Abstract
The sterol hormone, 1α,25-dihydroxyvitamin D3 (1α,25(OH)2D3), regulates gene expression and messenger RNA (mRNA) concentrations in zebrafish in vivo. Since mRNA concentrations and translation are influenced by micro-RNAs (miRNAs), we examined the influence of 1α,25(OH)2D3 on miRNA expression in zebrafish in vivo with whole transcriptome RNA sequencing, searched for miRNA binding sites in 1α,25(OH)2D3-sensitive genes, and performed correlation analyses between 1α,25(OH)2D3-sensitive miRNAs and mRNAs. In vehicle- and 1α,25(OH)2D3-treated, 7-day postfertilization larvae, between 282 and 295 known precursor miRNAs were expressed, and in vehicle- and 1α,25(OH)2D3-treated fish, between 83 and 122 novel miRNAs were detected. Following 1α,25(OH)2D3 treatment, 31 precursor miRNAs were differentially expressed (p<0.05). The differentially expressed miRNAs are predicted to potentially alter mRNAs for metabolic enzymes, transcription factors, growth factors, and Jak-STAT signaling. We verified the role of a 1α,25(OH)2D3-sensitive miRNA, miR125b, by demonstrating alterations in the concentrations of the mRNA of a 1α,25(OH)2D3-regulated gene, Cyp24a1, following transfection of renal cells with a miR125b miRNA mimic. Changes in the Cyp24a1 mRNA concentration by the miR125b miRNA mimic were associated with changes in the protein for Cyp24a1. Our data show that 1α,25(OH)2D3 regulates miRNA in zebrafish larvae in vivo and could thereby influence vitamin D-sensitive mRNA concentrations.
Introduction
The active form of vitamin D3, 1α,25-dihydroxyvitamin D3 (1α,25(OH)2D3), alters gene expression in the developing zebrafish in vivo.1 Initially restricted to a modest number of responding loci, by day 7 of treatment, the expression of approximately 2500 protein-coding genes is either increased or decreased in response to 1α,25(OH)2D3 when assessed by whole transcriptome RNA shotgun sequencing.1 To regulate gene expression in target tissues, 1α,25(OH)2D3 binds to a widely expressed receptor, the vitamin D receptor (VDR), and the ligand-bound receptor, either as a heterodimer with the retinoic acid X-receptor or as a homodimer binds vitamin D response elements (VDREs) in diverse genes,2–10 many of which encode proteins that alter calcium or phosphorus transport and bone mineralization or bone resorption.11–16 In Danio rerio, there are two VDR genes (VDRa and VDRb) that encode receptors with 453 (VDRA) and 422 (VDRB) amino acids with highly conserved DNA and ligand binding domains.17 Only the Danio VDRA plays a role in intestinal calcium transport.18 As described earlier, a large number of messenger RNAs (mRNAs) encoding proteins important in lipid, xenobiotic, and cell signaling pathways were shown to be regulated by 1α,25(OH)2D3 in developing zebrafish.1
The role of post-transcriptional regulation by micro-RNAs (miRNAs) has been established as a key contributory mechanism for gene expression in several model organisms for a variety of cellular processes.19–21 miRNAs, which represent 1%–2% of genes in worms, flies, and vertebrates,22 post-transcriptionally regulate gene expression in plants and vertebrates by binding to the 3′ untranslated regions of mRNAs19–21 and altering mRNA decay or translation.23 miRNAs are transcribed by RNA polymerase II24 (or sometimes RNA polymerase III)25 to give rise to long primary RNAs that are processed by the microprocessor complex (containing Drosha and DGCR8) to generate pre-miRNAs.26,27 Pre-miRNAs are cleaved by the cytoplasmic ribonuclease, Dicer, to generate mature miRNAs.28,29 One of the two strands of a miRNA is loaded into the RNA-silencing (RISC) complex before interaction with a target mRNA.30
Steroid hormones such as estrogens, progesterones, and androgens regulate miRNAs in cell culture and in vivo,31–39 and miRNAs regulate steroid hormone receptor concentrations within cells.31 There is a paucity of information regarding the regulation of miRNAs by the calcium regulating sterol, 1α,25(OH)2D3. The effects of 1α,25(OH)2D3 on miRNA expression have been examined in cells maintained in culture.40–51 Using microarrays, Alvarez-Diaz et al. recently demonstrated that 1α,25(OH)2D3 induces miRNA-22 expression in cultured colon cancer cells in a time-, dose- and VDR-dependent manner.40 Gocek et al. studied the effects of the exogenously added 1α,25(OH)2D3 on miRNA expression in human myeloid leukemia cells and demonstrated the induction of miR-32 in such cells.44 That study showed that miR-32 targets the 3′-untranslated region of the mRNA encoding the proapoptotic factor Bim, reducing its expression. Wang et al. examined the effects of 1α,25(OH)2D3 on miRNA expression in LNCap prostate cancer cells and demonstrated the regulation of a number of different miRNAs.49 Essa et al. showed recently that 1α,25(OH)2D3 and/or epigenetic drugs modulated VDR mRNA expression in 1α,25(OH)2D3-responsive and -resistant melanoma cell lines.43 Two very recent reports show that 1α,25(OH)2D3 regulates miRNAs in prostate tissue and in tumor xenografts in vivo,52,53 but no reports have demonstrated changes in miRNA expression in normal physiological states.
The effects of 1α,25(OH)2D3, steroids, and peptide hormones can be assessed by adding hormones to the zebrafish incubation medium54 and correlating changes in gene expression with organ, cartilage, and bone development. We previously examined 1α,25(OH)2D3-mediated gene expression in zebrafish embryos and larvae in vivo and showed that the sterol alters cartilage and bone development.1 To determine the influence of 1α,25(OH)2D3 on miRNA expression in vivo, we treated zebrafish embryos/larvae with 1α,25(OH)2D3 or vehicle and assessed miRNA expression at 7 days postfertilization (dpf), following addition of 1α,25(OH)2D3 or vehicle with whole transcriptome shotgun sequencing (RNA-seq). We chose 7 dpf larvae in which to examine changes in the miRNA expression, to establish a model system in which changes in miRNA expression could be correlated with ossification and cartilage development, both of which are established by this time.55,56 We now demonstrate that multiple miRNAs are altered by 1α,25(OH)2D3 in the developing zebrafish. These miRNAs potentially regulate multiple transcription factors, growth factors and growth factor binding proteins, peptide hormones, and various amino acid metabolic pathways (e.g., metabolic pathways for glycine, serine, threonine, and tryptophan). In addition, we demonstrate that miR125b alters Cyp24a1 mRNA expression in cultured cells.
Materials and Methods
Treatment of zebrafish with 1α,25(OH)2D3
Zebrafish embryos were obtained and cultured as described from natural mating of Segrest wild-type adult zebrafish in the Mayo Clinic Zebrafish Core Facility.57 One hundred forty zebrafish embryos were placed in a 20 mL embryo medium (pH 7.2) containing 1-phenyl-2-thiourea (PTU) (0.003% (w/v) and were maintained at 28°C–30°C. At 24 hours post-fertilization (1 dpf), 10 μL 1α,25(OH)2D3 in ethanol (gift of Dr. Milan Uskokovic, Hoffmann LaRoche, Nutley, NJ) was added to embryos maintained in 20 mL of fresh embryo medium (final concentration 1α,25(OH)2D3 300 pM). Control zebrafish were treated with 10 μL ethanol alone (vehicle controls). The medium containing either 300 pM 1α,25(OH)2D3 or vehicle was changed every 24 h. At 7 dpf, larvae were removed and immediately frozen at −80°C for RNA preparation. Thirty larvae were used for preparation of RNA. Four individual cDNA libraries from 1α,25(OH)2D3-treated fish and four individual libraries form vehicle-treated fish were prepared at 7 days.
