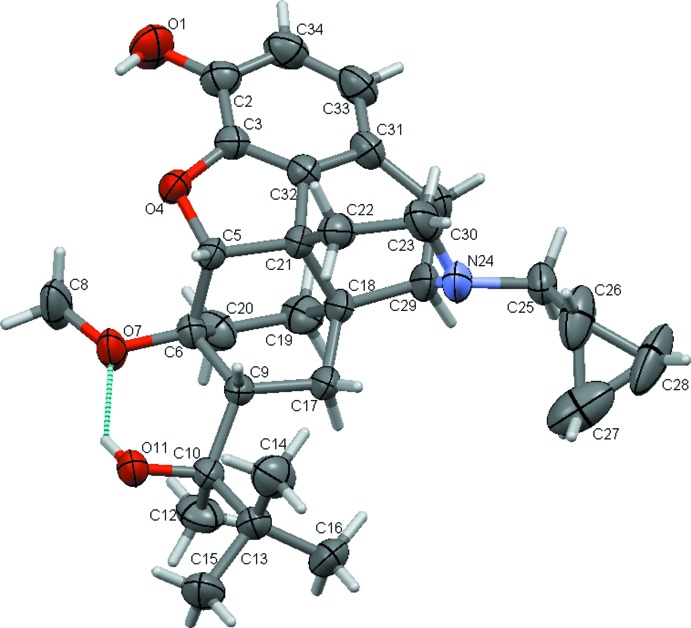
Abstract
In the crystal structure of a semi-synthetic opioid drug buprenorphine, C29H41NO4 {systematic name: (2S)-2-[(5R,6R,7R,14S)-9α-cyclopropylmethyl-3-hydroxy-6-methoxy-4,5-epoxy-6,14-ethanomorphinan-7-yl]-3,3-dimethylbutan-2-ol}, the cyclopropylmethyl group is disordered over two sites with an occupancy factor of 0.611 (3) for the major component. One of the hydroxy groups is involved in intramolecular O—H⋯O hydrogen bond. The other hydroxy group acts as a proton donor in an intermolecular O—H⋯O interaction that connects molecules into a zigzag chain along the b axis.
Related literature
For the crystal structure of buprenorphine hydrochloride, see: Flippen-Anderson et al. (1994 ▶); Kratochvil et al. (1994 ▶). For pharmacological information on buprenorphine, see: Weinberg et al. (1988 ▶); Huang et al. (2001 ▶). For the Kitaigorodskii packing coefficient, see: Kitajgorodskij (1973 ▶).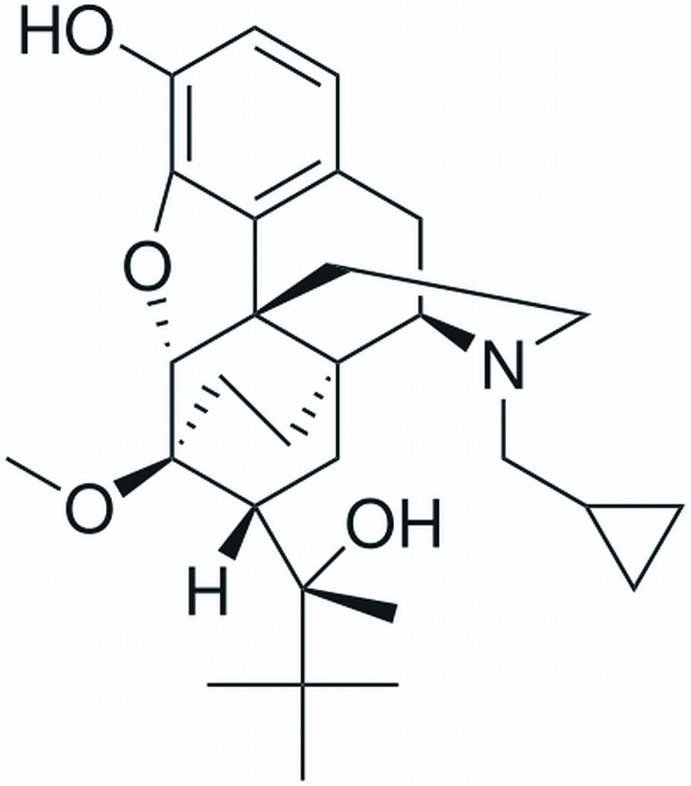
Experimental
Crystal data
C29H41NO4
M r = 467.63
Monoclinic,

a = 9.8154 (6) Å
b = 10.4283 (9) Å
c = 13.4508 (9) Å
β = 108.796 (5)°
V = 1303.37 (16) Å3
Z = 2
Mo Kα radiation
μ = 0.08 mm−1
T = 296 K
0.45 × 0.45 × 0.25 mm
Data collection
Bruker KappaCCD diffractometer
14483 measured reflections
4886 independent reflections
4312 reflections with I > 2σ(I)
R int = 0.026
Refinement
R[F 2 > 2σ(F 2)] = 0.052
wR(F 2) = 0.146
S = 1.04
4886 reflections
352 parameters
112 restraints
H atoms treated by a mixture of independent and constrained refinement
Δρmax = 0.32 e Å−3
Δρmin = −0.31 e Å−3
Data collection: COLLECT (Hooft, 1998 ▶); cell refinement: SCALEPACK (Otwinowski & Minor, 1997 ▶); data reduction: DENZO (Otwinowski & Minor, 1997 ▶) and SCALEPACK; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: Mercury (Macrae et al., 2008 ▶); software used to prepare material for publication: enCIFer (Allen et al., 2004 ▶).
Supplementary Material
Crystal structure: contains datablock(s) New_Global_Publ_Block, I. DOI: 10.1107/S1600536814009672/gk2610sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536814009672/gk2610Isup2.hkl
