Abstract
Purpose
Activating extrinsic apoptotic pathways targeting death receptors (DR) using agonistic antibodies or tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) is promising for cancer therapy. However, most pancreatic cancers are resistant to TRAIL therapy. The present studies aimed to identify combination therapies that enhance the efficacy of TRAIL therapy; and to investigate the underlying mechanisms.
Experimental Design
A xenograft model in nude mice was used to determine pancreatic cancer tumorigenesis and therapeutic efficacy of TRA-8, a monoclonal agonistic antibody for DR5. Pancreatic cancer cells were used to characterize mechanisms underlying poly(ADP-ribose) polymerase-1 (PARP-1) in regulating TRA-8-induced apoptosis in vitro.
Results
PARP-1 was found highly expressed in the TRA-8-resistant PANC-1 and Suit-2 cells, compared with TRA-8-sensitive BxPc-3 and MiaPaca-2. Inhibition of PARP-1 with a pharmacologic inhibitor sensitized PANC-1 and Suit2 cells to TRA-8 induced apoptosis in a dose-dependent manner. Furthermore, small interfering RNAs specifically knocking down PARP-1 markedly enhanced TRA-8-induced apoptosis in vitro, and augmented the efficacy of TRA-8 therapy on tumorigenesis in vivo. PARP-1 knockdown increased TRA-8-induced activation of caspase-8 in the death-induced signaling complex (DISC). Immuoprecipitation with DR5 antibody identified the recruitment of PARP-1 and PARP-1-mediated protein poly-ADP-ribosylation(pADPr) modification in the DR5-associated DISC. Further characterization revealed that PARP-1-mediated pADPr modification of caspase-8 inhibited caspase-8 activation, which may contribute to its function in regulating TRA-8 resistance.
Conclusions
Our studies not only provide novel molecular insights into the function of PARP-1 in regulating the extrinsic apoptosis machinery, but also support interventions combining PARP-1 inhibitors with death receptor agonists for pancreatic cancer therapy.
Keywords: Pancreatic Cancer, resistance, death receptor, apoptosis, PARP-1
INTRODUCTION
Pancreatic cancer is an important therapeutic challenge with nearly similar incidence and mortality rates. It is the most lethal cancer of the digestive system with a 5-year survival rate of 5% (1) . Due to the resistance of pancreatic cancer to chemotherapy and radiotherapy, current treatments for pancreatic cancer are generally not curative with modest benefit (2, 3). We and others have demonstrated that modulating death receptor-activated extrinsic apoptosis pathways may provide potential novel avenues for cancer therapy (4, 5). Upon ligand stimulation, death receptors (Fas or death receptor 4/5, DR4/5) recruit Fas-associated death domain (FADD) and the initiator caspases, caspase-8 or caspase-10, resulting in formation of the death-inducing signaling complex (DISC). The recruited caspase-8 or caspase-10 undergoes autocatalytic cleavage and activation to trigger the caspase cascade that ultimately leads to apoptosis (4, 5). We have previously reported that regulating the DISC formation promotes Fas death receptor-induced apoptosis of cancer cells, including pancreatic cancer cells (6-10).
Activating the DR4/5-mediated extrinsic apoptotic signaling pathway by tumor necrosis factor– related apoptosis-induced ligand (TRAIL) induces apoptosis of a variety of cancer cells, including melanoma, colon, lung, breast, kidney and pancreatic cancers (11, 12) . With virtually no toxicity for normal cells, recombinant TRAIL or agonistic antibodies specifically targeting DR4/5 are currently being tested in several phase I-III clinical trials (12-14). Although TRAIL is a promising anticancer agent, many cancer cells, including pancreatic cancer cells, develop resistance to TRAIL-induced apoptosis. The resistance of cancer cells to TRAIL-induced apoptosis has been associated with mutations of DR5 receptor (15), increased expression of the decoy death receptors DcR1 or DcR2(16, 17), decreased expression of apoptotic protein caspase-8 (18), and increased expression of a number of anti-apoptotic proteins, such as Bcl-2, Bcl-XL (19, 20) and the FLICE-like inhibitory protein (FLIP)(21, 22).
Combination therapy with other anticancer agents has been shown to enhance the efficacy of TRAIL-treatment for the resistant cancers (23-25). We and others have found that activating intrinsic apoptotic signaling pathways using gemcitabine (26), doxorubicin (27) or ionizing radiation (28), sensitizes resistant cancer cells to death receptors-induced apoptosis. These intrinsic apoptotic activators function via inducing mitochondria leakage or DNA damage (26-28).
The poly(ADP-ribose) polymerases (PARPs) are key to repairing damaged DNA by modifying proteins associated with DNA repair (29). Inhibition of PARPs results in accumulation of irreparable damaged DNA and facilitates triggering of the intrinsic apoptosis signaling pathway in cancer therapy (29). PARP-1 is the most abundant of PARPs, accounting for 75–90% of the PARPs (29). Inhibition of PARP-1 through siRNA knockdown or chemical inhibitors for PARP has been used to treat cancers with DNA repair deficiencies, such as BRCA-deficient breast and ovarian cancer (30) as well as BRCA2-associated pancreatic cancer (31). Additionally, combined use of PARP inhibitors with DNA damaging reagents or radiation sensitizes glioma, ovarian and pancreatic cancers to therapy (32-34).
The role of PARP-1 in death receptor-mediated extrinsic apoptotic signaling pathways is unknown. The present study provides the first evidence that PARP-1 inhibition sensitizes resistant pancreatic cancer cells to DR5-activated apoptosis and enhances the efficacy of DR5 antibody therapy for pancreatic cancers in a mouse tumor xenograft model. Furthermore, we have uncovered a novel mechanism of PARP-1 in regulating DR5-induced DISC recruitment and subsequent caspase-8 activation, which is independent of its known function in DNA repair. Our studies provide novel molecular insights into the function PARP-1 in the death receptor-activated apoptosis machinery, and support the use of combined therapy with DR5 agonists and PARP inhibitors for resistant pancreatic tumors.
MATERIALS AND METHODS
Cell culture, antibodies and reagents
The human pancreatic cancer cell lines BxPc-3, MiaPaCa-2, PANC-1 and Suit-2 were from the American Type Culture Collection (ATCC, Manassas, VA, USA). BxPc-3 and Suit-2 cells were grown in RPMI 1640; MiaPaCa-2 and PANC-1 cells were grown in DMEM (Invitrogen, Carlsbad, CA) supplemented with penicillin (5 units/ml), streptomycin (5 ug/ml), and 10% heat-inactivated fetal bovine serum.
DR5 agonist antibody TRA-8 was generated as previously described (35). All antibodies used were commercially available, including FADD (Millipore, Billerica, MA), caspase-3 and pADPr (Enzo Life, Plymouth Meeting, PA), caspase-8 (BD Bioscience, San Jose, CA and Cell Signaling Technology, Danvers, MA), PARP-1 and GAPDH (Santa Cruz Biotech, Santa Cruz, CA), DR5 (Prosci, Poway, CA).
Protein G agarose was from Invitrogen (Carlsbad, CA). Recombinant human PARP-1 protein was purchased from Enzo (Plymouth Meeting, PA). β-NAD was from Sigma-Aldrich (St. Louis, MO). Active human recombinant caspase-8 was from Research & Diagnostics (Minneapolis, MN). Full length mouse recombinant caspase-8 was from Abcam (Cambridge, MA)
Gene knockdown with lentivirus-delivered shRNA
Lentiviral constructs expressing a 21-nucleotide short hairpin RNA (shRNA) targeting human PARP-1 gene (GenBank accession number NM_001618.3) or human FADD gene (GenBankTM accession number NM_003824.3) were purchased from Open Biosystems (Huntsville, AL). The constructs were packaged into lentivirus-like particles pseudotyped with the vesicular stomatitis virus glycoprotein as we previously described (10). Transduction was performed by incubating cancer cells with lentivirus, and stably transduced cells were selected with puromycin (2μg/ml).