RNA preparation for RNA-seq and quantitative polymerase chain reaction
RNA was prepared as previously described, using RNA/protein spin columns (Clontech, Mountain View, CA).1 A lysis solution was added to 25–30 larvae that were lysed by passage successively through 21- and 27-gauge needles. Individual lysates were applied to RNA spin columns, and RNAs were eluted into nuclease-free water. Before library construction, the RNA quality was assessed by capillary electrophoresis against a reference size standard.
miRNA library preparation
We synthesized miRNA libraries from 1α,25(OH)2D3- or vehicle-treated zebrafish larvae using total RNA samples and a NEBNext® Multiplex Small RNA Kit (New England BioLabs, Ipswich, MA). Adaptors were ligated to the 3′ ends of the small noncoding RNAs present in 500 ng of total RNA. A complementary primer was annealed to the 3′ adaptor sequences followed by ligation of a 5′ RNA adaptor. A cDNA library was created by reverse transcriptase (Superscript III; Invitrogen, Carlsbad, CA) of the adaptor ligated and annealed small RNA population. The library was enriched by 15 cycles of polymerase chain reaction (PCR) employing a common 5′ primer and a 3′ primer containing one of the eight index primers (3′ adaptor complement; index sequences equivalent to Illumina TruSeq Small RNA sequences). The PCR products were subsequently purified. The libraries were assessed for miRNA products by Agilent Bioanalyzer DNA 1000 (Santa Clara, CA) analysis. The 130–160 bp region was quantitated to determine equimolar amounts of vehicle- and 1α,25(OH)2D3-treated sample libraries to pool. The pooled small RNA libraries were fractionated to extract a miRNA-enriched sample using 3% Pippin Prep (Sage Science, Beverly, MA) gel cassettes. It is unlikely that ribosomal, transfer, and sn/sno RNAs are present in the purified fraction used for analysis. The recovered fraction was purified and reconstituted in 10 μL of nuclease-free dH2O. Pooled miRNA fractions were assessed by a second Agilent DNA 1000 assay. A predominant peak at 140–150 bp indicated that miRNA modification and size selection steps were performed as expected. The final concentration of each library pool was determined. Libraries were loaded onto single end flow cells at concentrations of 8–10 pM to generate cluster densities of 700,000/mm2 following Illumina's standard protocol using the Illumina cBot cluster kit version 3. The flow cells were sequenced as two reads: read 1–51 cycles using the small RNA sequencing primer and an Index read to demultiplex the samples. Libraries were sequenced on an Illumina HiSeq 2000 using TruSeq SBS sequencing kit version 3 and SCS version 1.4.8 data collection software. Base calling was performed using Illumina's RTA version 1.12.4.2.
mRNA library preparation
mRNA-seq libraries were prepared from identical RNA samples as previously described using the mRNA v1 sample prep kit protocol (Illumina, San Diego, CA).1 Poly-A containing mRNA was purified using poly-T oligo magnetic beads. The purified mRNA was fragmented, RNA fragments were copied into first-strand cDNA, and second strand cDNA synthesis was carried out. The cDNA was purified, ends were repaired and phosphorylated after which, an A base was added to the 3′ ends of double-stranded DNA. Paired-end DNA adaptors (Illumina) with a single T-base overhang at the 3′ end were ligated to the double-stranded DNA and separated on a 2% agarose gel. DNA fragments of approximately 250–300 bp were excised, purified, and enriched by PCR. Sequencing was carried out as previously described.1
Quantitative PCR
Quantitative PCR (QPCR) was carried out using a Roche LightCycler 480 QPCR apparatus in 96-well QPCR plates (Roche Diagnostics Corp., Indianapolis, IN) using SYBR® Green master mix, Universal RT (Exiqon, Woburn, MA). RNA was isolated as described earlier.1 A Universal cDNA Synthesis Kit (Exiqon) was used to generate double-stranded DNA from 70 ng total RNA for each sample. We used predesigned locked nucleic acid LNA™-enhanced microRNA-specific QPCR primer sets from Exiqon for dre-let-7i (hsa-let-7i), dre-miR-100 (hsa-miR-100), dre-miR-96 (hsa-miR-96) (unchanging reference control), dre-miR-125b (hsa-miR-125b), U6 snRNA (additional reference control), or the custom LNA™ PCR primer set unique for zebrafish dre-miR-21. A reverse transcribed product was used to generate PCR products with each LNA primer pair. QPCR data were subjected to relative quantitation against dre-miR-96 using software supplied with the instrument.
Analysis of miRNAs
The generated single-end sequence reads were analyzed by the miRDeep2 tool.58 Known and novel miRNAs were interrogated for each sample, and their expression profiles were derived by counting the number of reads aligning to the genomic coordinates of all known and potential novel miRNAs predicted by the miRDeep tool.59 We consider a candidate to be a novel real miRNA if its miRDeep score is larger than 0. Specifically, for genomic regions aligned against sequencing reads, the miRDeep tool (1) investigates genomic DNA bracketing the alignments and computes their secondary RNA structure; (2) identifies plausible miRNA precursor sequences and scores their likelihood to be real miRNA precursors.60 We used a cutoff score value 0 in this study, since four samples were analyzed, and the estimated true positive rate on average is larger than 60%. In our standard bioinformatics workflow, snoRNA and other small RNAs are not assessed. For differential gene expression analysis between two conditions, we first eliminated miRNAs without any reads across all samples. Because scaling by total lane counts (e.g., reads per kilobase per million [gene counts/kilobase of exon for that gene/total counts of each biological replicate]×1,000,000) can bias estimates of differential expression, we used quantile-based normalization on read counts to determine if genes are differentially expressed61 using the negative binomial method62 requiring an adjusted p-value <0.01 controlled for multiple testing using the Benjamini and Hochberg correction.63 miRNAs with significantly differential expression (adjusted p-value <0.01) between two conditions were selected for downstream target prediction. Due to the lack of bioinformatics tools with statistically significant accuracy in predicting miRNA binding sites, we proposed a two-step computational approach to improve the prediction accuracy and to create an optimal framework for deciphering biological functions of miRNAs. First, we used the TargetScan algorithm64 to predict potential mRNAs for each differentially expressed miRNA. This algorithm predicts targets of vertebrate miRNAs by identifying mRNAs with conserved complementarity to the seed sequence of the miRNA. Second, we applied Pearson correlation analysis between differentially expressed mRNAs and differentially expressed miRNAs. We assessed statistical significance by using the q value of the false discovery rate (qFDR) as described by Storey and Tibshirani65 We defined the miRNA-mRNA correlation coefficient qFDR < 0.01 to be statistically significant. The common mRNA genes identified by both steps are treated as high-confidence targets for each miRNA. The biological effects of each miRNA, which occur through a variety of mechanisms,21 can subsequently be experimentally validated. The hypergeometric test was performed for enrichment analysis between the mRNAs that are correlated with one differentially expressed miRNA and the predicted mRNA targets for this miRNA. The FDR value was then calculated as described by Storey.66 Lists of predicted target genes for the differentially expressed miRNAs were used for pathway/network level analysis. We conducted pathway analysis of predicted target genes using DAVID (Database for Annotation, Visualization, and Integrated Discovery v6.7).67 An adjusted p-value less than 0.05 was used to assign biological significance. The goal was to investigate whether the predicted target genes were enriched with genes in canonical pathways in public databases. DAVID is a web resource consisting of an integrated biological knowledgebase and analytical tools that aim to extract and understand biological themes in large gene lists. It provides the gene functional classification as well as the identification (ID) conversion of the list of gene ID accessions to any other accession of choice. It also provides pathways from other databases such as Kyoto Encyclopedia of Genes and Genomes (KEGG),68 Panther,69 BioCarta (http://cgap.nci.nih.gov/Pathways/BioCarta_Pathways), and Reactome,70 and provides tissue expression and disease-specific information based on represented genes.
VDR element analysis of promoter regions of differentially expressed miRNAs
Promoter sequences of all differentially expressed miRNAs from −5000 to +5000 relative to the transcription start site were retrieved from the latest available zebrafish reference genome in the ENSEMBL database (Zv9 version) using customized Perl scripts. The match program71 provided by the TRANSFAC database in BIOBASE (www.biobase-international.com) was used to detect potential VDR binding sites in these sequences, using customized VDR profiles, in which four candidate VDR-related matrices from the MATRIX table were selected to minimize the rate of false positives. Matches exhibiting a matrix similarity scores above 0.9 were used.
VDR element analysis of promoter regions of differentially expressed mRNAs
Promoter sequences of all differentially expressed mRNAs from −5000 to +5000 relative to the transcription start site were retrieved from the latest available zebrafish reference genome in the ENSEMBL database (Zv9 version) using customized Perl scripts. Analysis was performed as described above to identify potential VDREs.
