Abstract
Lin28B is a RNA-binding protein that inhibits the let-7 microRNA family and acts as an oncogene in various human malignant diseases. Conversely, the members of let-7 family function as tumor suppressers and are often inactivated in cancers. The interaction of Lin28B/let-7 plays a crucial part of tumorigenesis. In this study, the authors examined the Lin28B expression using immunohistochemistry in 190 breast cancers and analyzed the correlation of Lin28B immunostaining and clinicopathological characteristics. Breast cancer patients previously diagnosed with invasive ductal carcinomas were enrolled in this study. All cases went through surgical procedures as the initial treatment. The characteristics of every case were collected, including tumor size, pathologic grade, metastatic lymphoid nodes, and estrogen receptor α (ERα), progesterone receptor (PR), and HER2 status. The immunostaining was scored by two independent investigators. Eighty-three (43.7%) of 190 cases showed positive expression of Lin28B. Lin28B immunostaining was increased in tumors compared with the adjacent tissues. Overexpression of Lin28B was linked to poor differentiation, advanced-stage disease, and Ki67-positive status (all p<0.05). Besides, Lin28B expression was significantly different among breast cancer subtypes. This study addresses the role of Lin28B in breast cancers and provides insight of its predictive effects in disease development.
Key words: : breast cancer, Ki67, Let-7, Lin28, Lin28B
Introduction
The RNA-binding protein Lin28B and its analogue Lin28A are key regulators of cellular transformation.1,2 So far, the Lin28/let7 axis is a well-established double-negative loop, promoting cell differentiation, cellular reprogramming, cell fate, and so on.1,3,4 Let-7s, a family of conservative microRNAs (miRNAs), are recognized as tumor suppressers which repress self-renewal and differentiation in both neoplastic and normal tissues.3 The members of let-7 activate tumorigenesis by derepressing the target oncogenes, such as MYC and RAS.3,5 Lin28 is primarily known for its post-transcriptional regulation of let-7s.1 Both Lin28A and Lin28B could selectively bind to let-7 precursors and inhibit their expression1,3,5
Lin28A and Lin28B perform different mechanisms in the blockage of let-7s and exhibit different expression profiles in tissues or cell lines, even though they share a high degree of similarity in nucleotide sequence identity and protein domains.1 Precursors of the let-7 miRNA family undergo series of processing steps to become mature biological units.1,6 In cell nucleoli, pri-miRNAs are cleaved into pre-miRNAs by a microprocessor containing the ribonuclease Drosha and cofactor DGCR8. Then, pre-miRNAs are processed into mature miRNAs, which are <22 nt length. Finally, the miRNAs are exported into the cytoplasm by Dicer.7 Evidence indicated that Lin28A bound to pre-let-7 and blocked the Dicer-mediated cleavage; whereas Lin28B stopped the Drosha-mediated cleavage through binding to the microprocessor complex.5 Consistent with these functional differences, Lin28A was revealed to localize in the cytoplasm, and Lin28B showed widespread expression in the nucleus and cytoplasm.1 In addition, Lin28B was highly expressed in the testis, placenta, fetal liver, several tumors, and cancer cell lines, whereas Lin28A showed limited expression in normal tissues.5 Their nonoverlapping localization in tissues or cell lines suggests that they are mutually exclusive in functions.5,8
Previous studies exploited that let-7s interacted with Lin28B to promote carcinogenesis.8,9 Others identified that Lin28B could drive diverse signaling pathways by directly regulating certain genes, including HMGA2, CCND2, IGF1R, and IGF2BP.5 Emerging proofs supported that Lin28B is a key regulator in normal or cancerous tissues by promoting cell proliferation, cell reprogramming, metabolism, migration, invasion, and even metastasis.2 Hence, Lin28B is thought to contribute to the behavior of advanced cancers with poor prognosis.10 Overexpression of Lin28B was observed in various tumor types, including lung cancer, colon cancer, cervical cancer, and breast cancer.2 Genome-wide association studies found that genetic variants of Lin28B were related with the timing of puberty and menarche.11 The age of menarche has been known as one of the reproductive risk factors associated with morbidity in cancers of the female reproductive system, especially breast cancers.12
There are a limited number of studies that pursued the expression of Lin28B in breast cancers. To determine the relationship between Lin28B expression and clinicopathological characteristics of breast cancer, the authors assessed semiquantitative protein expression analyses on formalin-fixed, paraffin-embedded primary tumors from 190 breast cancer patients.
