Abstract
The International Mouse Phenotyping Consortium (IMPC) plans to phenotype 20,000 single-gene knockout mice to gain an insight into gene function. Approximately 30% of these knockout mouse lines will be embryonic or perinatal lethal. The IMPC has selected three-dimensional (3D) imaging to phenotype these mouse lines at relevant stages of embryonic development in an attempt to discover the cause of lethality using detailed anatomical information. Rate of throughput is paramount as IMPC production centers have been given the ambitious task of completing this phenotyping project by 2021. Sifting through the wealth of data within high-resolution 3D mouse embryo data sets by trained human experts is infeasible at this scale. Here, we present a phenotyping pipeline that identifies statistically significant anatomical differences in the knockout, in comparison with the wild type, through a computer-automated image registration algorithm. This phenotyping pipeline consists of three analyses (intensity, deformation, and atlas based) that can detect missing anatomical structures and differences in volume of whole organs as well as on the voxel level. This phenotyping pipeline was applied to micro-CT images of two perinatal lethal mouse lines: a hypomorphic mutation of the Tcf21 gene (Tcf21-hypo) and a knockout of the Satb2 gene. With the proposed pipeline we were able to identify the majority of morphological phenotypes previously published for both the Tcf21-hypo and Satb2 mutant mouse embryos in addition to novel phenotypes. This phenotyping pipeline is an unbiased, automated method that highlights only those structural abnormalities that survive statistical scrutiny and illustrates them in a straightforward fashion.
Keywords: Micro-CT, Embryo, 3D, Imaging, Phenotyping
INTRODUCTION
The sequencing of the human genome has produced a vast resource of information; primarily, the location and chemical structure of each gene in the genome. At the present time, there is relatively little known of the role and function of each of the estimated 21,000 genes in the genome. Efforts to determine experimentally the relationship between genotype and expressed phenotype in the human individual are carried out in the mouse. This is mainly due to the 99% genetic homology between the two species and the development of established tools to manipulate the mouse genome.
A systematic approach to knock out each of the ∼20,000 genes in the mouse genome was proposed and funded in 2004 as a first step towards understanding gene function (Austin et al., 2004; Auwerx et al., 2004). This promoted the creation of the International Knockout Mouse Consortium (IKMC, www.knockoutmouse.org) in 2007 (International Mouse Knockout Consortium et al., 2007) with the goal of completing this systematic knockout mouse program by 2012. Through high-throughput gene trapping and gene targeting pipelines (Skarnes et al., 2011), >95% of this proposed work has now been completed (Bradley et al., 2012).
Creating and comprehensively phenotyping these 20,000 knockout mouse lines is an ambitious task taken on by the International Mouse Phenotyping Consortium (IMPC, www.mousephenotype.org) with the goal of phenotyping all mouse lines by 2021 (Brown and Moore, 2012). Of these knockout lines, ∼30% will be embryonic or perinatal lethal. Therefore, dedicated phenotyping pipelines are needed to investigate phenotypic abnormalities during embryonic development.
Three-dimensional (3D) imaging is an attractive option for phenotyping mouse embryos (Nieman et al., 2011; Norris et al., 2013). It has many advantages as a primary screen including whole-embryo coverage, the promise of computer-automated analysis, and future potential for data mining (Adams et al., 2013; Norris et al., 2013). As a result, 3D imaging has been deemed the most appropriate phenotyping assay for the primary screen of embryonic and perinatal lethal knockout mice (Adams et al., 2013). However, there is no consensus regarding how to analyze these data to identify anatomical phenotypes.
We have published numerous examples, primarily in the mouse brain, illustrating the value of image registration methods for identifying anatomical differences between groups of mice. Our image registration pipeline is a computer-automated algorithm in which 3D images are digitally morphed into a population average image, in which homologous points in anatomy are aligned. Image registration (Kovacevic, 2004; Lerch et al., 2011) is essential for qualitative visual assessment such that anatomical alignment is comparable between samples and it also lends itself to the statistical analysis of structural phenotypes between two groups of mice. Abnormal anatomical phenotypes were identified in the brains of mouse models of autism (Ellegood et al., 2013, 2011, 2010), schizophrenia (Clapcote et al., 2007), Alzheimer's disease (Lau et al., 2008) and Huntington's disease (Lerch et al., 2008) using image registration and deformation-based analysis.
Image registration of mature mouse embryos images acquired by magnetic resonance imaging (MRI) and optical projection tomography (OPT) (Sharpe et al., 2002) has been previously described (Cleary et al., 2011; Sussman et al., 2013; Zamyadi et al., 2010). In these studies, structural volume differences were identified between viable strains of mice using image registration methods. However, a pipeline to search out anatomical abnormalities in embryonic and perinatal lethal mouse knockout strains such as those produced by the IMPC has not yet been reported.
Ground work towards a phenotyping pipeline was published in Development where we investigated the natural variation in organ volumes in the embryonic day (E) 15.5 C57Bl/6 mouse embryo (Wong et al., 2012). We accomplished this by generating a novel 3D atlas of 48 segmented organs based on a consensus average micro-CT image of 35 individuals (Fig. 1). The present study is a natural extension of previous work; we outline a computer-automated pipeline that is able to identify abnormal mutant phenotypes in embryonic and perinatal lethal mice. The pipeline consists of three image analyses: intensity, deformation and atlas based. Here, we execute this three-pronged analysis pipeline on micro-CT images of E15.5 embryos that carry a hypomorphic allele for a transcription factor, Tcf21, and compared them with wild-type embryos (Quaggin et al., 1999). In addition, we conduct our analysis on the first perinatal lethal mouse knockout strain observed by the Davis, Toronto, Charles River and Chori (DTCC) branch of the IMPC, Satb2. The analysis is conducted at E15.5 to match the consensus gestational age for phenotyping late embryonic or perinatal lethal lines set by the IMPC. In both the Tcf21 and Satb2 analysis pipelines, we were able to recapitulate known anatomical phenotypes of these mutant strains as well as identify novel structural abnormalities found through our unbiased, whole body analysis.
Fig. 1.
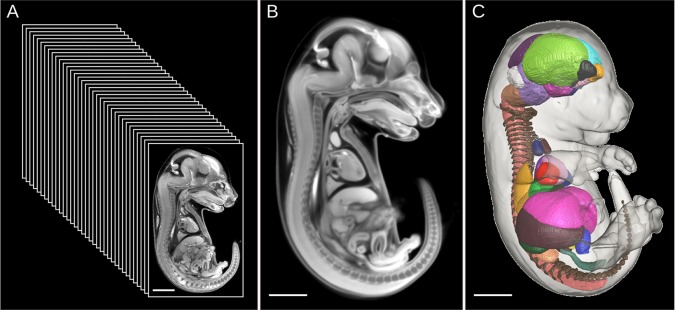
Generation of the 3D segmented mouse embryo atlas. (A-C) Thirty-five C57Bl/6 E15.5 mouse embryo micro-CT images (A) were morphed into a high quality representative population average atlas (B) through image registration. Forty-eight organ structures were manually painted through the 3D population average atlas. The resultant 3D mouse embryo segmented atlas (C) can be used to automatically measure organ volumes of new individual embryo images. Figure is adapted from our previous publication in Development (Wong et al., 2012).