Regulation of Cyp24A1 mRNA and protein by miR125b and miR21
The mouse Cyp24a1 gene was examined for the presence of mir125b and miR21 binding sites using the TRANFAC program. These miRNAs were shown to be increased 1.8- and 5.1-fold by 1α,25(OH)2D3. A binding site for mir125b was detected in the 3′ untranslated region (3′UTR) of the Cyp24a1 gene, but was absent in the case of miR21. Immortalized renal proximal tubular cells grown as previously described,72 were transfected with miRNA mimics for miR125b (sequence mmu-miR-125b-5p: 5′ UCCCUGAGACCCUAACUUGUGA 3′) and miR21a (sequence mmu-miR21a-5p: 5′ UAGCUUAUCAGACUGAUGUUGA 3′) or a control Dy574 miRNA mimic transfection control for 10 h using the Opti-MEM1 transfection reagent (DharmaFECT1) (Thermo Scientific, Pittsburg, PA). Following transfection of cells with the miRNA mimic for 10 h, the transfecting reagent was removed, a new medium was added, and the cells were grown for an additional 38 h. Cells were then harvested, and RNA and protein were isolated. Cyp24a1 RNA was quantitated using reverse transcription (RT)-PCR with appropriate primers (forward 5′ CCTGGGACACCATTTTCAA 3′, reverse 5′ TGCTGATAAATATCACAAAGGAAATC 3′).72 The Cyp24A1 protein was detected using western blotting methods with an antibody produced against recombinant Cyp24A1.73
Generation of mouse proximal tubule cells
Proximal tubule cells were isolated from the SV40 T-antigen mutant tsA58 ImmortoMouse (Charles River, Wilmington, MA) mouse kidney.74 The cortex from isolated kidneys of a 12-week-old female ImmortoMouse was harvested, minced, and digested with 1% collagenase type II (Worthington Biochemical Corporation, Lakewood, NJ) at 37°C. Cell suspensions were filtered through a nylon mesh, suspended in 45% Percoll, and centrifuged at 27,000 g for 15 min at 4°C. Isolated proximal tubule cells were grown on Matrigel-coated tissue culture plates, in DMEM/F12, 10% FBS, 100 U/mL penicillin G, 100 μg/mL streptomycin, and 1.2 mg/mL sodium bicarbonate. Either 6-well tissue culture plates (seeded at 1×105 cells/well) or 24-well tissue culture plates (seeded at 1×104 cells/well) were used for miRNA mimic experiments upon cell confluence.
Data sharing
All sequence data that were analyzed in this report have been deposited in the Gene Expression Omnibus.
Results
We show that known and novel miRNAs are expressed in 7 dpf zebrafish embryos in both vehicle- and 1α,25(OH)2D3-treated fish. Multiple miRNAs are repressed or induced by 1α,25(OH)2D3. Table 1 lists the detailed information of different categories of miRNAs for each sample treated with vehicle or 1α,25(OH)2D3. Between 287 and 293 known precursor miRNAs were expressed in 7 dpf zebrafish in the vehicle-treated group, and between 282 and 295 known precursor miRNAs were expressed in the 1α,25(OH)2D3-treated group. The detailed information of known precursor miRNAs in each sample is listed in Supplementary Table S1 (Supplementary Data are available online at www.liebertpub.com/zeb). Besides known miRNAs in the miRBase, novel miRNAs in each sample were detected using the miRDeep2 tool. In the vehicle-treated group, 266 unique novel miRNAs were detected in 7 dpf zebrafish larvae in at least one out of four replicates, while in the 1α,25(OH)2D3-treated group, 246 unique novel miRNAs were detected in at least one out of four replicates. Supplementary Tables S2a and b list the sequence of these novel miRNAs in each sample in either the vehicle- or the 1α,25(OH)2D3-treated group.
Table 1.
Summary Statistics of miRNA in Each Sample
| 7 dpf ET1 | 7 dpf ET2 | 7 dpf ET3 | 7 dpf ET4 | 7 dpf VD1 | 7 dpf VD2 | 7 dpf VD3 | 7 dpf VD4 | |
|---|---|---|---|---|---|---|---|---|
| Detected known miRNA | 291 | 293 | 292 | 287 | 282 | 283 | 287 | 295 |
| Novel miRNA | 122 | 105 | 110 | 102 | 87 | 83 | 101 | 117 |
| Expressed miRNA | 348 | 349 | 349 | 346 | 343 | 346 | 346 | 346 |
Detected known precursor miRNA: the number of known precursor miRNAs detected by miRDeep tool.
Novel miRNA: the number of novel miRNAs detected by miRDeep tool.
Expressed miRNA: the number of known miRNAs with no less than five mapped sequencing reads.
dpf, day postfertilization; ET, ethanol; VD, 1α,25(OH)2D3; miRNA, micro-RNA.
In Table 2 are listed miRNAs that are increased or decreased following 6 days of treatment with 1α,25(OH)2D3. We confirmed the changes in concentrations of these miRNAs by performing quantitative PCR specific for miRNAs. We observed changes in dre-let-7i (0.85-fold change by QPCR vs. 0.68 change by RNA-seq), dre-miR-21 (6.5-fold change by QPCR vs. 5.1-fold change by RNA-seq), dre-miR-100 (2.4-fold change by QPCR vs. 2.1-fold change by RNA-seq),), and dre-miR-125b (1.7-fold change by QPCR vs. 1.8 change by RNA-seq). Dre-miR-96 was used to normalize the data as this was found to be an invariant miRNA on initial sequencing. We further investigated whether these differentially expressed miRNAs are directly regulated by the 1α,25(OH)2D3 treatment by performing transcription factor binding site analysis for all differentially expressed miRNAs. We searched for four candidate VDR elements from the TRANSFAC database both upstream and downstream of the transcription start sites of these differentially expressed miRNAs. Eighteen out of 21 differentially expressed mature miRNAs have at least one potential VDR element site within −5000 to +5000 bps of their transcription start sites (Table 3).
Table 2.
Change in Expression of miRNAs Following Treatment of 7 dpf Zebrafish Larvae with Vehicle or 1α,25(OH)2D3
| Mature miRNA | Precursor miRNA | Fold change | p-Value |
|---|---|---|---|
| dre-miR-2190 | dre-miR-2190-1 | 6.38 | 0.02 |
| dre-miR-2190 | dre-miR-2190-2 | 6.38 | 0.02 |
| dre-miR-2190 | dre-miR-2190-3 | 6.38 | 0.02 |
| dre-miR-2190 | dre-miR-2190-4 | 6.38 | 0.02 |
| dre-miR-21 | dre-miR-21-1 | 5.07 | 0.9×10−50 |
| dre-miR-21 | dre-miR-21-2 | 5.06 | 0.9×10−50 |
| dre-miR-735 | dre-miR-735 | 2.69 | 0.02 |
| dre-miR-150 | dre-miR-150 | 2.50 | 1.92×10−16 |
| dre-miR-100 | dre-miR-100-2 | 2.13 | 0.002 |
| dre-miR-100 | dre-miR-100-1 | 2.13 | 0.002 |
| dre-miR-125b | dre-miR-125b-1 | 1.76 | 1.0×10−6 |
| dre-miR-125b | dre-miR-125b-2 | 1.76 | 1.0×10−6 |
| dre-miR-125b | dre-miR-125b3 | 1.75 | 1.0×10−6 |
| dre-miR-146a | dre-miR-146a | 1.74 | 0.02 |
| dre-miR-125c | dre-miR-125c | 1.6 | 0.00069 |
| dre-miR-17a* | dre-miR-17a-1 | 1.54 | 0.39 |
| dre-miR-let-7e | dre-miR-let- 7e | 1.52 | 0.021 |
| dre-miR-146b | dre-miR-146b | 1.52 | 0.003 |
| dre-miR-214 | dre-miR-214 | 1.48 | 0.009 |
| dre-miR-181c | dre-miR-181c | 1.44 | 0.032 |
| dre-miR-126a | dre-miR-126a | 1.41 | 0.009 |
| dre-miR-140* | dre-miR-140 | 1.37 | 0.02 |
| dre-miR-22a | dre-miR-22a | 1.35 | 0.02 |
| dre-miR-184 | dre-miR-184 | 0.70 | 0.013 |
| dre-miR-184 | dre-miR-184-2 | 0.70 | 0.013 |
| dre-miR-192 | dre-miR-192 | 0.70 | 0.04 |
| dre-miR-let-7i | dre-miR-let-7i | 0.67 | 0.003 |
| dre-miR-729 | dre-miR-729 | 0.64 | 0.0008 |
| dre-miR-459* | dre-miR-459 | 0.62 | 0.0007 |
| dre-miR-122 | dre-miR-122 | 0.52 | 7.64×10−7 |
| dre-miR-29a | dre-miR-29a | 0.44 | 0.04 |
miRNAs, which decreased following treatment with 1α,25(OH)2D3, are highlighted in gray.
1α,25(OH)2D3, 1α,25-dihydroxyvitamin D3.
Table 3.
