CNTD1 coordinates the maturation and designation of meiotic crossover sites from an excess pool of double-strand break intermediates by regulating dynamic changes in key protein complexes associated with these sites.
Abstract
Meiotic crossovers (COs) are crucial for ensuring accurate homologous chromosome segregation during meiosis I. Because the double-strand breaks (DSBs) that initiate meiotic recombination greatly outnumber eventual COs, this process requires exquisite regulation to narrow down the pool of DSB intermediates that may form COs. In this paper, we identify a cyclin-related protein, CNTD1, as a critical mediator of this process. Disruption of Cntd1 results in failure to localize CO-specific factors MutLγ and HEI10 at designated CO sites and also leads to prolonged high levels of pre-CO intermediates marked by MutSγ and RNF212. These data show that maturation of COs is intimately coupled to deselection of excess pre-CO sites to yield a limited number of COs and that CNTD1 coordinates these processes by regulating the association between the RING finger proteins HEI10 and RNF212 and components of the CO machinery.
Introduction
A small subset of the 200–300 double-strand breaks (DSBs) formed during early prophase of meiosis I in mouse spermatocytes is used to generate a highly regulated number of meiotic crossovers (COs; 20–30), with the excess DSBs being repaired as non-COs. The progressive differentiation process during prophase I that leads to CO formation can be observed cytologically by immunolocalization of conserved CO-promoting factors (Baker et al., 1996; Kneitz et al., 2000; Kolas et al., 2005; Holloway et al., 2008; Cole et al., 2012). During zygonema, localization of the meiosis-specific MutSγ heterodimer (MSH4/MSH5) to a subset of the initial DSBs reduces the pool of potential CO intermediates by half (Kneitz et al., 2000). MutSγ focus numbers subsequently decline as spermatocytes progress through pachynema, during which time MutLγ heterodimer (MLH1/MLH3) is recruited to a subset of these sites at a frequency and distribution corresponding to that of the final CO sites (Santucci-Darmanin et al., 2000). Another conserved pro-CO factor, RING finger protein RNF212 (orthologue of Caenorhabditis elegans ZHP-3), has been implicated in determining which MutSγ sites will mature into COs, likely by selective stabilization of pre-CO intermediates at sites where MutSγ and RNF212 colocalize (Reynolds et al., 2013). However, the initial number of MutSγ/RNF212 colocalization sites in early pachynema still significantly exceeds the final CO tally, implying that this proposed RNF212-driven mechanism is insufficient to account for the final number of COs. Thus, an additional level of regulation is required to eliminate the excess MutSγ/RNF212-marked sites not designated for a final CO fate. More recently, the putative ubiquitin E3 ligase, HEI10 (human enhancer of invasion-10; also known as CCNBP1 [cyclin B1–interacting protein 1]) has been demonstrated to play a significant role in this process in plants and mice, with loss of Hei10 resulting in persistent accumulation of MutSγ foci, and a failure to recruit MutLγ, leading to a failure to establish COs (Singh et al., 2007; Ward et al., 2007; Chelysheva et al., 2012; Wang et al., 2012; Qiao et al., 2014).
To investigate how a limited and tightly regulated number of COs are reliably generated from a substantial excess of initial recombination intermediates, we examined the role of CNTD1 (cyclin N-terminal domain–containing-1) during mouse meiosis. CNTD1 is the mammalian orthologue of C. elegans COSA-1 (CO site–associated 1), a cyclin-related protein that was recently shown to function in conjunction with MSH-4/MSH-5 and ZHP-3 in promoting meiotic COs (Yokoo et al., 2012). COSA-1 colocalizes with MSH-5 and ZHP-3 at presumptive CO sites in C. elegans and is proposed to function in a self-reinforcing mechanism to sequester CO-promoting factors at designated CO sites. Cntd1 transcripts are highly enriched in mouse and human testis (Skinner et al., 2008; Yokoo et al., 2012), and we show here that mouse CNTD1 is a critical regulator of this CO maturation and stabilization from meiotic CO precursors to mature COs. Loss of CNTD1 in mice results in severe meiotic disruption in late prophase I spermatocytes, resulting in drastically reduced CO numbers and subsequent infertility. Importantly, MutSγ and RNF212 focus frequency remains elevated well into late pachynema, and MLH1/MLH3 fail to load at any of these sites, suggesting that CNTD1 is essential for the final selection of MutSγ sites and the subsequent loading of MutLγ, two processes that are inextricably linked via their CNTD1 codependence.
Results and discussion
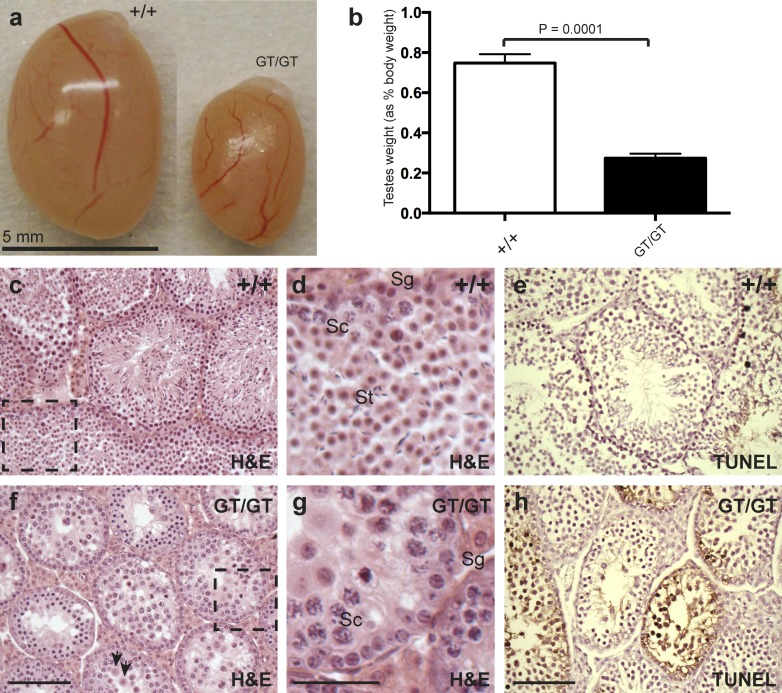
To examine the function of mouse Cntd1 in meiosis, we generated a mouse line with a modified Cntd1 gene trap allele that severely reduces or eliminates Cntd1 gene function (Cntd1GT; Fig. S1). Homozygous mutant mice (Cntd1GT/GT) are grossly similar to wild-type (WT) littermates, surviving into adulthood and exhibiting appropriate mating behavior. However, Cntd1GT/GT mutant males are sterile, showing significantly decreased testis size compared with WT and heterozygote males, and no epididymal spermatozoa (n = 5 WT, 7 heterozygote, and 18 mutant; Fig. 1, a and b; and not depicted), consistent with Cntd1 transcripts being highly enriched in mouse and human testis (Dezso et al., 2008; Thorrez et al., 2008). Cntd1GT/GT females are also sterile and exhibit meiotic phenotypes similar to those described herein (Fig. S2, n–s). Analysis of testis morphology revealed a loss of spermatozoa in the seminiferous tubules of Cntd1GT/GT males (Fig. 1, c, d, f, and g), whereas GCNA-1–associated spermatogonia and early spermatocytes were unaffected (not depicted). Accordingly, testes from Cntd1GT/GT males exhibit increased apoptosis of spermatocytes and no postmeiotic spermatids (TUNEL-positive cells in WT = mean of 4.14 per 20× view, n = 7; in mutant = mean of 48.83 per 20× view, n = 6; P = 0.0007; Fig. 1, e and h).