RESULTS
Intensity-based analysis: identifying gross differences
Once image registration is completed, it is inferred that in every mouse embryo image each homologous point in anatomy is co-aligned at the exact same location in space. However, if no homology in anatomy exists between embryo images, arising from a missing organ or situs inversus for example, the registration algorithm will not apply meaningful deformations in that local region of the image. Similarly, an organ for which the volume is grossly hyper- or hypoplastic in the mutant compared with their corresponding wild type will not register because the deformations required are larger than our constraints allow.
We can therefore attribute local mis-registration in the mouse embryo images to gross differences in mutant morphology. These phenotypes can be identified by calculating significant differences in image intensity post-registration. Specifically, the post-registration images are first intensity normalized and then blurred by a 0.1 mm full-width-half-maximum (FWHM) Gaussian kernel to reduce the contribution of random noise. The individual intensity images are fitted to a linear model by genotype to compute the t-statistic at every voxel within the image. The final output is a 3D t-statistic map subjected to a 5% false discovery rate (FDR) threshold that is superimposed onto the population average intensity image to present the locations of significant intensity differences.
Deformation-based analysis: identifying local volume and shape differences
The completed registration algorithm generates a deformation field for each mouse embryo image, which encodes all deformations needed to morph each image into the final population average image. The Jacobian determinant of the deformation field is a scalar measure of the amount by which each voxel in each image needs to expand or contract to fit into the population average image. This measure, in turn, corresponds to local volume at the voxel level. As in the intensity-based analysis, the Jacobian determinant of the deformation field is blurred by a 0.1 mm FWHM Gaussian kernel and then fitted to a linear model by genotype to calculate a t-statistic at every voxel. To normalize for overall embryo volume, the deformation fields used only include deformations subsequent to global scaling and shearing in the 12-parameter linear registration. A color-coded 3D t-statistic map, subjected to a 5% FDR threshold, is calculated and superimposed onto the population average intensity image to illustrate the location of local volume differences between genotypes.
Atlas-based analysis: identifying whole organ volume differences
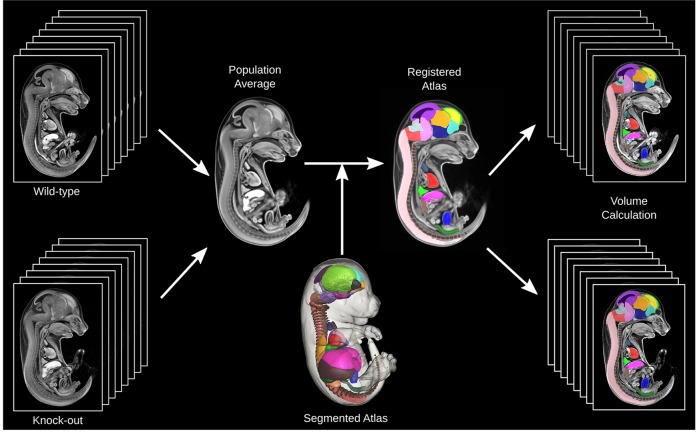
Fig. 2 is a schematic demonstrating the process for automatic volume measurements for each of the 48 anatomical structures within our previously published 3D segmented atlas (Wong et al., 2012). After image registration to produce a population average, the previously segmented 3D atlas is registered towards the population average image. The inverse deformation fields for each of the individual mouse embryos can be applied to the 3D segmented atlas such that all of the 48 segmented structures can be back propagated to each image. This allows for automatic volume measurements for these 48 organs for all input mouse embryo images. To normalize for embryo volume, each of the organ volumes is presented as a ratio of whole embryo volume. These can be fitted to a linear model by genotype and the t-statistic computed for each organ structure. A 5% FDR threshold was applied to control for the 48 independent statistical tests performed.
Fig. 2.
Schematic of the algorithm for identifying whole organ volume differences. Images from eight wild-type and eight knockout mouse embryos are registered into a population average image. The 3D segmented atlas (Wong et al., 2012) is then registered to this population average such that each segmented structure is resampled onto the population average space to provide a segmented population atlas. The segmented structures can then be back propagated onto the native images using the inverse of the transformations, which allows for automated volume measurements of the 48 structures.
Hypomorphic Tcf21 mutant
The result of the intensity-based analysis is presented in Fig. 3, revealing gross morphological differences between the wild-type and homozygous Tcf21-hypo embryos. Equivalent transverse sections through the thoracic cavity post-registration reveal a local mis-registration at the right middle lobe of the lung (Fig. 3A,B). This can be visualized better as a color map of the t-statistic of the image intensity overlaid onto the population average image with a FDR threshold of 5% (Fig. 3C). The pixels labeled blue in Fig. 3C are those that have significantly lower image intensity in the mutant population than in the wild type. This mis-registration corresponds to severe hypoplastic lungs in the Tcf21-hypo embryo. It is of note that this analysis is conducted on the whole mouse embryo volume, in an unbiased fashion. Significant differences are seen only in the right middle lobe and sparsely in the right accessory lobe of the lung. In contrast to the homozygous Tcf21-hypo embryos, no differences in intensity are found between the heterozygotes and the wild types.
Fig. 3.
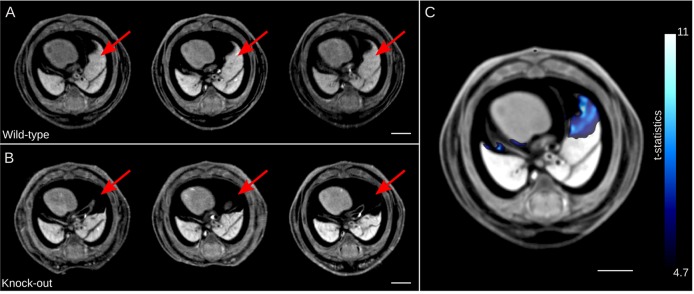
Analysis of image intensity differences in the Tcf21-hypo embryo images. (A,B) Three representative individual transverse sections of wild type (A) and hypomorphic Tcf21-hypo mutant (B) through the thoracic cavity post image registration are shown. The wild-type embryos (A) exhibit a normal middle lobe of the right lung morphology (red arrows). The Tcf21-hypo mouse embryos (B) are missing lung tissue at the corresponding locations (red arrows). (C) Highlighted in blue are significant intensity differences in the mutant. The color bar presents the t-statistic of which the minimum corresponds to a FDR threshold of 5%. Scale bars: 2 mm.
The hypoplastic phenotype of the remaining lobes of the lung in the Tcf21-hypo mutants is captured with both deformation (Fig. 4A) and atlas (Fig. 4B) based analyses. Fig. 4A presents a t-statistic map of the Jacobian determinant calculated for deformations between the Tcf21-hypo and wild-type populations superimposed onto a transverse and coronal section through the population average intensity image. The pixels labeled in blue are those areas that are significantly smaller in the mutant than in the wild type with a FDR threshold of 5%, whereas the pixels in red are areas of larger volume in the mutant. It is strikingly evident that all the lobes of the lung are flagged as smaller in the Tcf21-hypo mutant versus the wild type except for the right middle lobe and right accessory lobe owing to the mis-registration demonstrated in the first analysis (Fig. 3). In the transverse section, red areas reside in the thoracic cavity, which are larger in the mutant in response to the hypoplastic lung phenotype. Again, no local volume differences were found in the heterozygotes versus the wild type over the whole embryo volume using this analysis.
Fig. 4.