Statistics of miRNA Targets Derived from Target Scan and Pearson Correlation Approaches
| Name | Chr | Start | End | Strand | # of potential VDR binding sites |
|---|---|---|---|---|---|
| DRE-MIR-150 | chr3 | 32708295 | 32708357 | + | 2 |
| DRE-MIR-21 | chr10 | 28880713 | 28880774 | − | 3 |
| DRE-MIR-735 | chr24 | 39680979 | 39681041 | − | 5 |
| DRE-MIR-100 | chr5 | 31628647 | 31628704 | + | 3 |
| DRE-MIR-125B | chr15 | 20409343 | 20409405 | + | 4 |
| DRE-MIR-146A | chr13 | 11537689 | 11537750 | + | 5 |
| DRE-MIR-125C | chr15 | 29150075 | 29150136 | + | 1 |
| DRE-MIR-17A | chr1 | 2806111 | 2806172 | + | 5 |
| DRE-LET-7E | chr23 | 5478703 | 5478777 | − | 6 |
| DRE-MIR-146B | chr21 | 40390767 | 40390827 | − | 3 |
| DRE-MIR-214 | chr20 | 14792434 | 14792496 | + | 5 |
| DRE-MIR-181C | chr3 | 34283001 | 34283058 | + | 6 |
| DRE-MIR-126A | chr8 | 12065966 | 12066026 | − | 3 |
| DRE-MIR-140 | chr25 | 35855094 | 35855156 | + | 3 |
| DRE-MIR-29A | chr4 | 10671282 | 10671407 | − | 1 |
| DRE-MIR-459 | chr1 | 1275358 | 1275421 | + | 1 |
| DRE-MIR-729 | chr4 | 12649029 | 12649091 | + | 4 |
| DRE-LET-7I | chr25 | 1688209 | 1688287 | − | 0 |
| DRE-MIR-184 | chr18 | 25989802 | 25989866 | − | 0 |
| DRE-MIR-192 | chr10 | 27698862 | 27698925 | + | 0 |
| DRE-MIR-2190 | chr5 | 1190769 | 1190872 | − | 9 |
VDR, vitamin D receptor.
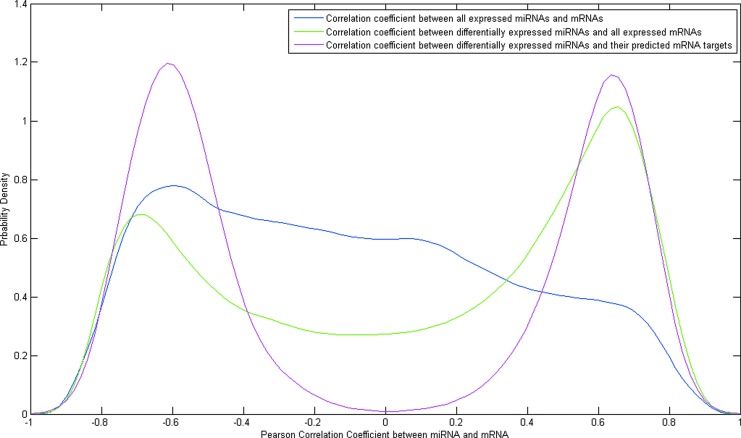
We identified genes that might potentially be altered by changes in miRNA amounts by performing TargetScan analysis, which identifies sequences in genes, to which a miRNA might bind, based on sequence specificity. The results of such analyses are shown in Supplementary Table S3. Supplementary Table S4 annotates the genes identified in Supplementary Table S3. Next, we analyzed the correlation between changes in miRNAs ascertained in the current study, and changes in expressed mRNAs measured by whole transcriptome analysis published by us earlier and deposited in the Gene Expression Omnibus (Accession #GSE38575). The results of such an analysis are shown in Table 4. The number of miRNAs common to both lists is considerably smaller than the number present in each list individually. Figure 1 shows the distribution plots of Pearson correlation coefficient values in different miRNA-mRNA groups. The correlations between differentially expressed miRNAs and their predicted mRNA targets are more enriched in larger absolute correlation coefficient values (i.e., either positively correlated or negatively correlated). To evaluate whether the mRNA lists generated from both correlation analysis and TargetScan prediction have a significant overlap, a hypergeometric test was performed for the results for each miRNA. A FDR value less than 0.1 indicated that our two-step computational approach identified potential mRNA targets with statistical significance.
Table 4.
Enriched Pathways in Predicted Messenger RNA Targets of Differentially Expressed micro-RNAs
| miRNA | Number of potential targets by TargetScan | Number of mRNAs correlated with each miRNA | Number of targets detected by both approaches |
|---|---|---|---|
| DRE-MIR-2190 | 468 | 895 | 10 |
| DRE-MIR-21 | 3590 | 1235 | 113 |
| DRE-MIR-735 | 2638 | 1331 | 77 |
| DRE-MIR-150 | 2240 | 1474 | 70 |
| DRE-MIR-100 | 163 | 45 | 0 |
| DRE-MIR-125B | 539 | 261 | 2 |
| DRE-MIR-146A | 2395 | 1200 | 88 |
| DRE-MIR-125C | 98 | 920 | 4 |
| DRE-MIR-17A* | 4345 | 1654 | 205 |
| DRE-LET-7E | 880 | 942 | 27 |
| DRE-MIR-146B | 469 | 154 | 2 |
| DRE-MIR-214 | 4559 | 1653 | 195 |
| DRE-MIR-181C | 0 | 460 | 0 |
| DRE-MIR-126A | 384 | 1210 | 6 |
| DRE-MIR-140* | 3327 | 1255 | 108 |
| DRE-MIR-29A | 2208 | 1555 | 104 |
| DRE-MIR-459* | 4699 | 1645 | 225 |
| DRE-MIR-729 | 2157 | 1485 | 82 |
| DRE-LET-7I | 123 | 1566 | 8 |
| DRE-MIR-184 | 801 | 1637 | 31 |
| DRE-MIR-192 | 2809 | 1655 | 123 |
Analysis was performed using the (DAVID) Database for Annotation, Visualization, and Integrated Discovery v6.7 program.
miRNA, micro-RNA; mRNAs, messenger RNAs.
FIG. 1.
The probability distribution of different zebrafish miRNA-mRNA groups whose expression is altered by exposure to 1α,25(OH)2D3 in vivo. The distribution of all miRNA-mRNA PCC shows a skewed normal distribution. The distribution of differentially expressed miRNAs and all expressed mRNAs demonstrate a two-peak distribution, and the distribution of differentially expressed miRNAs and their predicted mRNA targets as determined by a two-step computational approach has even higher peaks around −0.85 and 0.85, respectively. Results were confirmed by the hypergeometric test. 1α,25(OH)2D3, 1α,25-dihydroxyvitamin D3; miRNA, micro-RNA; mRNA, messenger RNA; PCC, Pearson correlation coefficient.
The expression of the Cyp24a1 gene is dramatically upregulated by 1α,25(OH)2D3. We determined whether the Cyp24a1 gene had binding sites for miRNAs that were upregulated by 1α,25(OH)2D3. An examination of the mouse Cyp24a1 gene revealed the presence of a mir125b-binding site in the 3′UTR of the gene. miR125b is increased 1.8-fold by 1α,25(OH)2D3. Immortalized renal proximal tubular cells transfected with the miRNA mimic for miR125b showed a 2.02-fold increase in Cyp24a1 mRNA assessed by RT-PCR (control 0.74±0.17 RU vs. 1.5±0.38 RU, p<0.011). Identical experiments performed with a miR21a mimic, which lacks a binding site in the Cyp24a1 gene, showed no changes in Cyp24a1 mRNA assessed by RT-PCR (control 0.88±0.34 RU vs. 0.91±0.41 RU, p=0.91). Assessment of transfection efficiency with Dy574 miRNA showed greater than 85% transfection efficiency. The Cyp24A1 protein, detected using western blotting methods, was increased following transfection of cells with the miRNA mimic for miR125b (control 31466.73±1139.52 [standard error of mean] vs. miR125b mimic treated 39943.47±2532.04, p=0.038), but not following transfection with the miRNA mimic for miR21a.
Analysis of the promoters of genes encoding mRNAs regulated by 1α,25(OH)2D3 demonstrated that 93% (3406 out of a total of 3665) of these genes had at least one VDRE in the region 5 kb before or after the transcription start site. Two hundred fifty-nine genes did not have a VDRE in this region. Of these genes, 50 had a miRNA binding site in the 3′ region of the cognate gene (Supplementary Table S5). The data suggest that these genes might be exclusively regulated through miRNA-dependent mechanisms.