Materials and Methods
Study subjects
Breast cancer tissues from 190 patients were collected at the Ganzhou Tumor Hospital, Jiangxi province between January 2008 and January 2013. Those patients who had neoadjuvant chemotherapy or had been treated elsewhere before coming to Ganzhou Tumor Hospital were excluded from the study. All cases were strictly diagnosed using histological and clinical criteria after surgical treatments. Only invasive ductal carcinoma was included in the study. Among 190 cases, 20 tumor samples were randomly selected for the assessments with the corresponding adjacent tissues. In addition, several samples of benign breast disease were also analyzed for comparison. The status of estrogen receptor α (ERα), progesterone receptor (PR), and Ki67 was determined by immunohistochemistry and collected from pathology reports. ERα and PR positivity was nuclear staining of 10% or more tumor cells. The authors evaluated the Ki67 labeling index using a cutoff value of 14%.13 HER2 immunoreactivity was scored for the intensity and the completeness of cell membrane staining (−, no staining; +, weak partial membranous staining in more than 10% tumor cells; ++, moderately complete membrane staining in more than 10% tumor cells; +++, strong complete membranous staining in more than 10% of tumor cells). HER2 (+++) was defined as positive, and the equivocal HER2 (++) was assessed by fluorescence in situ hybridization.14 Both HER2 (−) and (+) were described as negative. Based on 2013 St Gallen Consensus, subtypes of breast cancer (Luminal A, Luminal B, HER2 overexpression, and basal like) were defined by ERα, PR, Ki67, and HER2 status.15 Information, including age, menopausal status, and tumors, lymph nodes, and metastases (TNM) data, was obtained from clinical inquiry and relevant examinations. The TNM staging classification was determined by two different physicians according to the seventh edition Union for International Cancer Control (UICC) cancer staging systems. The study protocol was approved by the Ethics Committee of Jinling Hospital, Southern Medical University.
Immunohistochemistry of Lin28B
Tissues were fixed with 10% formalin at room temperature and were embedded in paraffin. The paraffin blocks were cut at 3 μm thickness. The sections were heated in 55°C–60°C for 2 hours, deparaffinized in xylene, hydrated through series of alcohol, and washed by phosphate-buffered saline solution (pH 7.4) three times. For antigen retrieval, the sections were heated with 10 mM citric buffer (pH 6.0) for 15 minutes. For endogenous peroxidase blockage, 3% H2O2 was added to each section and stayed for 5 minutes. To avoid nonspecific binding, 10% fetal bovine serum was used for each section for 10 minutes of incubation. The immunostaining began with the Lin28B (rabbit polyclonal, ab71415, 1:20; Abcam) as the primary antibody for 60 minutes of staining, continued with the MaxVision™ HRP-Polymer anti-Rabbit IHC Kit (KIT-5005; Maxim) for 15 minutes according to the manufacturer's protocol, and followed by diaminobenzidine for visualization. In addition, all sections were counterstained with hematoxylin. The testis tissue was used as positive control.
Evaluation of Lin28B immunoreactivity
Every section was evaluated and scored independently by two pathologists. The relative immunointensity was estimated by calculating the percentage of positive stained tumor cells (<10%, −; 10%–25%, +; 25%–50%, ++; >50%, +++). The positive expression was defined as the expression levels from “+” to “+++.”
Statistical analyses
Statistical analyses were conducted with SPSS 18.0 for windows (SPSS). The relationship between Lin28B expression and the various groups of clinicopathological characteristics was analyzed by the χ2 test or Fisher's exact test, where appropriate. The p<0.05 was considered as significance. Variables that showed significances with Lin28B expression were further examined by the Spearman rank correlation. The Wilcoxon signed-rank test was used for comparison of Lin28B expression between breast tumors and the corresponding adjacent tissues in 20 pairs of specimens.