CCDC reference: 1000182
Additional supporting information: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| O1—H1A⋯O11i | 0.88 (4) | 1.93 (4) | 2.798 (3) | 166 (3) |
| O11—H11A⋯O7 | 0.92 (3) | 1.81 (3) | 2.574 (2) | 139 (3) |
Symmetry code: (i)  .
.
supplementary crystallographic information
1. Comment
Buprenorphine is a semisynthetic opioid (Weinberg et al., 1988) that is used as a pain killer. The molecule has clearly defined hydrophilic and hydrophobic parts. In the latter part, static disorder for cyclopropylmethyl group is observed with the occupation of 0.612 (8) and 0.388 (8) for the two disordered sites. The minor component of the disordered part adopts a conformation that is similar to the one observed for buprenorphine hydrochloride salt (Flippen-Anderson et al., 1994; Kratochvil et al., 1994). In the major component the C29-N24-C25-C26 torsion angle equals to -155.4 (4)° whereas in the minor component the corresponding C29-N24-C25A-C26A angle is -72.2 (7)°. This disorder may result from a relatively loose packing of the crystal (Kitaigorodskii packing cooeficient of 0.65 (Kitajgorodskij, 1973)) that allows for some flexibility in the hydrophobic parts of the molecule.
2. Experimental
Suspension of 29.6 mg of buprenorphine in 200 ml of ethyl acetate was stirred at 25 °C for 14 days. After that time the liquid was separated from the solid and left for evaporation at room temperature. After several days colorless crystals (m.p. 492.5 K) appeared that were used for diffraction studies.
3. Refinement
All C-bound H-atoms were included in the geometrically determined positions with Uiso=1.2 Ueq(C). H atoms from the OH groups were located on a Fourier difference map and refined isotropically. In the absence of significant anomalous scattering effects, Friedel pairs were merged. The absolute configuration is known from the synthetic route. The cyclopropylmethyl group is disordered over two positions. To properly model the disordered fragment restrains were imposed on some bond lengths and anisotropic thermal parameters [DFIX, SADI and SIMU commands in SHELXL-97 (Sheldrick, 2008)].