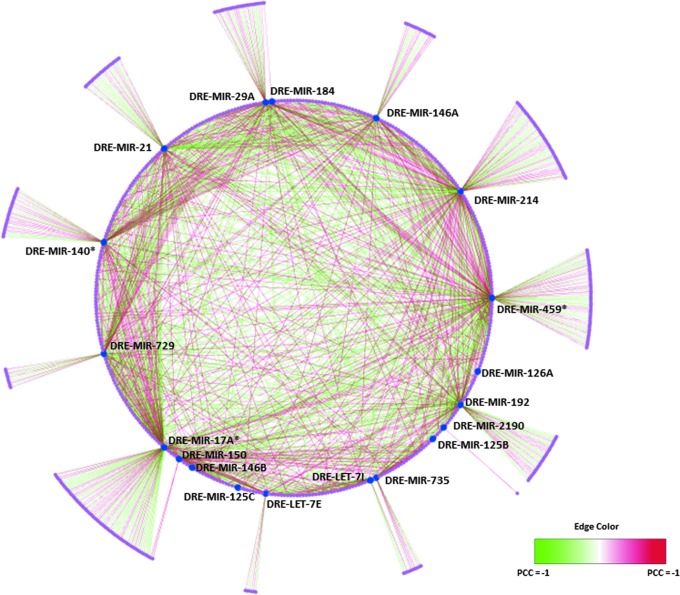
We analyzed the role of the altered miRNAs in different metabolic pathways. Shown in Figure 2 is the regulatory network containing miRNAs and their predicted candidate mRNA targets. The edge color denotes their correlation relationship. We observed a comparable number of both positive and negative correlation relationships between miRNAs and their putative mRNA targets. Although the canonical miRNA regulation model suggests that miRNAs repress the expression of their downstream target genes,22 it has been demonstrated in several studies that the miRNA regulatory network will generate complex expression patterns, causing either positive or negative correlations between miRNAs and their target genes.75–78 Competing endogenous RNAs, which sequester miRNAs, act as miRNA decoys and are likely to have additional effects on the miRNA function.79–82 Our results demonstrate that miRNAs and their target genes have not only negative correlation expression patterns but much more complex regulation mechanisms. The pathway and gene ontology enrichment analyses for the putative mRNA target list were performed by the DAVID program. Table 5 presents the enriched KEGG pathways in the putative mRNA targets. The pathways altered by miRNAs include those for glycine, serine, threonine, and tryptophan metabolism, and the Jak-STAT signaling pathway. We further investigated which KEGG pathways are enriched in putative mRNA targets for each differentially expressed miRNA. Supplementary Table S6 shows the KEGG pathways enriched in the putative mRNA targets (adjusted p-value less than 0.01) for each differentially expressed miRNA.
FIG. 2.
Overview of the regulatory networks containing differentially expressed miRNAs and their candidate mRNA targets in response to 1α,25(OH)2D3 in vivo. Blue nodes denote differentially expressed miRNAs, and red nodes denote their predicted mRNA targets. Edges denote the regulatory relationships between miRNAs and their predicted mRNA targets.
Table 5.
Enriched KEGG Pathways in the Putative mRNA Targets
| KEGG pathway term | Gene count | p-Value |
|---|---|---|
| Alanine, aspartate, and glutamate metabolism | 8 | 0.00075 |
| Jak-STAT signaling pathway | 14 | 0.00096 |
| Aminoacyl-tRNA biosynthesis | 8 | 0.0013 |
| p53 signaling pathway | 10 | 0.003 |
| Tryptophan metabolism | 7 | 0.0073 |
| Porphyrin metabolism | 5 | 0.043 |
| Glycine, serine, and threonine metabolism | 5 | 0.047 |
KEGG, Kyoto Encyclopedia of Genes and Genomes.
Discussion
Steroid hormones, and sterols such as vitamin D and its metabolites, alter biochemical processes and development by regulating gene expression and by altering mRNAs for a number of protein coding genes.1,5,7,11–16,83–97 In addition to regulating mRNA expression, steroids such as estrogen, progesterone, and androgen also regulate noncoding RNA, specifically, miRNA expression in cell culture and in vivo.31–39 By altering mRNA stability, miRNAs change the expression of coding mRNAs.
There is only limited information about how the active metabolite of vitamin D3, 1α,25(OH)2D3, regulates the expression of miRNAs in vivo. In previously published work, the effects of 1α,25(OH)2D3 on miRNA expression have been examined in cells maintained in culture.40–51 Two very recent reports show that 1α,25(OH)2D3 regulates miRNAs in prostate tissue and in tumor xenografts in vivo,52,53 but no reports have demonstrated changes in miRNA expression in normal physiological states. Furthermore, how the changes in miRNAs are associated with the changes in mRNAs in vivo is also unknown. We used a zebrafish model to assess the effects of 1α,25(OH)2D3 on miRNA expression for the following reasons: (1) miRNAs are expressed in developing zebrafish embryos and larvae and regulate several processes, including neural, cardiac, and fin development98–107; (2) zebrafish are readily treated with hormones and drugs1,54; (3) the effects of 1α,25(OH)2D3 on mRNA expression as a function of the developmental stage has been examined by our laboratory in the past and allow correlation between changes in miRNA expression and mRNA expression; and (4) the consequences of altered miRNA expression can be assessed morphologically because of the transparency of developing zebrafish.
We made several novel observations regarding miRNA expression in zebrafish larvae. First, in 7 dpf zebrafish larvae, many more miRNAs are expressed than in zebrafish at the earlier stages of development. Chen et al. showed that 154 distinct miRNAs were expressed in zebrafish embryos up to 2 dpf.98 Kloosterman et al. identified 139 known and 66 new miRNAs in 5 dpf zebrafish larvae.101 We show that in 7 dpf zebrafish, approximately 295 known miRNAs and 120 new, previously undescribed miRNAs, are expressed in vehicle-treated fish, with approximately an equal number being expressed in 1α,25(OH)2D3-treated zebrafish. We believe that the higher number of miRNAs expressed in 7 dpf zebrafish larvae compared with the number expressed at earlier stages is likely to be due to the increased complexity of organs and the need for fine tuning of mRNA expression rather than any methodological differences in library preparation or sequencing. The sequences and identities of these miRNAs are presented in the data and serve as a reference for expressed miRNAs at this stage of zebrafish larval development.
Second, we demonstrate that 1α,25(OH)2D3 regulates 31 separate precursor miRNAs following 6 days of treatment with hormone. Of these, eight are decreased, whereas 23 are increased. We noted that 18 of the 21 1α,25(OH)2D3-regulated mature miRNAs had VDREs in regions of DNA 5000 bp upstream or downstream of the transcription start site. We recognize that additional VDREs may be present in regions of DNA adjacent to the miRNA gene beyond those analyzed in this report, as Pike and colleagues have shown that VDREs are often located >10–100 kb from the transcriptional start sites of vitamin D-sensitive genes.85,108 Whether such VDREs are present at more distant locations is unknown. In addition, the impact of such VDREs on miRNA gene expression is unknown. However, our data do demonstrate the presence of authentic VDREs that are likely to be relevant to the regulation of miRNA gene expression in ∼85% of the 1α,25(OH)2D3-sensitive miRNAs. Of note, two vdr genes, vdra and vdrb, are present in the zebrafish genome.17 The VDRa is the longer of the two receptors with 37 additional NH2-terminal amino acids. Both VDRa and VDRb have highly similar DNA-binding domains that are similar to the DNA-binding domains of VDRs of mammalian origin. It is therefore likely that the receptors bind to similar DNA elements. The analysis in our report was carried out assuming that VDRa and VDRb bound similar DNA nucleotide elements. Further analyses to detect long-range VDREs and an examination of VDREs for binding affinities to the two VDRs are required in the future.
Third, we show that many genes are potentially regulated by these miRNAs when assessed by the Target Scan program. However, because we previously described changes in mRNA concentrations in response to 1α,25(OH)2D3, we can correlate changes in miRNAs with the changes in mRNAs following treatment with hormones. The changes can be statistically associated and by using a combined approach, genes regulated by changes in miRNA concentrations can be identified. Of great interest, many of the miRNAs shown to be altered, regulate 1α,25(OH)2D3-sensitive mRNAs. For example, miRNAs shown to be altered potentially regulate mRNAs for transcription factors (e.g., vdrb, runx2b, klf11, mycch, myccl1b), growth factors and growth factor binding proteins (igf2a, igf2b, igfbp2a, igfbp2b, tgfβ1a), leptin receptor (lepr), RNA encoding proteins involved in glycine, serine, threonine, alanine, aspartate, glutamate, and tryptophan metabolism, and JAK-STAT and p53 signaling. These results were further validated by analysis using DAVID.