Assessment of apoptosis
Apoptosis was determined by Annexin V-FITC and propidium iodide (PI) staining (BD Biosciences) and analyzed by flow cytometry (BD Biosciences).
Western blot analysis
Proteins were extracted, quantified with a BCA protein assay kit (Thermo Scientific, Waltham, MA), separated by SDS-PAGE and transferred to Immobilon P membranes (Millipore, Billerica, MA) as described previously (10). Membranes were blocked in 5% non-fat milk, incubated with primary antibodies overnight at 4°C and then horseradish peroxidase-conjugated secondary antibodies for 1 hour at room temperature. Signals were detected using the Immobilon Western chemiluminescent horseradish peroxidase substrate detection kit (Millipore).
Immunoprecipitation
Five hundred micrograms of extracted proteins were incubated with 1 μg of antibodies as indicated in figure legends for 1 hour, and subsequently incubated with 50 μl of 1:1 slurry of protein G-agarose beads overnight at 4°C. Beads were then washed and 20μl 2xLaemmli sample buffer was added to the beads followed by heating at 95°C for 5 minutes and chilling on ice. After brief centrifugation, proteins in the supernatant were analyzed by Western blotting with specific antibodies.
In Vitro pADPr modification and caspase-8 activation assays
Recombinant full length mouse Caspase-8 (300ng, Abcam) was immunoprecipitated using antibody for caspase-8. Immunoprecipitated beads were incubated in 100 μl reaction buffer (100 mM Tris-HCl [pH 8], 10 mM MgCl2, 1 mM dithiothreitol) with or without 100 ng of PARP-1 and 500 μM β-NAD for 30 minutes at 37°C and then washed 3 times.
For caspase-8 activation analysis, immunoprecipitated beads were incubated in 100 μl reaction buffer (20 mM HEPES [pH 7,5], 5 mM dithiothreitol) with or without 100 ng of recombinant active human caspase-8 (Research & Diagnostic) at 37°C for 60 minutes. Beads were then washed and 20μl 2×Laemmli sample buffer was added to the beads followed by heating at 95°C for 5 minutes and chilling on ice. After brief centrifugation, proteins in the supernatant were analyzed by Western blotting with specific antibodies.
Mouse xenograft model
The animal protocol was approved by the Institutional Animal Care and Use Committee at the University of Alabama at Birmingham, Birmingham, AL, USA. Male athymic nu/nu mice (6 weeks old, NCI-Frederick) were used for tumor inoculation. Briefly, control PANC-1 pancreatic cancer cells or PANC-1 cells with PARP-1 knockdown were inoculated subcutaneously into the flanks of mice (2×106 cells in 200 ml PBS/site). Three weeks following tumor inoculation, mice were divided into 2 groups (8 mice per group), mice in the treatment group were intraperitoneally injected with TRA-8 once a week for 6 weeks (200μg/mice), and mice in the control group were injected with 0.9% sodium chloride. Tumor size and body weight were measured every week and volumes were determined using the formula volume=length × width2/2. At the end of the experiment, tumors were removed from mice, half of each tumor was homogenized for Western blot analysis and the other half was fixed in 4% paraformaldehyde and embedded in paraffin.
TUNEL staining
TUNEL staining was performed on tumor sections (7 μm) (DeadEnd Fluorometric TUNEL System; Promega) to determine cell death, and DAPI staining (4',6-diamidine-2-phenylindole dihydrochloride) was used to identify nuclei. Stained specimens were examined microscopically (Leica M165 FC). For quantitative analysis, cell numbers were counted under a microscope (x 200). Four fields in each slide were counted and the percentage of apoptotic cells was determined.
Statistical analysis
Results are expressed as means ± SD. Differences between two groups were identified with Student's t-test. For multiple groups, one-way analysis of variance and Student-Newman-Keuls tests was performed to identify differences. Significance was defined as p < 0.05.
RESULTS
Inhibition of PARP-1 activity sensitizes resistant pancreatic cancer cells to TRA-8
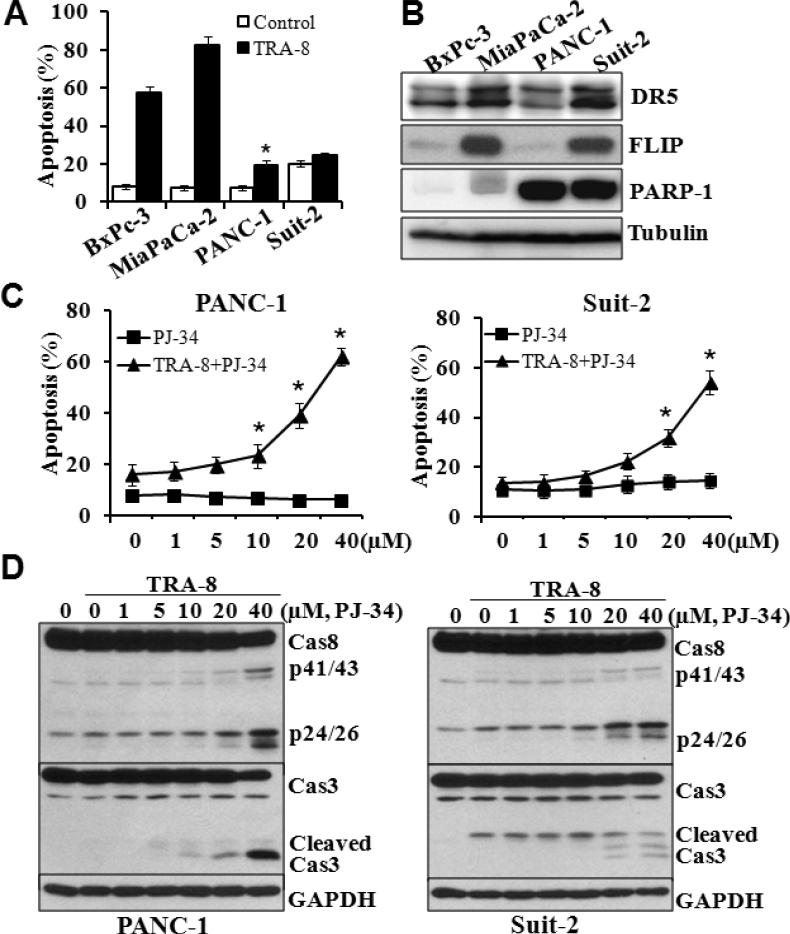
Using TRA-8, a monoclonal agonistic antibody specifically targeting DR5 (35), we determined death receptor-activated apoptosis in four pancreatic cell lines. BxPc-3 and MiaPaCa-2 cells were highly sensitive to TRA-8-induced apoptosis, whereas PANC-1 and Suit-2 cells are resistant to TRA-8 (Fig 1A). To understand the underlying mechanisms, we analyzed the expression of the receptor DR5, as well as the expression of an anti-apoptotic protein FLIP (Fig 1B) that has been shown to inhibit DR5-induced apoptosis (36). The expression of DR5 is higher in MiaPaCa-2 and Suit2 cells compared with that in BxPc-3 and PANC-1 cells, which was not correlated with their sensitivity to TRA-8-induced apoptosis. Similarly, the expression level of FLIP was not correlated to the resistance of the cells to TRA-8-induced apoptosis, suggesting that additional mechanisms may contribute to TRA-8 resistance.