Figures
Fig. 1.
Molecular structure and atom numbering scheme for buprenorphine. Displacement ellipsoids are shown at the 50% probability level. The minor position of the disordered part has been omitted for clarity.
Fig. 2.
Crystal packing diagram - view along the a axis. Hydrogen bonds are shown as blue lines.
Crystal data
| C29H41NO4 | F(000) = 508 |
| Mr = 467.63 | Dx = 1.192 Mg m−3 |
| Monoclinic, P21 | Melting point: 492.15 K |
| Hall symbol: P 2yb | Mo Kα radiation, λ = 0.71073 Å |
| a = 9.8154 (6) Å | Cell parameters from 6648 reflections |
| b = 10.4283 (9) Å | θ = 1.0–32.6° |
| c = 13.4508 (9) Å | µ = 0.08 mm−1 |
| β = 108.796 (5)° | T = 296 K |
| V = 1303.37 (16) Å3 | Block, colourless |
| Z = 2 | 0.45 × 0.45 × 0.25 mm |
Data collection
| Bruker KappaCCD diffractometer | 4312 reflections with I > 2σ(I) |
| Radiation source: fine-focus sealed tube | Rint = 0.026 |
| Horizonally mounted graphite crystal monochromator | θmax = 32.6°, θmin = 3.8° |
| Detector resolution: 9 pixels mm-1 | h = −14→14 |
| CCD scans | k = −15→15 |
| 14483 measured reflections | l = −20→20 |
| 4886 independent reflections |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.052 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.146 | H atoms treated by a mixture of independent and constrained refinement |
| S = 1.04 | w = 1/[σ2(Fo2) + (0.0762P)2 + 0.1599P] where P = (Fo2 + 2Fc2)/3 |
| 4886 reflections | (Δ/σ)max = 0.007 |
| 352 parameters | Δρmax = 0.32 e Å−3 |
| 112 restraints | Δρmin = −0.31 e Å−3 |
Special details
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | Occ. (<1) | |
| O1 | 0.79854 (19) | 0.6680 (2) | 0.14153 (17) | 0.0696 (5) | |
| H1A | 0.754 (4) | 0.621 (4) | 0.086 (3) | 0.078 (10)* | |
| C2 | 0.7100 (2) | 0.6599 (2) | 0.20083 (17) | 0.0492 (4) | |
| C3 | 0.59432 (19) | 0.57569 (18) | 0.18247 (14) | 0.0408 (3) | |
| O4 | 0.55939 (15) | 0.48055 (14) | 0.10755 (10) | 0.0442 (3) | |
| C5 | 0.44475 (17) | 0.40252 (17) | 0.12545 (12) | 0.0352 (3) | |
| H5A | 0.3618 | 0.4015 | 0.0609 | 0.042* | |
| C6 | 0.48701 (18) | 0.26326 (17) | 0.15977 (13) | 0.0378 (3) | |
| O7 | 0.51953 (16) | 0.19164 (15) | 0.07950 (13) | 0.0520 (4) | |
| C8 | 0.6421 (3) | 0.2234 (3) | 0.0510 (3) | 0.0751 (9) | |