We showed that the expression of the mRNA and protein of a highly 1α,25(OH)2D3-regulated gene, the Cyp24A1, is increased by the 1α,25(OH)2D3-regulated miRNA miR125b, in renal proximal tubular cells, thus demonstrating the importance of this miRNA in vitamin D-regulated gene expression. Whereas these results are not generalizable to the mRNAs of other genes that contain miRNA-binding sites, the data are consistent with the notion that 1α,25(OH)2D3-regulated miRNAs alter 1α,25(OH)2D3-sensitive mRNA expression. Although the canonical miRNA regulation model suggests that miRNAs repress the expression of their downstream target genes,22 it has been demonstrated in several studies that the miRNA regulatory network will generate complex expression patterns, causing either positive or negative correlations between miRNAs and their target genes.75–78 Several reports have suggested that miRNAs can positively regulate mRNA expression through binding to 3′ UTRs.109–113
In conclusion, we demonstrate that 1α,25(OH)2D3 alters miRNA expression in zebrafish in vivo. The changes in miRNAs correlate with the changes in mRNA expression. The data show complex regulation of gene expression by 1α,25(OH)2D3.
Supplementary Material
Acknowledgments
Supported by NIH grants AR-058003 and AR-60869 and a grant from the Marion and Ralph Falk Medical Trust. Additional funding was provided by an NIH Howard Temin Pathway to Independence Award in Cancer Research (4R00CA126184), the Grand Duchy of Luxembourg-Institute for Systems Biology Consortium, and the Camille-Dreyfus Teacher-Scholar Program (NDP).
Disclosure Statement
No competing financial interests exist.
References
- 1.Craig TA, Zhang Y, McNulty MS, et al. Research resource: whole transcriptome RNA sequencing detects multiple 1alpha,25-dihydroxyvitamin d3-sensitive metabolic pathways in developing zebrafish. Mol Endocrinol 2012;26:1630–1642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jurutka PW, Bartik L, Whitfield GK, et al. Vitamin D receptor: key roles in bone mineral pathophysiology, molecular mechanism of action, and novel nutritional ligands. J Bone Miner Res 2007;22Suppl 2:V2–V10 [DOI] [PubMed] [Google Scholar]
- 3.McDonnell DP, Mangelsdorf DJ, Pike JW, Haussler MR, O'Malley BW. Molecular cloning of complementary DNA encoding the avian receptor for vitamin D. Science 1987;235:1214–1217 [DOI] [PubMed] [Google Scholar]
- 4.Thompson PD, Jurutka PW, Haussler CA, Whitfield GK, Haussler MR. Heterodimeric DNA binding by the vitamin D receptor and retinoid X receptors is enhanced by 1,25-dihydroxyvitamin D3 and inhibited by 9-cis-retinoic acid. Evidence for allosteric receptor interactions. J Biol Chem 1998;273:8483–8491 [DOI] [PubMed] [Google Scholar]
- 5.Bortell R, Owen TA, Bidwell JP, et al. Vitamin D-responsive protein-DNA interactions at multiple promoter regulatory elements that contribute to the level of rat osteocalcin gene expression. Proc Natl Acad Sci U S A 1992;89:6119–6123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eisman JA, DeLuca HF. Intestinal 1,25-dihydroxyvitamin D3 binding protein: specificity of binding. Steroids 1977;30:245–257 [DOI] [PubMed] [Google Scholar]
- 7.Carlberg C, Bendik I, Wyss A, et al. Two nuclear signalling pathways for vitamin D. Nature 1993;361:657–660 [DOI] [PubMed] [Google Scholar]
- 8.Craig TA, Sommer S, Sussman CR, Grande JP, Kumar R. Expression and regulation of the vitamin D receptor in the zebrafish, Danio rerio. J Bone Miner Res 2008;23:1486–1496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jin CH, Pike JW. Human vitamin D receptor-dependent transactivation in Saccharomyces cerevisiae requires retinoid X receptor. Mol Endocrinol 1996;10:196–205 [DOI] [PubMed] [Google Scholar]
- 10.Umesono K, Murakami KK, Thompson CC, Evans RM. Direct repeats as selective response elements for the thyroid hormone, retinoic acid, and vitamin D3 receptors. Cell 1991;65:1255–1266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.DeLuca HF. Overview of general physiologic features and functions of vitamin D. Am J Clin Nutr 2004;80:1689S–96S [DOI] [PubMed] [Google Scholar]
- 12.Kumar R. Vitamin D and calcium transport. Kidney Int 1991;40:1177–1189 [DOI] [PubMed] [Google Scholar]
- 13.Haussler MR, Haussler CA, Bartik L, et al. Vitamin D receptor: molecular signaling and actions of nutritional ligands in disease prevention. Nutr Rev 2008;66:S98–S112 [DOI] [PubMed] [Google Scholar]
- 14.Cai Q, Chandler JS, Wasserman RH, Kumar R, Penniston JT. Vitamin D and adaptation to dietary calcium and phosphate deficiencies increase intestinal plasma membrane calcium pump gene expression. Proc Natl Acad Sci U S A 1993;90:1345–1349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wasserman RH, Smith CA, Brindak ME, et al. Vitamin D and mineral deficiencies increase the plasma membrane calcium pump of chicken intestine. Gastroenterology 1992;102:886–894 [DOI] [PubMed] [Google Scholar]
- 16.Christakos S, Dhawan P, Porta A, Mady LJ, Seth T. Vitamin D and intestinal calcium absorption. Mol Cell Endocrinol 2011;347:25–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bertrand S, Thisse B, Tavares R, et al. Unexpected novel relational links uncovered by extensive developmental profiling of nuclear receptor expression. PLoS Genet 2007;3:e188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lin CH, Su CH, Tseng DY, Ding FC, Hwang PP. Action of vitamin D and the receptor, VDRa, in calcium handling in zebrafish (Danio rerio). PLoS One 2012;7:e45650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ambros V. The functions of animal microRNAs. Nature 2004;431:350–355 [DOI] [PubMed] [Google Scholar]
- 20.Bartel DP, Chen CZ. Micromanagers of gene expression: the potentially widespread influence of metazoan microRNAs. Nat Rev Genet 2004;5:396–400 [DOI] [PubMed] [Google Scholar]
- 21.Ebert MS, Sharp PA. Roles for microRNAs in conferring robustness to biological processes. Cell 2012;149:515–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell 2009;136:215–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fabian MR, Sonenberg N, Filipowicz W. Regulation of mRNA translation and stability by microRNAs. Annu Rev Biochem 2010;79:351–379 [DOI] [PubMed] [Google Scholar]
- 24.Lee Y, Kim M, Han J, et al. MicroRNA genes are transcribed by RNA polymerase II. EMBO J 2004;23:4051–4060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Borchert GM, Lanier W, Davidson BL. RNA polymerase III transcribes human microRNAs. Nat Struct Mol Biol 2006;13:1097–1101 [DOI] [PubMed] [Google Scholar]
- 26.Gregory RI, Chendrimada TP, Shiekhattar R. MicroRNA biogenesis: isolation and characterization of the microprocessor complex. Methods Mol Biol 2006;342:33–47 [DOI] [PubMed] [Google Scholar]
- 27.Gregory RI, Yan KP, Amuthan G, et al. The Microprocessor complex mediates the genesis of microRNAs. Nature 2004;432:235–240 [DOI] [PubMed] [Google Scholar]
- 28.Hutvagner G, McLachlan J, Pasquinelli AE, Balint E, Tuschl T, Zamore PD. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science 2001;293:834–838 [DOI] [PubMed] [Google Scholar]
- 29.Ketting RF, Fischer SE, Bernstein E, Sijen T, Hannon GJ, Plasterk RH. Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes Dev 2001;15:2654–2659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chendrimada TP, Gregory RI, Kumaraswamy E, et al. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature 2005;436:740–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cochrane DR, Cittelly DM, Richer JK. Steroid receptors and microRNAs: relationships revealed. Steroids 2011;76:1–10 [DOI] [PubMed] [Google Scholar]
- 32.Bhat-Nakshatri P, Wang G, Collins NR, et al. Estradiol-regulated microRNAs control estradiol response in breast cancer cells. Nucleic Acids Res 2009;37:4850–4861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yu F, Yao H, Zhu P, et al. let-7 regulates self renewal and tumorigenicity of breast cancer cells. Cell 2007;131:1109–1123 [DOI] [PubMed] [Google Scholar]
- 34.Li H, Bian C, Liao L, Li J, Zhao RC. miR-17-5p promotes human breast cancer cell migration and invasion through suppression of HBP1. Breast Cancer Res Treat 2011;126:565–575 [DOI] [PubMed] [Google Scholar]