Figure 1. Inhibition of PARP-1 sensitizes TRAIL-resistant pancreatic cancer cells to TRA-8.
A. Apoptosis of pancreatic cancer cells, analyzed by Annexin V/PI staining, after exposure to TRA-8 (0.5μg/ml) for 24 hours (n=3, *p<0.01, **p<0.001 compared with control). B. Western blot analysis of the expression of Death Receptor 5 (DR5), c-FLIP, PARP-1 and tubulin in pancreatic cancer cells. C. Apoptosis of PANC-1 and Suit-2 cells that were exposed to increasing concentrations of PJ34, with or without of TRA-8 (0.5μg/ml) for 24 hours (n=3, *p<0.05). D. Western blot analysis of the expression of caspase-8 and caspase-3 in PANC-1 and Suit-2 cells that were exposed to increasing concentrations of PJ34 with TRA-8 (0.5μg/ml) for 8 hours. The expression of GAPDH was used as a loading control. Representative blots of three independent experiments are shown.
We found that the expression of PARP-1 was correlated with TRA-8 resistance of the pancreatic cancer cells. The expression of PARP-1 was markedly higher in the TRA-8-resistant cell PANC-1 and Suit-2 cells, compared with that in the TRA-8-sensitive BxPc-3 and MiaPaCa-2 cells (Fig 1B), suggesting a role of PARP-1 in TRA-8 resistance. Using a pharmacological inhibitor for PARPs, PJ-34, we found that inhibition of PARP activity enhanced TRA-8-induced apoptosis in PANC-1 and Suit-2 cells in a concentration-dependent manner (Fig 1C). Such an effect of PJ-34 was not due to its toxicity, as PJ-34 did not induce apoptosis at all concentrations used (0-40μM). Consistent with its effects on TRA-8-induced apoptosis, PJ-34 dramatically enhanced TRA-8-induced activation of caspase-8 and its downstream apoptotic effector, caspase-3 (Fig 1D).
Knockdown of PARP-1 sensitizes TRAIL resistant pancreatic cancer cells to TRA-8
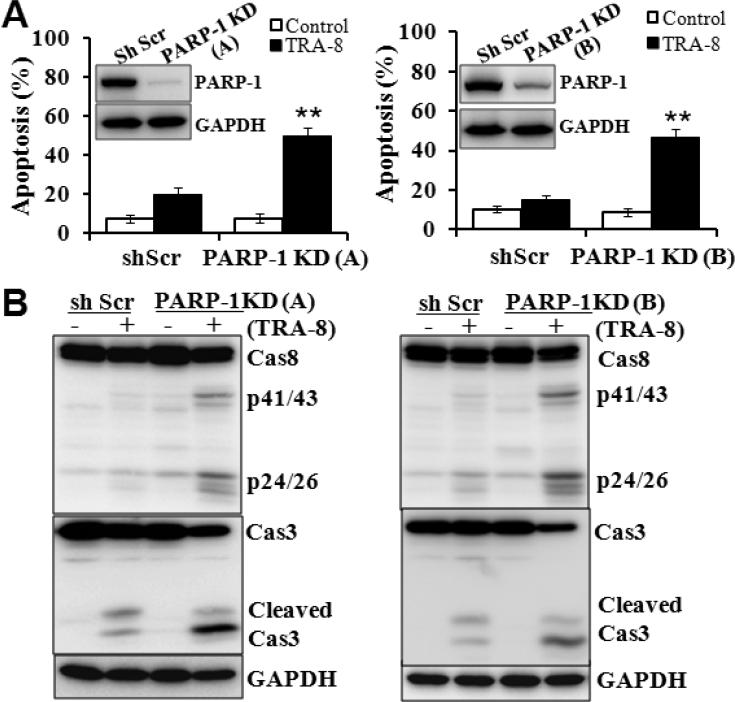
The specific role of PARP-1 in mediating TRA-8 resistance of pancreatic cancer cells was further determined using shRNAs specific for PARP-1. Two lines of PANC-1 cells with PARP-1 knockdown were generated using shRNAs specifically targeting different regions of the PARP-1 gene. Knockdown of PARP-1 by the two shRNAs was confirmed by Western blot analysis (Fig 2A, inserts). Knockdown of PARP-1 per se did not affect cell viability (Fig 2A, white bars), but significantly increased the sensitivity of PANC-1 cells to TRA-8 induced apoptosis (Fig 2A, black bars). Similar to the observations with PJ-34 (Fig 1D), PARP-1 knockdown increased TRA-8-induced activation of caspase-8 and caspase-3 (Fig 2B).
Figure 2. PARP-1 knockdown sensitizes TRAIL-resistant pancreatic cancer cells to TRA-8.
A. Apoptosis of control (shScr) or PARP-1 knockdown cells (PARP-1 KD) after exposure to TRA-8 (0.5μg/ml) for 24 hours (n=3 for each group, *p<0.001). Western blot analysis of the expression of PARP-1 in the control shScr cells and two lines of PARP-1 KD cells is shown in the inserts. B. Western blot analysis of the expression of caspase-8, caspase-3 and GAPDH in control shScr and two PARP-1 KD cells exposed to TRA-8 (0.5μg/ml) for 8 hours. Representative blots from three independent experiments are shown.
PARP-1 knockdown enhances efficacy of TRA-8 therapy in vivo
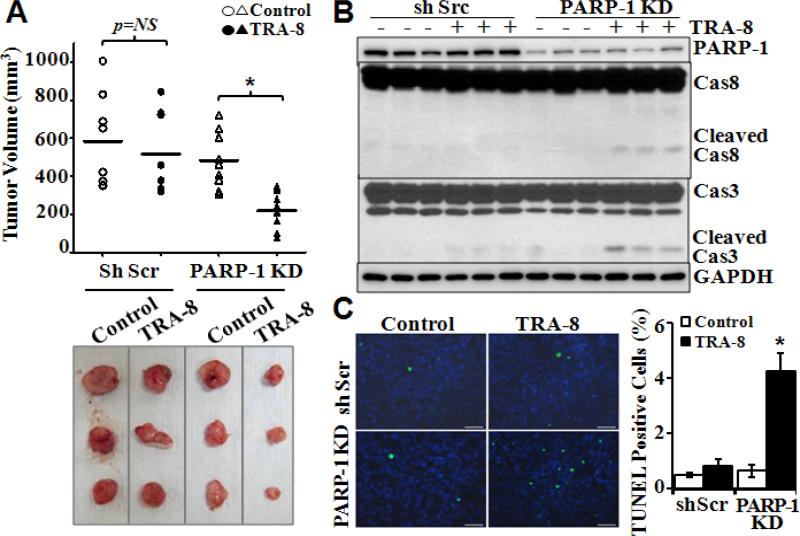
To determine the effects of PARP-1 knockdown on the efficacy of TRA-8 therapy on tumorigenesis in vivo, we characterized tumorigenesis and response to TRA-8 treatment of PARP-1 knockdown PANC-1 cells in a mouse xenograft model. PANC-1 cells with scrambled shRNA were used as controls. In the control groups, TRA-8 treatment did not reduce tumor size, further demonstrating resistance of these cancer cells to TRA-8 therapy (Fig 3A). In contrast, TRA-8 treatment effectively reduced tumor size of the PARP-1 knockdown cells (Fig 3A). Western blot analysis determined that the expression of PARP-1 was maintained at a lower level in xenograft tumors of PARP-1 knockdown cells, which was not affected by TRA-8 treatment (Fig 3B). Activation of caspase-8 and its downstream apoptosis effector caspase-3, indicated by cleaved caspase-8 and caspase-3, was demonstrated in PARP-1 knockdown tumors from the TRA-8-treated mice (Fig 3B). Furthermore, TUNEL staining of tumor sections demonstrated significant increases in cell death in the PARP-1 knockdown tumors from TRA-8-treated mice (Fig 3C).
