Abstract
Background
The five amino acid (AA) signature including isoleucine (Ile), leucine (Leu), valine (Val), tyrosine (Tyr), and phenylalanine (Phe) has been associated with incident diabetes and insulin resistance. We investigated whether this same AA signature, single nucleotide polymorphisms (SNPs) in genes in their catabolic pathway, were associated with development of impaired fasting glucose (IFG) after atenolol treatment.
Methods and Results
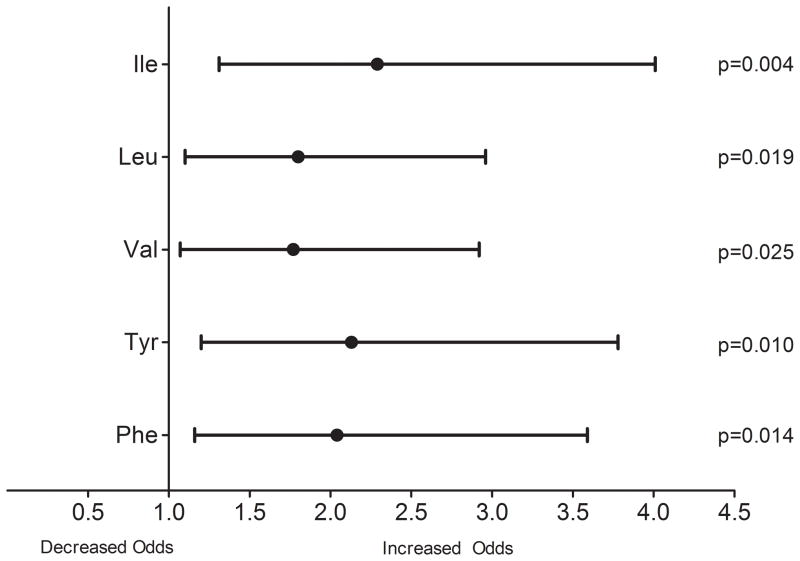
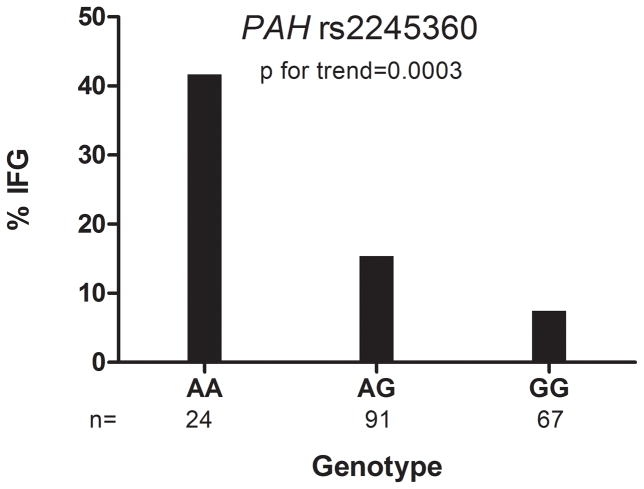
Among 234 European American participants enrolled in the Pharmacogenomic Evaluation of Antihypertensive Responses (PEAR) study and treated with atenolol for 9 weeks, we prospectively followed a nested cohort that had both metabolomics profiling and genotype data available, for the development of IFG. We assessed the association between baseline circulating levels of Ile, Leu, Val, Tyr and Phe, as well as SNPs in BCAT1 and PAH with development of IFG. All baseline AA levels were strongly associated with IFG development. Each increment in standard deviation of the five AAs was associated with the following odds ratio and 95% confidence interval for IFG based on fully adjusted model: Ile 2.29 (1.31–4.01), Leu 1.80 (1.10–2.96), Val 1.77 (1.07–2.92), Tyr 2.13 (1.20–3.78) and Phe 2.04 (1.16–3.59). The composite p value was 2x10−5. Those with PAH (rs2245360) AA genotype had the highest incidence of IFG (p for trend=0.0003).
Conclusions
Our data provide important insight into the metabolic and genetic mechanisms underlying atenolol associated adverse metabolic effects.