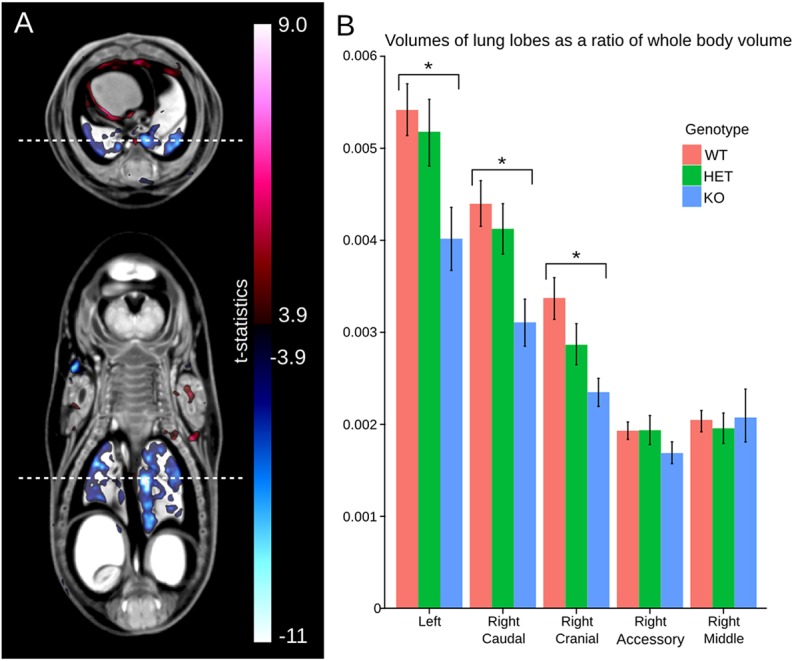
Lung volume differences exhibited in the Tcf21-hypo mutant. (A) A t-statistic map of the Jacobian determinant versus genotype superimposed onto a transverse (top) and coronal (bottom) section through the lung. The dashed white line corresponds to the location of the other image slice. The red color bar corresponds to areas larger in the Tcf21-hypo mutant versus the wild type and the blue color bar corresponds to areas smaller in the mutant. The minimum value on the color bar corresponds to an FDR threshold of 5%. (B) A bar graph of the volume of all five lobes of the lung as a ratio of whole embryo volume for the wild type, heterozygotes and Tcf21-hypo mutants calculated by atlas-based analysis. The error bars represent 95% confidence intervals. *FDR<5%.
Volumes as a ratio of whole embryo volume for each of the five lobes of the lung were calculated using the atlas-based analysis and are presented in Fig. 4B. The left, right caudal and right cranial lobes were significantly smaller in the homozygous Tcf21-hypo embryos compared with the wild type with an FDR threshold of 5%. However, there is no measured difference in overall volume of the right middle lobe and right accessory lobe between the wild type and homozygous Tcf21-hypo mutants. Again, the intensity-based analysis showed that the right middle lobe, and to a lesser extent the right accessory lobe, failed to register, prompting an inaccurate back propagation of the segmentation of these structures from the 3D-segmented atlas. Again, the heterozygotes did not have any significant structural volume differences compared with the wild type. However, it is observed that heterozygote volume measurements consistently lie between those of the wild type and those of the homozygous Tcf21-hypo mouse embryos, as would be expected.
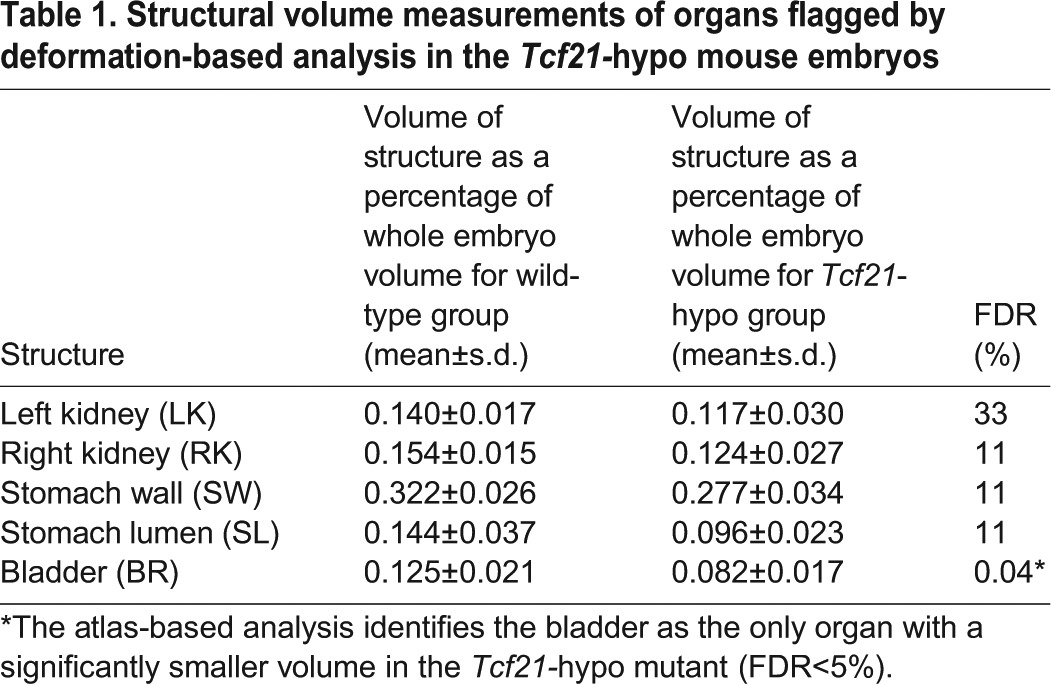
The largest t-statistic generated by the analysis of local volume differences coincides with an extremely small entrance to the esophagus and larynx in the homozygous Tcf21-hypo mutants (Fig. 5A,B). Traversing posteriorly down the esophagus, numerous punctate areas can be identified in blue, indicating a smaller esophagus in the Tcf21-hypo mutants (Fig. 5A). These volume differences continue into the stomach where both the stomach lumen and stomach wall are much smaller in the homozygous Tcf21-hypo mutants computed by deformation-based analysis (Fig. 5B,C). In addition, localized volumes in both kidneys are found to be significantly smaller by deformation-based analysis (Fig. 5B,C) as well as in the bladder (Fig. 5A,C). The stomach wall and lumen, right and left kidney, and bladder were evaluated using the atlas-based analysis to investigate whether there are whole-structure volume differences in these organs (Table 1). Using this method, only the bladder was flagged as significantly different (FDR<5%) in the Tcf21-hypo mutant.
Fig. 5.
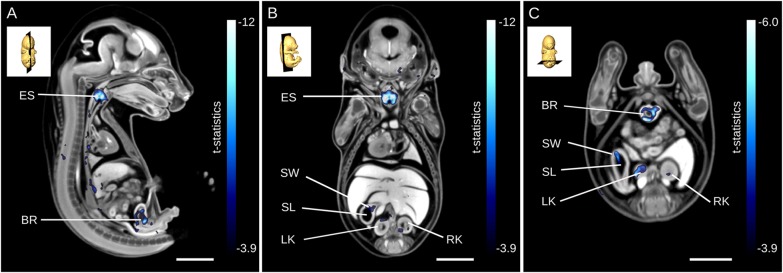
Deformation-based analysis identifying several anatomical phenotypes in the Tcf21-hypo mutant. (A-C) Results from deformation-based analysis presented for a mid-sagittal section through the esophagus (A), a coronal section through the kidneys (B) and an alternative transverse section through the kidneys (C). The blue color bar has a minimum that corresponds to an FDR threshold of 5% in. BR, bladder; ES, esophagus; LK, left kidney; RK, right kidney; SL, lumen; SW, stomach wall. Scale bars: 2 mm.
