SID-2 is required for environmental RNA interference. Proc Natl Acad Sci USA. 2007;104(25):10565–10570. doi: 10.1073/pnas.0611282104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Winter JF, et al. Caenorhabditis elegans screen reveals role of PAR-5 in RAB-11-recycling endosome positioning and apicobasal cell polarity. Nat Cell Biol. 2012;14(7):666–676. doi: 10.1038/ncb2508. [DOI] [PubMed] [Google Scholar]
- 20.Huang J, Brumell JH. Bacteria-autophagy interplay: A battle for survival. Nat Rev Microbiol. 2014;12(2):101–114. doi: 10.1038/nrmicro3160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lundquist EA. Small GTPases. WormBook. 2006:1–18. doi: 10.1895/wormbook.1.67.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen CC, et al. RAB-10 is required for endocytic recycling in the Caenorhabditis elegans intestine. Mol Biol Cell. 2006;17(3):1286–1297. doi: 10.1091/mbc.E05-08-0787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grant BD, Donaldson JG. Pathways and mechanisms of endocytic recycling. Nat Rev Mol Cell Biol. 2009;10(9):597–608. doi: 10.1038/nrm2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen W, Feng Y, Chen D, Wandinger-Ness A. Rab11 is required for trans-golgi network-to-plasma membrane transport and a preferential target for GDP dissociation inhibitor. Mol Biol Cell. 1998;9(11):3241–3257. doi: 10.1091/mbc.9.11.3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Casanova JE, et al. Association of Rab25 and Rab11a with the apical recycling system of polarized Madin-Darby canine kidney cells. Mol Biol Cell. 1999;10(1):47–61. doi: 10.1091/mbc.10.1.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hoekstra D, Tyteca D, van IJzendoorn SC. The subapical compartment: A traffic center in membrane polarity development. J Cell Sci. 2004;117(Pt 11):2183–2192. doi: 10.1242/jcs.01217. [DOI] [PubMed] [Google Scholar]
- 27.van Ijzendoorn SC. Recycling endosomes. J Cell Sci. 2006;119(Pt 9):1679–1681. doi: 10.1242/jcs.02948. [DOI] [PubMed] [Google Scholar]
- 28.Takahashi S, et al. Rab11 regulates exocytosis of recycling vesicles at the plasma membrane. J Cell Sci. 2012;125(Pt 17):4049–4057. doi: 10.1242/jcs.102913. [DOI] [PubMed] [Google Scholar]
- 29.Poteryaev D, Fares H, Bowerman B, Spang A. Caenorhabditis elegans SAND-1 is essential for RAB-7 function in endosomal traffic. Embo J. 2007;26(2):301–312. doi: 10.1038/sj.emboj.7601498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sasidharan N, et al. RAB-5 and RAB-10 cooperate to regulate neuropeptide release in Caenorhabditis elegans. Proc Natl Acad Sci USA. 2012;109(46):18944–18949. doi: 10.1073/pnas.1203306109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Welz T, Wellbourne-Wood J, Kerkhoff E. Orchestration of cell surface proteins by Rab11. Trends Cell Biol. 2014 doi: 10.1016/j.tcb.2014.02.004. 10.1016/j.tcb.2014.02.004. [DOI] [PubMed] [Google Scholar]
- 32.Bruce EA, Stuart A, McCaffrey MW, Digard P. Role of the Rab11 pathway in negative-strand virus assembly. Biochem Soc Trans. 2012;40(6):1409–1415. doi: 10.1042/BST20120166. [DOI] [PubMed] [Google Scholar]
- 33.Ma H, Croudace JE, Lammas DA, May RC. Expulsion of live pathogenic yeast by macrophages. Curr Biol. 2006;16(21):2156–2160. doi: 10.1016/j.cub.2006.09.032. [DOI] [PubMed] [Google Scholar]
- 34.Nicola AM, Robertson EJ, Albuquerque P, Derengowski LdaS, Casadevall A. Nonlytic exocytosis of Cryptococcus neoformans from macrophages occurs in vivo and is influenced by phagosomal pH. MBio. 2011;2(4) doi: 10.1128/mBio.00167-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Takeuchi H, Furuta N, Morisaki I, Amano A. Exit of intracellular Porphyromonas gingivalis from gingival epithelial cells is mediated by endocytic recycling pathway. Cell Microbiol. 2011;13(5):677–691. doi: 10.1111/j.1462-5822.2010.01564.x. [DOI] [PubMed] [Google Scholar]
- 36.Knodler LA, et al. Dissemination of invasive Salmonella via bacterial-induced extrusion of mucosal epithelia. Proc Natl Acad Sci USA. 2010;107(41):17733–17738. doi: 10.1073/pnas.1006098107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jeon MK, Klaus C, Kaemmerer E, Gassler N. Intestinal barrier: Molecular pathways and modifiers. World J Gastrointest Pathophysiol. 2013;4(4):94–99. doi: 10.4291/wjgp.v4.i4.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Didier ES, Weiss LM. Microsporidiosis: Current status. Curr Opin Infect Dis. 2006;19(5):485–492. doi: 10.1097/01.qco.0000244055.46382.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Checroun C, Wehrly TD, Fischer ER, Hayes SF, Celli J. Autophagy-mediated reentry of Francisella tularensis into the endocytic compartment after cytoplasmic replication. Proc Natl Acad Sci USA. 2006;103(39):14578–14583. doi: 10.1073/pnas.0601838103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Collins CA, et al. Atg5-independent sequestration of ubiquitinated mycobacteria. PLoS Pathog. 2009;5(5):e1000430. doi: 10.1371/journal.ppat.1000430. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.