Clinical Trial Registration
clinicaltrials.gov; Unique Identifier: NCT00246519
Keywords: amino acids, impaired glucose tolerance, pharmacogenetics, metabolomics, beta-blocker
Hypertension and being overweight or obese have previously been identified as risk factors for diabetes and frequently coexist.1, 2 Impaired fasting glucose (IFG), impaired glucose tolerance (IGT) and insulin resistance, important early components of cardiometabolic dysfunction, are also prevalent in those with hypertension 3 and significantly increase the risk for diabetes.4, 5 β blockers, although very effective blood pressure (BP) lowering agents, are associated with adverse metabolic effects, including hyperglycemia, IFG, and diabetes, all of which are associated with adverse cardiovascular consequences long-term.6–8 While the mechanistic underpinnings of β blocker associated adverse metabolic effects are incompletely understood, we have previously shown that risk for β blocker associated adverse metabolic effects is present in those with and without abdominal obesity.6
Identification of risk factors for diabetes has been a focus for decades, with the ultimate goal of developing strategies to delay or prevent onset of diabetes in those at highest risk. Based on data from observational and randomized clinical trials, clinical characteristics known to increase risk for drug induced diabetes include age, ethnicity, race, body mass index, hypertension, stroke, among many others.9
Metabolomics, a rapidly growing field that enables mapping of global biochemical changes associated with disease or treatment,10 has been used successfully to map pathways implicated in mechanisms of variation of response to drugs.11 Recently, metabolomics data from an observational study of the Framingham Offspring cohort identified a small cluster of essential amino acids (AAs), including baseline levels of three branched-chain amino acids (BCAA), isoleucine (IIe), leucine (Leu) and valine (Val), and two aromatic amino acids (AAA), phenylalanine (Phe) and tyrosine (Tyr), as metabolomic risk factors associated with a significant and independent increased risk for diabetes.12 This same 5 AA cluster has also been identified as a predictor of insulin resistance in young adults.13
While the clinical characteristics that predict risk for drug induced diabetes are similar to those that predict diabetes of other etiologies,14, 15 there are currently no data available to assess whether the same profile of BCAA and AAA that predicted diabetes and insulin resistance risk might also predict risk for drug induced alterations in glucose status. Likewise, while branched chain amino-acid transaminase 1, BCAT1 and phenylalanine hydroxylase, PAH, genes that catabolize BCAA and AA respectively, have been previously associated with diabetes16 and insulin resistance,17 there are no data regarding the associations of single nucleotide polymorphisms (SNPs) in these genes with drug induced metabolic changes. Therefore, in a nested cohort from the Pharmacogenomic Evaluation of Antihypertensive Responses (PEAR) study, we used a targeted “pharmacometabolomics informed pharmacogenomics” approach 18 to investigate whether baseline levels of the previously identified BCAA and AAA and SNPs in genes in their catabolic pathways, increase the odds of development of IFG, a phenotype associated with significant increased risk of diabetes and cardiovascular disease following short term exposure to atenolol.4, 19
Methods
PEAR is a prospective, randomized, parallel group, titration study undertaken to evaluate the pharmacogenomic determinants of the antihypertensive and adverse metabolic responses to atenolol and hydrochlorothiazide in hypertensive participants without a history of heart disease or diabetes. Details regarding study design and enrollment criteria have been previously published.20 The study is registered at clinicaltrials.gov, NCT00246519, http://clinicaltrials.gov/ct2/show/NCT00246519.
Study Population
PEAR participants were enrolled from primary care clinics at the University of Florida (Gainesville, FL), Emory University (Atlanta, GA), and the Mayo Clinic (Rochester, MN). Because we are seeking to extend findings generated in a primarily White population, this analysis includes a subset of European American men and women with mild-to-moderate essential hypertension, between the ages of 17–65, selected for metabolomics profiling and with available genotype information within the atenolol arm of PEAR (Supplement Figure 1). For metabolomics profiling, patients were divided into quartiles based on the DBP response to atenolol monotherapy. Equal numbers of patients were selected randomly within each response quartile using the PROC SURVEYSELECT procedure (simple random sampling without replacement). Prior to baseline measurements, those receiving treatment for hypertension at enrollment had all antihypertensive drugs discontinued for a median 27 (interquartile range [IQR] 19–34) days. Participants with IFG (glucose ≥ 100 mg/dl) at baseline were excluded from this analysis.
PEAR Protocol
Written informed consent was obtained voluntarily from all participants, and institutional review board approval was obtained from all study sites, which included University of Florida, Mayo Clinic and Emory University. Atenolol was initiated at 50mg, and titrated to 100mg, based on BP > 120/70 mmHg and tolerability. After at least 6 weeks on the final dose, response was determined, including BP measurements and laboratory-based assessments, and results described in this analysis come from this response assessment time point. PEAR is registered at ClinicalTrials.gov, #NCT00246519.
Laboratory Measurements
At baseline and after completion of atenolol monotherapy, fasting blood samples were collected for glucose and insulin. Insulin sensitivity status was calculated using the homeostatic model assessment – insulin resistance (HOMA-IR).21
Biochemical Assays
Glucose was measured in plasma, on an Hitachi 911 Chemistry Analyzer (Roche Diagnostics) at the central laboratory at the Mayo Clinic, Rochester, MN, using spectrophotometry by an automated enzymatic assay. Plasma insulin was measured using the Access Ultrasensitive Insulin immunoassay system (Beckman Instruments). All samples were tested in duplicate, and data reported are means of the duplicate samples.
Amino Acid Quantification
Plasma samples were transferred from the PEAR laboratory to the metabolomics core laboratory, University of California, Davis, where samples were extracted, derivatized and analyzed as reported previously in great detail.22, 23 Briefly, mass spectrometry used a Leco Pegasus IV time of flight (TOF) mass spectrometer with 280°C transfer line temperature, electron ionization at −70V and an ion source temperature of 250°C. Mass spectra were acquired from m/z 85–500 at 20 spectra/sec and 1750 V detector voltage. Quantitative data were normalized to the sum intensities of all known metabolites and used for statistical investigation.
Genotyping and Quality Control
To explore a potential functional mechanism underlying our observation that the 5 AA signature was associated with development of IFG following exposure to atenolol, we sought to investigate genes in common catabolic pathways. Because we had a limited population (n=184) with available genetic data and that did not have IFG at baseline, we used a candidate gene approach for this analysis. For the BCAAs, we chose to focus on BCAT1, which encodes the cytosolic form of the enzyme branched-chain amino acid transaminase. This enzyme catalyzes the reversible transamination of branched-chain alpha-keto acids to branched-chain L-amino acids essential for cell growth, and importantly, the aminotransferase step is the first step in the catabolic process for Ile, Leu and Val. For the AAs, we chose to focus on PAH, which encodes the enzyme phenylalanine hydroxylase. Phenylalanine hydroxylase catalyzes the hydroxylation of the aromatic side-chain of phenylalanine to generate tyrosine, and is the rate-limiting enzyme of the metabolic pathway that degrades excess phenylalanine.