Figure 3. PARP-1 knockdown enhances the efficacy of TRA-8 on tumorigenesis in mice.
A. Tumorigenesis of control shScr cells and PARP-1 KD cells in nude mouse model as described in “materials and methods”. Tumor volumes and representative tumors in each group at 6 weeks after treatment are shown. NS: no significance; n=8, *p<0.001. B. Western blot analysis of the expression of PARP-1, caspase-8, caspase-3 and GAPDH in isolated tumors. C. Cell death, analyzed by TUNEL staining, in tumors; and Quantitative analysis of TUNEL-positive cells as percentage of total cells in the tumor sections (n=5 tumors in each group, * p<0.001, bar=50 μm).
Activation of caspase-8 is responsible for PARP-1 knockdown-rendered TRA-8 sensitivity
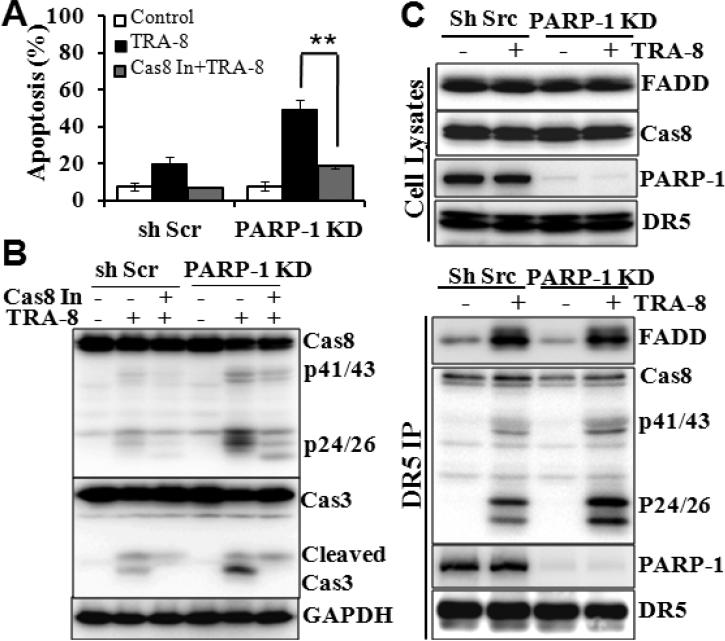
Because inhibition of PARP-1, by PJ-34 (Fig 1) or by shRNAs (Fig 2), increased TRA-8-induced activation of the upstream apoptosis initiator, caspase-8, we determined whether inhibition of caspase-8 may block the effects of PARP-1 inhibition. Z-IETD-FMK, a caspase-8 inhibitor, blocked TRA-8-induced apoptosis in the control PANC-1 cells. Moreover, inhibition of caspase-8 significantly reduced TRA-8-induced apoptosis in the PARP-1 knockdown cells (Fig 4A). Consistently, caspase-8 inhibition resulted in marked decreases in TRA-8-induced cleavage/activation of caspase-8 and its downstream caspase-3 in the PARP-1 knockdown cells (Fig 4 B).
Figure 4. Knockdown of PARP-1 sensitizes cells to TRA-8 through enhancing activation of caspase-8.
Control shScr cells or PARP-1 KD cells were exposed to TRA-8 (0.5μg/ml), with or without the caspase-8 inhibitor (Cas8 In, Z-IETD-FMK, 20μmol/L). A. Apoptosis, analyzed at 24 hours after treatment (n=3, **p<0.001). B. Western blot analysis of caspase-8, caspase-3 and GAPDH at 8 hours after treatment. C. Immunoprecipitation analysis of DR5-associated DISC upon TRA-8 stimulation (1μg/ml, 60 minutes). Western blot analysis shown the expression of FADD, caspase 8, PARP-1 and DR5 in cell lysates, and Western blot analysis of DR5-associated DISC recruitment of FADD, caspase-8, PARP-1. Representative blots from three independent experiments are shown.
Furthermore, knockdown of PARP-1 did not affect the expression of the death receptor DR5, the adaptor protein FADD, or the procaspase-8 (Fig 4C). As procaspase-8 is known to be proteolytically cleaved and activated in the DR5-induced DISC, we analyzed DR5-recrutied DISC proteins, including FADD and caspase-8, in response to TRA-8 stimulation. PARP-1 knockdown did not affect the recruitment of FADD into DR5-associated DISC in response to TRA-8 stimulation (Fig 4C). As expected, TRA-8 induced the recruitment and activation of caspase-8 into the DR5-associated DISC in the control cells (Fig 4C, shScr). TRA-8-induced DR5-recruitment of procaspase-8 appeared to be similar in the control and PARP-1 knockdown cells. However, increased activation of caspase-8 was demonstrated in the PARP-1 knockdown cells (Fig 4C, p41/43 and p24/26). Furthermore, PARP-1 was identified in the DR5-associated complex in the control cells under the basal condition, which was not affected by TRA-8 treatment (Fig 4C, PARP-1).
Activation of DR5 induces pADPr modification of DISC component proteins
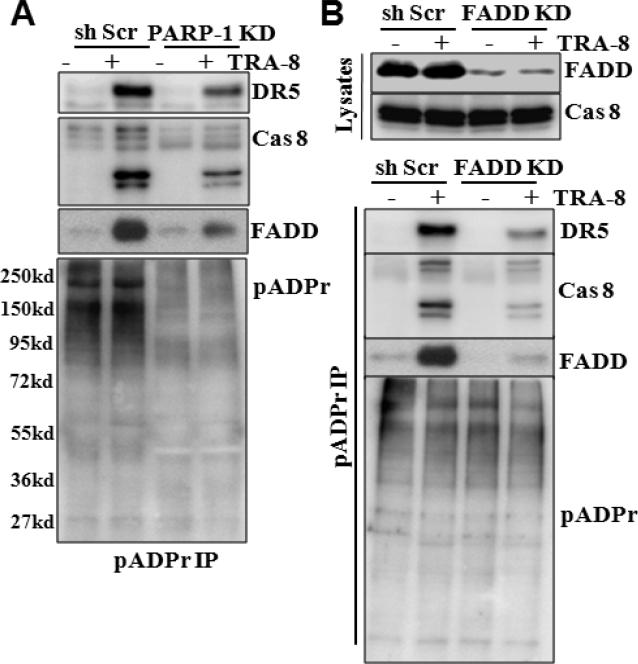
A key function of PARP-1 is to modify proteins by adding poly(ADP-ribose) (pADPr) chain to target proteins, which regulates the function of the modified proteins. To determine the function of PARP-1 in the DISC, we analyzed pADPr modification profile of the DR5-associated DISC proteins. Using anti-pADPr antibody, we determined that knockdown of PARP-1 significantly reduced overall protein pADPr modification in the PANC-1 cells (Fig 5A). TRA-8 treatment did not affect overall protein pADPr modification profile (Fig 5A, pADPr). However, increased pADPr modification on DISC components, DR5, FADD and caspase-8, was detected in the TRA-8-treated control cells (Fig 5A, shScr). Furthermore, PARP-1 knockdown decreased TRA-8-induced pADPr modification on DR5, FADD and caspase-8 (Fig 5A, PARP-1 KD).
Figure 5. Activation of DR5 induces pADPr modification of DISC proteins.