| H8A | 0.6489 | 0.1664 | −0.0032 | 0.113* | |
| H8B | 0.7270 | 0.2153 | 0.1112 | 0.113* | |
| H8C | 0.6338 | 0.3101 | 0.0258 | 0.113* | |
| C9 | 0.34273 (18) | 0.20874 (16) | 0.17199 (13) | 0.0355 (3) | |
| H9A | 0.2657 | 0.2508 | 0.1163 | 0.043* | |
| C10 | 0.3121 (2) | 0.06174 (16) | 0.15353 (14) | 0.0410 (3) | |
| O11 | 0.30844 (16) | 0.03344 (14) | 0.04772 (11) | 0.0463 (3) | |
| H11A | 0.401 (3) | 0.054 (3) | 0.050 (2) | 0.056 (7)* | |
| C12 | 0.4294 (3) | −0.0206 (2) | 0.2276 (2) | 0.0634 (6) | |
| H12A | 0.5217 | 0.0071 | 0.2256 | 0.095* | |
| H12B | 0.4145 | −0.1087 | 0.2060 | 0.095* | |
| H12C | 0.4259 | −0.0123 | 0.2978 | 0.095* | |
| C13 | 0.1567 (3) | 0.0210 (2) | 0.15323 (18) | 0.0512 (4) | |
| C14 | 0.0423 (3) | 0.1179 (3) | 0.0930 (3) | 0.0660 (7) | |
| H14A | −0.0506 | 0.0906 | 0.0941 | 0.099* | |
| H14B | 0.0418 | 0.1233 | 0.0216 | 0.099* | |
| H14C | 0.0641 | 0.2006 | 0.1256 | 0.099* | |
| C15 | 0.1171 (4) | −0.1091 (3) | 0.0959 (3) | 0.0722 (7) | |
| H15A | 0.0227 | −0.1344 | 0.0951 | 0.108* | |
| H15B | 0.1858 | −0.1729 | 0.1319 | 0.108* | |
| H15C | 0.1181 | −0.1006 | 0.0251 | 0.108* | |
| C16 | 0.1457 (4) | 0.0033 (3) | 0.2641 (2) | 0.0775 (9) | |
| H16A | 0.0492 | −0.0211 | 0.2587 | 0.116* | |
| H16B | 0.1696 | 0.0825 | 0.3023 | 0.116* | |
| H16C | 0.2114 | −0.0624 | 0.3005 | 0.116* | |
| C17 | 0.3285 (3) | 0.26244 (19) | 0.27611 (15) | 0.0467 (4) | |
| H17A | 0.2287 | 0.2835 | 0.2661 | 0.056* | |
| H17B | 0.3588 | 0.1978 | 0.3307 | 0.056* | |
| C18 | 0.4216 (2) | 0.38261 (19) | 0.30967 (13) | 0.0449 (4) | |
| C19 | 0.5770 (3) | 0.3344 (2) | 0.35075 (16) | 0.0557 (5) | |
| H19A | 0.5886 | 0.2774 | 0.4099 | 0.067* | |
| H19B | 0.6420 | 0.4063 | 0.3745 | 0.067* | |
| C20 | 0.6138 (2) | 0.2624 (2) | 0.26239 (17) | 0.0531 (5) | |
| H20A | 0.6965 | 0.3024 | 0.2507 | 0.064* | |
| H20B | 0.6392 | 0.1744 | 0.2840 | 0.064* | |
| C21 | 0.40045 (19) | 0.46967 (16) | 0.21299 (12) | 0.0366 (3) | |
| C22 | 0.2464 (2) | 0.5222 (2) | 0.17321 (18) | 0.0498 (4) | |
| H22A | 0.2390 | 0.5842 | 0.1180 | 0.060* | |
| H22B | 0.1805 | 0.4524 | 0.1433 | 0.060* | |
| C23 | 0.2020 (3) | 0.5858 (3) | 0.2599 (3) | 0.0722 (8) | |
| H23A | 0.2542 | 0.6656 | 0.2803 | 0.087* | |
| H23B | 0.1000 | 0.6053 | 0.2342 | 0.087* | |
