Abstract
Distinct properties of poly(ADP-ribose)—including its structural diversity, nucleation potential, and low complexity, polyvalent, highly charged nature—could contribute to organizing cellular architectures. Emergent data indicate that poly(ADP-ribose) aids in the formation of nonmembranous structures, such as DNA repair foci, spindle poles, and RNA granules. Informatics analyses reported here show that RNA granule proteins enriched for low complexity regions, which aid self-assembly, are preferentially modified by poly(ADP-ribose), indicating how poly(ADP-ribose) could direct cellular organization.
What is poly(ADP-ribose)?
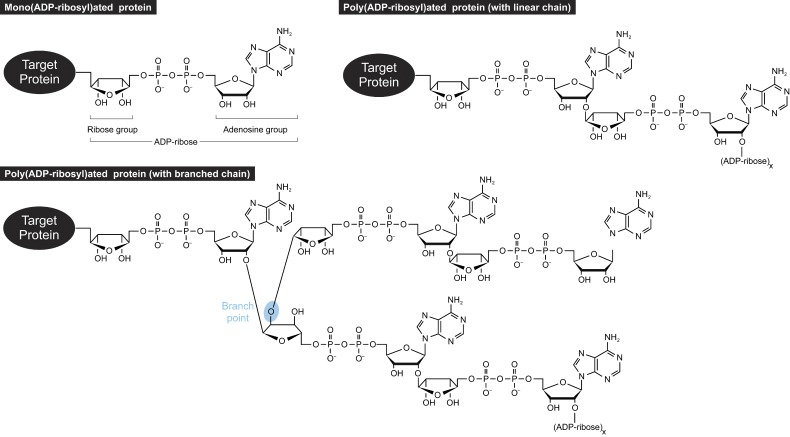
Poly(ADP-ribose), or simply PAR, is a polymer of two or more ADP-ribose units. This post-translational modification is added to proteins by ADP-ribosyltransferases, commonly known as poly(ADP-ribose) polymerases (PARPs; Hottiger et al., 2010; Gibson and Kraus, 2012). The covalent addition of multiple ADP-ribose units, or PARylation, can occur at different amino acids, including aspartate, glutamate, and lysine residues. To form the polymeric PAR, subunits are linked to one another via ribose–ribose bonds (Fig. 1; Miwa et al., 1979). Usually, the ribose group of one ADP-ribose unit is connected to the adenosine of the adjacent ADP-ribose unit. Occasionally, the nonadenosine ribose groups from neighboring ADP-ribose units can be linked, resulting in branching of the polymer (Miwa et al., 1981).
Figure 1.
Protein ADP-ribosylation. Shown here is mono(ADP-ribosyl)ation and poly(ADP-ribosyl)ation with linear or branched chains.
Regulation of poly(ADP-ribose) formation
In humans, there are 17 PARPs that share homologous domains, some with catalytic activities that transfer the ADP-ribose moiety from NAD+ to specific amino acid residues on substrate proteins, or to ADP-ribose itself. Six of them are predicted to add multiple ADP-ribose units whereas nine are predicted to only add single units (Table 1; Hottiger et al., 2010; Gibson and Kraus, 2012). Subtle changes within the catalytic domain determine not only the number of ADP-ribose subunits to be added, but also how these subunits are connected. Whereas PARP-1 is known to generate branching once every 20–50 ADP-ribose units (Alvarez-Gonzalez and Jacobson, 1987), PARP-5a has no such activities detectable (Rippmann et al., 2002). It is not currently known what controls the branching frequency or the length limit of PAR.
Table 1.
Activities and localization of PARPs
| Gene name | Alternative name | Activity | Localization |
| PARP-1 | ARTD1 | PARylation (branching) | Nucleus, nucleoli |
| PARP-2 | ARTD2 | PARylation | Cytoplasmic puncta, nuclear puncta, nucleoli |
| PARP-3 | ARTD3 | MARylation/PARylation? | Cytoplasmic puncta, nuclear puncta |
| PARP-4 | ARTD4 | PARylation | Cytoplasmic puncta and nucleus |
| PARP-5a | ARTD5, TNKS | PARylation | Cytoplasmic puncta, centrosome, stress granules, spindle pole |
| PARP-5b | ARTD6, TNKS2 | PARylation | Cytoplasmic puncta, spindle |
| PARP-6 | ARTD17 | MARylation? | Cytoplasmic puncta |
| PARP-7 | ARTD14, tiPARP | MARylation? | Cytoplasmic puncta, nuclear puncta |
| PARP-8 | ARTD16 | MARylation? | Cytoplasmic puncta, centrosome, nuclear envelope, spindle pole |
| PARP-9 | ARTD9, BAL1 | Inactive | Cytoplasm, plasma membrane, nucleus |
| PARP-10 | ARTD10 | MARylation | Cytoplasmic puncta |
| PARP-11 | ARTD11 | MARylation? | Cytoplasmic puncta, nuclear puncta, centriole |
| PARP-12 | ARTD12, ZC3HDC1 | MARylation | Cytoplasmic puncta, Golgi, stress granules |
| PARP-13 | ARTD13, ZC3HAV1, ZAP | Inactive | Cytoplasmic puncta, stress granules |
| PARP-14 | ARTD8, BAL2, CoaSt6 | MARylation | Cytoplasmic puncta, nuclear puncta, focal adhesion, stress granules |
| PARP-15 | ARTD7, BAL3 | MARylation? | Stress granules |
| PARP-16 | ARTD15 | MARylation | Cytoplasmic puncta, reticular |
PARylation, poly(ADP-ribosyl)ation; MARylation, mono(ADP-ribosyl)ation. Predicted activities are indicated with question marks. Localization data are based on Meder et al. (2005); Leung et al. (2011); and Vyas et al. (2013).