A. pADPr modification of DISC proteins. Control (shScr) and PARP-1 KD cells were exposed to TRA-8 (1μg/ml) for 60 minutes. Immunoprecipitation was performed with pADPr antibody, and Western blotting analysis was performed to determined pADPr-modification of DR5, caspase-8 and FADD. B. The effects of FADD knockdown on pADPr-modification of DISC proteins. Expression of FADD in the control (shScr) and FADD KD cells was determined by Western blot analysis. Immunoprecipitation performed with pADPr antibody and Western blot analysis of pADPr-modified DR5, caspase-8 and FADD. Representative blots from three independent experiments are shown.
To further determine that pADPr modification of DR5, FADD and caspase-8 occurred in the DR5-associated DISC, we knocked down FADD, the adaptor protein of DISC formation, with shRNA specifically targeting FADD (Fig 5B). Knockdown of FADD did not affect the overall protein pADPr modification profile, but decreased TRA-8-induced pADPr modification on DR5 and caspase-8 (Fig 5B, pADPr), indicating that TRA-8-induced pADPr modification on DR5 and caspase-8 is dependent on FADD-mediated DISC formation (Fig 5B).
pADPr modification inhibits caspase-8 activation
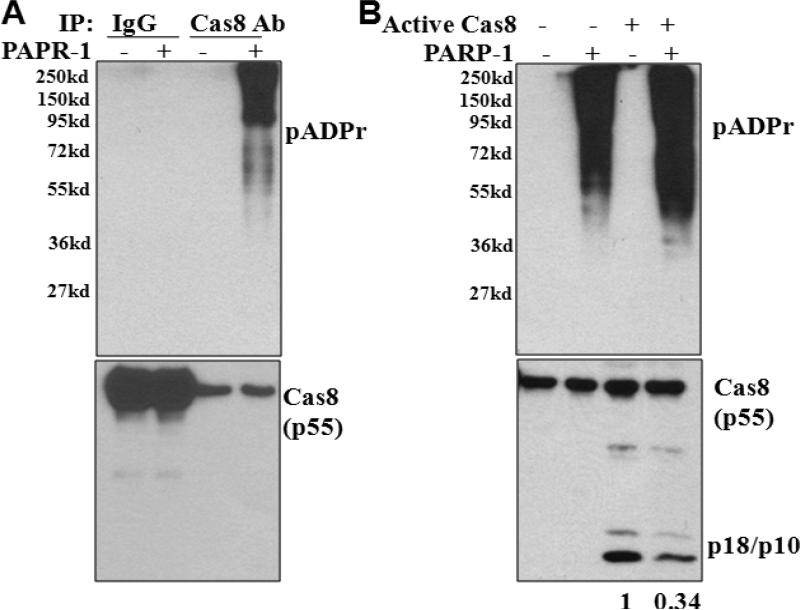
Using purified recombinant PARP-1 and caspase-8 proteins, a direct effect of PARP-1 on pADPr modification of caspase-8 was demonstrated (Fig 6A). Furthermore, the effect of pADPr modification of caspase-8 on its activation was characterized. In the absence of PARP-1, active caspase-8 cleaved procaspase-8 into cleaved p10/18 fragments (Fig 6B). However, decreased caspase-8 cleavage was observed when PARP-1 was added (last lane, Fig 6B), indicating that pADPr modification of caspase-8 inhibited caspase-8 activation.
Figure 6. pADPr modification of caspase-8 decreases caspase-8 activation.
A. pADPr modification of caspsae-8 by recombinant PARP-1 in vitro. Recombinant caspase-8 was incubated with IgG (control) or caspase-8 antibody (Cas8 Ab) conjugated to protein G-agarose beads in the presence of absence of PARP-1 protein. After washes, proteins on the beads were subject to SDS-PAGE and Western blot analysis for pADPr-modification (pADPr). B. pADPr modification inhibits cleavage/activation of caspase-8 in vitro. Caspase-8 was conjugated to protein G-agarose beads by Cas8 Ab and incubated with/without PARP-1 recombinant protein as A, and then exposed to addition of active caspase-8. Western blot analysis determines the effects of PARP-1-mediated pADPr-modification on caspase-8 cleavage. The numbers shown on the bottom depict relative density of cleaved caspase-8 (p18/p10). Representative blots from three independent experiments are shown.
DISCUSSION
TRAIL therapies have shown low toxicity in patients in several clinical trials (12-14). However, therapeutic effects of TRAIL agonists as a monotherapy have been limited due to tumor resistance (12-14). Experimental studies have postulated multiple mechanisms that are responsible for TRAIL resistance, including low expression of the death receptors DR4/DR5, high expression of decoy receptor DcR1/DcR2, and increased expression of anti-apoptotic molecular such as FLIP (12). However, a consistent correlation between the expression of DR4/DR5 and TRAIL sensitivity of cancer cells has not been determined (12). We demonstrated in the present study that the expression of DR5 was not correlated with the sensitivity of pancreatic cancer cells to DR5 agonistic antibody TRA-8-induced apoptosis. TRA-8 is a monoclonal antibody that specifically targets DR5, but not DR4 or the decoy receptors DcR1 and DcR2 (35), thus the resistance of the pancreatic cancer cells to TRA-8-induced apoptosis is independent of the expression of the decoy receptors. Additionally, the expression level of the anti-apoptotic protein FLIP was not correlated to TRA-8 resistance of the pancreatic cancer cells, although we and others have demonstrated a protective effect of FLIP on death receptor-induced apoptosis in several cancer cells (6, 21, 22) .
We found that the expression of PARP-1 contributed to the TRA-8 resistance of pancreatic cancer cells. As the predominant member of the PARP family that regulates protein pADPr modification, PARP-1 is an important regulator of the DNA base excision repair pathway and is essential for the maintenance of genomic integrity and cell survival in response to genotoxic insults (37, 38). PARP-1 expression is elevated in several cancer cells (39) and is associated with adverse outcomes of breast and ovarian cancers (39, 40). Consistently, we found that the expression of PARP-1 was increased in two pancreatic cell lines, PANC-1 and Suit-2 cells, which exhibited resistance to TRA-8-induced apoptosis. Inhibition of PARP-1, using a pharmacological inhibitor or shRNA, rendered the resistant cells more sensitive to TRA-8-induced apoptosis, demonstrating the engagement of PARP-1 in TRA-8-resistent pancreatic cancer cells.
Consistent with its essential role in DNA repair, PARP-1 antagonists, used as a mono-therapy for tumors with DNA repair deficiencies or in combination with DNA damage-inducing agents, increase accumulation of damaged DNA that triggers intrinsic apoptotic pathways (31-34). In pancreatic cancer, PARP inhibitors enhance the effects of chemotherapy with DNA alkylating agents or radiation therapy, both via promoting DNA damage-induced intrinsic apoptotic signaling pathways. The present studies have provided the first evidence that PARP-1 inhibition alone did not induce apoptosis of pancreatic cancer cells, but enhanced TRA-8-induced apoptosis, suggesting that PARP-1 regulates TRA-8-induced extrinsic apoptosis via a mechanism independent of its function in DNA repair.
The expression of PARP-1 was identified in both nucleus and cytoplasm of the PANC-1 and Suit-2 cells (Data not shown). PARPs are mainly localized in the nucleus, where they execute the DNA repair function (29). Recently, PARP-1 has also been identified in the cytoplasm of neurons (41) and smooth muscle cells (42), as well as in the mitochondria of fibroblasts and HeLa cells (43). In breast cancer, expression of cytoplasmic PARPs is correlated with tumor aggressiveness, higher risk for relapse and death (44). However, the function of cytoplasmic PARP-1 in cancer is not clear. PARP-1 is capable of modifying proteins via pADPr (29, 32). Chang et al. have shown that pADPr modification of the spindle pole protein NuMA in the cytoplasm by PARP-1 is required for assembly, maintenance of bipolar spindle structure as well as the function of the mitotic spindle (45). Our studies demonstrated the presence of PARP-1 in a new cellular location in pancreatic cancer cells, the death receptor-associated DISC (Figure 4C). The presence of PARP-1 in the DR5-associated complex enhanced pADPr modification of DISC proteins. An association between PARP-1 and DR5 was demonstrated under basal conditions (Fig 4C, DR5/PARP-1), however, no pADPr modification on DR5 was present (Fig 5A, DR5). pADPr modification of the DISC complex, DR5/Caspase8/FADD, was identified in response to TRA-8 treatment (Fig 5A) and was found to be dependent on the DISC-recruitment of FADD (Fig 5B). Therefore, PARP-1 may not directly modify DR5 at basal conditions, but regulates the pADPr modification of the DR5-associated DISC complex, and thus modulating downstream apoptotic signaling.