| N24 | 0.2327 (3) | 0.5006 (2) | 0.3518 (2) | 0.0752 (7) | |
| C25 | 0.1960 (6) | 0.5433 (5) | 0.4459 (4) | 0.0546 (11) | 0.612 (8) |
| H25A | 0.2198 | 0.6332 | 0.4601 | 0.065* | 0.612 (8) |
| H25B | 0.2495 | 0.4937 | 0.5070 | 0.065* | 0.612 (8) |
| C25A | 0.1310 (9) | 0.5706 (7) | 0.3988 (7) | 0.0579 (17) | 0.388 (8) |
| H25C | 0.1742 | 0.6493 | 0.4331 | 0.069* | 0.388 (8) |
| H25D | 0.0407 | 0.5908 | 0.3450 | 0.069* | 0.388 (8) |
| C26 | 0.0370 (5) | 0.5228 (7) | 0.4219 (4) | 0.0817 (19) | 0.612 (8) |
| H26 | −0.0280 | 0.5633 | 0.3586 | 0.098* | 0.612 (8) |
| C27 | −0.0006 (12) | 0.3835 (9) | 0.4492 (6) | 0.101 (3) | 0.612 (8) |
| H27A | −0.0864 | 0.3432 | 0.4024 | 0.122* | 0.612 (8) |
| H27B | 0.0784 | 0.3255 | 0.4820 | 0.122* | 0.612 (8) |
| C28 | −0.0193 (12) | 0.4978 (15) | 0.5104 (10) | 0.111 (4) | 0.612 (8) |
| H28A | −0.1169 | 0.5251 | 0.5018 | 0.133* | 0.612 (8) |
| H28B | 0.0473 | 0.5075 | 0.5811 | 0.133* | 0.612 (8) |
| C26A | 0.1090 (8) | 0.4783 (8) | 0.4756 (5) | 0.068 (2) | 0.388 (8) |
| H26A | 0.1901 | 0.4331 | 0.5253 | 0.082* | 0.388 (8) |
| C27A | −0.0235 (18) | 0.407 (2) | 0.4081 (16) | 0.143 (6) | 0.388 (8) |
| H27C | −0.0687 | 0.4359 | 0.3366 | 0.171* | 0.388 (8) |
| H27D | −0.0275 | 0.3148 | 0.4182 | 0.171* | 0.388 (8) |
| C28A | −0.031 (2) | 0.492 (3) | 0.495 (2) | 0.129 (7) | 0.388 (8) |
| H28C | −0.0410 | 0.4516 | 0.5570 | 0.155* | 0.388 (8) |
| H28D | −0.0822 | 0.5725 | 0.4755 | 0.155* | 0.388 (8) |
| C29 | 0.3847 (3) | 0.4674 (2) | 0.39174 (18) | 0.0652 (7) | |
| H29A | 0.3989 | 0.4147 | 0.4546 | 0.078* | |
| C30 | 0.4872 (4) | 0.5871 (3) | 0.4255 (2) | 0.0727 (8) | |
| H30A | 0.4337 | 0.6566 | 0.4434 | 0.087* | |
| H30B | 0.5656 | 0.5651 | 0.4884 | 0.087* | |
| C31 | 0.5502 (3) | 0.6346 (2) | 0.34350 (17) | 0.0512 (4) | |
| C32 | 0.51126 (19) | 0.57300 (18) | 0.24722 (14) | 0.0407 (3) | |
| C33 | 0.6662 (3) | 0.7197 (2) | 0.36339 (19) | 0.0595 (5) | |
| H33A | 0.6944 | 0.7668 | 0.4254 | 0.071* | |
| C34 | 0.7396 (2) | 0.7347 (2) | 0.2915 (2) | 0.0567 (5) | |
| H34A | 0.8112 | 0.7968 | 0.3042 | 0.068* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| O1 | 0.0602 (9) | 0.0785 (13) | 0.0795 (12) | −0.0284 (9) | 0.0357 (9) | −0.0159 (10) |