PARylation is reversible and two broad classes of enzymes are responsible for PAR degradation (Table 2)—the first cleaves the ribose–ribose bonds between ADP-ribose subunits and the second removes the terminal ADP-ribose groups from modified proteins. The primary enzyme that involves PAR degradation is known as poly(ADP-ribose) glycohydrolase (PARG; Hatakeyama et al., 1986). Though PARG can efficiently cleave the ribose–ribose bonds within PAR chain, it cannot remove the terminal ADP-ribose groups from PARylated substrates, thereby generating mono(ADP-ribosyl)ated proteins (Slade et al., 2011). Though less efficient than PARG, ADP-ribosylhydrolase 3 (ARH3) can also cleave the ribose–ribose bonds within PAR in vitro and in cells (Mueller-Dieckmann et al., 2006; Oka et al., 2006; Niere et al., 2012). Recently, three enzymes were identified in humans that break the covalent bonds between terminal ADP-ribose groups and acidic residues on modified proteins—MacroD1, MacroD2, and TARG1 (Jankevicius et al., 2013; Rosenthal et al., 2013; Sharifi et al., 2013). These three enzymes all possess a macrodomain that recognizes single ADP-ribose units, but among these three members, TARG1 is unique for its ability to also reduce PARylation by removing the whole PAR chain specifically at glutamate–ADP-ribose ester bonds (Sharifi et al., 2013).
Table 2.
Properties and localization of PAR degrading enzymes
| Gene name | Alternative name | Cleave ribose–ribose bonds? | Cleave between ADP-ribose and acidic residues? | Localization |
| PARG | + | − | Cytoplasm, nucleus, mitochondria | |
| ARH3 | ADPRHL2 | + | Not tested | Cytoplasm, nucleus, mitochondria |
| MacroD1 | LRP16 | − | + | Cytoplasm, nucleus |
| MacroD2 | C20orf133 | − | + | Cytoplasm, nucleus |
| TARG1 | C6orf130, OARD1 | − | + | Cytoplasm, nucleus |
ARH3, ADP-ribosylhydrolase 3; PARG, poly(ADP-ribose) glycohydrolase; TARG1, terminal ADP-ribose protein glycohydrolase 1.
Poly(ADP-ribose) in cellular organization
DNA repair complexes.
The founding member of the ADP-ribosyltransferase family PARP-1 assembles DNA repair complexes whereby PARP-1 auto-PARylates itself at sites of DNA damage, thereby recruiting proteins by noncovalent interactions to repair DNA (Gibson and Kraus, 2012). In addition, recent results show that PARylation is required during genotoxic stress for subnuclear relocalization of RNA-binding proteins (Jungmichel et al., 2013). For example, splicing factor THRAP3 accumulation to nuclear speckles is dependent on PARP-1 catalytic activity, as is the relocalization of the dual RNA/DNA-binding protein TAF15 to nucleoli (Jungmichel et al., 2013). Though the functional significances of these latter phenomena remain to be characterized, these data highlight the critical role of PAR in reorganizing cellular architectures during genotoxic stress.
Stress granules.
During heat shock, viral infection, and other stress conditions, stalled translation complexes aggregate to form cytoplasmic RNA-rich structures called stress granules (SGs; Kedersha et al., 2013). These cytoplasmic structures are enriched with PAR, five PARPs, and two cytoplasmic PARG isoforms (Leung et al., 2011). Overexpression of cytoplasmic PARG isoforms inhibits the formation of SGs, whereas overexpression of SG-localized PARPs induces SG formation even in the absence of stress. Consistently, PARG knockdown results in the delay of SG disassembly when stress is relieved. These data taken together suggest that the precise balance of PAR synthesis and degradation regulates the structural integrity of SGs (Leung et al., 2011, 2012).
Nucleolus.
In Drosophila, nearly half of nuclear PARP-1 and PAR are localized in steady-state nucleoli (Boamah et al., 2012), where ribosomal RNA transcription and processing occur. Genetic depletion of PARP-1 results in the disintegration of nucleoli, separating the ribosomal DNA compartment from processing enzymes (Boamah et al., 2012). Similar subcellular mislocalization phenotypes are observed when fly larvae are treated with the general PARP inhibitor 3-aminobenzamide, suggesting that the enzymatic activity of PARP-1 is required for the nucleolar structural integrity (Boamah et al., 2012). Consistently, PARG knockout flies with excess endogenous amounts of PAR have additional nucleoli with ultrastructural abnormalities (Boamah et al., 2012). Such PAR-dependent reorganization of nuclear structures renders PARP-1 and PARG mutants inefficient in ribosome production (Boamah et al., 2012). Thus, a proper level of PAR is critical not only for the structural integrity, but also the functions of cellular organelles.
Mitotic spindles.
During mitosis, when many nonmembranous structures disintegrate, PAR is involved in the formation of bipolar spindles (Chang et al., 2004). PAR in the spindle appears to exchange very slowly with the neighboring environment. Addition of PARG or anti-PAR antibodies to isolated spindles in vitro results in their rapid breakdown. Though several PARPs including PARP-1 were identified in mammalian spindle structures (Table 1), only PARylation mediated by PARP-5a, which makes linear chains, is required for both the structure and function of spindles (Chang et al., 2005). Thus, the chemical structures of PAR dictated by specific PARPs could potentially control how cellular architectures are formed.