Genotypes for SNPs in the BCAT1 and PAH genes were obtained from the Omni1M quad GWAS Beadchip. The Omni1M quad is a GWAS chip that used the Infinium II assay and genotypes were called using BeadStudio software and GenTrain2 calling algorithm. Procedures for quality Control and principal component (PC) analysis for determination of population substructure are described in detail in the supplementary materials. After QC procedures, minor allele frequency filter of >0.05, and linkage disequilibrium and monogenic SNP pruning, a total of 96 SNPs, 69 SNPs in BCAT1 and 27 SNPs in PAH, were analyzed in the genetic association study.
Primary Outcome
We examined the association between baseline plasma metabolite level of Ile, Leu, Val, Tyr and Phe, as well as SNP genotypes and development of IFG, defined as a new occurrence of fasting glucose ≥ 100 mg/dl24, following treatment with atenolol monotherapy. After a 12 hour fast, glucose was measured in all participants at baseline, prior to initiation of atenolol, and again at the end of atenolol therapy. Mean duration of treatment with atenolol was 9 weeks. Participants with a fasting glucose ≥ 100 mg/dl at baseline were excluded from this study. Participants with a baseline fasting glucose < 100 mg/dl and a fasting glucose ≥ 100 mg/dl at the end of atenolol treatment were considered to have developed IFG. Participants with a baseline fasting glucose < 100 mg/dl and a fasting glucose that remained < 100 mg/dl at the end of atenolol treatment were considered to have not developed IFG.
Statistical Analyses
Frequencies and percentages were calculated for categorical baseline characteristics and were compared by using Chi-square tests between those who did and did not develop IFG. Means and standard deviations or medians and interquartile ranges were calculated for continuous baseline characteristics and were compared using t-tests or Wilcoxon rank sum tests, respectively, between those who did and did not develop IFG. Change and percent change in fasting glucose, baseline glucose, baseline insulin and baseline AA levels were log-transformed due to non-normal distribution and compared across case/control groups. We utilized a regression model to assess the association between baseline AA level with the percent change in glucose, adjusted for age, gender, BMI, baseline glucose and insulin, and HOMA IR, in the entire cohort with metabolomics data available (n=150). Whether a participant developed IFG, as a binary outcome variable, was fitted by multiple logistic regression models. In separate analyses, each baseline plasma AA level was entered in the logistic model individually as a primary risk predictor. Because the distribution of those who did and did not develop IFG was significantly different by gender and previous research has shown that plasma amino acid levels differ significantly by gender,25 we conducted a subgroup analysis where each baseline plasma AA level as a primary risk predictor of IFG development was tested separately by gender. The values of AA level were standardized (mean=0 and standard deviation=1) so that the regression coefficients are interpreted as odds ratio per standard deviation.12 Odds ratios for AA level predicting IFG were estimated. For logistic regression analyses, we tested four separate models: 1) unadjusted, 2) adjusted for baseline age, sex and BMI, 3) model two plus fasting glucose and fasting insulin and 4) model three plus HOMA-IR, to test the effect of insulin resistance. We evaluated the composite effect of the AAs by combining the individual p values from the model that included HOMA-IR. Since the BCAA and AAA were all strongly correlated (r = 0.56–0.91, p<0.0001 for all comparisons), an extension to Fisher’s combination for correlated tests was performed, for testing significance of the composite p value.26 For amino acid analyses, P values less than 0.05 were considered statistically significant.
For pharmacogenomics analyses, deviations from Hardy Weinberg Equilibrium (HWE) were assessed using Fisher’s Exact Test with alpha=0.05. The associations of the BCAT1 and PAH SNPs and IFG were evaluated using logistic regression, adjusting for age, gender, baseline glucose, insulin and HOMA (model 4 above), and the first 2 PCs for ancestry. To account for multiple comparisons, alpha levels for the SNP associations were set based on the total number of SNPs included in the analysis, which was 96: 0.05/96 = 0.0005. SNP genotype QC and genetic association analyses were conducted in PLINK.27 All other analyses were performed by using SAS v9.3 (SAS institute, Cary, NC).
Results
Pharmacometabolomic Analyses
Among the 234 participants randomized to the atenolol group in the PEAR study, metabolomic profiling was conducted on 150 (Supplement Figure 1). Of those, 122 did not have IFG at baseline and are the focus of the metabolomics analysis. A total of 24 participants developed IFG during an average 9 weeks of treatment with atenolol. Baseline characteristics of the participants included in the metabolomic analysis are summarized in Table 1 and do not differ from the entire atenolol cohort (data not shown). While those who developed IFG were more likely to be men, the cohort was similar in age, body mass index and systolic and diastolic BP, in those who did and did not develop IFG. Baseline fasting glucose was higher (Table 1), and change in glucose following treatment with atenolol was significantly greater among those who developed IFG than those who did not (mean 13.5 mg/dl vs. 2.1 mg/dl, p<0.0001) (Supplement Figure 2A). Median baseline plasma levels of Ile, Leu, Val and Phe were significantly higher in those who developed IFG than those who did not, while there was no difference in baseline levels of Tyr (Supplement Figure 2B).