Our studies revealed that PARP-1 modified caspase-8; and determined a novel function of PARP-1 in regulating pADPr modification of caspase-8, thereby inhibiting caspase-8 activation. Such an observation is consistent with previous studies demonstrating that pADPr modification inhibits protein function (46, 47). For instance, pADPr modification of p65 NF-kappaB decreases its interaction with the nuclear export factor and thus retains it in the nucleus (46). Similarly, pADPr modification of the ATPase imitation switch protein inhibits ATPase activity by reducing its binding affinity with its nucleosomal substrate (47). The activation of procaspase 8 is believed to occur in the death receptors-activated DISC, which recruits procaspase-8 to form homodimers that provide the proximity necessary to initiate cleavage and activation of procaspase-8 (48-50). Therefore, the DISC structure provides a platform for the oligomerization of procaspase-8 that allows two procaspase-8 homodimers to be in close proximity. By the same token, the presence of PARP-1 in the DR5-associated complex provides the proximity for PARP-1 to modify (pro)caspase-8 in the DISC. This concept is supported by several lines of evidence: 1) pADPr modification of caspase-8 was identified in the DR5-associated DISC induced by TRA-8; 2) inhibition of DISC formation by FADD knockdown blocked pADPr modification on caspase-8; 3) recombinant PARP-1 induced pADPr modification of caspase-8 and inhibited caspase-8 cleavage/ activation directly. We found PARP-1 inhibited the cleavage of procaspase-8 into the p18/p10 active form (Fig 6B). The amino acids that are critical for the cleavage of caspase-8 into the p18/p10 active form are the aspartate residues at 216, 374 and 384 (48, 49). Because aspartate is one of the PARP-1 targeting modification residues (29), pADPr modification on these caspase-8 aspartate residues may contribute to the inhibition of caspase-8 cleavage and activation. In support of this concept, when PARP-1 was inhibited, by a pharmacological inhibitor or shRNA, TRA-8-induced activation of caspase-8 was increased, and TRA-8-induced apoptosis was enhanced. This is also supported by the observation that caspase-8 inhibitor blocked the effect of PARP-1 inhibition on TRA-8-induced apoptosis.
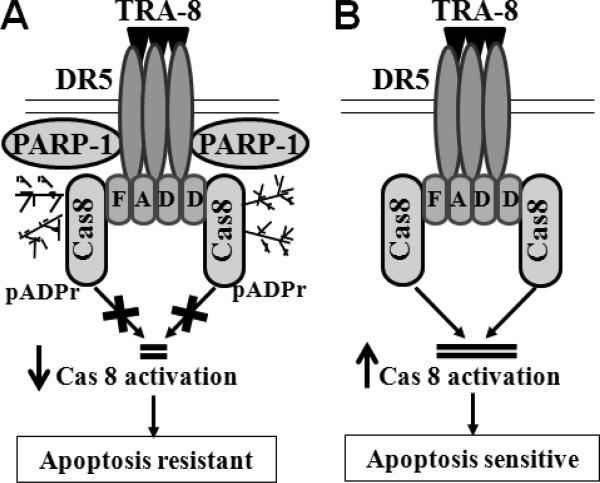
In summary, we have found that PARP-1 knockdown sensitized resistant pancreatic cancer cells to TRA-8-induced apoptosis in vitro and enhanced the efficacy of TRA-8 therapy of pancreatic tumors in vivo. PARP-1 was identified in the DR5-associated complex, which induced pADPr modification of caspase-8 in the TRA-8-activated DR5-associated DISC and modulated caspase-8 activation. Therefore, inhibition of PARP-1 blocks pADRr modification of caspase-8, which facilitates DR5-mediated activation of caspase-8 in the DISC, and thus sensitizing tumor cells to TRA-8-induced apoptosis (Fig 7). These studies demonstrate a novel function of PARP-1 in the death receptor-activated apoptotic signaling machinery, which provides important molecular insights into the mechanisms underlying resistance of pancreatic cancer to TRAIL therapy. Results from the present studies support interventions combining PARP-1 inhibitors with death receptor agonists for pancreatic cancer therapy.
Figure 7. A Schematic model of PARP-1 regulation of DR5-mediated apoptosis.
A. In response to TRA-8, DR5-bound PARP-1 modifies DR5-recruited caspase-8 (Cas8), which inhibits DISC-associated cleavage and activation of caspsae-8. Therefore, high expression of PARP-1 renders tumor cells resistant to TRA-8-induced apoptosis. B. Inhibition of PARP-1 blocks pADRr modification of caspase-8, which facilitates DR5-mediated activation of caspsae-8 in the DISC, and therefore sensitizing tumor cells to TRA-8-induced apoptosis.
STATEMENT OF TRANSLATIONAL RELEVANCE.
The present studies explore the potential use of combination therapies that enhance the efficacy of TRAIL therapy of resistant pancreatic cancer, a fatal disease with very limited therapeutic options. Using a pancreatic cancer xenograft model in nude mice, we have demonstrated that PARP-1 knockdown sensitizes the resistant pancreatic cancer cells to TRA-8-induced apoptosis and thus enhances the efficacy of TRA-8 therapy. PARP inhibitors are already used in Clinical Trials for different tumors because of its known function in DNA repair. The studies presented here highlight a novel function of PARP-1 in regulating the extrinsic apoptosis machinery that contributes to pancreatic cancer resistance, and support interventions combining PARP-1 inhibitors with death receptor agonists for pancreatic cancer therapy. Thus these studies set up a stage for a possibility of clinical applications of the combination therapy in pancreatic cancer patients.
Acknowledgments
This work was supported in part by UAB Department of Pathology start-up funds to YC and a VA Merit Review Award to JMM.