Principles of poly(ADP-ribose) in cellular organization
Chain length of PAR specifies which proteins it binds.
Cellular architectures are commonly built by cross-linking proteins in a noncovalent manner via protein–protein interactions (e.g., prion-generated amyloid fibers) or protein–RNA interactions (many RNA granules). In the latter case, a single RNA can serve as a scaffold that allows the binding of multiple RNA-binding proteins. Analogous to RNA, PAR can also serve as a scaffold for multiple proteins to bind in a noncovalent manner (Gibson and Kraus, 2012; Leung et al., 2012; Krietsch et al., 2013). Intriguingly, the ability of a protein to bind to PAR is dependent on the chain length. For example, DNA repair factors XPA, DEK, and Chk1 exhibit a strong preference for long PAR chain length (>40mer), whereas other DNA repair factors p53 and WRN, as well as histone H1, can efficiently bind to short polymers (Fahrer et al., 2007, 2010; Min et al., 2013; Popp et al., 2013). Such length-dependent PAR binding also plays a regulatory role as demonstrated by Chk1, whereby purified PAR of >65mers but not 30mers activates its autophosphorylation in vitro (Min et al., 2013). Though helical structures have been proposed for longer PAR polymer (Minaga and Kun, 1983), how length and branching affect PAR structure remains unclear. Notably, each ADP-ribose subunit is ∼0.5 kD; as ADP-ribose units are rapidly polymerized, the resultant PAR could present itself as a sizeable structure. For instance, PAR polymers can be up to 67 residues with two branch points per molecule after DNA damage, and polymers of 244 residues with 6 branch points upon heat shock (Alvarez-Gonzalez and Jacobson, 1987). Such sizeable nature of PAR makes it a likely candidate for building scaffolds of cellular architectures.
Nucleation of cellular architectures through PARylation?
An emerging concept in cellular self-organization is that nonmembranous structures are formed through phase transition (Hyman and Simons, 2012; Weber and Brangwynne, 2012). As cellular systems continuously seek a state with minimal free energy, it is sometimes favorable for certain protein components to aggregate into locally distinct domains (also known as “droplet” phase). The phase transition is usually triggered by an increase in local concentration of certain types of proteins, particularly those with multi-domains and/or with intrinsically disordered, low complexity sequences (i.e., repetitive amino acid sequence; Huntley and Golding, 2002). Recently, McKnight and colleagues reported that RNA granules can be seeded in vitro by a chemical with β-stranded–like structure (Han et al., 2012; Kato et al., 2012). Incubation of cell or tissue lysate with this chemical selectively precipitates ∼170 proteins commonly found in RNA granules. Many of these proteins contain low complexity regions, which are necessary and sufficient for the formation of RNA granules in vitro and in cells. These recent data are consistent with previous findings demonstrating that repetitive amino acid sequences in prion or QN-rich domain are critical for RNA granule assembly in yeast and human cells (Gilks et al., 2004; Decker et al., 2007; Reijns et al., 2008).
But what triggers the local concentration of proteins in cells? One possibility is nucleation through polynucleotides as proposed for RNA in the high-order assembly of FUS protein or nuclear bodies (Teixeira et al., 2005; Shevtsov and Dundr, 2011; Schwartz et al., 2013), and for DNA in the recruitment of the C-terminal domain of RNA polymerase II (Kwon et al., 2013). So, could this third naturally occurring polynucleotide, PAR, play a similar role for nucleating cellular structures? Intriguingly, when cross-comparing the list of selectively precipitated RNA granule proteins from McKnight and colleagues (Han et al., 2012) with three independent proteomic datasets of identified PARylated proteins, the overlap is highly statistically significant by Fisher’s exact test (p-value = 2.14 × 10−20 [Zhang et al., 2013], 1.8 × 10−20 [Gagné et al., 2008], and 1.27 × 10−24 [Jungmichel et al., 2013]; see Table S1). Statistical significance was also observed with a list of known RNA granule components from Kato et al. (2012) (Table S1). Among the list of 170 proteins, 28 proteins have shown to be ADP-ribosylated at glutamate and aspartate residues, and it appears that, with this limited set of data, the low complexity region is generally depleted of ADP-ribosylated aspartate but enriched for ADP-ribosylated glutamate (see Table S1 for statistical analyses).
One potential function for ADP-ribosylation is to facilitate local concentration of proteins near low complexity regions, thereby passing the critical threshold amount of protein interactions needed to form “droplet” phase. Compared with the other two polynucleotides, one distinct feature of PAR is that it is a protein modification, where the polymer size can be enzymatically regulated on the timescale of seconds to minutes (Wielckens et al., 1983; Alvarez-Gonzalez and Althaus, 1989). Therefore, one might hypothesize that the PAR polymers synthesized on proteins could become instantaneous “seeding” platforms to recruit large numbers of proteins by noncovalent interactions (Fig. 2). These recruited proteins can then be oligomerized via low complexity regions to form cellular structures. The advantage of PAR being a protein modification is that it provides a regulated point of nucleation upon certain stress conditions (e.g., DNA damage or stress granule formation). Given that proteins bind to PAR in a length-dependent manner, the chain length might also serve as a timer for recruiting specific proteins to assemble cellular structures in a defined temporal order. As stress is relieved, PAR can be degraded and structures disassembled.