- 35.Zhang B, Pan X, Cobb GP, Anderson TA. microRNAs as oncogenes and tumor suppressors. Dev Biol 2007;302:1–12 [DOI] [PubMed] [Google Scholar]
- 36.Pan Q, Luo X, Chegini N. Differential expression of microRNAs in myometrium and leiomyomas and regulation by ovarian steroids. J Cell Mol Med 2008;12:227–240 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 37.Ribas J, Ni X, Haffner M, et al. miR-21: an androgen receptor-regulated microRNA that promotes hormone-dependent and hormone-independent prostate cancer growth. Cancer Res 2009;69:7165–7169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tessel MA, Krett NL, Rosen ST. Steroid receptor and microRNA regulation in cancer. Curr Opin Oncol 2010;22:592–597 [DOI] [PubMed] [Google Scholar]
- 39.Gupta A, Caffrey E, Callagy G, Gupta S. Oestrogen-dependent regulation of miRNA biogenesis: many ways to skin the cat. Biochem Soc Trans 2012;40:752–758 [DOI] [PubMed] [Google Scholar]
- 40.Alvarez-Diaz S, Valle N, Ferrer-Mayorga G, et al. MicroRNA-22 is induced by vitamin D and contributes to its antiproliferative, antimigratory and gene regulatory effects in colon cancer cells. Hum Mol Genet 2012;21:2157–2165 [DOI] [PubMed] [Google Scholar]
- 41.Cristobo I, Larriba MJ, de los Rios V, Garcia F, Munoz A, Casal JI. Proteomic analysis of 1alpha,25-dihydroxyvitamin D3 action on human colon cancer cells reveals a link to splicing regulation. J Proteomics 2011;75:384–397 [DOI] [PubMed] [Google Scholar]
- 42.Essa S, Denzer N, Mahlknecht U, et al. VDR microRNA expression and epigenetic silencing of vitamin D signaling in melanoma cells. J Steroid Biochem Mol Biol 2010;121:110–113 [DOI] [PubMed] [Google Scholar]
- 43.Essa S, Reichrath S, Mahlknecht U, Montenarh M, Vogt T, Reichrath J. Signature of VDR miRNAs and epigenetic modulation of vitamin D signaling in melanoma cell lines. Anticancer Res 2012;32:383–389 [PubMed] [Google Scholar]
- 44.Gocek E, Wang X, Liu X, Liu CG, Studzinski GP. MicroRNA-32 upregulation by 1,25-dihydroxyvitamin D3 in human myeloid leukemia cells leads to Bim targeting and inhibition of AraC-induced apoptosis. Cancer Res 2011;71:6230–6239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Guan H, Liu C, Chen Z, et al. 1,25-Dihydroxyvitamin D3 up-regulates expression of hsa-let-7a-2 through the interaction of VDR/VDRE in human lung cancer A549 cells. Gene 2013;522:142–146 [DOI] [PubMed] [Google Scholar]
- 46.Kasiappan R, Shen Z, Tse AK, et al. 1,25-Dihydroxyvitamin D3 suppresses telomerase expression and human cancer growth through microRNA-498. J Biol Chem 2012;287:41297–41309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Komagata S, Nakajima M, Takagi S, Mohri T, Taniya T, Yokoi T. Human CYP24 catalyzing the inactivation of calcitriol is post-transcriptionally regulated by miR-125b. Mol Pharmacol 2009;76:702–709 [DOI] [PubMed] [Google Scholar]
- 48.Maier S, Daroqui MC, Scherer S, et al. Butyrate and vitamin D3 induce transcriptional attenuation at the cyclin D1 locus in colonic carcinoma cells. J Cell Physiol 2009;218:638–642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang WL, Chatterjee N, Chittur SV, Welsh J, Tenniswood MP. Effects of 1alpha,25 dihydroxyvitamin D3 and testosterone on miRNA and mRNA expression in LNCaP cells. Mol Cancer 2011;10:58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang WL, Welsh J, Tenniswood M. 1,25-Dihydroxyvitamin D3 modulates lipid metabolism in prostate cancer cells through miRNA mediated regulation of PPARA. J Steroid Biochem Mol Biol 2013;136:247–251 [DOI] [PubMed] [Google Scholar]
- 51.Wang X, Gocek E, Liu CG, Studzinski GP. MicroRNAs181 regulate the expression of p27Kip1 in human myeloid leukemia cells induced to differentiate by 1,25-dihydroxyvitamin D3. Cell Cycle 2009;8:736–741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Giangreco AA, Vaishnav A, Wagner D, et al. Tumor suppressor microRNAs, miR-100 and -125b, are regulated by 1,25-dihydroxyvitamin D in primary prostate cells and in patient tissue. Cancer Prev Res (Phila) 2013;6:483–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Padi SK, Zhang Q, Rustum YM, Morrison C, Guo B. MicroRNA-627 mediates the epigenetic mechanisms of vitamin D to suppress proliferation of human colorectal cancer cells and growth of xenograft tumors in mice. Gastroenterology 2013;145:437–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fleming A, Sato M, Goldsmith P. High-throughput in vivo screening for bone anabolic compounds with zebrafish. J Biomol Screen 2005;10:823–831 [DOI] [PubMed] [Google Scholar]
- 55.Schilling TF, Kimmel CB. Musculoskeletal patterning in the pharyngeal segments of the zebrafish embryo. Development 1997;124:2945–2960 [DOI] [PubMed] [Google Scholar]
- 56.Mabee PM, Olmstead KL, Cubbage CC. An experimental study of intraspecific variation, developmental timing, and heterochrony in fishes. Evolution 2000;54:2091–2106 [DOI] [PubMed] [Google Scholar]
- 57.Westerfield M. The Zebrafish Book: A Guide for the Laboratory Use of Zebrafish (Danio rerio), 4th edition. Eugene: University of Oregon Press, 2000 [Google Scholar]
- 58.Yang X, Li L. miRDeep-P: a computational tool for analyzing the microRNA transcriptome in plants. Bioinformatics (Oxford, England) 2011;27:2614–2615 [DOI] [PubMed] [Google Scholar]
- 59.Ambros V, Bartel B, Bartel DP, et al. A uniform system for microRNA annotation. RNA 2003;9:277–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Friedlander MR, Chen W, Adamidi C, et al. Discovering microRNAs from deep sequencing data using miRDeep. Nat Biotechnol 2008;26:407–415 [DOI] [PubMed] [Google Scholar]
- 61.Bullard JH, Purdom E, Hansen KD, Dudoit S. Evaluation of statistical methods for normalization and differential expression in mRNA-Seq experiments. BMC Bioinformatics 2010;11:94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol 2010;11:R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B 1995;57:289–300 [Google Scholar]
- 64.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 2005;120:15–20 [DOI] [PubMed] [Google Scholar]
- 65.Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci U S A 2003;100:9440–9445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Storey JD. A direct approach to false discovery rates. J R Stat Soc B 2002;64:479–498 [Google Scholar]
- 67.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 2009;4:44–57 [DOI] [PubMed] [Google Scholar]
- 68.Kanehisa M, Araki M, Goto S, et al. KEGG for linking genomes to life and the environment. Nucleic Acids Res 2008;36:D480–D484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mi H, Guo N, Kejariwal A, Thomas PD. PANTHER version 6: protein sequence and function evolution data with expanded representation of biological pathways. Nucleic Acids Res 2007;35:D247–D252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Joshi-Tope G, Gillespie M, Vastrik I, et al. Reactome: a knowledgebase of biological pathways. Nucleic Acids Res 2005;33:D428–D432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kel AE, Gossling E, Reuter I, Cheremushkin E, Kel-Margoulis OV, Wingender E. MATCH (TM): a tool for searching transcription factor binding sites in DNA sequences. Nucleic Acids Res 2003;31:3576–3579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ryan ZC, Ketha H, McNulty MS, et al. Sclerostin alters serum vitamin D metabolite and fibroblast growth factor 23 concentrations and the urinary excretion of calcium. Proc Natl Acad Sci U S A 2013;110:6199–6204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kumar R, Schaefer J, Grande JP, Roche PC. Immunolocalization of calcitriol receptor, 24-hydroxylase cytochrome P-450, and calbindin D28k in human kidney. Am J Physiol 1994;266:F477–F485 [DOI] [PubMed] [Google Scholar]
- 74.Cunningham R, Steplock D, Wang F, et al. Defective parathyroid hormone regulation of NHE3 activity and phosphate adaptation in cultured NHERF-1−/− renal proximal tubule cells. J Biol Chem 2004;279:37815–37821 [DOI] [PubMed] [Google Scholar]