Table 1.
Baseline characteristics of the cohort with metabolomics data, according to IFG status
| Developed IFG (n=24) | Did Not Develop IFG (n=98) | |
|---|---|---|
| Characteristic | ||
| European American (n, %) | 24, 100 | 98, 100 |
| Women (n,%) | 6, 25* | 62, 63* |
| Current smoker (n,%) | 3, 13 | 13, 13 |
| Mean age, years | 52.5 (9) | 49.6 (10) |
| Mean BMI, kg/m2 | 30.5 (4.5) | 30.3 (6.2) |
| Mean waist circ., cm | 98.5 (11) | 96.6 (13) |
| Mean SBP, mmHg | 143 (9) | 145(10) |
| Mean DBP, mmHg | 92 (5) | 92 (6) |
| Median Fasting glucose, mg/dL | 93 (86.0–98.0)* | 88.3 (83.5–91.5)* |
| Median Fasting insulin, μIU/mL | 8.3 (4.8–11.2) | 6.6 (4.9–9.6) |
| Mean Potassium, meq/L | 4.5 (0.40) | 4.3 (0.34) |
| Median HOMA IR | 1.86 (1.06–2.56) | 1.42 (1.05–1.97) |
Continuous variables that are normally distributed (age, BMI, waist circumference, SBP and DBP) are presented as mean (standard deviation). Continuous variables such as glucose, insulin and HOMA are not normally distributed and are presented as median (interquartile range). Categorical variables were presented as number and percentages. BMI=body mass index, SBP=systolic blood pressure, DBP=diastolic blood pressure, HOMA-IR=homeostatic model assessment-insulin resistance.
denotes a p value < 0.05 by using Chi-square or Wilcoxon rank sum test to compare frequency between those who did and did not develop IFG.
Baseline levels of all five AAs were significantly associated with percent change in glucose, even after adjustment for age, sex, BMI, baseline glucose and insulin and HOMA-IR. The beta coefficients of the baseline five AAs are shown in Table 2.
Table 2.
Association between baseline and percent change in glucose following treatment with atenolol. Beta coefficients were from multiple regression adjusted for baseline glucose, baseline insulin, HOMA-IR, gender, age and body mass index (BMI).
| Beta Coefficient (SE) | P value | ||
|---|---|---|---|
|
| |||
| Baseline AA | Ile | 0.31 (0.08) | 0.0003 |
| Leu | 0.28 (0.08) | 0.001 | |
| Val | 0.24 (0.08) | 0.004 | |
| Tyr | 0.21 (0.068) | 0.01 | |
| Phe | 0.18 (0.08) | 0.03 | |
SE= standard error, Ile=isoleucine, Leu=leucine, Val=valine, Tyr=tyrosine, Phe=phenylalanine
Association of amino acids with impaired fasting glucose
Association between baseline AA level (per SD increment) and odds for development of IFG for models 1–4 in the overall cohort is summarized in Supplement Table 1. Results from Model 4 showed the following odds ratio and 95% confidence interval for IFG: Ile 2.29 (1.31–4.01), Leu 1.80 (1.10–2.96), Val 1.77 (1.07–2.92), Tyr 2.13 (1.20–3.78) and Phe 2.04 (1.16–3.59). (Figure 1). When the p values from the individual AAs were combined, the composite p value was 2x10−5 for Model 4, indicating a very strong association between this five AA metabolite profile and odds for IFG following atenolol treatment. In subgroup analysis of odds for IFG according to gender, with the covariates from Model 4, in women (n=6 developed IFG and n=62 did not develop IFG), the ORs ranged from 1.18–3.67, however none of the associations reach statistical significance likely due to the small number of women that developed IFG. In men (n=18 developed IFG and n=36 did not develop IFG), the ORs ranged from 1.75–2.47, and baseline levels of Ile, Tyr and Phe were associated with significantly increased adjusted odds for developing IFG. Data summarizing adjusted odds for IFG according to gender are presented in Supplement Table 2.
Figure 1.
Odds ratio and 95% Confidence Intervals (per standard deviation) for baseline amino acid level and development of impaired fasting glucose (IFG) from logistic regressions adjusted for age, gender, BMI, baseline fasting glucose and insulin and HOMA-IR (model 4), n=122. BMI = Body Mass Index, HOMA-IR=Homeostatic Model Assessment – Insulin Resistance, Ile=isoleucine, Leu=leucine, Val=valine, Tyr=tyrosine, Phe=phenylalanine
Pharmacogenetic Analysis
Patients with the PAH rs2245360 AA genotype had the highest incidence of IFG (41.7%), compared with patients with AG genotype (15.2%) and GG genotype (7.4%), p for trend=0.0003, which achieved statistical significance (Figure 2). In analyses of odds per allele for developing IFG after atenolol monotherapy, PAH rs2245360 had adjusted ORs of 3.51, 95% confidence interval 1.62–7.63, p = 0.0015. None of the BCAT1 SNPs tested achieved Bonferroni corrected statistical significance for development of IFG.
Figure 2.