Figure 2.
Model of PAR-mediated RNA granule formation. PARylation could facilitate the local concentration of RNA granule proteins with low complexity regions, thereby promoting the oligomerization of these protein regions to form RNA granules.
Alternatively, this sizeable modification when directly added at the low complexity region can potentially disrupt oligomerization. As demonstrated for phosphorylation, the addition of post-translational modifications at a low complexity region can alter existing protein–protein interactions and shift the equilibrium of phase transition for structure formation (Han et al., 2012; Li et al., 2012; Kwon et al., 2013). As we are still at the very early stage in identifying ADP-ribosylated sites by mass spectrometry, future biophysical and biochemical studies will allow us to further pinpoint the roles of specific ADP-ribosylation sites in the formation of RNA granules and other cellular structures.
Highly charged PAR competes with other polynucleotides for protein binding.
Apart from PARylated substrate, another class of key constituents in PAR-mediated cellular architecture is PAR-binding proteins. One critical property of PAR is its highly negative charges contributed by the two phosphate groups of each ADP-ribose subunit (Fig. 1), which are intrinsically inclined to bind to positively charged protein surfaces (e.g., those that bind DNA and RNA). In fact, the highly negative charge of PAR is able to effectively compete for histone binding to DNA that has only one phosphate group in each subunit (Realini and Althaus, 1992). The noncovalent PAR-mediated association with histones is so rigorous that it withstands phenol partitioning, strong acid, detergents, and high salt (Realini and Althaus, 1992). However, when considering equivalent numbers of negative charges, PAR is 100–1,000 times more potent than other naturally occurring polyanions, such as poly(A) and heparin, in dissociating histone–DNA complexes (Realini and Althaus, 1992). These experimental data suggest that the interaction between histone–PAR is more than electrostatic and has led to the “histone shuttle” hypothesis, where auto-PARylation of PARP-1 at DNA damage sites results in a transient and reversible dissociation of histones from DNA (Realini and Althaus, 1992). The resulting nucleosomal unfolding potentially allows the access of DNA-binding proteins for repair. Once DNA damage is repaired, PAR is degraded and histones reassociate with DNA.
Given that PAR-binding proteins and PARylated substrates are also enriched for RNA-binding proteins (Gagné et al., 2008; Jungmichel et al., 2013; Zhang et al., 2013), it is feasible that PAR can compete with RNA for protein binding. As stress granules are proposed sites for sorting mRNAs (Kedersha et al., 2013) and PAR is enriched in these cytoplasmic RNA granules (Leung et al., 2011), it will be of interest to test whether the competition between PAR and RNA for protein binding is involved in remodeling of ribonucleoprotein complexes upon stress. Notably, some PAR-binding domains can also bind poly(A) in vitro (Neuvonen and Ahola, 2009), whereas some RNA-binding domains bind equally well with PAR (Kalisch et al., 2012; Krietsch et al., 2013), suggesting the possibility of competition between RNA and PAR binding in cells.
Chemically, PAR sequence complexity is relatively low with homo-polymeric stretches of adenosine-containing units. Analogous to the low complexity sequence observed in proteins, the repeating ADP-ribose units may allow for the formation of a coherent cellular structure by recruiting multiple proteins via PAR-binding domains (Kalisch et al., 2012; Krietsch et al., 2013). For example, macrodomains recognize terminal ADP-ribose units, PBZ domains recognize adjacent ADP-riboses, and WWE domains recognize iso-ADP-riboses (Fig. 3). Based on peptide library screening, more than 500 proteins are predicted to bind to PAR (Pleschke et al., 2000; Gagné et al., 2008). Though there is no significant overlap between McKnight and colleagues’ list of RNA granule proteins with predicted PAR binders, statistically significant overlap is observed with characterized PAR-binding proteins (Krietsch et al., 2013; p-value = 8.13 × 10−7; see Table S2 for statistical analyses). However, the statistical significance only holds if the analyses included those PAR-binding proteins that are also PARylated substrates. In fact, all the PAR-binding proteins found in the McKnight list are also PARylated substrates. These data suggest that PARylation rather than PAR binding is a critical determinant for granule formation. Potentially, these proteins that are both PARylated and PAR binding may act as hubs for mediating cellular structures where PARs act as spokes (Fig. 2, white proteins).
Figure 3.
ADP-ribose recognition by protein domains. The WWE domain recognizes the smallest structural unit of PAR, iso-ADP-ribose, the macrodomain recognizes the terminal ADP-ribose groups, and the PBZ domain can recognize adjacent ADP-ribose groups.
Intriguingly, recent data also suggest that PAR can compete with negatively charged phosphopeptides. For example, PAR-binding pockets were identified in FHA and BRCT domains that respectively recognize phospho-threonine and phospho-serine (Li et al., 2013). This new form of competition opens the possibility that PAR can reorganize specific post-translational forms of proteins in cellular structures and alter signaling pathway responses.
Predictions and prospects
Technical development.