Results
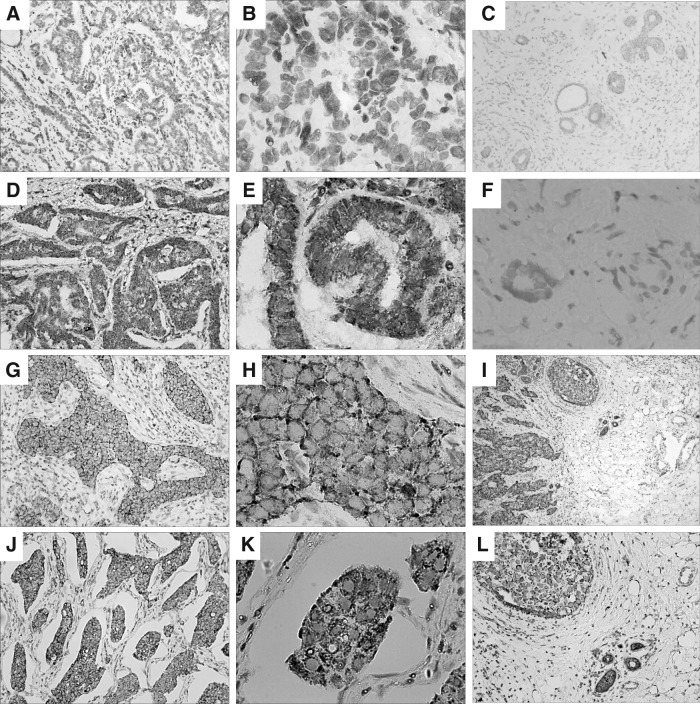
Clinicopathological features of 190 breast cancer cases are summarized in Table 1. Briefly, 83 of 190 (43.7%) cases showed positive expression of Lin28B in immunohistochemistry analyses, whereas the remaining 107 (56.3%) cases showed negative expression. The correlations between Lin28 expression and various groups are shown in Table 1. For further dissection, the authors analyzed the paired specimens of tumors and adjacent tissues. Lin28B immunostaining was increased in tumors compared with the adjacent tissues (p=0.002, Wilcoxon signed-rank test). In the Lin28B-positive cancer, the adjacent tissue apparently showed a relatively lower or negative expression (Table 2). Lin28B was predominantly stained in the cytoplasm, but a few sections presented a combination expression of the nucleus and cytoplasm (Fig. 1). Besides cancerous tissue samples, the authors also conducted Lin28B immunohistochemistry in several tissue samples of benign breast disease (Fig. 1). Consistent with previous studies, all of them showed negative expression.1
Table 1.
Correlation Between Clinicopathogical Features and Lin28B Expression in 190 Breast Cancer Cases
| Lin28B expression | |||||||
|---|---|---|---|---|---|---|---|
| Clinicopathological features | na(%) | Negative (%) | Positive (%) | χ2 | pb | r | pc |
| All cases | 190 (100) | 107 (56.3) | 83 (43.7) | ||||
| Age | 0.021 | 0.884 | |||||
| ≤46.5 | 95 (50.0) | 54 (28.4) | 41 (21.6) | ||||
| >46.5 | 95 (50.0) | 53 (27.9) | 42 (22.1) | ||||
| Menopausal status | 0.229 | 0.632 | |||||
| Premenopausal | 120 (63.2) | 66 (34.7) | 54 (28.4) | ||||
| Postmenopausald | 70 (36.8) | 41 (21.6) | 29 (15.3) | ||||
| Tumor size (cm) | 4.729 | 0.094 | |||||
| <2 | 53 (27.9) | 31 (16.3) | 22 (11.6) | ||||
| 2–5 | 123 (64.7) | 72 (37.9) | 51 (26.8) | ||||
| >5 | 14 (7.4) | 4 (2.1) | 10 (5.3) | ||||
| Histological differentiation | 13.790 | 0.001 | 0.216 | 0.003 | |||
| Well | 61 (32.1) | 39 (20.5) | 22 (11.6) | ||||
| Moderate | 81 (42.6) | 52 (27.4) | 29 (15.3) | ||||
| Poor | 48 (25.3) | 16 (8.4) | 32 (16.8) | ||||
| TNM staging | 9.213 | 0.002 | 0.220 | 0.002 | |||
| I–II | 97 (51.1) | 65 (34.2) | 32 (16.8) | ||||
| III–IV | 93 (48.9) | 42 (22.1) | 51 (26.8) | ||||
| Metastatic lymph nodes | 30.996 | <0.0001 | 0.359 | <0.0001 | |||
| 0 | 72 (37.9) | 59 (31.1) | 13 (6.8) | ||||
| 1–3 | 45 (23.7) | 18 (9.5) | 27 (14.2) | ||||
| ≥4 | 73 (38.4) | 30 (15.8) | 43 (22.6) | ||||
| ER | 0.160 | 0.899 | |||||
| Negative | 86 (45.3) | 48 (25.3) | 38 (20.0) | ||||
| Positive | 104 (54.7) | 59 (31.1) | 45 (23.7) | ||||
| PR | 2.307 | 0.129 | |||||
| Negative | 92 (48.4) | 57 (30.0) | 35 (18.4) | ||||
| Positive | 98 (51.6) | 50 (26.3) | 48 (25.3) | ||||
| Her-2 | 1.264 | 0.261 | |||||
| Negative | 147 (77.4) | 86 (45.3) | 61 (32.1) | ||||
| Positive | 43 (22.6) | 21 (11.1) | 22 (11.6) | ||||
| Ki67 | 13.268 | <0.0001 | 0.037 | 0.677 | |||
| Negative | 76 (40.0) | 55 (28.9) | 21 (11.1) | ||||
| Positive | 92 (60.0) | 52 (27.4) | 62 (32.6) | ||||
| Subtype | 20.450 | <0.0001 | |||||
| Luminal A | 52 (27.4) | 41 (21.6) | 11 (5.8) | ||||
| Luminal B | 59 (31.1) | 22 (11.6) | 37 (19.5) | ||||
| Her-2 overexpression | 29 (15.3) | 14 (12.8) | 15 (7.9) | ||||
| Basal like | 50 (26.3) | 30 (15.8) | 20 (10.5) | ||||
Number of cases in each group.