| C2 | 0.0464 (9) | 0.0461 (10) | 0.0558 (10) | −0.0073 (8) | 0.0174 (8) | −0.0025 (8) |
| C3 | 0.0458 (8) | 0.0371 (8) | 0.0408 (7) | −0.0032 (7) | 0.0158 (6) | −0.0005 (6) |
| O4 | 0.0529 (7) | 0.0451 (7) | 0.0428 (6) | −0.0126 (6) | 0.0267 (5) | −0.0058 (5) |
| C5 | 0.0413 (7) | 0.0379 (7) | 0.0308 (6) | −0.0052 (6) | 0.0177 (5) | −0.0017 (6) |
| C6 | 0.0422 (7) | 0.0368 (7) | 0.0390 (7) | −0.0003 (6) | 0.0197 (6) | −0.0058 (6) |
| O7 | 0.0526 (7) | 0.0521 (8) | 0.0643 (8) | −0.0096 (6) | 0.0368 (6) | −0.0220 (7) |
| C8 | 0.0686 (13) | 0.0789 (17) | 0.103 (2) | −0.0219 (14) | 0.0626 (14) | −0.0399 (16) |
| C9 | 0.0454 (8) | 0.0309 (6) | 0.0366 (7) | 0.0013 (6) | 0.0219 (6) | −0.0002 (6) |
| C10 | 0.0550 (9) | 0.0303 (7) | 0.0450 (8) | 0.0014 (6) | 0.0264 (7) | 0.0003 (6) |
| O11 | 0.0568 (7) | 0.0429 (7) | 0.0489 (6) | −0.0055 (6) | 0.0304 (6) | −0.0113 (5) |
| C12 | 0.0773 (15) | 0.0390 (10) | 0.0713 (14) | 0.0109 (10) | 0.0204 (12) | 0.0095 (10) |
| C13 | 0.0661 (11) | 0.0370 (8) | 0.0646 (11) | −0.0087 (8) | 0.0406 (10) | −0.0049 (8) |
| C14 | 0.0492 (11) | 0.0595 (14) | 0.0966 (18) | −0.0044 (10) | 0.0339 (12) | −0.0078 (13) |
| C15 | 0.0888 (18) | 0.0478 (12) | 0.097 (2) | −0.0240 (12) | 0.0537 (16) | −0.0190 (13) |
| C16 | 0.117 (2) | 0.0613 (15) | 0.0815 (16) | −0.0256 (16) | 0.0692 (17) | −0.0056 (13) |
| C17 | 0.0724 (12) | 0.0366 (8) | 0.0426 (8) | −0.0043 (8) | 0.0348 (8) | −0.0019 (7) |
| C18 | 0.0703 (11) | 0.0386 (8) | 0.0337 (7) | −0.0049 (8) | 0.0277 (7) | −0.0026 (6) |
| C19 | 0.0773 (14) | 0.0489 (10) | 0.0340 (8) | 0.0003 (10) | 0.0082 (9) | 0.0052 (8) |
| C20 | 0.0517 (10) | 0.0477 (10) | 0.0543 (10) | 0.0058 (8) | 0.0093 (8) | −0.0023 (9) |
| C21 | 0.0461 (8) | 0.0340 (7) | 0.0359 (7) | −0.0009 (6) | 0.0216 (6) | −0.0008 (6) |
| C22 | 0.0485 (9) | 0.0411 (9) | 0.0671 (12) | 0.0013 (8) | 0.0289 (9) | 0.0010 (8) |
| C23 | 0.0728 (14) | 0.0496 (12) | 0.117 (2) | −0.0031 (11) | 0.0626 (15) | −0.0192 (14) |
| N24 | 0.1076 (17) | 0.0555 (11) | 0.0982 (16) | −0.0207 (12) | 0.0827 (15) | −0.0271 (11) |
| C25 | 0.072 (3) | 0.0528 (19) | 0.052 (2) | 0.0074 (19) | 0.038 (2) | −0.0066 (17) |
| C25A | 0.068 (4) | 0.056 (3) | 0.060 (4) | 0.022 (3) | 0.034 (3) | 0.003 (3) |
| C26 | 0.065 (3) | 0.127 (5) | 0.064 (2) | 0.040 (3) | 0.035 (2) | 0.012 (3) |
