Short summary
Mutant RAS-driven tumorigenesis was thought for decades to arise independently of wild-type RAS isoforms, but recent evidence points to their involvement. In this issue of Cancer Cell, Grabocka et al. report how loss of wild-type RAS alters oncogenic signaling and dampens the DNA-damage response, thereby promoting tumor progression and chemosensitivity.
Preview
It has been over thirty years since constitutively active mutant forms of KRAS, HRAS and NRAS were shown to transform cells in culture, thus suggesting their ability to drive tumorigenesis autonomously. Indeed, it is now known that mutations in these genes contribute to loss of growth control in approximately 30% of all human cancers (Pylayeva-Gupta et al., 2011). However, detailed analyses of the growth factor signaling pathways impacted by RAS later demonstrated that wild-type (WT) RAS isoforms playa significant role in the transformative abilities of oncogenic RAS mutants (Huang et al., 1993; Lim et al., 2008; Young et al., 2013) but the molecular mechanism remained unknown. In this issue of Cancer Cell, Grabocka et al. provide a major advance in understanding the relationship between oncogenic RAS and WT RAS isoforms in tumorigenesis by elucidating how expression of WT RAS isoforms affect both tumor progression and chemotherapeutic sensitivity by modulating the DNA damage response (Grabocka et al, 2014).
To examine the contribution of WT RAS isoforms in promoting tumorigenesis in KRAS-driven tumors, Grabocka et al. first use WT KRAS and mutant KRAS (G12D) pancreatic and colon carcinoma cells engineered for inducible suppression of WT HRAS and NRAS expression. They show that silencing of HRAS or NRAS in mutant-KRAS cells increases MAPK-RSK and PI3K-AKTsignaling and delays progression through G2/M phase, but has no effect on cells expressing WT KRAS. Furthermore, no substantial change in this delay was observed when both HRAS and NRAS were concurrently depleted, suggesting that HRAS and NRAS function within a single module to regulate oncogenic KRAS signaling. Together with previous studies, these results demonstrate that HRAS and NRAS work to limit oncogenic signaling, which in turn leads to cell cycle delays.
The delayed cell cycle progression and mitotic defects observed upon WT RAS isoform suppression are consistent with the well-established effect of oncogenic signaling on genomic instability and cell cycle checkpoint activation. Oncogenic RAS expression, like most oncogenes, causes replication stress, which is defined as the DNA damage response (DDR) associated with perturbed S phase progression and leads to the activation of the ATR and ATM kinases (Halazonetis et al., 2008). Engagement of the DDR initially acts as a barrier to tumorigenesis by inducing cell cycle arrest, senescence, or apoptosis. However, as tumors evolve, attenuation or loss of specific components of the DDR, such as p53, suppresses these outcomes, thus affording tumor progression. CHK1, a checkpoint kinase operating directly downstream of ATR, has been shown in some cases to be inhibited by growth factor signaling pathways through phosphorylation on S280, which prevents CHK1 activation via phosphorylation of S317 and S345 by ATR (King et al., 2004). Therefore, S280 phosphorylation of CHK1 is one mechanism among many by which oncogenic stress-induced checkpoint response can be compromised. However, checkpoint abrogation can be a double-edged sword, allowing cell cycle progression while further promoting genomic instability.
Grabocka et al. demonstrate that depletion of HRAS or NRAS leads to an increase in inhibitory phosphorylation of CHK1 at S280 and a decrease of phospho-CHK1 at S317 and S345 without affecting the ATM-CHK2 signaling pathway. Consistent with checkpoint mitigation after suppression of WT RAS isoforms, KRAS mutant cells failed to block mitotic entry soon after exogenous DNA damage. Because the long-term effects of checkpoint abrogation can be genome destabilizing, these results are in agreement with the mitotic defects and increased phosphorylation of the histone variant H2AX (γH2AX) when WT RAS isoforms were suppressed in mutant RAS-transformed cells. These findings indicate that oncogene-enforced limitation of DNA damage checkpoint control may promote additional genomic instability in affected tumors (Figure 1).