Figure 1.
Cntd1GT/GT males show a severe infertility phenotype. (a and b) Cntd1GT/GT testes are 64% smaller than testes of WT and heterozygote (not depicted) littermates (WT = 0.75% total body weight ± 0.04, n = 5; heterozygote = 0.74% ± 0.11, n = 7; mutant = 0.27% ± 0.02, n = 7; t test, P = 0.0001) and have no epididymal spermatozoa (not depicted). Error bars show SDs. (c–h) Cntd1GT/GT testes show abnormal morphology and increased cell death. Hematoxylin and eosin (H&E) staining of WT (c and d) and Cntd1GT/GT (f and g) testis sections reveals differences in testis morphology, with the Cntd1GT/GT males showing decreased cellularity in the lumen of the tubules, and the absence of later spermatogenic stages. Arrows indicate cells at metaphase undergoing apoptosis. Higher magnification images of insets in c and f are shown in d and g, respectively; Sg, spermatogonia; Sc, prophase I spermatocytes; St, postmeiotic spermatids. TUNEL labeling of testis sections from WT (e) and Cntd1GT/GT males (h) reveals an increase in cells undergoing apoptosis in the Cntd1GT/GT testis (44 TUNEL-positive cells per 20× view number). Bars: (c, e, f, and h) 100 µm; (d and g) 50 µm.
The presence of metaphase-stage spermatocytes (Fig. 1 f, arrows) in Cntd1GT/GT males distinguishes them from mutants lacking proteins critical for meiotic recombination initiation, synapsis, and/or early steps in DSB repair (e.g., Spo11−/−, Sycp3−/−, and Dmc1−/−), in which spermatocytes arrest before pachynema (Pittman et al., 1998; Yoshida et al., 1998; Baudat et al., 2000; Yuan et al., 2000; Kolas et al., 2004). Instead, the Cntd1GT/GT mutant phenotype is reminiscent of meiotic CO-defective mutants, such as Mlh1−/−, Mlh3−/−, Hei10mei4/mei4, and Rnf212−/−, whose spermatocytes display normal homologue pairing and initial DSB processing and are able to develop beyond pachynema but fail to form appropriate numbers of COs (Edelmann et al., 1996; Eaker et al., 2002; Lipkin et al., 2002; Ward et al., 2007; Strong and Schimenti, 2010; Reynolds et al., 2013).
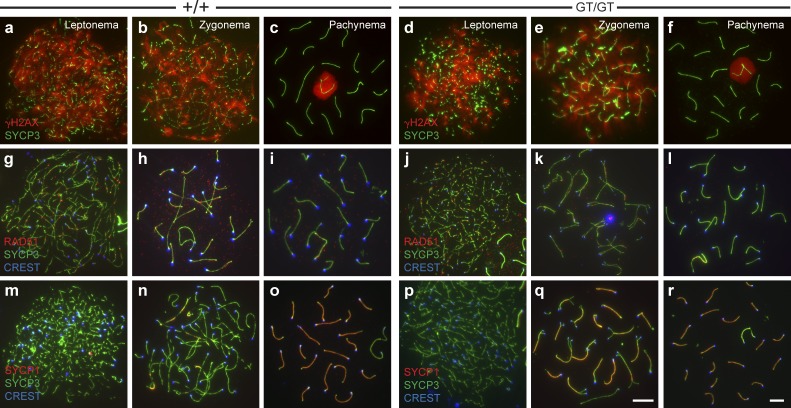
To assess prophase I progression in Cntd1GT/GT spermatocytes, chromosome spreads were stained with various antibodies to visualize the substages of prophase I. DSB induction and repair were assessed by staining for phosphorylated histone H2AX (γ-H2AX) and strand exchange protein RAD51, and these did not differ between WT and Cntd1GT/GT spermatocytes (Fig. 2, a–l; Moens et al., 1997; Hunter and Kleckner, 2001; Hunter et al., 2001; Mahadevaiah et al., 2001). Furthermore, no obvious differences were observed in the percentages of meiocytes from different substages, as assessed by the status of the synaptonemal complex components SYCP3 and SYCP1 (n = 3 mice from each genotype; Fig. 2, m–r; Schmekel et al., 1996; Schalk et al., 1998). Analysis of prophase I stages observed under the microscope revealed no difference in the proportion of cells at each substage between Cntd1+/+ and Cntd1GT/GT males (P = 0.94, χ2 analysis; n = 204 and 202 cells, respectively). Collectively, these data indicate that early recombination and synapsis events are normal in Cntd1GT/GT males.
Figure 2.
Early synapsis and DSB repair markers are appropriately localized in Cntd1GT/GT spermatocytes. (a–r) Meiotic chromosome spreads from WT (a–c, g–i, and m–o) and Cntd1GT/GT males (d–f, j–l, and p–r) were stained with antibodies to SYCP3, γ-H2AX (a–f), RAD51 (g–l), SYCP1 (m–r), and centromere marker CREST. γ-H2AX localizes to DSBs in early prophase I stage leptonema and zygonema (a, b, d, and e) before becoming sequestered to the sex body at pachynema (c and f). RAD51 accumulates in high numbers on leptotene and zygotene chromosomes (g, h, j, and k) before being removed at most sites by pachynema (i and l). SYCP1 accumulates along synapsed regions of the chromosomes during zygonema (n and q, yellow regions) and, by pachynema, has localized along the entire length of the fully synapsed autosomes (o and r). No differences in localization of these markers was observed between WT and Cntd1GT/GT males, as demonstrated by quantitation of RAD51 focus frequency at zygonema (83 ± 24.6 and 78 ± 23.0, respectively; mean ± SD) and pachynema (17 ± 6.8 and 19 ± 5.8, respectively; mean ± SD). The RAD51 focus counts at each stage were not statistically significant between genotypes (P = 0.80 and P = 0.69 for zygonema and pachynema, respectively). Bars, 10 µm.