If PAR is critically involved in cellular organization, how can we test this model? Until recently, it has been difficult to identify PARylation sites (Chapman et al., 2013; Zhang et al., 2013). This is because PAR is heterogeneous in length and therefore no unique mass signatures are associated with this modification for mass spectrometry identification. Now given that many endogenous sites have been identified on glutamate and aspartate residues (Zhang et al., 2013), site-directed mutagenesis studies can be performed to test the importance of how PAR mediates cellular structures. However, as ADP-ribosylation can occur at other residues such as cysteine, lysine, and arginine (Cervantes-Laurean et al., 1997), a more comprehensive proteomics tool is urgently needed in the field. Another challenge is to purify a sufficient quantity of PAR of defined length and structure for in vitro biochemistry experiments. Currently, it is possible to make milligram quantities of small- to medium-sized polymer of defined length (Tan et al., 2012), but tools for enriching branched polymers are lacking. Similarly, techniques for chemical synthesis of PAR attached to defined amino acids are still in early development (Moyle and Muir, 2010; Kistemaker et al., 2013). It will be of interest to see whether specific PAR structures can seed the formation of cellular structures in vitro.
The intricate relationships between mono– and poly–ADP-ribosylation.
Systematic analyses of the ADP-ribosyltransferase/PARP family have begun to unravel the complexity of regulation by this family, where some members add only single ADP-ribose units (Hottiger et al., 2010; Vyas et al., 2013). As demonstrated by a recent study for auto-PARylation of PARP-1, transfer of ADP-ribose units can be mediated sequentially by more than one enzyme (Mao et al., 2011), where the initial ADP-ribose was conjugated at a lysine residue by a non-PARP enzyme SIRT6. Whether PARylation can be mediated through sequential actions of ADP-ribosyltransferases that add single and multiple ADP-ribose units remains unclear. As many ADP-ribosyltransferases/PARPs have been identified within the same nonmembranous cellular structures (e.g., nucleoli, stress granules, and spindle poles), these structures thus serve as useful models to investigate their individual roles and the interplay among various members. However, even though homologues of PARP and PARG have been identified in animals, plants, fungi, and protists (Otto et al., 2005; Briggs and Bent, 2011; Slade et al., 2011), it should be noted that not all eukaryotes, notably budding yeast, possess these enzymes, but they still can form similar cellular structures. It is therefore important to figure out what and how PAR offers additional enzymatic regulation in the formation and maintenance of these structures.
Therapeutics.
Finally, the persistence of nonmembranous cellular structures such as stress granules has been shown to be associated with the pathology of cancers and neurodegenerative diseases, where therapeutic modulation of their structural integrity by small molecular kinase inhibitors is currently being explored (Wippich et al., 2013; Kim et al., 2014). As PARP inhibitors are clinically tested in treating cancers and neurodegenerative diseases (Jagtap and Szabó, 2005; Garber, 2013), understanding the principles of poly(ADP-ribose)-mediated organization could potentially have a wide impact on therapy paradigms.
Online supplemental material
Datasets involved and statistical analyses are detailed in Tables S1 and S2. Table S1 and S2 focus on comparisons of RNA granule proteins with PARylated proteins and PAR-binding proteins, respectively. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201402114/DC.
Supplementary Material
Acknowledgments
I would like to thank members of the Leung laboratory for lively and stimulating discussions; Drs. Phil Sharp, Joe Gall, Ted Dawson, Paul Chang, Mike Matunis, and Roy Parker for insightful comments on the manuscript; Dr. Arjun Bhutkar for informatics discussion; Drs. Dmitri Filippov and Scott Bailey in PAR structure discussions; and Drs. Barry Zirkin and Tun-Han Leung for technical support.
The author declares no competing financial interests.
Footnotes
Abbreviations used in this paper:
- PAR
- poly(ADP-ribose)
- PARG
- poly(ADP-ribose) glycohydrolase
- PARP
- poly(ADP-ribose) polymerase
References
- Alvarez-Gonzalez R., Althaus F.R. 1989. Poly(ADP-ribose) catabolism in mammalian cells exposed to DNA-damaging agents. Mutat. Res. 218:67–74 10.1016/0921-8777(89)90012-8 [DOI] [PubMed] [Google Scholar]
- Alvarez-Gonzalez R., Jacobson M.K. 1987. Characterization of polymers of adenosine diphosphate ribose generated in vitro and in vivo. Biochemistry. 26:3218–3224 10.1021/bi00385a042 [DOI] [PubMed] [Google Scholar]