Footnotes
Disclosure: None
REFERENCES
- 1.Hartman DJ, Krasinskas AM. Assessing treatment effect in pancreatic cancer. Arch Pathol Lab Med. 2012;136:100–9. doi: 10.5858/arpa.2011-0144-RA. [DOI] [PubMed] [Google Scholar]
- 2.Moore MJ, Goldstein D, Hamm J, Figer A, Hecht JR, Gallinger S, et al. Erlotinib plus gemcitabine compared with gemcitabine alone in patients with advanced pancreatic cancer: a phase III trial of the National Cancer Institute of Canada Clinical Trials Group. J Clin Oncol. 2007;25:1960–6. doi: 10.1200/JCO.2006.07.9525. [DOI] [PubMed] [Google Scholar]
- 3.Conroy T, Desseigne F, Ychou M, Bouche O, Guimbaud R, Becouarn Y, et al. FOLFIRINOX versus gemcitabine for metastatic pancreatic cancer. N Engl J Med. 2011;364:1817–25. doi: 10.1056/NEJMoa1011923. [DOI] [PubMed] [Google Scholar]
- 4.Wilson NS, Dixit V, Ashkenazi A. Death receptor signal transducers: nodes of coordination in immune signaling networks. Nat Immunol. 2009;10:348–55. doi: 10.1038/ni.1714. [DOI] [PubMed] [Google Scholar]
- 5.Yang A, Wilson NS, Ashkenazi A. Proapoptotic DR4 and DR5 signaling in cancer cells: toward clinical translation. Curr Opin Cell Biol. 2010;22:837–44. doi: 10.1016/j.ceb.2010.08.001. [DOI] [PubMed] [Google Scholar]
- 6.Chen Y, Xu J, Jhala N, Pawar P, Zhu ZB, Ma L, et al. Fas-mediated apoptosis in cholangiocarcinoma cells is enhanced by 3,3′-diindolylmethane through inhibition of AKT signaling and FLICE-like inhibitory protein. Am J Pathol. 2006;169:1833–42. doi: 10.2353/ajpath.2006.060234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jing G, Yuan K, Liang Q, Sun Y, Mao X, McDonald JM, et al. Reduced CaM/FLIP binding by a single point mutation in c-FLIP(L) modulates Fas-mediated apoptosis and decreases tumorigenesis. Lab Invest. 2012;92:82–90. doi: 10.1038/labinvest.2011.131. [DOI] [PubMed] [Google Scholar]
- 8.Pawar P, Ma L, Byon CH, Liu H, Ahn EY, Jhala N, et al. Molecular mechanisms of tamoxifen therapy for cholangiocarcinoma: role of calmodulin. Clin Cancer Res. 2009;15:1288–96. doi: 10.1158/1078-0432.CCR-08-1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pawar PS, Micoli KJ, Ding H, Cook WJ, Kappes JC, Chen Y, et al. Calmodulin binding to cellular FLICE-like inhibitory protein modulates Fas-induced signalling. Biochem J. 2008;412:459–68. doi: 10.1042/BJ20071507. [DOI] [PubMed] [Google Scholar]
- 10.Yuan K, Jing G, Chen J, Liu H, Zhang K, Li Y, et al. Calmodulin mediates Fas-induced FADD-independent survival signaling in pancreatic cancer cells via activation of Src-extracellular signal-regulated kinase (ERK). J Biol Chem. 2011;286:24776–84. doi: 10.1074/jbc.M110.202804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dimberg LY, Anderson CK, Camidge R, Behbakht K, Thorburn A, Ford HL. On the TRAIL to successful cancer therapy? Predicting and counteracting resistance against TRAIL-based therapeutics. Oncogene. 2013;32:1341–50. doi: 10.1038/onc.2012.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Holoch PA, Griffith TS. TNF-related apoptosis-inducing ligand (TRAIL): a new path to anti-cancer therapies. Eur J Pharmacol. 2009;625:63–72. doi: 10.1016/j.ejphar.2009.06.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang S. TRAIL: a sword for killing tumors. Curr Med Chem. 2010;17:3309–17. doi: 10.2174/092986710793176285. [DOI] [PubMed] [Google Scholar]
- 14.Wiezorek J, Holland P, Graves J. Death receptor agonists as a targeted therapy for cancer. Clin Cancer Res. 2010;16:1701–8. doi: 10.1158/1078-0432.CCR-09-1692. [DOI] [PubMed] [Google Scholar]
- 15.Shin MS, Kim HS, Lee SH, Park WS, Kim SY, Park JY, et al. Mutations of tumor necrosis factor-related apoptosis-inducing ligand receptor 1 (TRAIL-R1) and receptor 2 (TRAIL-R2) genes in metastatic breast cancers. Cancer Res. 2001;61:4942–6. [PubMed] [Google Scholar]
- 16.Sanlioglu AD, Karacay B, Koksal IT, Griffith TS, Sanlioglu S. DcR2 (TRAIL-R4) siRNA and adenovirus delivery of TRAIL (Ad5hTRAIL) break down in vitro tumorigenic potential of prostate carcinoma cells. Cancer Gene Ther. 2007;14:976–84. doi: 10.1038/sj.cgt.7701087. [DOI] [PubMed] [Google Scholar]
- 17.Sheikh MS, Huang Y, Fernandez-Salas EA, El-Deiry WS, Friess H, Amundson S, et al. The antiapoptotic decoy receptor TRID/TRAIL-R3 is a p53-regulated DNA damage-inducible gene that is overexpressed in primary tumors of the gastrointestinal tract. Oncogene. 1999;18:4153–9. doi: 10.1038/sj.onc.1202763. [DOI] [PubMed] [Google Scholar]
- 18.Hopkins-Donaldson S, Bodmer JL, Bourloud KB, Brognara CB, Tschopp J, Gross N. Loss of caspase-8 expression in highly malignant human neuroblastoma cells correlates with resistance to tumor necrosis factor-related apoptosis-inducing ligand-induced apoptosis. Cancer Res. 2000;60:4315–9. [PubMed] [Google Scholar]
- 19.Hinz S, Trauzold A, Boenicke L, Sandberg C, Beckmann S, Bayer E, et al. Bcl-XL protects pancreatic adenocarcinoma cells against. Oncogene. 2000;19:5477–86. doi: 10.1038/sj.onc.1203936. [DOI] [PubMed] [Google Scholar]
- 20.Sinicrope FA, Penington RC, Tang XM. Tumor necrosis factor-related apoptosis-inducing ligand-induced apoptosis is inhibited by Bcl-2 but restored by the small molecule Bcl-2 inhibitor, HA 14-1, in human colon cancer cells. Clin Cancer Res. 2004;10:8284–92. doi: 10.1158/1078-0432.CCR-04-1289. [DOI] [PubMed] [Google Scholar]
- 21.Chen F, Guo J, Zhang Y, Zhao Y, Zhou N, Liu S, et al. Knockdown of c-FLIP(L) enhanced AD5-10 anti-death receptor 5 monoclonal antibody-induced apoptosis in human lung cancer cells. Cancer Sci. 2009;100:940–7. doi: 10.1111/j.1349-7006.2009.01119.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Geserick P, Drewniok C, Hupe M, Haas TL, Diessenbacher P, Sprick MR, et al. Suppression of cFLIP is sufficient to sensitize human melanoma cells to TRAIL- and CD95L-mediated apoptosis. Oncogene. 2008;27:3211–20. doi: 10.1038/sj.onc.1210985. [DOI] [PubMed] [Google Scholar]
- 23.Bai J, Sui J, Demirjian A, Vollmer CM, Jr., Marasco W, Callery MP. Predominant Bcl-XL knockdown disables antiapoptotic mechanisms: tumor necrosis factor-related apoptosis-inducing ligand-based triple chemotherapy overcomes chemoresistance in pancreatic cancer cells in vitro. Cancer Res. 2005;65:2344–52. doi: 10.1158/0008-5472.CAN-04-3502. [DOI] [PubMed] [Google Scholar]