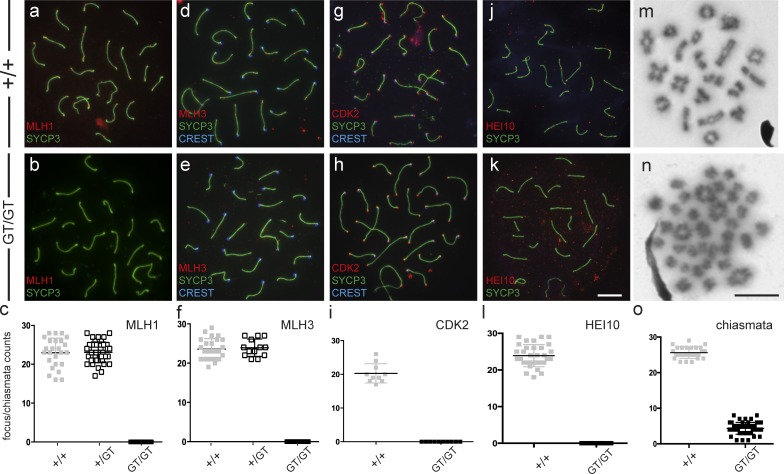
Despite the success of synapsis and formation of early recombination intermediates, Cntd1GT/GT mutants are severely defective in meiotic CO formation. During late pachynema in WT spermatocytes, maturing COs are visualized as sites of accumulation of MLH1 and MLH3, together comprising the MutLγ heterodimer, and CDK2 (which also localizes at telomeres; Fig. 3, a–i; Ashley et al., 2001; Marcon and Moens, 2003; Kolas et al., 2005; Cohen et al., 2006). In Cntd1GT/GT males, no MLH1 or MLH3 foci were observed on pachytene chromosomes compared with the mean of 23.0 ± 3.8 (n = 25 WT, 41 heterozygote, and 25 mutant, from two individuals of each genotype) MLH1 foci and 23.6 ± 2.7 (n = 25 WT, 13 heterozygote, and 25 mutant, from two individuals of each genotype) MLH3 foci found in WT spermatocyte spreads (Fig. 3, a–f). CO-associated CDK2 foci were also absent in the Cntd1GT/GT mutant, but CDK2 localization at telomeres persisted (n = 10, from two individuals of each genotype; Fig. 3, g–i). The failure to load MLH1, MLH3, and CDK2 indicates that crossing over through the canonical meiotic CO pathway is severely disrupted in Cntd1GT/GT males. Accordingly, the number of chiasmata resulting from COs was also substantially reduced in diakinesis-stage spermatocytes in Cntd1GT/GT males (Fig. 3, m–o), to only 17% of the WT chiasmata count (n = 26 WT and 52 mutant, from two individuals of each genotype). The presence of residual chiasmata suggests that some COs may be produced in the Cntd1GT/GT mutant even when MutLγ does not accumulate at repair sites, consistent with previous observations of residual MutLγ-independent COs and chiasmata in Mlh3−/− males (Kolas et al., 2005; Svetlanov et al., 2008).
Figure 3.
Absence of late prophase markers of meiotic CO sites and severe reduction in chiasmata in Cntd1GT/GT spermatocytes. (a–l) Late pachytene spermatocyte spreads from WT (a, d, g, and j) and Cntd1GT/GT males (b, e, h, and k) stained with antibodies against SYCP3, MLH1 (a and b), MLH3 (d and e), CDK2 (g and h), HEI10 (j and k), and centromere marker CREST (d, e, g, and h). c, f, i, and l show quantitation of MLH1, MLH3, CDK2, and HEI10 focus frequencies in individual nuclei from WT (filled gray boxes), Cntd1GT/+ heterozygous (open boxes), and Cntd1GT/GT mutant (filled black boxes) spermatocytes; only nontelomere-associated CDK2 foci were included. For quantitation of HEI10 foci, only foci associated with SYCP3-labeled chromosome cores were scored. Counts were as follows: MLH1 foci in WT, Cntd1GT/+, and Cntd1GT/GT = 23.0 ± 3.8, 23.1 ± 2.7, and 0, n = 25, 41, and 25, respectively; MLH3 foci in WT, Cntd1GT/+, and Cntd1GT/GT = 23.6 ± 2.7, 23.9 ± 2.2, and 0, n = 25, 13, and 25, respectively; CDK2 foci in WT and Cntd1GT/GT = 20.3 ± 2.8 and 0, n = 10 and 10, respectively; HEI10-foci in WT and Cntd1GT/GT = 23.9 ± 3.0 and 0, n = 32 and 10, respectively. No significant differences between WT and Cntd1GT/+ spermatocytes were observed for either MLH1 or MLH3 foci (P = 0.9 and P = 0.7, respectively); thus, in contrast to the Hei10 and Rnf212 loci (Reynolds et al., 2013; Qiao et al., 2014), the Cntd1 locus does not appear to be haploinsufficient. (m and n) Chromosome spreads from WT (m) and Cntd1GT/GT (n) diakinesis-stage spermatocytes. (o) Quantitation showed an 83% drop in chiasmata in the Cntd1GT/GT males compared with WT (WT = 25.62 ± 0.32, n = 26; mutant = 4.35 ± 0.23, n = 52). Data are means ± SD. Bars, 10 µm.
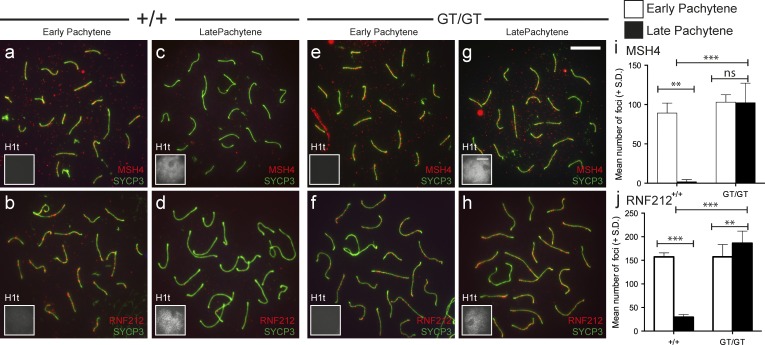
Given the absence of late CO markers in Cntd1GT/GT spermatocytes, we assessed the status of intermediate steps in the progressive differentiation of meiotic recombination sites. Specifically, we examined localization of MSH4 (a component of the pre-CO complex MutSγ; Kneitz et al., 2000) and the predicted small ubiquitin-like modifier E3 ligase RNF212 (Reynolds et al., 2013) on chromosome spreads from early and late pachytene spermatocytes. In WT controls, MSH4 foci were abundant at early pachynema (89.2 ± 5.9 foci per nucleus, n = 5) and had declined precipitously by late pachynema (2 ± 0.9, n = 9; Fig. 4 i). Similarly, RNF212 foci in WT controls were abundant during early pachynema (157.3 ± 3.5, n = 6) and had declined substantially by late pachynema (29.7 ± 1.9, n = 9; Fig. 4 j). Similar dynamics of MSH4 and RNF212 localization were observed in Mlh3−/− males, indicating that progressive reduction in numbers of foci harboring these components does not require loading of the MutLγ complex (Fig. S2, a–d). In contrast, Cntd1GT/GT spermatocytes had abundant MSH4 and RNF212 foci during early pachynema, but the numbers of MSH4 and RNF212 foci did not decline in late pachynema, remaining elevated above 100 foci per cell as seen in early pachynema (MSH4 early pachynema = 103.1 ± 3.3, n = 8; late pachynema = 102.3 ± 7.9, n = 10; RNF212 early pachynema = 157.5 ± 10.6, n = 6; and late pachynema = 186.5 ± 6.8, n = 14; Fig. 4, e–j). The frequencies of MSH4 and RNF212 foci in late pachytene spermatocytes from Cntd1GT/GT mutants were therefore significantly higher than in WT late pachytene spermatocytes (both P < 0.0001). Intriguingly, the numbers of late pachytene RNF212 foci in the Cntd1GT/GT mutant were also significantly higher than the numbers of early pachytene foci in either the mutant or the WT (P = 0.04 and P = 0.0013, respectively), indicating that RNF212 focus numbers continue to increase during pachytene progression in the absence of CNTD1.