- Boamah E.K., Kotova E., Garabedian M., Jarnik M., Tulin A.V. 2012. Poly(ADP-Ribose) polymerase 1 (PARP-1) regulates ribosomal biogenesis in Drosophila nucleoli. PLoS Genet. 8:e1002442 10.1371/journal.pgen.1002442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs A.G., Bent A.F. 2011. Poly(ADP-ribosyl)ation in plants. Trends Plant Sci. 16:372–380 10.1016/j.tplants.2011.03.008 [DOI] [PubMed] [Google Scholar]
- Cervantes-Laurean D., Jacobson E.L., Jacobson M.K. 1997. Preparation of low molecular weight model conjugates for ADP-ribose linkages to protein. Methods Enzymol. 280:275–287 10.1016/S0076-6879(97)80119-X [DOI] [PubMed] [Google Scholar]
- Chang P., Jacobson M.K., Mitchison T.J. 2004. Poly(ADP-ribose) is required for spindle assembly and structure. Nature. 432:645–649 10.1038/nature03061 [DOI] [PubMed] [Google Scholar]
- Chang P., Coughlin M., Mitchison T.J. 2005. Tankyrase-1 polymerization of poly(ADP-ribose) is required for spindle structure and function. Nat. Cell Biol. 7:1133–1139 10.1038/ncb1322 [DOI] [PubMed] [Google Scholar]
- Chapman J.D., Gagné J.-P., Poirier G.G., Goodlett D.R. 2013. Mapping PARP-1 auto-ADP-ribosylation sites by liquid chromatography-tandem mass spectrometry. J. Proteome Res. 12:1868–1880 10.1021/pr301219h [DOI] [PubMed] [Google Scholar]
- Decker C.J., Teixeira D., Parker R. 2007. Edc3p and a glutamine/asparagine-rich domain of Lsm4p function in processing body assembly in Saccharomyces cerevisiae. J. Cell Biol. 179:437–449 10.1083/jcb.200704147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahrer J., Kranaster R., Altmeyer M., Marx A., Bürkle A. 2007. Quantitative analysis of the binding affinity of poly(ADP-ribose) to specific binding proteins as a function of chain length. Nucleic Acids Res. 35:e143 10.1093/nar/gkm944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahrer J., Popp O., Malanga M., Beneke S., Markovitz D.M., Ferrando-May E., Bürkle A., Kappes F. 2010. High-affinity interaction of poly(ADP-ribose) and the human DEK oncoprotein depends upon chain length. Biochemistry. 49:7119–7130 10.1021/bi1004365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagné J.-P., Isabelle M., Lo K.S., Bourassa S., Hendzel M.J., Dawson V.L., Dawson T.M., Poirier G.G. 2008. Proteome-wide identification of poly(ADP-ribose) binding proteins and poly(ADP-ribose)-associated protein complexes. Nucleic Acids Res. 36:6959–6976 10.1093/nar/gkn771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garber K. 2013. PARP inhibitors bounce back. Nat. Rev. Drug Discov. 12:725–727 10.1038/nrd4147 [DOI] [PubMed] [Google Scholar]
- Gibson B.A., Kraus W.L. 2012. New insights into the molecular and cellular functions of poly(ADP-ribose) and PARPs. Nat. Rev. Mol. Cell Biol. 13:411–424 10.1038/nrm3376 [DOI] [PubMed] [Google Scholar]
- Gilks N., Kedersha N.L., Ayodele M., Shen L., Stoecklin G., Dember L.M., Anderson P. 2004. Stress granule assembly is mediated by prion-like aggregation of TIA-1. Mol. Biol. Cell. 15:5383–5398 10.1091/mbc.E04-08-0715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han T.W., Kato M., Xie S., Wu L.C., Mirzaei H., Pei J., Chen M., Xie Y., Allen J., Xiao G., McKnight S.L. 2012. Cell-free formation of RNA granules: bound RNAs identify features and components of cellular assemblies. Cell. 149:768–779 10.1016/j.cell.2012.04.016 [DOI] [PubMed] [Google Scholar]
- Hatakeyama K., Nemoto Y., Ueda K., Hayaishi O. 1986. Purification and characterization of poly(ADP-ribose) glycohydrolase. Different modes of action on large and small poly(ADP-ribose). J. Biol. Chem. 261:14902–14911 [PubMed] [Google Scholar]
- Hottiger M.O., Hassa P.O., Lüscher B., Schüler H., Koch-Nolte F. 2010. Toward a unified nomenclature for mammalian ADP-ribosyltransferases. Trends Biochem. Sci. 35:208–219 10.1016/j.tibs.2009.12.003 [DOI] [PubMed] [Google Scholar]
- Huntley M.A., Golding G.B. 2002. Simple sequences are rare in the Protein Data Bank. Proteins. 48:134–140 10.1002/prot.10150 [DOI] [PubMed] [Google Scholar]
- Hyman A.A., Simons K. 2012. Cell biology. Beyond oil and water—phase transitions in cells. Science. 337:1047–1049 10.1126/science.1223728 [DOI] [PubMed] [Google Scholar]
- Jagtap P., Szabó C. 2005. Poly(ADP-ribose) polymerase and the therapeutic effects of its inhibitors. Nat. Rev. Drug Discov. 4:421–440 10.1038/nrd1718 [DOI] [PubMed] [Google Scholar]
- Jankevicius G., Hassler M., Golia B., Rybin V., Zacharias M., Timinszky G., Ladurner A.G. 2013. A family of macrodomain proteins reverses cellular mono-ADP-ribosylation. Nat. Struct. Mol. Biol. 20:508–514 10.1038/nsmb.2523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jungmichel S., Rosenthal F., Altmeyer M., Lukas J., Hottiger M.O., Nielsen M.L. 2013. Proteome-wide identification of poly(ADP-Ribosyl)ation targets in different genotoxic stress responses. Mol. Cell. 52:272–285 10.1016/j.molcel.2013.08.026 [DOI] [PubMed] [Google Scholar]