p for χ2 test.
p for Spearman rank correlation.
Postmenopausal status for natural menopause.
ER, estrogen receptor; PR, progesterone receptor; TNM, tumors, lymph nodes, and metastases.
Table 2.
Expression of Lin28B in Breast Invasive Ductal Carcinomas and Adjacent Tissues
| Numbera | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | pb |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Invasive ductal carcinoma | − | − | + | +++ | − | +++ | ++ | − | − | − | +++ | − | + | ++ | +++ | ++ | − | + | ++ | +++ | |
| Adjacent tissue | − | − | − | − | − | + | − | − | − | − | ++ | − | − | − | ++ | − | − | − | − | + | 0.002 |
Each sample of breast cancer was collected with the corresponding adjacent tissue.
p for Wilcoxon signed-rank test.
FIG. 1.
Lin28B expression in breast invasive ductal carcinoma by immunohistochemical staining. (A, B) Well-differentiated tumor cells scored as Lin28B (−). (D, E) Lin28B staining was scored as (+) in well-differentiated tumor cells. (G, H) Lin28B staining was scored as (++) in moderately differentiated tumor cells. (J, K) Lin28B staining was scored as (+++) in poorly differentiated tumor cells. (C, F) Benign breast disease was negative expression of Lin28B. (I, L) Breast cancer tissue in the upper left portion was scored as (++), and adjacent tissue in the lower right portion was scored as (+/−). (D, E, G–L) All presented staining in the cytoplasm. Magnification: 40× in (I), 100× in (A, C, D, G, J, and L), 400× in (B, E, F, H, and K).
Clinical characteristics
In all, 190 patients with a median age of 46.5 years (range, 26–87 years) were included in the final analyses. Among these cases, 120 (63.2%) patients were premenopause and 70 (36.8%) patients were postmenopause. According to the UICC cancer staging systems seventh edition stage grouping criteria, 97 (51.1%) cases were stage I–II, and 93 (48.9%) were stage III–IV. There were 72 (37.9%) cases without metastatic lymphoid nodes, 45 (23.7%) with more than one and less than three metastatic lymphoid nodes, and 73 (38.4%) with more than four metastatic lymphoid nodes (Table 1). The proportion of Lin28B-positive cases was higher in stage III–IV disease and multiple lymph nodes metastasis (p<0.05 for both). In the Spearman rank correlation, both TNM staging and metastatic lymph nodes showed a positive correlation with Lin28B expression (r=0.220 and 0.359, respectively, p<0.05 for both) (Table 1).
Pathological characteristics
In histological differentiation, 61 (32.1%) cases were well differentiated, 81 (42.6%) cases were moderately differentiated, and 48 (25.3%) cases were poorly differentiated. Eighty-six (45.3%) cases, ERα negative; 104 (54.7%) cases, ERα positive; 92 (48.4%) cases, PR negative; 98 (51.6%) cases, PR positive; 43 (22.6%) cases, HER2 positive; 147 (77.4%), HER2 negative; 92 (60.0%), Ki67 positive; 76 (40.0%) cases, Ki67 negative (Table 1). In breast cancer subtypes, 52 (27.4%) cases were Luminal A; 59 (31.1%) cases were Luminal B; 29 (15.3%) cases were HER2 overexpression; and 50 (26.3%) cases were basal like. The distribution of Lin28B expression was significantly different among histological differentiation, Ki67 status, and intrinsic subtypes (p<0.05 for all). Compared with Ki67-negative cases, Ki67-positve cases had a significantly higher proportion of positive expression of Lin28B. As expected, similar results were seen between the Luminal A and Luminal B subtypes, which were mostly determined by Ki67 status. Additionally, histological differentiation and Ki67 labeling index presented a positive association with Lin28B expression in Spearman rank correlation (r=0.216 and 0.037, respectively, p<0.05 for both) (Table 1). No significant differences with Lin28B expression were found in ERα, PR, and HER2 status.