Figure 4.
CO-promoting proteins MSH4 and RNF212 fail to be removed from chromosome cores in Cntd1GT/GT spermatocytes. (a–h) WT (a–d) and Cntd1GT/GT mutant (e–h) spermatocytes at both early (a, b, e, and f) and late (c, d, g, and h) pachynema, stained with antibodies against SYCP3 and MSH4 (a, c, e, and g) or RNF212 (b, d, f, and h). (i and J) MSH4 (i) and RNF212 (j) foci were quantified at each stage. Insets show H1t staining as a staging indicator (H1t is only evident in mid- to late pachynema). Counts are as follows: MSH4 early pachynema in WT and Cntd1GT/GT mutants, 89.20 ± 5.69 and 103.1 ± 3.35, respectively. MSH4 late pachynema in WT and Cntd1GT/GT mutants, 1.89 ± 0.93 and 102.3 ± 7.87, respectively. RNF212 in early pachynema in WT and Cntd1GT/GT mutants, 157.3 ± 3.5 and 157.5 ± 10.6, respectively. RNF212 in late pachynema in WT and Cntd1GT/GT mutants, 29.7 ± 1.9 and 186.5 ± 6.8, respectively. **, P < 0.05; ***, P < 0.0001; Mann–Whitney U test). Bars: (g applies to a–h) 10 µm; (H1T insets) 20 µm.
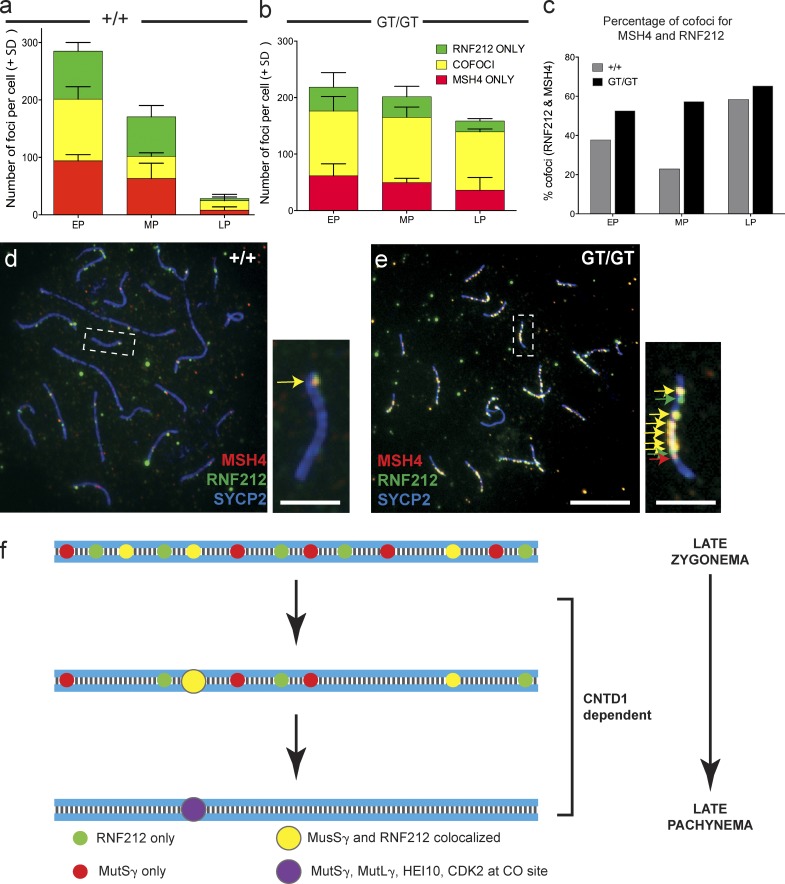
In addition to quantitating MSH4 and RNF212 foci individually in spermatocytes spreads from adult testes (Fig. 4, i and j), we also quantitated colocalization of MSH4 and RNF212 on meiotic chromosome cores in early, mid-, and late pachytene spermatocytes (n = 7 for each substage; Fig. 5). As previously reported, only a subset of RNF212 foci colocalized with MSH4 foci in WT spermatocytes at early pachynema, and both total foci and MSH4/RNF212 cofoci declined in number during pachytene progression, albeit the proportion of cofoci increased between early and late pachynema (cofoci = 38% of total in early pachynema and 58% in late pachynema, n = 16; Fig. 5, a, c, and d; Reynolds et al., 2013). In contrast, although the proportion of MSH4/RNF212 cofoci during early pachynema was similar between Cntd1GT/GT spermatocytes and WT spermatocytes, both the numbers of foci and the proportion of cofoci remained high throughout pachynema in the Cntd1GT/GT mutants (total foci in WT, 284 in early pachynema vs. 28 in late pachynema; total foci in Cntd1GT/GT mutants, 218 in early pachynema vs.158 in late pachynema; cofoci in WT, 107 in early pachynema vs. 17 in late pachynema; and cofoci in Cntd1GT/GT mutants, 114 in early pachynema vs. 103 in late pachynema; n = 10; Fig. 4, b and d; and Fig. 5, a–e), and the numbers of cofoci were significantly higher at midpachynema (P < 0.01) and late pachynema (P < 0.001) in Cntd1GT/GT mutants compared with WT (Fig. 5 c). These data indicate that CNTD1 is not required either for loading of RNF212 or MutSγ or for the association of RNF212 with MutSγ-specified recombination intermediates but is required for progressive removal of these proteins from excess recombination sites through pachynema.
Figure 5.