| C27 | 0.074 (4) | 0.132 (7) | 0.089 (5) | −0.023 (4) | 0.014 (3) | 0.043 (5) |
| C28 | 0.094 (5) | 0.174 (9) | 0.095 (5) | 0.058 (6) | 0.074 (5) | 0.054 (5) |
| C26A | 0.074 (4) | 0.085 (5) | 0.063 (4) | 0.029 (4) | 0.047 (3) | 0.020 (4) |
| C27A | 0.082 (8) | 0.177 (14) | 0.174 (14) | −0.044 (9) | 0.048 (10) | −0.034 (12) |
| C28A | 0.099 (10) | 0.180 (15) | 0.119 (12) | 0.023 (11) | 0.051 (8) | 0.036 (12) |
| C29 | 0.114 (2) | 0.0501 (12) | 0.0533 (10) | −0.0199 (13) | 0.0572 (13) | −0.0161 (10) |
| C30 | 0.115 (2) | 0.0607 (14) | 0.0589 (12) | −0.0255 (15) | 0.0515 (13) | −0.0260 (12) |
| C31 | 0.0679 (12) | 0.0413 (9) | 0.0486 (10) | −0.0064 (9) | 0.0245 (9) | −0.0119 (8) |
| C32 | 0.0480 (8) | 0.0356 (8) | 0.0402 (7) | −0.0018 (7) | 0.0166 (6) | −0.0023 (6) |
| C33 | 0.0744 (14) | 0.0455 (10) | 0.0571 (11) | −0.0099 (10) | 0.0190 (10) | −0.0178 (9) |
| C34 | 0.0542 (10) | 0.0452 (10) | 0.0675 (13) | −0.0122 (9) | 0.0154 (9) | −0.0108 (10) |
Geometric parameters (Å, º)
| O1—C2 | 1.358 (3) | C17—H17B | 0.9700 |
| O1—H1A | 0.88 (4) | C18—C19 | 1.530 (3) |
| C2—C3 | 1.393 (3) | C18—C21 | 1.544 (2) |
| C2—C34 | 1.397 (3) | C18—C29 | 1.545 (3) |
| C3—C32 | 1.372 (2) | C19—C20 | 1.544 (3) |
| C3—O4 | 1.377 (2) | C19—H19A | 0.9700 |
| O4—C5 | 1.470 (2) | C19—H19B | 0.9700 |
| C5—C6 | 1.540 (2) | C20—H20A | 0.9700 |
| C5—C21 | 1.548 (2) | C20—H20B | 0.9700 |
| C5—H5A | 0.9800 | C21—C32 | 1.494 (2) |
| C6—O7 | 1.431 (2) | C21—C22 | 1.533 (3) |
| C6—C20 | 1.532 (3) | C22—C23 | 1.522 (3) |
| C6—C9 | 1.583 (2) | C22—H22A | 0.9700 |
| O7—C8 | 1.415 (3) | C22—H22B | 0.9700 |
| C8—H8A | 0.9600 | C23—N24 | 1.472 (4) |
| C8—H8B | 0.9600 | C23—H23A | 0.9700 |
| C8—H8C | 0.9600 | C23—H23B | 0.9700 |
| C9—C17 | 1.556 (2) | N24—C29 | 1.456 (4) |
| C9—C10 | 1.567 (2) | N24—C25 | 1.492 (4) |
| C9—H9A | 0.9800 | C25—C26 | 1.504 (7) |
| C10—O11 | 1.443 (2) | C25—H25A | 0.9700 |
| C10—C12 | 1.522 (3) | C25—H25B | 0.9700 |
| C10—C13 | 1.582 (3) | C26—C28 | 1.487 (9) |
| O11—H11A | 0.92 (3) | C26—C27 | 1.572 (9) |
| C12—H12A | 0.9600 | C26—H26 | 0.9800 |
| C12—H12B | 0.9600 | C27—C28 | 1.493 (10) |
| C12—H12C | 0.9600 | C27—H27A | 0.9700 |
| C13—C14 | 1.534 (4) | C27—H27B | 0.9700 |