From the genotyped population, incidence of impaired fasting glucose according to the top PAH SNP, n=184. IFG=impaired fasting glucose.
Conclusions
We have shown, for the first time, that baseline plasma levels of Ile, Leu, Val, Tyr and Phe, as well as a gene in a catabolic pathway, are strongly associated with increased odds of developing IFG in hypertensive participants, treated relatively short-term (9 weeks) with atenolol, a commonly prescribed β blocker. Importantly, IFG is an independent predictor of diabetes,4 and adding measures of anthropometrics and/or insulin resistance status into prediction models is often more informative for assessing diabetes risk.28 Here, we show that baseline levels of 4 of the 5 AAs investigated are significantly different in those who do and do not develop IFG, and the AAs can further inform a prediction model for drug induced IFG, an early metabolic risk phenotype in the diabetes continuum. Our data extend recent findings from the Framingham Offspring study,12 an observational study primarily in Whites, that showed a strong association between these AAs and incident diabetes.
Overweight, obesity, glucose intolerance and insulin resistance have been closely linked through many important biochemical and regulatory pathways.29 Knowledge that higher blood concentrations of Ile, Leu, Val, Tyr and Phe are elevated in people with obesity, insulin resistance or diabetes is not new.30 The continued rise in prevalence of risk conditions for diabetes in the last 2 decades has increased the need to better understand all relevant underlying pathways. Recent developments in metabolite profiling or metabolomics, have provided insight into additional biochemical pathways that play an important role in glucose and insulin regulation.31 In a study comparing lean and obese individuals, BCAA and AAA were recently recognized as a metabolite cluster strongly associated with insulin sensitivity.32 Additionally, BCAA have been associated with coronary artery disease 33 and diabetes.12 We have further confirmed BCAA as a significant predictor for atenolol-induced IFG, in a population of middle-aged, otherwise healthy, hypertensive individuals, providing additional evidence for its utility as an important risk biomarker. In an analysis stratified by gender, Huffman, et al, showed that a cluster of large neutral AA’s (Leu/Ile ratio, Val, Phe, Try, proline and histidine) along with uric acid was a significant independent predictor of insulin sensitivity in both men and women.25 In our subgroup analysis evaluating the 5 AA signature stratified by gender, we confirmed an association with increased odds for IFG in men, and saw similar, though non-significant trends in women. Our inability to confirm an association in women is likely due to the small number of women who developed IFG in our study, and warrants further investigation in a larger cohort of women treated with atenolol.
Importantly, evidence also exists suggesting that these AAs may play a causal role in metabolic dysregulation. A study evaluating the effect of a high fat diet with or without BCAA supplementation in rats demonstrated that the high fat plus BCAA diet resulted in a higher rate of insulin resistance than the high fat diet alone.32 Similarly, a small study of healthy men who were infused a solution containing 18 AAs showed that AAs impair both insulin-mediated suppression of glucose production and insulin-stimulated glucose disposal in skeletal muscle.34 Together, these data suggest that these metabolites contribute to the development of metabolic dysregulation and are not just innocent bystanders in the metabolic disease continuum.35 Our data from a relatively overweight/obese hypertensive population suggest that treatment with atenolol may be another environmental exposure that has interactions between BCAA, AAA and glucose regulation, as we observed significantly increased odds for IFG, an important early predictor for diabetes, associated with all amino acids tested.
PAH, on chromosome 12 (12q22-q24.2) has been associated with altered metabolic status. In the African American cohort from the Insulin Resistance Atherosclerosis Family Study, strong linkage between the 12q22-24.2 region (PAH) and acute insulin response to glucose was observed.17 Linkage to fasting insulin has previously been reported near 12q23 in the Amish as well.36 Our observation of a significant difference in development of IFG according to PAH rs2245360 genotype suggests this gene may play an important functional role in metabolic risk.
β-adrenergic blockers have long been recommended as first-line therapy for the treatment of hypertension, especially in patients with coronary artery disease.37 However, β-blockers have been implicated in altering glucose homeostasis, primarily through inhibition of pancreatic insulin secretion and promoting insulin resistance. β-receptor selectivity appears to play a role in the degree of downstream metabolic effects, which include not only glucose increases but also weight gain and dyslipidemia. Nonselective and higher-dose selective agents result in the largest adverse metabolic changes.38 Newer β-blockers including nebivolol and carvedilol appear to minimally affect glucose homeostasis and improve insulin sensitivity.39, 40 While the mechanisms underlying this differential effect on glucose remain unclear, they likely extend beyond β blockade. Atenolol, the β blocker used to treat participants in PEAR is cardioselective, and the 100mg dose used is the usual dose used to treat hypertension. We observe that AAs and PAH, a gene previously associated with insulin sensitivity, are also associated with atenolol-associated IFG. These data suggest the underlying mechanisms of drug-induced and primary dysmetabolism may be the same. Drugs may be an environmental trigger in patients with metabolic risk factors.