Persistent colocalization of MSH4 and RNF212 in late pachynema in Cntd1GT/GT spermatocytes. (a–e) Quantitation of foci containing only MSH4, only RNF212, or both MSH4 and RNF212 (a–c) associated with chromosome cores in spermatocytes from Cntd1+/+ (a and d) and Cntd1GT/GT (b and e) males. Graph a shows progressive diminution of overall focus numbers in WT spermatocytes, accompanied by an increased proportion of cofoci in late pachynema (LP) relative to early pachynema (EP). MP, midpachynema. Graph b shows that both focus numbers and the proportion of cofoci are maintained at high levels throughout pachynema in Cntd1GT/GT spermatocytes (a and b, n = 7 for each substage). The percentages of only cofoci are provided in graph c for both genotypes. The frequency of cofoci was significantly higher in Cntd1GT/GT spermatocytes compared with WT at mid- and late pachynema (Mann–Whitney U test, P < 0.01 and P < 0.001, respectively). For this quantitation, Cntd1+/+ and Cntd1GT/GT spermatocyte spreads were stained with antibodies against SYCP2, RNF212, and MSH4 to quantitate the frequency of RNF212/MSH4 colocalization (yellow) specifically associated with the meiotic chromosome cores. The quantitation of foci in graphs a and b is provided for each individual protein (in their respective fluorochrome colors), along with the cofocus counts (in yellow to reflect the merge of the red and green cofocus counts). d and e show example images of late pachytene spreads from Cntd1+/+ and Cntd1GT/GT testes, respectively, with dashed boxes indicating the specific chromosomes shown in the associated magnifications. Bars: (d and e) 10 µm; (insets) 5 µm. (f) Model for designation of CO sites through pachynema of prophase I, through selection by MutSγ and RNF212, and ultimate CO promotion by MutLγ.
Collectively, our data indicate that mouse CNTD1, like its C. elegans orthologue COSA-1 (Yokoo et al., 2012), is an important factor for maturation of meiotic COs. Moreover, our data provide new insight regarding how CO maturation is ultimately restricted to a small subset of potential sites during mammalian meiosis. Persistence of high levels of RNF212/MutSγ cofoci in Cntd1GT/GT spermatocytes indicates that colocalization of RNF212 and MutSγ, although undoubtedly required for CO maturation, is not sufficient to recruit CDK2 or MutLγ to prospective CO sites. Thus, these data demonstrate that selective stabilization of MutSγ by RNF212 cannot alone explain CO site selection. Instead, our data indicate requirements for both (a) an additional CO differentiation step that subsequently enables loading of MutLγ and (b) an active deselection process that destabilizes or removes excess pre-CO intermediates to achieve the final outcome. Moreover, our work supports a model in which recruitment of CDK2 and MutLγ and deselection of excess RNF212/MutSγ-marked sites are intimately coupled events, with CNTD1 playing a key role in integrating these processes. The data presented are consistent with CNTD1 functioning predominantly either to promote installation of MutLγ, to induce the removal of MutSγ/RNF212, or both. Interestingly, the 17% incidence of residual chiasmata observed in Cntd1GT/GT mice is higher than the 3% residual chiasmata seen in Rnf212−/− mice (Reynolds et al., 2013) or the 10% residual chiasmata observed in Mlh3−/− mice (Kolas et al., 2005; Svetlanov et al., 2008). Furthermore, residual chiasma levels correlate with the numbers of MutSγ foci observed at mid-pachynema in these meiotic mutants (Rnf212−/−, reduced; Mlh3−/−, normal; Cntd1GT/GT, elevated). This correlation raises the possibility that MutSγ may be responsible for promoting all meiotic COs, including those derived from recombination intermediates processed in the absence of MutLγ.
The identity of CNTD1 as a member of the cyclin superfamily suggests that it may accomplish these tasks by partnering with a CDK subunit to form a CNTD1–CDK protein kinase complex. Unfortunately, rigorous testing of two custom antibodies and seven commercially available antibodies raised against CNTD1 epitopes has failed to identify any reliable reagents for visualizing the CNTD1 protein, precluding any confident localization or colocalization of CNTD1 with key interactors, such as putative CDK partners. However, several additional lines of evidence implicate HEI10, a RING finger protein that functions as an E3 ubiquitin ligase in vitro (Toby et al., 2003), as a likely functional partner and candidate substrate for CNTD1 in regulating CO site selection and CO maturation. First, HEI10 has both a predicted cyclin-interacting motif (RXL) and multiple consensus CDK phosphorylation motifs ([S/T]P; Fig. S3) and can be phosphorylated in vitro by purified cyclin B/CDC2 (Toby et al., 2003). Moreover, the meiotic defects observed in Hei10mei4/mei4 mutant mice, which contain an in-frame deletion that eliminates the RXL motif (Ward et al., 2007), are distinct from other known meiotic mutants but closely parallel the meiotic defects observed in the Cntd1GT/GT mutant (Ward et al., 2007; Qiao et al., 2014). Finally, the HEI10 protein localizes at designated CO sites in late pachytene spermatocytes (Qiao et al., 2014), and this localization is lost in the Cntd1GT/GT mutant (Fig. 3, j–l). Whereas we detected 23.9 ± 0.5 synaptonemal complex–associated HEI10 foci per nucleus in WT late pachytene spermatocytes, synaptonemal complex–associated HEI10 foci were absent in Cntd1GT/GT spermatocytes (n = 32 WT and 10 mutant). Together, these data indicate that CNTD1 and HEI10 collaborate to promote CO maturation and deselection of excess pre-CO sites and suggest that association with and/or phosphorylation by a putative CNTD1-dependent CDK complex may be required for HEI10 function.
A previous study has identified two potential CDK subunits that might partner with CNTD1 to comprise a dedicated meiotic CNTD1–CDK complex that promotes CO progression: CDK4, which is abundant on chromosomes early in pachynema (>150 foci) and then declines steadily during pachytene progression in spermatocytes from WT mice (Fig. S2, e–h and m), and CDK2, which localizes specifically at CO-designated sites during mid- to late pachynema (Fig. 3, g–i; Ashley et al., 2001). We found that CDK4 foci are not only present but persist at high levels throughout pachynema in the Cntd1GT/GT mutant, similar to the persistent localization seen for RNF212 and MutSγ (n = 37 WT and 47 mutant; Fig. S2, i–m). This contrasts with the absence of CO-associated CDK2 foci in mutant spermatocytes, collectively making CDK2 a more likely candidate CNTD1 partner. Together with our evidence that CNTD1 acts in conjunction with HEI10, these data support the conclusion that CNTD1 likely functions at CO-designated sites.
In principle, the coordinate regulation of CO maturation and deselection of excess CO-eligible sites could simply reflect dependence of these two processes on CNTD1 as a common regulator (Fig. 5 f). Consistent with the possibility that CNTD1 might function directly in the deselection process, both RNF212 and the MutSγ heterodimer are plausible candidate CDK substrates (Fig. S3). Alternatively, CNTD1 may function specifically to promote designation/maturation of selected CO intermediates, with removal of RNF212 and MutSγ from other sites occurring as a secondary downstream consequence. This type of functional coupling could reflect a feedback network in which potential intermediates are retained until the cell senses that one event per chromosome pair has been successfully designated for CO maturation, which in turn triggers a change in state that leads to removal of the excess intermediates. This is an attractive scenario, as it provides a means to constrain CO number while at the same time guaranteeing formation of the obligatory CO needed to ensure successful chromosome segregation.