Table 1.
Structural volume measurements of organs flagged by deformation-based analysis in the Tcf21-hypo mouse embryos
Satb2 mutant
Gross morphological phenotypes can be identified by visually comparing a mid-sagittal section of the population average image of eight wild-type Satb2 mouse embryos (Fig. 6A) and eight knockout mouse embryos (Fig. 6B). Immediately apparent is a shorter tongue and lower jaw in the mutant, as well as a cleft palate. These phenotypes are highlighted using both intensity (Fig. 6C) and deformation (Fig. 6D) based analyses. The voxel-based intensity analysis shows the cleft palate phenotype in blue as well as an area of mis-registration due to a shorter tongue (Fig. 6C, blue) and due to a wider base of the tongue (Fig. 6C, red). The voxel-based Jacobian determinant analysis reveals large craniofacial abnormalities in the Satb2 knockout due to a much shorter jaw and tongue (Fig. 6D, blue). It is of note that neither analysis identified any morphological differences outside of the craniofacial region. In addition, none of the 48 structures in the segmented 3D mouse embryo atlas was found to be significantly different in volume in the knockout compared with wild type.
Fig. 6.
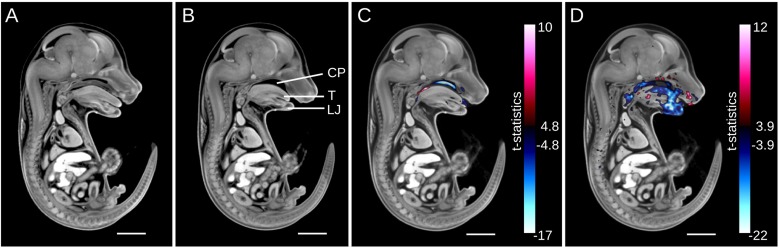
Gross phenotypes of the Satb2 knockout demonstrated by intensity- and deformation-based analyses. (A,B) A population average image of the Satb2 wild-type (A) and knockout (B) mouse embryos demonstrate visually evident phenotypes, such as a cleft palate (CP), shorter lower jaw (LJ) and shorter tongue (T). (C) Intensity-based analysis highlights the cleft palate in blue. Red and blue correspond to areas of brighter and darker image intensity in the knockout, respectively. (D) Deformation-based analysis shows a much smaller tongue and jaw in blue. Red and blue correspond to areas of larger and smaller volumes in the knockout, respectively. The minimum of the color bars corresponds to a FDR threshold of 5%. Scale bars: 2 mm.
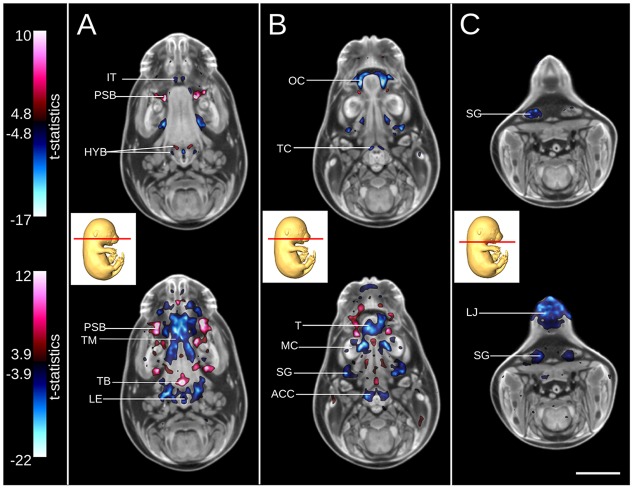
Transverse sections through the tongue (Fig. 7A), mandible (Fig. 7B) and lower jaw (Fig. 7C) demonstrate the sensitivity of both the intensity- and deformation-based analyses in identifying detailed mutant phenotypes. Intensity-based analysis (Fig. 7A, top) reveals missing primordium of the incisor teeth (Fig. 7A, top, blue), bulges in the palatal shelf mesenchyme (Fig. 7A, top, red), as well gross differences to the cartilage primordium of the greater and lesser horn of the hyoid bone. For the same slice, deformation-based analysis (Fig. 7A, bottom) shows volumes differences indicative of palatal shelf bulges and a larger base of the tongue (Fig. 7A, bottom, red), as well as significantly smaller intrinsic muscles of the tongue, entrance to the larynx and epiglottis. In a more posterior slice, the results of the intensity-based analysis (Fig. 7B, top) shows a much larger oral cavity in the mutant due to the cleft palate and shorter tongue, as well as the missing primordium of the thyroid cartilage (Fig. 7B, top, blue). For the same slice, the deformation-based analysis (Fig. 7B, bottom) reveals a smaller tongue, Meckel's cartilage, submandibular gland and the primordium of both the arytenoid and cricoid cartilages. Lastly, for a section through the lower jaw (Fig. 7C, top), image intensity differences are identified in the submandibular gland. Results of deformation-based analysis for the same section (Fig. 7C, bottom) also shows a smaller local volume of the submandibular gland as well as a much shorter lower jaw.
Fig. 7.
Subtle phenotypes of the Satb2 knockout demonstrated by intensity- and deformation-based analyses. (A-C) Transverse sections through the tongue (A), mandible (B) and lower jaw (C). Intensity-based analysis illustrating image intensity differences between the Satb2 wild type and knockout (top) and deformation-based analysis identifying local volume differences for the same sections (bottom). For intensity-based analysis (top), red and blue correspond to areas of brighter or darker image intensity in the knockout, respectively. For deformation-based analysis (bottom), red and blue correspond to areas of larger and smaller local volumes in the knockout, respectively. The minimum value for each color bar corresponds with a FDR threshold of 5%. ACC, arytenoid and cricoid cartilages; HYB, cartilage primordium of greater and lesser horn of hyoid bone; IT, incisor teeth; LE, opening to larynx and epiglottis; LJ, lower jaw; MC, Meckel's cartilage; OC, oral cavity; PSB, palatal shelf bulges; SG, submandibular gland; T, tongue; TB, base of tongue; TC, thyroid cartilage; TM, intrinsic muscle of tongue. Scale bar: 2 mm.
DISCUSSION
Concordance of results with previous literature
Tcf21 belongs to the type 2 basic helix-loop-helix transcription factor family and is essential for the development of various organs including heart, lung, kidney, gonads and spleen. The morphological phenotypes of conventional Tcf21 knockout mice were well described previously (Quaggin et al., 1999). Conventional Tcf21 knockout mice die just after birth because of respiratory failure and tetralogy of Fallot. The lungs of Tcf21 knockout mice are severely hypoplastic, lack mature alveoli and type II pneumocytes and show defects in proximodistal differentiation of the airway epithelium (Quaggin et al., 1999). Additionally, kidneys of Tcf21 knockout mice show a profound defect in nephron differentiation, exhibiting immature podoytes and glomeruli.