There are a few limitations of our study that are worthy of mention. First, while we targeted our analysis on a metabolite cluster previously identified for risk of diabetes in a population of Whites, we conducted our analyses only in a cohort of European Americans. Because our cohorts is relatively small, particularly for analyses by gender, these results should be replicated in other populations with hypertension, untreated and treated with β blockers and other drugs that possess adverse metabolic risks, for confirmation. Second, while we did observe a significant association with IFG in our cohort treated with atenolol, we cannot exclude that other metabolites are also playing a role. Third, our findings were identified among European American hypertensive individuals, treated with atenolol and as such are only generalizable among similar patient populations. However, use of β blockers in general, and atenolol specifically is highly prevalent. In 2010, more than 36 million prescriptions were filled for atenolol or atenolol combinations in the US.41 Lastly, further investigation is warranted to 1) determine whether this 5 amino acid signature is causal in the development of IFG or is simply a marker of insulin resistance and impaired beta cell function and 2) confirm our pharmacogenetic association and to extend this finding to other antihypertensive agents associated with hyperglycemia, including thiazide diuretics and other race and ethnic groups at high risk for metabolic dysfunction.
In conclusion, we have extended the previous findings associating BCAA and AAAs with incident diabetes, to atenolol-induced IFG development. Our findings are important as they suggest novel biomarkers for the identification of those individuals at risk of developing antihypertensive treatment-induced diabetes. They may also provide insights to help better understand the mechanisms of β-blocker-induced dysglycemia. While pharmacogenomics has shown to be informative with regard to drug:gene interactions for β blocker BP response,42 this study represents one of the first to employ a targeted pharmacometabolomic investigation combined with a pharmacogenomics investigation informed by the pharmacometabolomic findings for the antihypertensive drug-induced dysglycemia phenotype. With the prevalence of overweight, obesity, insulin resistance, hypertension and diabetes on the rise not only in the US, but worldwide, understanding as many aspects of the mechanistic underpinnings of drug induced adverse metabolic effects as possible is important. Pharmacometabolomics is a new, but rapidly growing field that has promise in defining pathways implicated in mechanisms of variation of response to therapies and compliments information gained from a pharmacogenomics approach. Together, they may be of immense value as the focus for drug therapy moves towards a personalized approach.
Supplementary Material
While it has long been known that over one hundred commonly prescribed medications can adversely affect glucose levels, the underlying mechanisms associated with this dysglycemia are not well understood. Our observation that a 5-amino acid signature previously associated with diabetes is also strongly associated with atenolol-induced impaired fasting glucose, even after adjustment for standard biochemical measures of insulin resistance, suggests this to be an important diabetes risk factor. With diabetes prevalence continuing to increase, recent emphasis on early identification of those at greatest risk has been an important prevention strategy. Metabolomic signatures such as the 5 amino acids we describe may become a biomarker which could be used in the clinical setting to identify individuals at increased risk for diabetes and to recognize those who would benefit from treatment with medications not associated with dysglycemia, where possible. Results from this study provide incentive to test the clinical utility of this strategy.
Acknowledgments
Funding Sources: This work was funded by the Pharmacometabolomics Research Network (RC2 GM092729) and the NIH Pharmacogenomics Research Network (U01 GM074492). Additional funding includes: K23 HL086558 (RMC-D), K23 HL091120 (ALB), and grants from the NIH National Center for Research Resources to the University of Florida (UL1 TR000064), Emory University (UL1 TR000454), and Mayo Clinic (UL1 TR000135).
Footnotes
This study was presented at the American Heart Association 2012 Scientific Sessions meeting, Los Angeles, California, in abstract form.
Conflict of Interest Disclosures: RK-D holds patents in the metabolomics field. All others have none.
References
- 1.Roger VL, Go AS, Lloyd-Jones DM, Benjamin EJ, Berry JD, Borden WB, et al. Subcommittee AHASCaSS. Heart disease and stroke statistics--2012 update: A report from the american heart association. Circulation. 2012;125:e2–e220. doi: 10.1161/CIR.0b013e31823ac046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bombelli M, Facchetti R, Sega R, Carugo S, Fodri D, Brambilla G, et al. Impact of body mass index and waist circumference on the long-term risk of diabetes mellitus, hypertension, and cardiac organ damage. Hypertension. 2011;58:1029–1035. doi: 10.1161/HYPERTENSIONAHA.111.175125. [DOI] [PubMed] [Google Scholar]