- 24.Chawla-Sarkar M, Bauer JA, Lupica JA, Morrison BH, Tang Z, Oates RK, et al. Suppression of NF-kappa B survival signaling by nitrosylcobalamin sensitizes neoplasms to the anti-tumor effects of Apo2L/TRAIL. J Biol Chem. 2003;278:39461–9. doi: 10.1074/jbc.M306111200. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 25.Shrader M, Pino MS, Lashinger L, Bar-Eli M, Adam L, Dinney CP, et al. Gefitinib reverses TRAIL resistance in human bladder cancer cell lines via inhibition of AKT-mediated X-linked inhibitor of apoptosis protein expression. Cancer Res. 2007;67:1430–5. doi: 10.1158/0008-5472.CAN-06-1224. [DOI] [PubMed] [Google Scholar]
- 26.Jing G, Yuan K, Turk AN, Jhala NC, Arnoletti JP, Zhang K, et al. Tamoxifen enhances therapeutic effects of gemcitabine on cholangiocarcinoma tumorigenesis. Lab Invest. 2011;91:896–904. doi: 10.1038/labinvest.2011.60. [DOI] [PubMed] [Google Scholar]
- 27.El-Zawahry A, McKillop J, Voelkel-Johnson C. Doxorubicin increases the effectiveness of Apo2L/TRAIL for tumor growth inhibition of prostate cancer xenografts. BMC Cancer. 2005;5:2. doi: 10.1186/1471-2407-5-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shankar S, Singh TR, Srivastava RK. Ionizing radiation enhances the therapeutic potential of TRAIL in prostate cancer in vitro and in vivo: Intracellular mechanisms. Prostate. 2004;61:35–49. doi: 10.1002/pros.20069. [DOI] [PubMed] [Google Scholar]
- 29.Gibson BA, Kraus WL. New insights into the molecular and cellular functions of poly(ADP-ribose) and PARPs. Nat Rev Mol Cell Biol. 2012;13:411–24. doi: 10.1038/nrm3376. [DOI] [PubMed] [Google Scholar]
- 30.Fong PC, Boss DS, Yap TA, Tutt A, Wu P, Mergui-Roelvink M, et al. Inhibition of poly(ADP-ribose) polymerase in tumors from BRCA mutation carriers. N Engl J Med. 2009;361:123–34. doi: 10.1056/NEJMoa0900212. [DOI] [PubMed] [Google Scholar]
- 31.Fogelman DR, Wolff RA, Kopetz S, Javle M, Bradley C, Mok I, et al. Evidence for the efficacy of Iniparib, a PARP-1 inhibitor, in BRCA2-associated pancreatic cancer. Anticancer Res. 2011;31:1417–20. [PubMed] [Google Scholar]
- 32.Mangerich A, Burkle A. How to kill tumor cells with inhibitors of poly(ADP-ribosyl)ation. Int J Cancer. 2011;128:251–65. doi: 10.1002/ijc.25683. [DOI] [PubMed] [Google Scholar]
- 33.Melisi D, Ossovskaya V, Zhu C, Rosa R, Ling J, Dougherty PM, et al. Oral poly(ADP-ribose) polymerase-1 inhibitor BSI-401 has antitumor activity and synergizes with oxaliplatin against pancreatic cancer, preventing acute neurotoxicity. Clin Cancer Res. 2009;15:6367–77. doi: 10.1158/1078-0432.CCR-09-0910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Porcelli L, Quatrale AE, Mantuano P, Leo MG, Silvestris N, Rolland JF, et al. Optimize radiochemotherapy in pancreatic cancer: PARP inhibitors a new therapeutic opportunity. Mol Oncol. 2012 Oct 29; doi: 10.1016/j.molonc.2012.10.002. pii: S1574-7891(12)00100-7. doi: 10.1016/j.molonc.2012.10.002.[Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ichikawa K, Liu W, Zhao L, Wang Z, Liu D, Ohtsuka T, et al. Tumoricidal activity of a novel anti-human DR5 monoclonal antibody without hepatocyte cytotoxicity. Nat Med. 2001;7:954–60. doi: 10.1038/91000. [DOI] [PubMed] [Google Scholar]
- 36.Zhang L, Fang B. Mechanisms of resistance to TRAIL-induced apoptosis in cancer. Cancer Gene Ther. 2005;12:228–37. doi: 10.1038/sj.cgt.7700792. [DOI] [PubMed] [Google Scholar]
- 37.Trucco C, Oliver FJ, de MG, Menissier-de MJ. DNA repair defect in poly(ADP-ribose) polymerase-deficient cell lines. Nucleic Acids Res. 1998;26:2644–9. doi: 10.1093/nar/26.11.2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.de Murcia JM, Niedergang C, Trucco C, Ricoul M, Dutrillaux B, Mark M, et al. Requirement of poly(ADP-ribose) polymerase in recovery from DNA damage in mice and in cells. Proc Natl Acad Sci U S A. 1997;94:7303–7. doi: 10.1073/pnas.94.14.7303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yelamos J, Farres J, Llacuna L, Ampurdanes C, Martin-Caballero J. PARP-1 and PARP-2: New players in tumour development. Am J Cancer Res. 2011;1:328–46. [PMC free article] [PubMed] [Google Scholar]
- 40.Goncalves A, Finetti P, Sabatier R, Gilabert M, Adelaide J, Borg JP, et al. Poly(ADP-ribose) polymerase-1 mRNA expression in human breast cancer: a meta-analysis. Breast Cancer Res Treat. 2011;127:273–81. doi: 10.1007/s10549-010-1199-y. [DOI] [PubMed] [Google Scholar]
- 41.Cookson MR, Ince PG, Usher PA, Shaw PJ. Poly(ADP-ribose) polymerase is found in both the nucleus and cytoplasm of human CNS neurons. Brain Res. 1999;834:182–5. doi: 10.1016/s0006-8993(99)01559-0. [DOI] [PubMed] [Google Scholar]
- 42.Ertsey R, Chapin CJ, Kitterman JA, Scavo LM. Ontogeny of poly(ADP-ribose) polymerase-1 in lung and developmental implications. Am J Respir Cell Mol Biol. 2004;30:853–61. doi: 10.1165/rcmb.2003-0248OC. [DOI] [PubMed] [Google Scholar]
- 43.Rossi MN, Carbone M, Mostocotto C, Mancone C, Tripodi M, Maione R, et al. Mitochondrial localization of PARP-1 requires interaction with mitofilin and is involved in the maintenance of mitochondrial DNA integrity. J Biol Chem. 2009;284:31616–24. doi: 10.1074/jbc.M109.025882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.von MG, Muller BM, Loibl S, Budczies J, Hanusch C, Darb-Esfahani S, et al. Cytoplasmic poly(adenosine diphosphate-ribose) polymerase expression is predictive and prognostic in patients with breast cancer treated with neoadjuvant chemotherapy. J Clin Oncol. 2011;29:2150–7. doi: 10.1200/JCO.2010.31.9079. [DOI] [PubMed] [Google Scholar]
- 45.Chang P, Jacobson MK, Mitchison TJ. Poly(ADP-ribose) is required for spindle assembly and structure. Nature. 2004;432:645–9. doi: 10.1038/nature03061. [DOI] [PubMed] [Google Scholar]
- 46.Zerfaoui M, Errami Y, Naura AS, Suzuki Y, Kim H, Ju J, et al. Poly(ADP-ribose) polymerase-1 is a determining factor in Crm1-mediated nuclear export and retention of p65 NF-kappa B upon TLR4 stimulation. J Immunol. 2010;185:1894–902. doi: 10.4049/jimmunol.1000646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sala A, La RG, Burgio G, Kotova E, Di GD, Collesano M, et al. The nucleosome-remodeling ATPase ISWI is regulated by poly-ADP-ribosylation. PLoS Biol. 2008;6:e252. doi: 10.1371/journal.pbio.0060252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhao Y, Sui X, Ren H. From procaspase-8 to caspase-8: revisiting structural functions of caspase-8. J Cell Physiol. 2010;225:316–20. doi: 10.1002/jcp.22276. [DOI] [PubMed] [Google Scholar]
- 49.Lavrik IN, Krammer PH. Regulation of CD95/Fas signaling at the DISC. Cell Death Differ. 2012;19:36–41. doi: 10.1038/cdd.2011.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Muzio M, Stockwell BR, Stennicke HR, Salvesen GS, Dixit VM. An induced proximity model for caspase-8 activation. J Biol Chem. 1998;273:2926–30. doi: 10.1074/jbc.273.5.2926. [DOI] [PubMed] [Google Scholar]