- Kalisch T., Amé J.-C., Dantzer F., Schreiber V. 2012. New readers and interpretations of poly(ADP-ribosyl)ation. Trends Biochem. Sci. 37:381–390 10.1016/j.tibs.2012.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato M., Han T.W., Xie S., Shi K., Du X., Wu L.C., Mirzaei H., Goldsmith E.J., Longgood J., Pei J., et al. 2012. Cell-free formation of RNA granules: low complexity sequence domains form dynamic fibers within hydrogels. Cell. 149:753–767 10.1016/j.cell.2012.04.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N.L., Ivanov P., Anderson P. 2013. Stress granules and cell signaling: more than just a passing phase? Trends Biochem. Sci. 38:494–506 10.1016/j.tibs.2013.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H.-J., Raphael A.R., LaDow E.S., McGurk L., Weber R.A., Trojanowski J.Q., Lee V.M.-Y., Finkbeiner S., Gitler A.D., Bonini N.M. 2014. Therapeutic modulation of eIF2α phosphorylation rescues TDP-43 toxicity in amyotrophic lateral sclerosis disease models. Nat. Genet. 46:152–160 10.1038/ng.2853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kistemaker H.A.V., van Noort G.J.V.D.H., Overkleeft H.S., van der Marel G.A., Filippov D.V. 2013. Stereoselective ribosylation of amino acids. Org. Lett. 15:2306–2309 10.1021/ol400929c [DOI] [PubMed] [Google Scholar]
- Krietsch J., Rouleau M., Pic E., Ethier C., Dawson T.M., Dawson V.L., Masson J.-Y., Poirier G.G., Gagné J.-P. 2013. Reprogramming cellular events by poly(ADP-ribose)-binding proteins. Mol. Aspects Med. 34:1066–1087 10.1016/j.mam.2012.12.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon I., Kato M., Xiang S., Wu L., Theodoropoulos P., Mirzaei H., Han T., Xie S., Corden J.L., McKnight S.L. 2013. Phosphorylation-regulated binding of RNA polymerase II to fibrous polymers of low-complexity domains. Cell. 155:1049–1060 10.1016/j.cell.2013.10.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung A.K.L., Vyas S., Rood J.E., Bhutkar A., Sharp P.A., Chang P. 2011. Poly(ADP-ribose) regulates stress responses and microRNA activity in the cytoplasm. Mol. Cell. 42:489–499 10.1016/j.molcel.2011.04.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung A., Todorova T., Ando Y., Chang P. 2012. Poly(ADP-ribose) regulates post-transcriptional gene regulation in the cytoplasm. RNA Biol. 9:542–548 10.4161/rna.19899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M., Lu L.-Y., Yang C.-Y., Wang S., Yu X. 2013. The FHA and BRCT domains recognize ADP-ribosylation during DNA damage response. Genes Dev. 27:1752–1768 10.1101/gad.226357.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P., Banjade S., Cheng H.-C., Kim S., Chen B., Guo L., Llaguno M., Hollingsworth J.V., King D.S., Banani S.F., et al. 2012. Phase transitions in the assembly of multivalent signalling proteins. Nature. 483:336–340 10.1038/nature10879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao Z., Hine C., Tian X., Van Meter M., Au M., Vaidya A., Seluanov A., Gorbunova V. 2011. SIRT6 promotes DNA repair under stress by activating PARP1. Science. 332:1443–1446 10.1126/science.1202723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meder V.S., Boeglin M., de Murcia G., Schreiber V. 2005. PARP-1 and PARP-2 interact with nucleophosmin/B23 and accumulate in transcriptionally active nucleoli. J. Cell Sci. 118:211–222 10.1242/jcs.01606 [DOI] [PubMed] [Google Scholar]
- Min W., Bruhn C., Grigaravicius P., Zhou Z.-W., Li F., Krüger A., Siddeek B., Greulich K.-O., Popp O., Meisezahl C., et al. 2013. Poly(ADP-ribose) binding to Chk1 at stalled replication forks is required for S-phase checkpoint activation. Nat Commun. 4:2993 10.1038/ncomms3993 [DOI] [PubMed] [Google Scholar]
- Minaga T., Kun E. 1983. Probable helical conformation of poly(ADP-ribose). The effect of cations on spectral properties. J. Biol. Chem. 258:5726–5730 [PubMed] [Google Scholar]
- Miwa M., Saikawa N., Yamaizumi Z., Nishimura S., Sugimura T. 1979. Structure of poly(adenosine diphosphate ribose): identification of 2′-[1″-ribosyl-2″-(or 3″-)(1″’-ribosyl)]adenosine-5′,5″,5″’-tris(phosphate) as a branch linkage. Proc. Natl. Acad. Sci. USA. 76:595–599 10.1073/pnas.76.2.595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miwa M., Ishihara M., Takishima S., Takasuka N., Maeda M., Yamaizumi Z., Sugimura T., Yokoyama S., Miyazawa T. 1981. The branching and linear portions of poly(adenosine diphosphate ribose) have the same alpha(1 leads to 2) ribose-ribose linkage. J. Biol. Chem. 256:2916–2921 [PubMed] [Google Scholar]