Discussion
The Lin28/let-7 axis is considered as the central maintenance of cell proliferation, cell growth, metabolism, and so on.3 Similar to Lin28A, Lin28B promotes cellular transformation and induces pluripotent stem cells in vitro and in vivo.1 By now, both of them discovered overexpression in advanced disease in multiple cancer types.1 This study elucidated the correlation between Lin28B expression and invasive ductal breast carcinoma according to immunohistochemistry.5 Notably, the authors found that Lin28B expression significantly associated with histological differentiation, TNM staging, and number of metastatic lymph nodes. Higher levels of expression were observed in tumors with poor histological differentiation, stage III–IV, and multiple metastatic lymph nodes. All these three features indicated advanced disease with aggressive characteristics and poor prognosis.
Recent findings undertook similar investigation of Lin28B expression on multiple types of cancer.16–18 It is proved that Lin28B overexpression was associated with tumor recurrence and poor overall survival in colon adenocarcinomas.19 Hamano et al. found that high expression of Lin28B correlated with lymph node metastasis and poor prognosis in esophagus cancer.20 It is therefore rational to link Lin28B overexpression to aggressive features and poor outcomes of cancer, and conclude the possible consequence on increasing proliferation and invasiveness. Results of this study in breast cancer were in accordance with these previously reported researches.
Subsequently, the current study also pointed out that Ki67 status was positively related to Lin28B expression. Ki67 is a marker of high proliferation and has a predictive and prognostic value for clinical practice.21 The difference between Luminal B and Luminal A breast cancer is largely dependent on Ki67 status.15 The prognosis of Luminal B is relatively poor than the one of Luminal A.22,23 Given the facts mentioned above, it is convincible to infer that Lin28B could be a prognosis factor of breast cancer. Sakurai et al. studied Lin28A and Lin28B expression in breast cancer and found that the expression of several miRNAs of the let-7 family was inversely correlated with Lin28. Meanwhile, they also found relevance between Lin28B expression and Ki67.24 Moreover, HER2 overexpression is also regarded as an aggressive clinical characteristic in breast cancer.25 It has been found that Lin28A was overexpressed in HER2-positive breast cancers.1 It is reported that genetic variants of Lin28A could disturb the Lin28/let-7 axis in breast cancer.26 Furthermore, Lin28A could enhance HER2 expression by binding to HER2 mRNA.25 As for Lin28B, triple-negative tumors were related with its overexpression in populations of European descent.1,2 However, the authors failed to capture a higher scale of Lin28B-positive expression in triple-negative breast cancers in the current study. Larger studies in a different population with long-term follow-up are needed to validate these findings.
In conclusion, this study provided evidences that Lin28B, a homologue of Lin28A, showed correlation with clinicopathological characteristics in breast cancers. Lin28B could be a predictive and prognostic marker of breast cancer. Additional studies will be necessary to investigate the roles of Lin28B in response to various treatments and clinical outcomes.
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (No. 81272252) and the Natural Science Foundation of Jiangsu Province (No. BK2011656) (to X.G.).
Disclosure Statement
No potential conflicts of interest were disclosed.