- 3.Hayden MR, Sowers JR. Treating hypertension while protecting the vulnerable islet in the cardiometabolic syndrome. J Am Soc Hypertens. 2008;2:239–266. doi: 10.1016/j.jash.2007.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nichols GA, Hillier TA, Brown JB. Normal fasting plasma glucose and risk of type 2 diabetes diagnosis. Am J Med. 2008;121:519–524. doi: 10.1016/j.amjmed.2008.02.026. [DOI] [PubMed] [Google Scholar]
- 5.Tabák AG, Jokela M, Akbaraly TN, Brunner EJ, Kivimäki M, Witte DR. Trajectories of glycaemia, insulin sensitivity, and insulin secretion before diagnosis of type 2 diabetes: An analysis from the whitehall ii study. Lancet. 2009;373:2215–2221. doi: 10.1016/S0140-6736(09)60619-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cooper-DeHoff RM, Wen S, Beitelshees AL, Zineh I, Gums JG, Turner ST, et al. Impact of abdominal obesity on incidence of adverse metabolic effects associated with antihypertensive medications. Hypertension. 2010;55:61–68. doi: 10.1161/HYPERTENSIONAHA.109.139592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Messerli FH, Bangalore S, Julius S. Risk/benefit assessment of beta-blockers and diuretics precludes their use for first-line therapy in hypertension. Circulation. 2008;117:2706–2715. doi: 10.1161/CIRCULATIONAHA.107.695007. discussion 2715. [DOI] [PubMed] [Google Scholar]
- 8.Elliott WJ, Meyer PM. Incident diabetes in clinical trials of antihypertensive drugs: A network meta-analysis. Lancet. 2007;369:201–207. doi: 10.1016/S0140-6736(07)60108-1. [DOI] [PubMed] [Google Scholar]
- 9.Bakris G, Stockert J, Molitch M, Zhou Q, Champion A, Bacher P, et al. Risk factor assessment for new onset diabetes: Literature review. Diabetes Obes Metab. 2009;11:177–187. doi: 10.1111/j.1463-1326.2008.00925.x. [DOI] [PubMed] [Google Scholar]
- 10.Kaddurah-Daouk R, Kristal BS, Weinshilboum RM. Metabolomics: A global biochemical approach to drug response and disease. Annu Rev Pharmacol Toxicol. 2008;48:653–683. doi: 10.1146/annurev.pharmtox.48.113006.094715. [DOI] [PubMed] [Google Scholar]
- 11.Trupp M, Zhu H, Wikoff WR, Baillie RA, Zeng ZB, Karp PD, et al. Metabolomics reveals amino acids contribute to variation in response to simvastatin treatment. PLoS One. 2012;7:e38386. doi: 10.1371/journal.pone.0038386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang TJ, Larson MG, Vasan RS, Cheng S, Rhee EP, McCabe E, et al. Metabolite profiles and the risk of developing diabetes. Nat Med. 2011;17:448–453. doi: 10.1038/nm.2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Würtz P, Soininen P, Kangas AJ, Rönnemaa T, Lehtimäki T, Kähönen M, et al. Branched-chain and aromatic amino acids are predictors of insulin resistance in young adults. Diabetes Care. 2013;36:648–655. doi: 10.2337/dc12-0895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cooper-Dehoff R, Cohen JD, Bakris GL, Messerli FH, Erdine S, Hewkin AC, et al. Predictors of development of diabetes mellitus in patients with coronary artery disease taking antihypertensive medications (findings from the International Verapamil sr-Trandolapril Study [INVEST]) Am J Cardiol. 2006;98:890–894. doi: 10.1016/j.amjcard.2006.04.030. [DOI] [PubMed] [Google Scholar]
- 15.Lindholm LH, Ibsen H, Borch-Johnsen K, Olsen MH, Wachtell K, Dahlöf B, et al. Risk of new-onset diabetes in the losartan intervention for endpoint reduction in hypertension study. J Hypertens. 2002;20:1879–1886. doi: 10.1097/00004872-200209000-00035. [DOI] [PubMed] [Google Scholar]
- 16.Rampersaud E, Damcott CM, Fu M, Shen H, McArdle P, Shi X, et al. Identification of novel candidate genes for type 2 diabetes from a genome-wide association scan in the old order amish: Evidence for replication from diabetes-related quantitative traits and from independent populations. Diabetes. 2007;56:3053–3062. doi: 10.2337/db07-0457. [DOI] [PubMed] [Google Scholar]
- 17.Rich SS, Bowden DW, Haffner SM, Norris JM, Saad MF, Mitchell BD, et al. Identification of quantitative trait loci for glucose homeostasis: The insulin resistance atherosclerosis study (iras) family study. Diabetes. 2004;53:1866–1875. doi: 10.2337/diabetes.53.7.1866. [DOI] [PubMed] [Google Scholar]
- 18.Ji Y, Hebbring S, Zhu H, Jenkins GD, Biernacka J, Snyder K, et al. Glycine and a glycine dehydrogenase (gldc) snp as citalopram/escitalopram response biomarkers in depression: Pharmacometabolomics-informed pharmacogenomics. Clin Pharmacol Ther. 2011;89:97–104. doi: 10.1038/clpt.2010.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Levitzky YS, Pencina MJ, D’Agostino RB, Meigs JB, Murabito JM, Vasan RS, et al. Impact of impaired fasting glucose on cardiovascular disease: The framingham heart study. J Am Coll Cardiol. 2008;51:264–270. doi: 10.1016/j.jacc.2007.09.038. [DOI] [PubMed] [Google Scholar]
- 20.Johnson JA, Boerwinkle E, Zineh I, Chapman AB, Bailey K, Cooper-DeHoff RM, et al. Pharmacogenomics of antihypertensive drugs: Rationale and design of the pharmacogenomic evaluation of antihypertensive responses (pear) study. Am Heart J. 2009;157:442–449. doi: 10.1016/j.ahj.2008.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wallace TM, Levy JC, Matthews DR. Use and abuse of homa modeling. Diabetes Care. 2004;27:1487–1495. doi: 10.2337/diacare.27.6.1487. [DOI] [PubMed] [Google Scholar]