- Moyle P.M., Muir T.W. 2010. Method for the synthesis of mono-ADP-ribose conjugated peptides. J. Am. Chem. Soc. 132:15878–15880 10.1021/ja1064312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller-Dieckmann C., Kernstock S., Lisurek M., von Kries J.P., Haag F., Weiss M.S., Koch-Nolte F. 2006. The structure of human ADP-ribosylhydrolase 3 (ARH3) provides insights into the reversibility of protein ADP-ribosylation. Proc. Natl. Acad. Sci. USA. 103:15026–15031 10.1073/pnas.0606762103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuvonen M., Ahola T. 2009. Differential activities of cellular and viral macro domain proteins in binding of ADP-ribose metabolites. J. Mol. Biol. 385:212–225 10.1016/j.jmb.2008.10.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niere M., Mashimo M., Agledal L., Dölle C., Kasamatsu A., Kato J., Moss J., Ziegler M. 2012. ADP-ribosylhydrolase 3 (ARH3), not poly(ADP-ribose) glycohydrolase (PARG) isoforms, is responsible for degradation of mitochondrial matrix-associated poly(ADP-ribose). J. Biol. Chem. 287:16088–16102 10.1074/jbc.M112.349183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oka S., Kato J., Moss J. 2006. Identification and characterization of a mammalian 39-kDa poly(ADP-ribose) glycohydrolase. J. Biol. Chem. 281:705–713 10.1074/jbc.M510290200 [DOI] [PubMed] [Google Scholar]
- Otto H., Reche P.A., Bazan F., Dittmar K., Haag F., Koch-Nolte F. 2005. In silico characterization of the family of PARP-like poly(ADP-ribosyl)transferases (pARTs). BMC Genomics. 6:139 10.1186/1471-2164-6-139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pleschke J.M., Kleczkowska H.E., Strohm M., Althaus F.R. 2000. Poly(ADP-ribose) binds to specific domains in DNA damage checkpoint proteins. J. Biol. Chem. 275:40974–40980 10.1074/jbc.M006520200 [DOI] [PubMed] [Google Scholar]
- Popp O., Veith S., Fahrer J., Bohr V.A., Bürkle A., Mangerich A. 2013. Site-specific noncovalent interaction of the biopolymer poly(ADP-ribose) with the Werner syndrome protein regulates protein functions. ACS Chem. Biol. 8:179–188 10.1021/cb300363g [DOI] [PMC free article] [PubMed] [Google Scholar]
- Realini C.A., Althaus F.R. 1992. Histone shuttling by poly(ADP-ribosylation). J. Biol. Chem. 267:18858–18865 [PubMed] [Google Scholar]
- Reijns M.A.M., Alexander R.D., Spiller M.P., Beggs J.D. 2008. A role for Q/N-rich aggregation-prone regions in P-body localization. J. Cell Sci. 121:2463–2472 10.1242/jcs.024976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rippmann J.F., Damm K., Schnapp A. 2002. Functional characterization of the poly(ADP-ribose) polymerase activity of tankyrase 1, a potential regulator of telomere length. J. Mol. Biol. 323:217–224 10.1016/S0022-2836(02)00946-4 [DOI] [PubMed] [Google Scholar]
- Rosenthal F., Feijs K.L.H., Frugier E., Bonalli M., Forst A.H., Imhof R., Winkler H.C., Fischer D., Caflisch A., Hassa P.O., et al. 2013. Macrodomain-containing proteins are new mono-ADP-ribosylhydrolases. Nat. Struct. Mol. Biol. 20:502–507 10.1038/nsmb.2521 [DOI] [PubMed] [Google Scholar]
- Schwartz J.C., Wang X., Podell E.R., Cech T.R. 2013. RNA seeds higher-order assembly of FUS protein. Cell Rep. 5:918–925 10.1016/j.celrep.2013.11.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharifi R., Morra R., Appel C.D., Tallis M., Chioza B., Jankevicius G., Simpson M.A., Matic I., Ozkan E., Golia B., et al. 2013. Deficiency of terminal ADP-ribose protein glycohydrolase TARG1/C6orf130 in neurodegenerative disease. EMBO J. 32:1225–1237 10.1038/emboj.2013.51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevtsov S.P., Dundr M. 2011. Nucleation of nuclear bodies by RNA. Nat. Cell Biol. 13:167–173 10.1038/ncb2157 [DOI] [PubMed] [Google Scholar]
- Slade D., Dunstan M.S., Barkauskaite E., Weston R., Lafite P., Dixon N., Ahel M., Leys D., Ahel I. 2011. The structure and catalytic mechanism of a poly(ADP-ribose) glycohydrolase. Nature. 477:616–620 10.1038/nature10404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan E.S., Krukenberg K.A., Mitchison T.J. 2012. Large-scale preparation and characterization of poly(ADP-ribose) and defined length polymers. Anal. Biochem. 428:126–136 10.1016/j.ab.2012.06.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira D., Sheth U., Valencia-Sanchez M.A., Brengues M., Parker R. 2005. Processing bodies require RNA for assembly and contain nontranslating mRNAs. RNA. 11:371–382 10.1261/rna.7258505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vyas S., Chesarone-Cataldo M., Todorova T., Huang Y.-H., Chang P. 2013. A systematic analysis of the PARP protein family identifies new functions critical for cell physiology. Nat Commun. 4:2240 10.1038/ncomms3240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber S.C., Brangwynne C.P. 2012. Getting RNA and protein in phase. Cell. 149:1188–1191 10.1016/j.cell.2012.05.022 [DOI] [PubMed] [Google Scholar]
- Wielckens K., George E., Pless T., Hilz H. 1983. Stimulation of poly(ADP-ribosyl)ation during Ehrlich ascites tumor cell “starvation” and suppression of concomitant DNA fragmentation by benzamide. J. Biol. Chem. 258:4098–4104 [PubMed] [Google Scholar]
- Wippich F., Bodenmiller B., Trajkovska M.G., Wanka S., Aebersold R., Pelkmans L. 2013. Dual specificity kinase DYRK3 couples stress granule condensation/dissolution to mTORC1 signaling. Cell. 152:791–805 10.1016/j.cell.2013.01.033 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Wang J., Ding M., Yu Y. 2013. Site-specific characterization of the Asp- and Glu-ADP-ribosylated proteome. Nat. Methods. 10:981–984 10.1038/nmeth.2603 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.