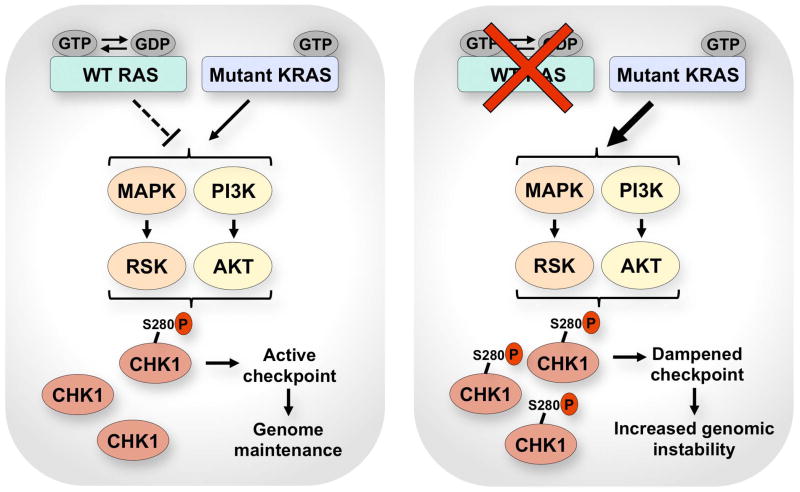
Figure 1. Regulation of the DNA damage response by wild-type RAS in mutant KRAS cells.
WT RAS antagonizes mutant KRAS signaling, thereby limiting the inhibitory phosphorylation of CHK1 at S280 from MAPK-RSK and PI3K-AKT signaling. Uninhibited pools of CHK1 afford an active DNA damage checkpoint response and some degree of genome maintenance. Suppressing WT RAS hyperactivates mutant KRAS signaling, which increases inhibitory phosphorylation of CHK1 at S280. CHK1 inhibition dampens the checkpoint and increases genomic instability.
Although the analysis described above goes far to explain the mechanism of cell cycle perturbations through CHK1 inhibition, it raises important questions on the mechanism by which WT RAS isoforms promote CHK1 S280 phosphorylation. Previous reports have shown that both the MAPK-RSK and PI3K-AKT pathways can cause CHK1 S280 phosphorylation (King et al., 2004; Ray-David et al., 2012). Grabocka et al. demonstrate that both of these pathways, MAPK-RSK and PI3K-AKT, are activated and involved in S280 phosphorylation upon suppression of WT RAS isoforms in KRASG12D-expressing cells and that each pathway contributes to CHK1 S280 phosphorylation. Similar to cultured cells, depletion of WT HRAS in mutant KRAS tumor xenografts also resulted in hyperactivation of the MAPK-RSK and PI3K-AKT pathways as well as repression of CHK1 activity following exposure to DNA-damaging chemotherapeutic agents. Thus, the silencing WT HRAS and NRAS in mutant KRAS cells leads to CHK1 S280 phosphorylation through hyperactivation of both the MAPK-RSK and PI3K-AKT pathways (Figure 1).
Mutation or deletion of p53 is well known to imbuea pro-survival quality to cancer cells by hampering induction of apoptosis from DNA-damaging chemotherapy; however, the opposite effect is observed in response to DNA-damaging agents when most other checkpoint genes are compromised. Indeed, genomic instability is exacerbated by checkpoint failure when cells with damaged DNA enter mitosis. With this feature of checkpoint failure in mind, the authors then queried whether cells with dampened checkpoint activity due to CHK1 S280 phosphorylation would be particularly sensitive to DNA damaging chemotherapies. Accordingly, they found that combining knockdown of WT HRAS with irinotecan caused an increase in cell death and tumor regression compared to either treatment alone. Because suppression of WT RAS isoforms sensitized tumors to a standard DNA-damaging treatment, these results may have significant value in the design of novel treatments for mutant KRAS-associated cancers. In summary, Grabocka et al. have now demonstrated that silencing of WT HRAS or NRAS in mutant KRAS cells significantly influences cancer biology in a way that will facilitate the design of individualized treatments.