- 22.Fiehn O, Wohlgemuth G, Scholz M, Kind T, Lee do Y, Lu Y, et al. Quality control for plant metabolomics: Reporting msi-compliant studies. Plant J. 2008;53:691–704. doi: 10.1111/j.1365-313X.2007.03387.x. [DOI] [PubMed] [Google Scholar]
- 23.Scholz M, Fiehn O. Setupx--a public study design database for metabolomic projects. Pac Symp Biocomput. 2007:169–180. [PubMed] [Google Scholar]
- 24.American Diabetes Association. Standards of medical care in diabetes--2014. Diabetes Care. 2014;37 (Suppl 1):S14–80. doi: 10.2337/dc14-S014. [DOI] [PubMed] [Google Scholar]
- 25.Huffman KM, Shah SH, Stevens RD, Bain JR, Muehlbauer M, Slentz CA, et al. Relationships between circulating metabolic intermediates and insulin action in overweight to obese, inactive men and women. Diabetes Care. 2009;32:1678–1683. doi: 10.2337/dc08-2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Galwey NW. A new measure of the effective number of tests, a practical tool for comparing families of non-independent significance tests. Genet Epidemiol. 2009;33:559–568. doi: 10.1002/gepi.20408. [DOI] [PubMed] [Google Scholar]
- 27.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. Plink: A tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lyssenko V, Almgren P, Anevski D, Perfekt R, Lahti K, Nissén M, et al. Predictors of and longitudinal changes in insulin sensitivity and secretion preceding onset of type 2 diabetes. Diabetes. 2005;54:166–174. doi: 10.2337/diabetes.54.1.166. [DOI] [PubMed] [Google Scholar]
- 29.Kim JA, Wei Y, Sowers JR. Role of mitochondrial dysfunction in insulin resistance. Circ Res. 2008;102:401–414. doi: 10.1161/CIRCRESAHA.107.165472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Felig P, Marliss E, Cahill GF. Plasma amino acid levels and insulin secretion in obesity. N Engl J Med. 1969;281:811–816. doi: 10.1056/NEJM196910092811503. [DOI] [PubMed] [Google Scholar]
- 31.Newgard CB. Interplay between lipids and branched-chain amino acids in development of insulin resistance. Cell Metab. 2012;15:606–614. doi: 10.1016/j.cmet.2012.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Newgard CB, An J, Bain JR, Muehlbauer MJ, Stevens RD, Lien LF, et al. A branched-chain amino acid-related metabolic signature that differentiates obese and lean humans and contributes to insulin resistance. Cell Metab. 2009;9:311–326. doi: 10.1016/j.cmet.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shah SH, Bain JR, Muehlbauer MJ, Stevens RD, Crosslin DR, Haynes C, et al. Association of a peripheral blood metabolic profile with coronary artery disease and risk of subsequent cardiovascular events. Circ Cardiovasc Genet. 2010;3:207–214. doi: 10.1161/CIRCGENETICS.109.852814. [DOI] [PubMed] [Google Scholar]
- 34.Tremblay F, Marette A. Amino acid and insulin signaling via the mtor/p70 s6 kinase pathway. A negative feedback mechanism leading to insulin resistance in skeletal muscle cells. J Biol Chem. 2001;276:38052–38060. doi: 10.1074/jbc.M106703200. [DOI] [PubMed] [Google Scholar]
- 35.Shah SH, Svetkey LP, Newgard CB. Branching out for detection of type 2 diabetes. Cell Metab. 2011;13:491–492. doi: 10.1016/j.cmet.2011.04.003. [DOI] [PubMed] [Google Scholar]
- 36.Hsueh WC, Mitchell BD, Aburomia R, Pollin T, Sakul H, Gelder Ehm M, et al. Diabetes in the old order amish: Characterization and heritability analysis of the amish family diabetes study. Diabetes Care. 2000;23:595–601. doi: 10.2337/diacare.23.5.595. [DOI] [PubMed] [Google Scholar]
- 37.Chobanian AV, Bakris GL, Black HR, Cushman WC, Green LA, Izzo JL, et al. Seventh report of the joint national committee on prevention, detection, evaluation, and treatment of high blood pressure. Hypertension. 2003;42:1206–1252. doi: 10.1161/01.HYP.0000107251.49515.c2. [DOI] [PubMed] [Google Scholar]
- 38.Jacob S, Rett K, Henriksen EJ. Antihypertensive therapy and insulin sensitivity: Do we have to redefine the role of beta-blocking agents? Am J Hypertens. 1998;11:1258–1265. doi: 10.1016/s0895-7061(98)00141-1. [DOI] [PubMed] [Google Scholar]
- 39.Bakris GL, Fonseca V, Katholi RE, McGill JB, Messerli FH, Phillips RA, et al. Metabolic effects of carvedilol vs metoprolol in patients with type 2 diabetes mellitus and hypertension: A randomized controlled trial. JAMA. 2004;292:2227–2236. doi: 10.1001/jama.292.18.2227. [DOI] [PubMed] [Google Scholar]
- 40.Celik T, Iyisoy A, Kursaklioglu H, Kardesoglu E, Kilic S, Turhan H, et al. Comparative effects of nebivolol and metoprolol on oxidative stress, insulin resistance, plasma adiponectin and soluble p-selectin levels in hypertensive patients. J Hypertens. 2006;24:591–596. doi: 10.1097/01.hjh.0000209993.26057.de. [DOI] [PubMed] [Google Scholar]
- 41.2010 top 200 generic drugs by total prescription. 2011. 2012 http://drugtopics.modernmedicine.com/drugtopics/data/articlestandard//drugtopics/252011/727243/article.pdf.
- 42.Shin J, Johnson JA. Pharmacogenetics of beta-blockers. Pharmacotherapy. 2007;27:874–887. doi: 10.1592/phco.27.6.874. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.