In the current study, we used a new hypomorphic allele for Tcf21 that expresses only 30% of Tcf21 transcripts compared with wild type to produce the Tcf21-hypo mutants. Morphological analysis of the homozygous Tcf21-hypo mice showed mild hypoplasia in lungs and kidneys, an intermediate phenotype compared with that of wild type and the conventional knockout mice (data not shown). The Tcf21-hypo mutants are perinatal lethal as well. We are able to recapitulate the known hypoplastic lung phenotype automatically by using three analysis techniques. The right middle lobe and accessory lobe are found to be severely hypoplastic through intensity-based analysis; the remaining three lung lobes are significantly smaller in the Tcf21-hypo mice after both deformation- and atlas-based analyses. Additionally, local areas in both the right and left kidney were found to be significantly smaller in the Tcf21-hypo embryos using deformation-based morphometry.
Whole-embryo analysis allows for truly unbiased anatomical phenotyping and its value is evident in the identification of novel esophagus, larynx, stomach and bladder phenotypes identified in Tcf21-hypo embryos. The differences begin at the entrance of the esophagus, continue down the esophagus and into the stomach where the deformation-based analysis reveals a significantly smaller stomach lumen and wall. Tcf21 is expressed in the smooth muscle layer of the gastrointestinal tract (Maezawa et al., 2012) and may be required for proper development. Additionally, both the deformation- and atlas-based analyses identified that the bladder is significantly smaller in the Tcf21-hypo embryo. This may be a downstream effect of renal failure (Quaggin et al., 1999) and hypoplasticity.
The heterozygous Tcf21-hypo embryos did not show any significant differences using any of the three analyses compared with the wild-type group. This result is expected and further validates the efficacy of the presented method and its ability to control for false positives.
In humans, genetic mutations in chromosomal region 2q32-q33 are associated with craniofacial malformations, including cleft palate. Satb2 is a candidate cleft palate gene in the 2q32 locus where haploinsufficiency has been seen to interrupt its function (Britanova et al., 2006; FitzPatrick et al., 2003). The role of Satb2 in craniofacial patterning was investigated in gene knockout mice by Dobreva et al. Their study involved extensive embryonic anatomical phenotyping using Alcian Blue/Alizarin Red staining of skeletal preparations, Hematoxylin-Eosin staining and scanning electron micrographs (Dobreva et al., 2006). Our automated analysis on micro-CT images of the Satb2 knockout identified the majority of anatomical phenotypes presented by Dobreva et al. that are observable at the E15.5 time point. We were able to confirm a shortened mandible, smaller and shorter tongue, cleft palate, malformations of the lesser horns and the body of the hyoid bone, and missing thyroid cartilage and incisor teeth in the Satb2 knockout mouse embryo. Additionally, we were able to identify the presence of bulges developed by the palatal shelve mesenchyme that was revealed through scanning electron micrographs in the study by Dobreva et al. The sensitivity of our analyses also reveals novel phenotypes, such as significantly smaller Meckel's cartilage, smaller submandibular glands, smaller entrance to the larynx and epiglottis, and smaller arytenoid and cricoid cartilages.
Benefits of the phenotyping image registration pipeline
We strongly believe that when phenotyping mouse embryos using 3D imaging, the bare minimum for image processing is an affine image registration of the data. This ensures that for qualitative comparison each mouse embryo data set is analyzed at corresponding locations in anatomy. However, it is difficult to interpret subtle volumetric differences when qualitatively comparing two-dimensional sections one at a time through a 3D image set. In addition, qualitative assessment of thousands of 3D embryo images for the IMPC initiative is tedious, low-throughput and prone to human error and bias.
The ability to identify image intensity differences is essential when phenotyping embryonic or perinatal lethal mouse knockout strains in which gross volume differences or missing structures are the reasons for premature death. This is evident in both homozygous Tcf21-hypo and Satb2 homozygous null mouse embryos, in which a severely hypoplastic right middle lobe of the lung and cleft palate, respectively, are sure contributors to perinatal lethality. In addition, subtle non-volumetric differences in the Satb2 knockout, such as missing incisor teeth and thyroid cartilage as well as alterations to the hyoid bone, are identified.
Deformation-based analysis is advantageous because it performs statistical analysis of local volume and shape differences at every element of the 3D data set. It often supports the findings of the atlas-based analysis and also adds local anatomical phenotypes to areas not included in the 3D segmented atlas. The atlas-based analysis did not identify any structural volume differences in the Satb2 mutant; however, the deformation-based analysis highlighted many craniofacial abnormalities, including the visually evident shorter tongue and lower jaw as well as the subtle finding of bulges in the palatal shelves.
The atlas-based analysis is a fully automatic way to segment and measure structural volumes of any given mouse embryo image. Alternatively, each structure for each embryo would need to be manually painted, which would not be feasible for high-throughput phenotyping projects. The atlas published in Development by Wong et al., for example, required 400 h of work by a trained expert (Wong et al., 2012). This analysis is limited in its breadth though; despite the use of the most comprehensive mouse embryo atlas to date, it still includes only 48 structures that cover approximately half of the entire embryo volume. More structures should be incorporated in the future. However, labeling every voxel is impossible owing to the structural variability of organs such as the intestines.
We believe that the three analyses presented should be performed in concert. Each is independent of the others such that they can be executed in parallel once image registration is complete. Despite their respective benefits and disadvantages, we do not propose a set hierarchy in the execution of the three analyses because in the case of a primary screen there is no a priori knowledge of the type of phenotype that may be present. However, in terms of the readout it might be best to interpret the intensity-based analysis first because it flags gross phenotypes, which are likely to be drivers for embryonic lethality, and flags false negatives in the deformation- and atlas-based analyses.
This automated pipeline is not restricted to micro-CT data. Registration of 3D mouse embryo images acquired by MRI (Cleary et al., 2011; Sussman et al., 2013; Zamyadi et al., 2010), and OPT have previously been published (Anderson et al., 2013; Sussman et al., 2013). Another imaging modality performing image acquisition for the IMPC is high resolution episcopic microscopy (HREM), which generates images with resolution of the order of 1 µm isotropic (Weninger et al., 2006). It will be interesting to see how image registration handles much more defined micro-tissue structures relayed by HREM imaging.
With the described computer processing power, memory and image resolution of the 3D images, an analysis of eight wild-type and eight knockout mouse embryos can be achieved within 24 h. A comparable computer cluster to that presented here is approachable by individual IMPC production centers with an estimated cost of $8000 per unit. The entire imaging analysis pipeline, including the image registration, does not involve manual intervention of any kind and leverages powerful computer processing that exists today. The final read-out for each analysis is simple, easily interpretable and can be easily uploaded to the IMPC data coordination center (DCC) with sufficient image compression.
Accuracy and limitations of the phenotyping pipeline
We have conducted a rigorous study on the sensitivity of our registration methods when applied to brain MRI images (van Eede et al., 2013). A known, induced volume change was applied to a set of MRI brain images and we analyzed how well we can recover this change. Once multiple comparisons are controlled using the FDR method, it was found that our voxel-by-voxel deformation-based analysis incurred a very low average rate of false positive voxels (0.17%). The validity of the results of the intensity- and deformation-based analyses is also evident in our analysis of the Satb2 mutant, shown in Fig. 7, in which most of the intensity and volume differences are bilateral. This demonstrates that the differences identified are not random, but survive statistical scrutiny in a symmetrical fashion. The FDR method for flagging phenotypes ensures that the probability of a false positive phenotype is <5%. However, there are no assurances of the validity of a false negative or the designation of a ‘no’ phenotype. The accuracy of our back-propagation of segmented structures using the atlas-based analysis is demonstrated in our previous study, in which each lobe of the lungs of individual mouse embryos were accurately back-propagated from the segmented atlas (Wong et al., 2012). However, inaccuracies in the volume measurements and back-propagation can occur in the liver because its location and lower boundary are dependent on the random patterning of the intestines (Wong et al., 2012).