- 75.Tang T, Kumar S, Shen Y, et al. Adverse interactions between micro-RNAs and target genes from different species. Proc Natl Acad Sci U S A 2010;107:12935–12940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wang L, Oberg AL, Asmann YW, et al. Genome-wide transcriptional profiling reveals microRNA-correlated genes and biological processes in human lymphoblastoid cell lines. PLoS One 2009;4:e5878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Huang GT, Athanassiou C, Benos PV. mirConnX: condition-specific mRNA-microRNA network integrator. Nucleic Acids Res 2011;39:W416–W423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lu J, Clark AG. Impact of microRNA regulation on variation in human gene expression. Genome Res 2012;22:1243–1254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Cesana M, Cacchiarelli D, Legnini I, et al. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell 2011;147:358–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Karreth FA, Tay Y, Perna D, et al. In vivo identification of tumor- suppressive PTEN ceRNAs in an oncogenic BRAF-induced mouse model of melanoma. Cell 2011;147:382–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sumazin P, Yang X, Chiu HS, et al. An extensive microRNA-mediated network of RNA-RNA interactions regulates established oncogenic pathways in glioblastoma. Cell 2011;147:370–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tay Y, Kats L, Salmena L, et al. Coding-independent regulation of the tumor suppressor PTEN by competing endogenous mRNAs. Cell 2011;147:344–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Leff SE, Rosenfeld MG, Evans RM. Complex transcriptional units: diversity in gene expression by alternative RNA processing. Annu Rev Biochem 1986;55:1091–1117 [DOI] [PubMed] [Google Scholar]
- 84.Tsai MJ, O'Malley BW. Molecular mechanisms of action of steroid/thyroid receptor superfamily members. Annu Rev Biochem 1994;63:451–486 [DOI] [PubMed] [Google Scholar]
- 85.Kim S, Yamazaki M, Zella LA, et al. Multiple enhancer regions located at significant distances upstream of the transcriptional start site mediate RANKL gene expression in response to 1,25-dihydroxyvitamin D3. J Steroid Biochem Mol Biol 2007;103:430–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ozono K, Liao J, Kerner SA, Scott RA, Pike JW. The vitamin D-responsive element in the human osteocalcin gene. Association with a nuclear proto-oncogene enhancer. J Biol Chem 1990;265:21881–21888 [PubMed] [Google Scholar]
- 87.Ozono K, Sone T, Pike JW. The genomic mechanism of action of 1,25-dihydroxyvitamin D3. J Bone Miner Res 1991;6:1021–1027 [DOI] [PubMed] [Google Scholar]
- 88.Sone T, Kerner S, Pike JW. Vitamin D receptor interaction with specific DNA. Association as a 1,25-dihydroxyvitamin D3-modulated heterodimer. J Biol Chem 1991;266:23296–23305 [PubMed] [Google Scholar]
- 89.Chen KS, DeLuca HF. Cloning of the human 1 alpha,25-dihydroxyvitamin D-3 24-hydroxylase gene promoter and identification of two vitamin D-responsive elements. Biochim Biophys Acta 1995;1263:1–9 [DOI] [PubMed] [Google Scholar]
- 90.Darwish HM, DeLuca HF. Analysis of binding of the 1,25-dihydroxyvitamin D3 receptor to positive and negative vitamin D response elements. Arch Biochem Biophys 1996;334:223–234 [DOI] [PubMed] [Google Scholar]
- 91.Malinen M, Ryynanen J, Heinaniemi M, Vaisanen S, Carlberg C. Cyclical regulation of the insulin-like growth factor binding protein 3 gene in response to 1alpha,25-dihydroxyvitamin D3. Nucleic Acids Res 2011;39:502–512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Polly P, Carlberg C, Eisman JA, Morrison NA. Identification of a vitamin D3 response element in the fibronectin gene that is bound by a vitamin D3 receptor homodimer. J Cell Biochem 1996;60:322–333 [DOI] [PubMed] [Google Scholar]
- 93.Sinkkonen L, Malinen M, Saavalainen K, Vaisanen S, Carlberg C. Regulation of the human cyclin C gene via multiple vitamin D3-responsive regions in its promoter. Nucleic Acids Res 2005;33:2440–2451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Turunen MM, Dunlop TW, Carlberg C, Vaisanen S. Selective use of multiple vitamin D response elements underlies the 1 alpha,25-dihydroxyvitamin D3-mediated negative regulation of the human CYP27B1 gene. Nucleic Acids Res 2007;35:2734–2747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wu Y, Craig TA, Lutz WH, Kumar R. Identification of 1 alpha,25-dihydroxyvitamin D3 response elements in the human transforming growth factor beta 2 gene. Biochemistry 1999;38:2654–2660 [DOI] [PubMed] [Google Scholar]
- 96.Im HJ, Craig TA, Pittelkow MR, Kumar R. Characterization of a novel hexameric repeat DNA sequence in the promoter of the immediate early gene, IEX-1, that mediates 1alpha,25-dihydroxyvitamin D(3)-associated IEX-1 gene repression. Oncogene 2002;21:3706–3714 [DOI] [PubMed] [Google Scholar]
- 97.Veenstra TD, Fahnestock M, Kumar R. An AP-1 site in the nerve growth factor promoter is essential for 1, 25-dihydroxyvitamin D3-mediated nerve growth factor expression in osteoblasts. Biochemistry 1998;37:5988–5994 [DOI] [PubMed] [Google Scholar]
- 98.Chen PY, Manninga H, Slanchev K, et al. The developmental miRNA profiles of zebrafish as determined by small RNA cloning. Genes Dev 2005;19:1288–1293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Choi PS, Zakhary L, Choi WY, et al. Members of the miRNA-200 family regulate olfactory neurogenesis. Neuron 2008;57:41–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Giraldez AJ, Cinalli RM, Glasner ME, et al. MicroRNAs regulate brain morphogenesis in zebrafish. Science 2005;308:833–838 [DOI] [PubMed] [Google Scholar]
- 101.Kloosterman WP, Steiner FA, Berezikov E, et al. Cloning and expression of new microRNAs from zebrafish. Nucleic Acids Res 2006;34:2558–2569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kloosterman WP, Wienholds E, Ketting RF, Plasterk RH. Substrate requirements for let-7 function in the developing zebrafish embryo. Nucleic Acids Res 2004;32:6284–6291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Lim LP, Glasner ME, Yekta S, Burge CB, Bartel DP. Vertebrate microRNA genes. Science 2003;299:1540. [DOI] [PubMed] [Google Scholar]
- 104.Thatcher EJ, Paydar I, Anderson KK, Patton JG. Regulation of zebrafish fin regeneration by microRNAs. Proc Natl Acad Sci U S A 2008;105:18384–18389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Wienholds E, Kloosterman WP, Miska E, et al. MicroRNA expression in zebrafish embryonic development. Science 2005;309:310–311 [DOI] [PubMed] [Google Scholar]
- 106.Wienholds E, Koudijs MJ, van Eeden FJ, Cuppen E, Plasterk RH. The microRNA-producing enzyme Dicer1 is essential for zebrafish development. Nat Genet 2003;35:217–218 [DOI] [PubMed] [Google Scholar]
- 107.Zhao Y, Samal E, Srivastava D. Serum response factor regulates a muscle-specific microRNA that targets Hand2 during cardiogenesis. Nature 2005;436:214–220 [DOI] [PubMed] [Google Scholar]
- 108.Pike JW, Meyer MB. Fundamentals of vitamin D hormone-regulated gene expression. J Steroid Biochem Mol Biol 2013;pii: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Place RF, Li LC, Pookot D, Noonan EJ, Dahiya R. MicroRNA-373 induces expression of genes with complementary promoter sequences. Proc Natl Acad Sci U S A 2008;105:1608–1613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Murphy AJ, Guyre PM, Pioli PA. Estradiol suppresses NF-kappa B activation through coordinated regulation of let-7a and miR-125b in primary human macrophages. J Immunol 2010;184:5029–5037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ma F, Liu X, Li D, et al. MicroRNA-466l upregulates IL-10 expression in TLR-triggered macrophages by antagonizing RNA-binding protein tristetraprolin-mediated IL-10 mRNA degradation. J Immunol 2010;184:6053–6059 [DOI] [PubMed] [Google Scholar]
- 112.Ghosh T, Soni K, Scaria V, Halimani M, Bhattacharjee C, Pillai B. MicroRNA-mediated up-regulation of an alternatively polyadenylated variant of the mouse cytoplasmic {beta}-actin gene. Nucleic Acids Res 2008;36:6318–6332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Tsai NP, Lin YL, Wei LN. MicroRNA mir-346 targets the 5′-untranslated region of receptor-interacting protein 140 (RIP140) mRNA and up-regulates its protein expression. Biochem J 2009;424:411–418 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.