Although the authors’ findings increase our mechanistic understanding of how WT and mutant RAS isoforms interact to promote tumor progression and modulate responses to DNA-damaging chemotherapies, interesting questions remain. For example, the effect of the WT KRAS allele on tumorigenesis driven by the mutant KRAS allele on signaling and the DDR have not yet been determined and are relevant given that the WT KRAS allele is often less expressed or completely lost in mutant KRAS-driven cancer cells. Because recent findings suggest both a tumor suppressive and promoting role for expression of the WT KRAS allele (Zhang et al., 2001; Matallanas et al., 2011), it is not immediately apparent how expression of the WT KRAS allele will effect oncogenic KRAS-transformed cells. Furthermore, it is important to note that oncogenic RAS has been associated with increased, not decreased, CHK1 activity (Halazonetis et al., 2008; Gilad et al., 2010). In these cases, CHK1 activity may be slightly stimulated by oncogenic stress, but only to suboptimal levels that are insufficient to counter the frequency of replication abnormalities produced by oncogene expression, leading to increased replication fork collapse and genomic instability (Gilad et al., 2010). Therefore, these studies predict great potential for ATR and CHK1 inhibitors as treatments for KRAS-driven cancers through their ability to further reduce ATR-CHK1 signaling to levels that are toxic (Gilad et al., 2010). Clearly, Grabocka et al. have provided novel insight into the role of WT RAS isoforms in regulating the DDR, providing a fresh look at an old research problem that will undoubtedly stimulate new discoveries for decades to come.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Grabocka, et al. this issue of Cancer Cell. [Google Scholar]
- Gilad O, Nabet BY, Ragland RL, Schoppy DW, Smith KD, Durham AC, Brown EJ. Combining ATR suppression with oncogenic Ras synergistically increases genomic instability, causing synthetic lethality or tumorigenesis in a dosage-dependent manner. Cancer Res. 2010;70:9693–9702. doi: 10.1158/0008-5472.CAN-10-2286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halazonetis TD, Gorgoulis VG, Bartek J. An oncogene-induced DNA damage model for cancer development. Science. 2008;319:1352–5. doi: 10.1126/science.1140735. [DOI] [PubMed] [Google Scholar]
- Huang DC, Marshall CJ, Hancock JF. Plasma membrane-targeted ras GTPase-activating protein is a potent suppressor of p21ras function. Mol Cell Biol. 1993;13:2420–31. doi: 10.1128/mcb.13.4.2420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King FW, Skeen J, Hay N, Shtivelman E. Inhibition of Chk1 by activated PKB/Akt. Cell Cycle. 2004;3:634–7. [PubMed] [Google Scholar]
- Lim KH, Ancrile BB, Kashatus DF, Counter CM. Tumour maintenance is mediated by eNOS. Nature. 2008;452:646–649. doi: 10.1038/nature06778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matallanas D, Romano D, Al-Mulla F, O’Neill E, Al-Ali W, Crespo P, Doyle B, Nixon C, Sansom O, Drosten M, et al. Mutant K-Ras activation of the proapoptotic MST2 pathway is antagonized by wild-type K-Ras. Mol Cell. 2011;44:893–906. doi: 10.1016/j.molcel.2011.10.016. [DOI] [PubMed] [Google Scholar]
- Pylayeva-Gupta Y, Grabocka E, Bar-Sagi D. Nat Rev Cancer. 2011;11:761–774. doi: 10.1038/nrc3106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray-David H, Romeo Y, Lavoie G, Deleris P, Tcherkezian J, Galan JA, Roux PP. RSK promotes G2 DNA damage checkpoint silencing and participates in melanoma chemoresistance. Oncogene. 2012;32:634–7. doi: 10.1038/onc.2012.472. [DOI] [PubMed] [Google Scholar]
- Young A, Lou D, McCormick F. Oncogenic and wild-type Ras play divergent roles in the regulation of mitogen-activated protein kinase signaling. Cancer Discov. 2013;3:112–23. doi: 10.1158/2159-8290.CD-12-0231. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Wang Y, Vikis HG, Johnson L, Liu G, Li J, Anderson MW, Sills RC, Hong HL, Devereux TR, et al. Wildtype Kras2 can inhibit lung carcinogenesis in mice. Nat Genet. 2001;29:25–33. doi: 10.1038/ng721. [DOI] [PubMed] [Google Scholar]