Our analysis pipeline assumes that any potential anatomical phenotype in the mutant is either fully penetrant or possessed by most of the knockout mouse embryos. It is common for knockout populations to have partially penetrant phenotypes and our three analysis tools will not report such a phenotype because it would not survive statistical scrutiny. This promotes a need for a method of statistical analysis to compare each mutant individual with a wild-type population. One method could be to analyze the number of standard deviations the knockout embryo is from the mean volume, Jacobian determinant, or image intensity of the wild-type population.
Our analyses have been successfully tested at the E15.5 developmental stage, one of the two stages in the IMPC pipeline. We account for the variability in developmental age (due to the unknown time of conception) by normalizing all our volumetric results by overall embryo size. This reduces the variability by as much as 50% depending on the structure (Wong et al., 2012). It is feasible to register embryos that bound the E15.5 stage because from E14.5 to E16.5 the mouse embryo is no longer undergoing organogenesis, only growth. Differences in volume and shape due to maturing organs exhibited by mouse embryos around E15.5 can still be registered. In a previous publication we demonstrated the capability of the image registration algorithm to register 35 mouse embryos that had an overall volume of 361.60±41.44 mm3 (±10%) (Wong et al., 2012).
At the other developmental stage to be phenotyped by the IMPC, E9.5, rapid organogenesis is occurring within hours and a simple size normalization will not suffice. At E9.5, mouse embryo samples should be somite matched; however, the amount of breeding required to reach 16 somite-matched mutants and wild-type mouse embryos is cost prohibitive. Furthermore, a segmented atlas approach would be difficult at this younger time point because there are very few volumetric structures that can be demarcated and segmented out at this stage. A wild-type atlas through time would be beneficial to be able to compare any given knockout mouse embryo with its age-matched wild type. This four-dimensional atlas could be interpolated throughout gestation making it possible to stage a mouse embryo by organ to discriminate structures that are growth retarded or developing normally.
The ability to detect a difference per organ depends on the inherent variability in that organ's volume and shape in the data sets. In our previous publication, we calculated that the average variance in 35 C57BL/6 mouse embryos was ∼10% per structure (Wong et al., 2012). A power analysis showed that with a sample size of eight wild types and eight knockouts, we could detect a 9% and 14% volume difference in the brain and liver, respectively. There is an asymptote in the power analysis whereby adding more samples does not increase the sensitivity, and we have chosen a sample size of eight because it is the breaking point of that function. Conversely, if the sample size is reduced the sensitivity drops exponentially.
Mouse production is the predominant consumer of both time and money in the IMPC pipeline. At modest sample sizes (n=1-3), manual annotation by trained experts can outpace mouse production and would not be the rate-limiting step. However, for quantitative and statistical analysis, large sample sizes of eight are needed and at that scale manual annotation becomes both tedious and expensive. A combination of both human experts and computational algorithms will no doubt be used to make best use of the abundance of structural data provided by 3D imaging.
Outlook
Large-scale phenotyping screens, like that of the IMPC, require an executable pipeline that is high-throughput, standardized between phenotyping centers and sensitive enough to identify phenotypic hits. Ex vivo 3D imaging promises to satisfy these requirements for the primary screen of embryonic lethal mouse lines identified by the IMPC. Imaging modalities such as OPT, MRI and micro-CT are capable of acquiring data for one knockout mouse line (eight wild types and eight knockouts) per day through parallel imaging techniques (Schneider et al., 2004; Wong et al., 2012; Zhang et al., 2010). Furthermore, 3D imaging can be standardized. The same micro-CT imaging system can be purchased and used at each phenotyping center and OPT systems can be easily installed at each center using an open-source imaging portal we have recently published and support (Wong et al., 2013a). 3D imaging generates a wealth of morphological data over the whole embryo volume. With high image resolution, as fine as 1 µm when using HREM, therein lies enough anatomical data for a high-hit rate for morphological phenotypes. However, the promise of 3D imaging as a primary screen for embryonic lethal phenotypes is limited without imaging analysis software to identify mutant phenotypes computationally. Here, we introduced a computer-automated three-level analysis based on image registration methods that can flag both subtle and gross morphological differences between wild-type and knockout mouse embryos. Analysis of each embryonic lethal line can be completed within 24 h with relatively affordable computational hardware and the presented software is open source and can be used at each phenotyping center as well. The development of this automated image analysis pipeline is a substantial advancement in how 3D imaging can be used as a primary screen for embryonic lethal lines.
MATERIALS AND METHODS
Sample preparation
A new hypomorphic allele for Tcf21 (Tcf21-hypo) was implemented in which loxP and Frt sites were inserted into the promoter lesion of Tcf21 gene by homologous recombination that resulted in a massive decrease of Tcf21 transcript production. Three groups of samples were included in this analysis: eight wild-type, eight heterozygous and eight homozygous Tcf21-hypo embryos collected from five litters. The background strain of these mice is a mixture of B6/CD1/129. The Satb2 mouse embryos were split into two groups as per the DTCC embryonic lethal screen: eight wild-type and eight homozygous null mouse embryos collected from five litters (Adams et al., 2013). The background strain of the Satb2 cohort is C57Bl/6N, which is the genetic background of all IMPC knockout lines.
For this study, the mice were mated and detection of a vaginal plug the following morning was considered to be 0.5 days post coitum (dpc). At 15.5 dpc, pregnant females were sacrificed by cervical dislocation. The mouse embryos were then dissected and fixed in 4% paraformaldehyde overnight at 4°C. The mouse embryos were then transferred to PBS for storage until potassium tri-iodide (Lugol) staining. Each sample was immersed in 15 ml stock Lugol solution (L6146, Sigma-Aldrich) overnight (17±1 h) (Wong et al., 2012). The samples were then embedded in 1% agarose (Bioshop) in 11-mm centrifuge tubes (Beckman Instruments) prior to micro-CT imaging. The Satb2 mouse embryos underwent an additional tissue stabilization protocol, which was developed to minimize tissue shrinkage due to Lugol staining as previously described (Wong et al., 2012, 2013b). The embryo tissue is cross-linked through formaldehyde fixation and macromolecules are linked to a hydrogel monomer creating a gel mesh that preserves tissue geometry (Chung et al., 2013; Wong et al., 2013b).
Micro-CT imaging
For each mouse embryo, 3D datasets were acquired using a Skyscan 1172 high-resolution micro-CT scanner (Bruker). With the X-ray source at 100 kVp and 100 µA and the use of a 0.5 mm aluminium filter, each specimen was rotated 360° around the vertical axis, generating 1200 views in 5 h. These image projections were reconstructed into digital cross-sections using the Feldkamp algorithm (Feldkamp and Davis, 1984) for cone beam CT. The resulting 3D data block contained 2000×1000×1000 voxels of 13.4 µm and was resampled to match the resolution (28 µm) and data size of the 3D segmented atlas previously published (Wong et al., 2012). The down-sampling of the 3D images was performed as the 2× decrease in voxel size allowed for an 8× reduction in the amount of RAM and processing time for the imaging analysis pipeline.