Despite demonstrating a conserved role for CNTD1/COSA-1 orthologues in promoting CO maturation, our analysis of Cntd1GT/GT mice has also revealed substantial plasticity in the regulatory circuits governing meiotic CO progression across species. Whereas MutSγ-marked pre-CO intermediates persist in Cntd1 mutant mice, implying that CNTD1 is required for their removal, MutSγ foci are lost in C. elegans cosa-1 mutants (Yokoo et al., 2012), indicating an apparently opposite role for COSA-1 in promoting formation or stabilization of early pre-CO intermediates. Furthermore, whereas CNTD1 collaborates with HEI10 in the mouse and is required for installation of both HEI10 and MutLγ at CO sites, the nematode lacks both HEI10 and MutLγ (Chelysheva et al., 2012) and instead retains MutSγ at CO sites during late pachynema and diplonema (Yokoo et al., 2012). We speculate that the presence of CNTD1/COSA-1 in the ancestral metazoan lineage enabled evolution of two coordinated regulatory modules involving distinct RING finger proteins: an RNF212/ZHP-3–dependent module governing MutSγ stability at potential CO sites and an HEI10-dependent module promoting installation of MutLγ at designated CO sites. We hypothesize that reconfiguration of the first module during nematode evolution rendered MutLγ expendable in worms, resulting in coordinate loss of the second module. Conversely, retention and coupling of the two modules (RNF212 and HEI10; Fig. 5 f) in the mouse may provide a means for “safe transfer” of CO intermediates from a protected, MutSγ-bound state to a MutLγ-bound state that promotes their resolution.
Materials and methods
Animals
Cntd1 transgenic mice were generated from the embryonic stem cell line EPD0190_3_E03: Cntd1tm1e(KOMP)Wtsi (obtained from the University of California, Davis Knockout Mouse Repository), which contains a gene trap cassette (FRT-lacZ-loxP-neo-FRT-loxP) in the first intron of mouse Cntd1 gene. A Spo11-Cre mouse line (Lyndaker et al., 2013) was crossed with these Cntd1 transgenic mice to remove the neo cassette. Genotyping of Cntd1 animals was performed using the following PCR primer pairs: CNTD1lacZ_forward (5′-CGACTCCTGGAGCCCGTCAG-3′) and CNTD1loxP_1 reverse (5′-GCGCGCCGTTTAAACATAACT-3′), which detect a 420-bp fragment from the mutant allele; CNTD1WT forward (5′-CTGACATTCGCTCTCGTTTCC-3′) and CNTD1WT reverse (5′-CGGCTGACAAAAGGTTTGG-3′), which produce a 520-bp band in the WT allele; and CNTD1neo_forward (5′-TTCTTCTGAGCGGGACTCTG-3′). Fertility tests were performed by mating Cntd1 males aged between 8 and 10 wk with WT adult females. Mating was determined by the presence of a copulation plug the next morning. Pregnancy was confirmed either by gentle abdominal palpation after gestation day 11 or the delivery of litters. Experimental animals were used under the strict guidance and approval of the Cornell University Institutional Animal Care and Use Committee.
Histology and immunohistochemistry
Testes from 4- or 8-wk-old mice were fixed in Bouin’s fixative for 6 h at room temperature or 10% formalin overnight at 4°C and then washed in 70% ethanol. Fixed and paraffin-embedded tissues were sectioned at 4 µm. Hematoxylin and eosin staining, TUNEL staining, and GCNA-1 staining were performed as described previously (Holloway et al., 2011), the former using ApopTag peroxidase kit (EMD Millipore) and the latter using Vectastain reagents (Vector Laboratories).
Sperm counts
The cauda epididymides were removed from adult mice and placed in prewarmed DMEM containing 4% bovine serum albumin. Each epididymis was squeezed with tweezers to extrude the sperm and then incubated at 32°C/5% CO2 for 20 min. A 20-µl aliquot of the sperm suspension was resuspended in 480 µl of 10% formalin, and the sperm cells were counted.
Chromosome analysis and immunofluorescence
Prophase I chromosome preparations and immunofluorescence were performed using previously described techniques (Peters et al., 1997; Kolas et al., 2005). In brief, testes were removed and decapsulated into hypotonic sucrose extraction buffer (containing 1.7% sucrose) and left on ice for 0.5–1 h. Tubules were chopped on glass depression slides in a bubble of 0.03% sucrose and added to slides coated in 1% paraformaldehyde. For analysis of female chromosome spreads, ovaries were removed from embryonic day 18.5 to day 0.5 postpartum females, briefly soaked in HEB, minced in 0.03% sucrose, and added to a bubble of paraformaldehyde on a well slide. Slides were slow dried and subjected to immunofluorescence analyses. The primary antibodies used were either generated in our laboratory, commercially available, or gifts from other laboratories (Kolas et al., 2005). They were mouse SYCP1 (1:1,000; against full-length protein), rabbit SYCP3 (1:5,000; against full-length protein), mouse SYCP3 (1:5,000; against full-length protein), guinea pig SYCP2 (1:5,000; a gift from J. Wang, University of Pennsylvania, Philadelphia, PA), CREST human autoimmune serum (1:30,000), mouse γ-H2AX (1:5,000; EMD Millipore), rabbit RAD51 (1:300; EMD Millipore), rabbit MSH4 or MSH5 (1:100; both obtained from Abcam), monoclonal human MLH1 (1:100; BD), rabbit CDK2 and CDK4 (both used at 1:100; Santa Cruz Biotechnology, Inc.), polyclonal rabbit MLH3 (1:300; Lipkin et al., 2002), mouse monoclonal CCNB1IP1/HEI10 (1:100; Abcam), and rabbit and guinea pig RNF212 (1:200 and 1:20, respectively; Reynolds et al., 2013). Secondary antibodies used were goat anti–mouse Alexa Fluor 488, goat anti–rabbit Alexa Fluor 555, goat anti–human Alexa Fluor 647, and goat anti–guinea pig Alexa Fluor 647 (all 1:1,000; Invitrogen).
Spermatocyte diakinesis spread preparations
Diakinesis chromosome preparations were prepared as described previously (Evans et al., 1964; Uroz et al., 2008), with a slight modification (Holloway et al., 2010). Slides were stained in Giemsa solution for 3 min, washed, and air dried.
Image acquisition
All slides were visualized using a microscope (Axio Imager.Z1; Carl Zeiss) under a 20×, 0.5 NA EC Plan Neofluar air immersion (Carl Zeiss) or 63×, 1.4 NA Plan Apochromat oil immersion differential interference contrast (Carl Zeiss) magnifying objective at room temperature. Images were captured on a charge-coupled device camera (AxioCam MRm; Carl Zeiss). The fluorochromes used were Alexa Fluor labeled with Cy3, Cy5, or FITC. Images were captured with a cooled charged-coupled device camera (AxioCam MRm) and processed using AxioVision software (version 4.7.2; Carl Zeiss).