References
- 1.Piskounova E, Polytarchou C, Thornton JE, et al. Lin28A and Lin28B inhibit let-7 microRNA biogenesis by distinct mechanisms. Cell 2011;147:1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Viswanathan SR, Powers JT, Einhorn W, et al. Lin28 promotes transformation and is associated with advanced human malignancies. Nat Genet 2009;41:843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nam Y, Chen C, Gregory RI, et al. Molecular basis for interaction of let-7 microRNAs with Lin28. Cell 2011;147:1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhou J, Ng SB, Chng WJ. LIN28/LIN28B: An emerging oncogenic driver in cancer stem cells. Int J Biochem Cell Biol 2013;45:973. [DOI] [PubMed] [Google Scholar]
- 5.Viswanathan SR, Daley GQ. Lin28: A microRNA regulator with a macro role. Cell 2010;140:445. [DOI] [PubMed] [Google Scholar]
- 6.Paroo Z, Ye X, Chen S, et al. Phosphorylation of the human microRNA-generating complex mediates MAPK/Erk signaling. Cell 2009;139:112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rybak A, Fuchs H, Smirnova L, et al. A feedback loop comprising lin-28 and let-7 controls pre-let-7 maturation during neural stem-cell commitment. Nat Cell Biol 2008;10:987. [DOI] [PubMed] [Google Scholar]
- 8.Thornton JE, Gregory RI. How does Lin28 let-7 control development and disease? Trends Cell Biol 2012;22:474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhu H, Shyh-Chang N, Segre AV, et al. The Lin28/let-7 axis regulates glucose metabolism. Cell 2011;147:81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Elks CE, Perry JR, Sulem P, et al. Thirty new loci for age at menarche identified by a meta-analysis of genome-wide association studies. Nat Genet 2010;42:1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cousminer DL, Berry DJ, Timpson NJ, et al. Genome-wide association and longitudinal analyses reveal genetic loci linking pubertal height growth, pubertal timing and childhood adiposity. Hum Mol Genet 2013;22:2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Anderson KN, Schwab RB, Martinez ME. Reproductive risk factors and breast cancer subtypes: A review of the literature. Breast Cancer Res Treat 2014;144:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cheang MC, Chia SK, Voduc D, et al. Ki67 index, HER2 status, and prognosis of patients with luminal B breast cancer. J Natl Cancer Inst 2009;101:736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jacobs TW, Gown AM, Yaziji H, et al. Specificity of HercepTest in determining HER-2/neu status of breast cancers using the United States Food and Drug Administration-approved scoring system. J Clin Oncol 1999;17:1983. [DOI] [PubMed] [Google Scholar]
- 15.Goldhirsch A, Winer EP, Coates AS, et al. Personalizing the treatment of women with early breast cancer: Highlights of the St. Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer 2013. Ann Oncol 2013;24:2206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guo Y, Chen Y, Ito H, et al. Identification and characterization of lin-28 homolog B (LIN28B) in human hepatocellular carcinoma. Gene 2006;384:51. [DOI] [PubMed] [Google Scholar]
- 17.El-Khairi R, Parnaik R, Duncan AJ, et al. Analysis of LIN28A in early human ovary development and as a candidate gene for primary ovarian insufficiency. Mol Cell Endocrinol 2012;351:264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.West JA, Viswanathan SR, Yabuuchi A, et al. A role for Lin28 in primordial germ-cell development and germ-cell malignancy. Nature 2009;460:909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.King CE, Cuatrecasas M, Castells A, et al. LIN28B promotes colon cancer progression and metastasis. Cancer Res 2011;71:4260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hamano R, Miyata H, Yamasaki M, et al. High expression of Lin28 is associated with tumour aggressiveness and poor prognosis of patients in oesophagus cancer. Br J Cancer 2012;106:1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fasching PA, Heusinger K, Haeberle L, et al. Ki67, chemotherapy response, and prognosis in breast cancer patients receiving neoadjuvant treatment. BMC Cancer 2011;11:486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goldhirsch A, Wood WC, Coates AS, et al. Strategies for subtypes—dealing with the diversity of breast cancer: Highlights of the St. Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer 2011. Ann Oncol 2011;22:1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reis-Filho JS, Pusztai L. Gene expression profiling in breast cancer: Classification, prognostication, and prediction. Lancet 2011;378:1812. [DOI] [PubMed] [Google Scholar]
- 24.Sakurai M, Miki Y, Masuda M, et al. LIN28: A regulator of tumor-suppressing activity of let-7 microRNA in human breast cancer. J Steroid Biochem Mol Biol 2012;131:101. [DOI] [PubMed] [Google Scholar]
- 25.Feng C, Neumeister V, Ma W, et al. Lin28 regulates HER2 and promotes malignancy through multiple mechanisms. Cell Cycle 2012;11:2486. [DOI] [PubMed] [Google Scholar]
- 26.Chen AX, Yu KD, Fan L, et al. Germline genetic variants disturbing the Let-7/LIN28 double-negative feedback loop alter breast cancer susceptibility. PLoS Genet 2011;7:e1002259. [DOI] [PMC free article] [PubMed] [Google Scholar]