Image registration
Each mouse embryo image (n=16; eight wild type and eight mutant) is morphed into a population average image such that the resulting transformations can be used for our three-stage analysis pipeline (Fig. 2). The computer-automated algorithm has been described previously (Kovacevic, 2004; Wong et al., 2012; Zamyadi et al., 2010). Briefly, each mouse embryo image is subjected to a six-parameter, linear registration (three translations, three rotations) towards a model embryo such that every image is in the same location in space and has the same orientation. An additional 12-parameter, linear registration (three translations, three rotations, three scales and three shears) is conducted in a pairwise manner (16×15 registrations) to normalize differences in embryo size. The average of the 15 transforms per mouse embryo image is calculated and the resultant transform is applied to each respective image. A final average image is then calculated from the average of the linearly transformed images. Following this, an iterative six-generation multiscale non-linear registration algorithm is applied whereby each mouse embryo image is registered towards the average of the linear registrations, and then subsequently towards the average image of the previous non-linear registration step. The registration pipeline code is written in Perl, utilizes MNI AUTOREG tools (Collins et al., 1994) and is fully computer automated. A more comprehensive description of the algorithm can be found here at https://wiki.mouseimaging.ca/display/MICePub/MiceBuildModelAlgorithm. The image registration was computed using a cluster of ten computers with the following hardware specifications: Dell PowerEdge 1950s with two quadcore Intel(R) Xeon(R) E5450 processors at 3.00 GHz and 64 GB of RAM running on the linux distribution Ubuntu 10.04.
Image analysis pipeline
For each of the three image analyses, a phenotype is flagged if the calculated value in the mutant group is statistically abnormal in comparison with the variability inherent to the wild-type group. We invoke the FDR to control for multiple comparisons inherent to each analysis (Genovese et al., 2002). We employ an FDR threshold of 5% to flag mutant phenotypes, ensuring that there is a 5% maximum likelihood that the identified phenotype is a false positive. The image analyses are computed on the outputs of the image registration algorithm using the free statistical software R.
Acknowledgements
We thank the DTCC consortium, especially the Toronto Centre of Phenogenomics, for performing the lethality screen, dissections and genotyping of the Satb2 knockout mouse embryos.
Footnotes
Competing interests
The authors declare no competing financial interests.
Author contributions
M.D.W. prepared the mouse embryos for imaging, acquired the image data sets, optimized the image registration, performed the statistical analysis and wrote the manuscript. M.D.W. and R.M.H. designed the experiments. R.M.H. supervised the study and edited the manuscript. Y.M. bred, dissected and genotyped the Tcf-21 hypo mutant mouse embryos and edited the manuscript. J.P.L. consulted on image registration and statistical analysis and edited the manuscript.
Funding
This work was supported by the National Institutes of Health [OD011185 to R.M.H and U54HG006364 to R.M.H.] and by Genome Canada. Deposited in PMC for release after 12 months.
References
- Adams D., Baldock R., Bhattacharya S., Copp A. J., Dickinson M., Greene N. D. E., Henkelman M., Justice M., Mohun T., Murray S. A., et al. (2013). Bloomsbury report on mouse embryo phenotyping: recommendations from the IMPC workshop on embryonic lethal screening. Dis. Model. Mech. 6, 571-579 10.1242/dmm.011833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson G. A., Wong M. D., Yang J., Henkelman R. M. (2013). 3D imaging, registration, and analysis of the early mouse embryonic vasculature. Dev. Dyn. 242, 527-538 10.1002/dvdy.23947 [DOI] [PubMed] [Google Scholar]
- Austin C. P., Battey J. F., Bradley A., Bucan M., Capecchi M., Collins F. S., Dove W. F., Duyk G., Dymecki S., Eppig J. T., et al. (2004). The knockout mouse project. Nat. Genet. 36, 921-924 10.1038/ng0904-921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auwerx J., Avner P., Baldock R., Ballabio A., Balling R., Barbacid M., Berns A., Bradley A., Brown S., Carmeliet P., et al. (2004). The European dimension for the mouse genome mutagenesis program. Nat. Genet. 36, 925-927 10.1038/ng0904-925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley A., Anastassiadis K., Ayadi A., Battey J. F., Bell C., Birling M.-C., Bottomley J., Brown S. D., Bürger A., Bult C. J., et al. (2012). The mammalian gene function resource: the International Knockout Mouse Consortium. Mamm. Genome 23, 580-586 10.1007/s00335-012-9422-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britanova O., Depew M. J., Schwark M., Thomas B. L., Miletich I., Sharpe P., Tarabykin V. (2006). Satb2 haploinsufficiency phenocopies 2q32-q33 deletions, whereas loss suggests a fundamental role in the coordination of jaw development. Am. J. Hum. Genet. 79, 668-678 10.1086/508214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown S. D. M., Moore M. W. (2012). The International Mouse Phenotyping Consortium: past and future perspectives on mouse phenotyping. Mamm. Genome 23, 632-640 10.1007/s00335-012-9427-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung K., Wallace J., Kim S.-Y., Kalyanasundaram S., Andalman A. S., Davidson T. J., Mirzabekov J. J., Zalocusky K. A., Mattis J., Denisin A. K., et al. (2013). Structural and molecular interrogation of intact biological systems. Nature 497, 332-337 10.1038/nature12107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clapcote S. J., Lipina T. V., Millar J. K., Mackie S., Christie S., Ogawa F., Lerch J. P., Trimble K., Uchiyama M., Sakuraba Y., et al. (2007). Behavioral phenotypes of Disc1 missense mutations in mice. Neuron 54, 387-402 10.1016/j.neuron.2007.04.015 [DOI] [PubMed] [Google Scholar]
- Cleary J. O., Modat M., Norris F. C., Price A. N., Jayakody S. A., Martinez-Barbera J. P., Greene N. D. E., Hawkes D. J., Ordidge R. J., Scambler P. J., et al. (2011). Magnetic resonance virtual histology for embryos: 3D atlases for automated high-throughput phenotyping. NeuroImage 54, 769-778 10.1016/j.neuroimage.2010.07.039 [DOI] [PubMed] [Google Scholar]
- Collins D. L., Neelin P., Peters T. M., Evans A. C. (1994). Automatic 3D intersubject registration of MR volumetric data in standardized Talairach space. J. Comput. Assist. Tomogr. 18, 192-205 10.1097/00004728-199403000-00005 [DOI] [PubMed] [Google Scholar]