Online supplemental material
Fig. S1 provides details regarding the genetrap (GT) allele for Cntd1 that was used in the current work. Fig. S2 shows chromosome spread images from WT, Cntd1GT/GT, and Mlh3−/− male and female mice stained with antibodies against various proteins involved in synapsis and recombination. Fig. S3 shows predicted cyclin-binding motifs and CDK phosphorylation sites in HEI10, RNF212, and MutSγ. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201401122/DC1.
Supplementary Material
Acknowledgments
We thank Peter Borst for his assistance with mouse handling and maintenance, Melissa Toledo for assistance with genotyping, Dr. Neil Hunter (University of California, Davis, Davis, CA) for providing anti-RNF212 antibodies, and Jeremy Wang (University of Pennsylvania) for providing anti-SYCP2 antibodies.
This work was supported by National Institutes of Health grants R01HD041012 to P.E. Cohen, R00HD065870 to J.K. Holloway, and R01GM67268 to A.M. Villeneuve.
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- CO
- crossover
- DSB
- double-strand break
- WT
- wild type
References
- Ashley T., Walpita D., de Rooij D.G. 2001. Localization of two mammalian cyclin dependent kinases during mammalian meiosis. J. Cell Sci. 114:685–693 [DOI] [PubMed] [Google Scholar]
- Baker S.M., Plug A.W., Prolla T.A., Bronner C.E., Harris A.C., Yao X., Christie D.M., Monell C., Arnheim N., Bradley A., et al. 1996. Involvement of mouse Mlh1 in DNA mismatch repair and meiotic crossing over. Nat. Genet. 13:336–342 10.1038/ng0796-336 [DOI] [PubMed] [Google Scholar]
- Baudat F., Manova K., Yuen J.P., Jasin M., Keeney S. 2000. Chromosome synapsis defects and sexually dimorphic meiotic progression in mice lacking Spo11. Mol. Cell. 6:989–998 10.1016/S1097-2765(00)00098-8 [DOI] [PubMed] [Google Scholar]
- Chelysheva L., Vezon D., Chambon A., Gendrot G., Pereira L., Lemhemdi A., Vrielynck N., Le Guin S., Novatchkova M., Grelon M. 2012. The Arabidopsis HEI10 is a new ZMM protein related to Zip3. PLoS Genet. 8:e1002799 10.1371/journal.pgen.1002799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen P.E., Pollack S.E., Pollard J.W. 2006. Genetic analysis of chromosome pairing, recombination, and cell cycle control during first meiotic prophase in mammals. Endocr. Rev. 27:398–426 10.1210/er.2005-0017 [DOI] [PubMed] [Google Scholar]
- Cole F., Kauppi L., Lange J., Roig I., Wang R., Keeney S., Jasin M. 2012. Homeostatic control of recombination is implemented progressively in mouse meiosis. Nat. Cell Biol. 14:424–430 10.1038/ncb2451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dezso Z., Nikolsky Y., Sviridov E., Shi W., Serebriyskaya T., Dosymbekov D., Bugrim A., Rakhmatulin E., Brennan R.J., Guryanov A., et al. 2008. A comprehensive functional analysis of tissue specificity of human gene expression. BMC Biol. 6:49 10.1186/1741-7007-6-49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaker S., Cobb J., Pyle A., Handel M.A. 2002. Meiotic prophase abnormalities and metaphase cell death in MLH1-deficient mouse spermatocytes: insights into regulation of spermatogenic progress. Dev. Biol. 249:85–95 10.1006/dbio.2002.0708 [DOI] [PubMed] [Google Scholar]
- Edelmann W., Cohen P.E., Kane M., Lau K., Morrow B., Bennett S., Umar A., Kunkel T., Cattoretti G., Chaganti R., et al. 1996. Meiotic pachytene arrest in MLH1-deficient mice. Cell. 85:1125–1134 10.1016/S0092-8674(00)81312-4 [DOI] [PubMed] [Google Scholar]
- Evans E.P., Breckon G., Ford C.E. 1964. An air-drying method for meiotic preparations from mammalian testes. Cytogenetics. 3:289–294 10.1159/000129818 [DOI] [PubMed] [Google Scholar]
- Holloway J.K., Booth J., Edelmann W., McGowan C.H., Cohen P.E. 2008. MUS81 generates a subset of MLH1-MLH3-independent crossovers in mammalian meiosis. PLoS Genet. 4:e1000186 10.1371/journal.pgen.1000186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holloway J.K., Morelli M.A., Borst P.L., Cohen P.E. 2010. Mammalian BLM helicase is critical for integrating multiple pathways of meiotic recombination. J. Cell Biol. 188:779–789 10.1083/jcb.200909048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holloway J.K., Mohan S., Balmus G., Sun X., Modzelewski A., Borst P.L., Freire R., Weiss R.S., Cohen P.E. 2011. Mammalian BTBD12 (SLX4) protects against genomic instability during mammalian spermatogenesis. PLoS Genet. 7:e1002094 10.1371/journal.pgen.1002094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter N., Kleckner N. 2001. The single-end invasion: an asymmetric intermediate at the double-strand break to double-holliday junction transition of meiotic recombination. Cell. 106:59–70 10.1016/S0092-8674(01)00430-5 [DOI] [PubMed] [Google Scholar]
- Hunter N., Börner G.V., Lichten M., Kleckner N. 2001. Gamma-H2AX illuminates meiosis. Nat. Genet. 27:236–238 10.1038/85781 [DOI] [PubMed] [Google Scholar]
- Kneitz B., Cohen P.E., Avdievich E., Zhu L., Kane M.F., Hou H., Jr, Kolodner R.D., Kucherlapati R., Pollard J.W., Edelmann W. 2000. MutS homolog 4 localization to meiotic chromosomes is required for chromosome pairing during meiosis in male and female mice. Genes Dev. 14:1085–1097 [PMC free article] [PubMed] [Google Scholar]
- Kolas N.K., Yuan L., Hoog C., Heng H.H., Marcon E., Moens P.B. 2004. Male mouse meiotic chromosome cores deficient in structural proteins SYCP3 and SYCP2 align by homology but fail to synapse and have possible impaired specificity of chromatin loop attachment. Cytogenet. Genome Res. 105:182–188 10.1159/000078188 [DOI] [PubMed] [Google Scholar]
- Kolas N.K., Svetlanov A., Lenzi M.L., Macaluso F.P., Lipkin S.M., Liskay R.M., Greally J., Edelmann W., Cohen P.E. 2005. Localization of MMR proteins on meiotic chromosomes in mice indicates distinct functions during prophase I. J. Cell Biol. 171:447–458 10.1083/jcb.200506170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipkin S.M., Moens P.B., Wang V., Lenzi M., Shanmugarajah D., Gilgeous A., Thomas J., Cheng J., Touchman J.W., Green E.D., et al. 2002. Meiotic arrest and aneuploidy in MLH3-deficient mice. Nat. Genet. 31:385–390 [DOI] [PubMed] [Google Scholar]