- Dobreva G., Chahrour M., Dautzenberg M., Chirivella L., Kanzler B., Fariñas I., Karsenty G., Grosschedl R. (2006). SATB2 is a multifunctional determinant of craniofacial patterning and osteoblast differentiation. Cell 125, 971-986 10.1016/j.cell.2006.05.012 [DOI] [PubMed] [Google Scholar]
- Ellegood J., Pacey L. K., Hampson D. R., Lerch J. P., Henkelman R. M. (2010). Anatomical phenotyping in a mouse model of fragile X syndrome with magnetic resonance imaging. NeuroImage 53, 1023-1029 10.1016/j.neuroimage.2010.03.038 [DOI] [PubMed] [Google Scholar]
- Ellegood J., Lerch J. P., Henkelman R. M. (2011). Brain abnormalities in a Neuroligin3 R451C knockin mouse model associated with autism. Autism Res. 4, 368-376 10.1002/aur.215 [DOI] [PubMed] [Google Scholar]
- Ellegood J., Babineau B. A., Henkelman R. M., Lerch J. P., Crawley J. N. (2013). Neuroanatomical analysis of the BTBR mouse model of autism using magnetic resonance imaging and diffusion tensor imaging. NeuroImage 70, 288-300 10.1016/j.neuroimage.2012.12.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldkamp L., Davis L. C., Kress J. W. (1984). Practical cone-beam tomography. J. Opt. Soc. Am. A. 1, 612-619 10.1364/JOSAA.1.000612 [DOI] [Google Scholar]
- FitzPatrick D. R., Carr I. M., McLaren L., Leek J. P., Wightman P., Williamson K., Gautier P., McGill N., Hayward C., Firth H., et al. (2003). Identification of SATB2 as the cleft palate gene on 2q32-q33. Hum. Mol. Genet. 12, 2491-2501 10.1093/hmg/ddg248 [DOI] [PubMed] [Google Scholar]
- Genovese C. R., Lazar N. A., Nichols T. (2002). Thresholding of statistical maps in functional neuroimaging using the false discovery rate. NeuroImage 15, 870-878 10.1006/nimg.2001.1037 [DOI] [PubMed] [Google Scholar]
- International Mouse Knockout Consortium; Collins F. S., Rossant J., Wurst W. (2007). A mouse for all reasons. Cell 128, 9-13 10.1016/j.cell.2006.12.018 [DOI] [PubMed] [Google Scholar]
- Kovacevic N. (2004). A three-dimensional MRI atlas of the mouse brain with estimates of the average and variability. Cereb. Cortex 15, 639-645 10.1093/cercor/bhh165 [DOI] [PubMed] [Google Scholar]
- Lau J. C., Lerch J. P., Sled J. G., Henkelman R. M., Evans A. C., Bedell B. J. (2008). Longitudinal neuroanatomical changes determined by deformation-based morphometry in a mouse model of Alzheimer's disease. NeuroImage 42, 19-27 10.1016/j.neuroimage.2008.04.252 [DOI] [PubMed] [Google Scholar]
- Lerch J. P., Carroll J. B., Spring S., Bertram L. N., Schwab C., Hayden M. R., Henkelman R. M. (2008). Automated deformation analysis in the YAC128 Huntington disease mouse model. NeuroImage 39, 32-39 10.1016/j.neuroimage.2007.08.033 [DOI] [PubMed] [Google Scholar]
- Lerch J. P., Sled J. G., Henkelman R. M. (2011). MRI phenotyping of genetically altered mice. Methods Mol. Biol. 711, 349-361 10.1007/978-1-61737-992-5_17 [DOI] [PubMed] [Google Scholar]
- Maezawa Y., Binnie M., Li C., Thorner P., Hui C.-C., Alman B., Taketo M. M., Quaggin S. E. (2012). A new cre driver mouse line, tcf21/pod1-cre, targets metanephric mesenchyme. PLoS ONE 7, e40547 10.1371/journal.pone.0040547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieman B. J., Wong M. D., Henkelman R. M. (2011). Genes into geometry: imaging for mouse development in 3D. Curr. Opin. Genet. Dev. 21, 638-646 10.1016/j.gde.2011.08.009 [DOI] [PubMed] [Google Scholar]
- Norris F. C., Wong M. D., Greene N. D. E., Scambler P. J., Weaver T., Weninger W. J., Mohun T. J., Henkelman R. M., Lythgoe M. F. (2013). A coming of age: advanced imaging technologies for characterising the developing mouse. Trends Genet. 29, 700-711 10.1016/j.tig.2013.08.004 [DOI] [PubMed] [Google Scholar]
- Quaggin S. E., Schwartz L., Cui S., Igarashi P., Deimling J., Post M., Rossant J. (1999). The basic-helix-loop-helix protein pod1 is critically important for kidney and lung organogenesis. Development 126, 5771-5783 [DOI] [PubMed] [Google Scholar]
- Schneider J. E., Böse J., Bamforth S. D., Gruber A. D., Broadbent C., Clarke K., Neubauer S., Lengeling A., Bhattacharya S. (2004). Identification of cardiac malformations in mice lacking Ptdsr using a novel high-throughput magnetic resonance imaging technique. BMC Dev. Biol. 4, 16 10.1186/1471-213X-4-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharpe J., Ahlgren U., Perry P., Hill B., Ross A., Hecksher-Sørensen J., Baldock R., Davidson D. (2002). Optical projection tomography as a tool for 3D microscopy and gene expression studies. Science 296, 541-545 10.1126/science.1068206 [DOI] [PubMed] [Google Scholar]
- Skarnes W. C., Rosen B., West A. P., Koutsourakis M., Bushell W., Iyer V., Mujica A. O., Thomas M., Harrow J., Cox T., et al. (2011). A conditional knockout resource for the genome-wide study of mouse gene function. Nature 474, 337-342 10.1038/nature10163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sussman D., van Eede M., Wong M. D., Adamson S. L., Henkelman M. (2013). Effects of a ketogenic diet during pregnancy on embryonic growth in the mouse. BMC Pregnancy Childbirth 13, 109 10.1186/1471-2393-13-109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Eede M. C., Scholz J., Chakravarty M. M., Henkelman R. M., Lerch J. P. (2013). Mapping registration sensitivity in MR mouse brain images. NeuroImage 82, 226-236 10.1016/j.neuroimage.2013.06.004 [DOI] [PubMed] [Google Scholar]
- Weninger W. J., Geyer S. H., Mohun T. J., Rasskin-Gutman D., Matsui T., Ribeiro I., Costa L. D. F., Izpisúa-Belmonte J. C., Müller G. B. (2006). High-resolution episcopic microscopy: a rapid technique for high detailed 3D analysis of gene activity in the context of tissue architecture and morphology. Anat. Embryol. 211, 213-221 10.1007/s00429-005-0073-x [DOI] [PubMed] [Google Scholar]
- Wong M. D., Dorr A. E., Walls J. R., Lerch J. P., Henkelman R. M. (2012). A novel 3D mouse embryo atlas based on micro-CT. Development 139, 3248-3256 10.1242/dev.082016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong M. D., Dazai J., Walls J. R., Gale N. W., Henkelman R. M. (2013a). Design and implementation of a custom built optical projection tomography system. PLoS ONE 8, e73491 10.1371/journal.pone.0073491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong M. D., Spring S., Henkelman R. M. (2013b). Structural stabilization of tissue for embryo phenotyping using micro-CT with iodine staining. PLoS ONE 8, e84321 10.1371/journal.pone.0084321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamyadi M., Baghdadi L., Lerch J. P., Bhattacharya S., Schneider J. E., Henkelman R. M., Sled J. G. (2010). Mouse embryonic phenotyping by morphometric analysis of MR images. Physiol. Genomics 42A, 89-95 10.1152/physiolgenomics.00091.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Schneider J. E., Portnoy S., Bhattacharya S., Henkelman R. M. (2010). Comparative SNR for high-throughput mouse embryo MR microscopy. Magn. Reson. Med. 63, 1703-1707 10.1002/mrm.22352 [DOI] [PubMed] [Google Scholar]