- Lyndaker A.M., Lim P.X., Mleczko J.M., Diggins C.E., Holloway J.K., Holmes R.J., Kan R., Schlafer D.H., Freire R., Cohen P.E., Weiss R.S. 2013. Conditional inactivation of the DNA damage response gene Hus1 in mouse testis reveals separable roles for components of the RAD9-RAD1-HUS1 complex in meiotic chromosome maintenance. PLoS Genet. 9:e1003320 10.1371/journal.pgen.1003320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahadevaiah S.K., Turner J.M., Baudat F., Rogakou E.P., de Boer P., Blanco-Rodríguez J., Jasin M., Keeney S., Bonner W.M., Burgoyne P.S. 2001. Recombinational DNA double-strand breaks in mice precede synapsis. Nat. Genet. 27:271–276 10.1038/85830 [DOI] [PubMed] [Google Scholar]
- Marcon E., Moens P. 2003. MLH1p and MLH3p localize to precociously induced chiasmata of okadaic-acid-treated mouse spermatocytes. Genetics. 165:2283–2287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moens P.B., Chen D.J., Shen Z., Kolas N.K., Tarsounas M., Heng H.H., Spyropoulos B. 1997. Rad51 immunocytology in rat and mouse spermatocytes and oocytes. Chromosoma. 106:207–215 10.1007/s004120050241 [DOI] [PubMed] [Google Scholar]
- Peters A.H., Plug A.W., van Vugt M.J., de Boer P. 1997. A drying-down technique for the spreading of mammalian meiocytes from the male and female germline. Chromosome Res. 5:66–68 10.1023/A:1018445520117 [DOI] [PubMed] [Google Scholar]
- Pittman D.L., Cobb J., Schimenti K.J., Wilson L.A., Cooper D.M., Brignull E., Handel M.A., Schimenti J.C. 1998. Meiotic prophase arrest with failure of chromosome synapsis in mice deficient for Dmc1, a germline-specific RecA homolog. Mol. Cell. 1:697–705 10.1016/S1097-2765(00)80069-6 [DOI] [PubMed] [Google Scholar]
- Qiao H., Prasada Rao H.B., Yang Y., Fong J.H., Cloutier J.M., Deacon D.C., Nagel K.E., Swartz R.K., Strong E., Holloway J.K., et al. 2014. Antagonistic roles of ubiquitin ligase HEI10 and SUMO ligase RNF212 regulate meiotic recombination. Nat. Genet. 46:194–199 10.1038/ng.2858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds A., Qiao H., Yang Y., Chen J.K., Jackson N., Biswas K., Holloway J.K., Baudat F., de Massy B., Wang J., et al. 2013. RNF212 is a dosage-sensitive regulator of crossing-over during mammalian meiosis. Nat. Genet. 45:269–278 10.1038/ng.2541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santucci-Darmanin S., Walpita D., Lespinasse F., Desnuelle C., Ashley T., Paquis-Flucklinger V. 2000. MSH4 acts in conjunction with MLH1 during mammalian meiosis. FASEB J. 14:1539–1547 10.1096/fj.14.11.1539 [DOI] [PubMed] [Google Scholar]
- Schalk J.A., Dietrich A.J., Vink A.C., Offenberg H.H., van Aalderen M., Heyting C. 1998. Localization of SCP2 and SCP3 protein molecules within synaptonemal complexes of the rat. Chromosoma. 107:540–548 10.1007/s004120050340 [DOI] [PubMed] [Google Scholar]
- Schmekel K., Meuwissen R.L., Dietrich A.J., Vink A.C., van Marle J., van Veen H., Heyting C. 1996. Organization of SCP1 protein molecules within synaptonemal complexes of the rat. Exp. Cell Res. 226:20–30 10.1006/excr.1996.0198 [DOI] [PubMed] [Google Scholar]
- Singh M.K., Nicolas E., Gherraby W., Dadke D., Lessin S., Golemis E.A. 2007. HEI10 negatively regulates cell invasion by inhibiting cyclin B/Cdk1 and other promotility proteins. Oncogene. 26:4825–4832 10.1038/sj.onc.1210282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skinner A.M., O’Neill S.L., Grompe M., Kurre P. 2008. CXCR4 induction in hematopoietic progenitor cells from Fanca−/−, -c−/−, and -d2−/− mice. Exp. Hematol. 36:273–282 10.1016/j.exphem.2007.11.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strong E.R., Schimenti J.C. 2010. Evidence implicating CCNB1IP1, a RING domain-containing protein required for meiotic crossing over in mice, as an E3 SUMO ligase. Genes (Basel). 1:440–451 10.3390/genes1030440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svetlanov A., Baudat F., Cohen P.E., de Massy B. 2008. Distinct functions of MLH3 at recombination hot spots in the mouse. Genetics. 178:1937–1945 10.1534/genetics.107.084798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorrez L., Van Deun K., Tranchevent L.C., Van Lommel L., Engelen K., Marchal K., Moreau Y., Van Mechelen I., Schuit F. 2008. Using ribosomal protein genes as reference: a tale of caution. PLoS ONE. 3:e1854 10.1371/journal.pone.0001854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toby G.G., Gherraby W., Coleman T.R., Golemis E.A. 2003. A novel RING finger protein, human enhancer of invasion 10, alters mitotic progression through regulation of cyclin B levels. Mol. Cell. Biol. 23:2109–2122 10.1128/MCB.23.6.2109-2122.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uroz L., Rajmil O., Templado C. 2008. Premature separation of sister chromatids in human male meiosis. Hum. Reprod. 23:982–987 10.1093/humrep/dem427 [DOI] [PubMed] [Google Scholar]
- Wang K., Wang M., Tang D., Shen Y., Miao C., Hu Q., Lu T., Cheng Z. 2012. The role of rice HEI10 in the formation of meiotic crossovers. PLoS Genet. 8:e1002809 10.1371/journal.pgen.1002809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward J.O., Reinholdt L.G., Motley W.W., Niswander L.M., Deacon D.C., Griffin L.B., Langlais K.K., Backus V.L., Schimenti K.J., O’Brien M.J., et al. 2007. Mutation in mouse hei10, an e3 ubiquitin ligase, disrupts meiotic crossing over. PLoS Genet. 3:e139 10.1371/journal.pgen.0030139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoo R., Zawadzki K.A., Nabeshima K., Drake M., Arur S., Villeneuve A.M. 2012. COSA-1 reveals robust homeostasis and separable licensing and reinforcement steps governing meiotic crossovers. Cell. 149:75–87 10.1016/j.cell.2012.01.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida K., Kondoh G., Matsuda Y., Habu T., Nishimune Y., Morita T. 1998. The mouse RecA-like gene Dmc1 is required for homologous chromosome synapsis during meiosis. Mol. Cell. 1:707–718 10.1016/S1097-2765(00)80070-2 [DOI] [PubMed] [Google Scholar]
- Yuan L., Liu J.G., Zhao J., Brundell E., Daneholt B., Höög C. 2000. The murine SCP3 gene is required for synaptonemal complex assembly, chromosome synapsis, and male fertility. Mol. Cell. 5:73–83 10.1016/S1097-2765(00)80404-9 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.