| C13—C16 | 1.541 (3) | C28—H28A | 0.9700 |
| C13—C15 | 1.546 (3) | C28—H28B | 0.9700 |
| C14—H14A | 0.9600 | C29—C30 | 1.576 (4) |
| C14—H14B | 0.9600 | C29—H29A | 0.9800 |
| C14—H14C | 0.9600 | C30—C31 | 1.511 (3) |
| C15—H15A | 0.9600 | C30—H30A | 0.9700 |
| C15—H15B | 0.9600 | C30—H30B | 0.9700 |
| C15—H15C | 0.9600 | C31—C32 | 1.384 (3) |
| C16—H16A | 0.9600 | C31—C33 | 1.400 (3) |
| C16—H16B | 0.9600 | C33—C34 | 1.388 (4) |
| C16—H16C | 0.9600 | C33—H33A | 0.9300 |
| C17—C18 | 1.531 (3) | C34—H34A | 0.9300 |
| C17—H17A | 0.9700 | ||
| C2—O1—H1A | 103 (2) | C18—C19—H19A | 109.8 |
| O1—C2—C3 | 125.0 (2) | C20—C19—H19A | 109.8 |
| O1—C2—C34 | 119.08 (19) | C18—C19—H19B | 109.8 |
| C3—C2—C34 | 115.77 (19) | C20—C19—H19B | 109.8 |
| C32—C3—O4 | 113.03 (15) | H19A—C19—H19B | 108.2 |
| C32—C3—C2 | 121.17 (17) | C6—C20—C19 | 111.52 (17) |
| O4—C3—C2 | 125.50 (17) | C6—C20—H20A | 109.3 |
| C3—O4—C5 | 107.60 (12) | C19—C20—H20A | 109.3 |
| O4—C5—C6 | 115.14 (14) | C6—C20—H20B | 109.3 |
| O4—C5—C21 | 106.95 (13) | C19—C20—H20B | 109.3 |
| C6—C5—C21 | 108.25 (12) | H20A—C20—H20B | 108.0 |
| O4—C5—H5A | 108.8 | C32—C21—C22 | 112.84 (15) |
| C6—C5—H5A | 108.8 | C32—C21—C18 | 106.10 (14) |
| C21—C5—H5A | 108.8 | C22—C21—C18 | 110.72 (15) |
| O7—C6—C20 | 111.26 (15) | C32—C21—C5 | 101.87 (13) |
| O7—C6—C5 | 111.72 (14) | C22—C21—C5 | 112.53 (15) |
| C20—C6—C5 | 109.76 (15) | C18—C21—C5 | 112.33 (14) |
| O7—C6—C9 | 108.39 (13) | C23—C22—C21 | 112.4 (2) |
| C20—C6—C9 | 113.49 (15) | C23—C22—H22A | 109.1 |
| C5—C6—C9 | 101.90 (13) | C21—C22—H22A | 109.1 |
| C8—O7—C6 | 119.87 (16) | C23—C22—H22B | 109.1 |
| O7—C8—H8A | 109.5 | C21—C22—H22B | 109.1 |
| O7—C8—H8B | 109.5 | H22A—C22—H22B | 107.8 |
| H8A—C8—H8B | 109.5 | N24—C23—C22 | 110.4 (2) |
| O7—C8—H8C | 109.5 | N24—C23—H23A | 109.6 |
| H8A—C8—H8C | 109.5 | C22—C23—H23A | 109.6 |
| H8B—C8—H8C | 109.5 | N24—C23—H23B | 109.6 |
| C17—C9—C10 | 115.25 (14) | C22—C23—H23B | 109.6 |
| C17—C9—C6 | 107.89 (14) | H23A—C23—H23B | 108.1 |
| C10—C9—C6 | 117.87 (13) | C29—N24—C23 | 111.19 (18) |
| C17—C9—H9A | 104.8 | C29—N24—C25 | 104.9 (3) |
| C10—C9—H9A | 104.8 | C23—N24—C25 | 119.5 (3) |
| C6—C9—H9A | 104.8 | C29—N24—C25A | 133.6 (4) |
| O11—C10—C12 | 107.66 (16) | C23—N24—C25A | 94.3 (4) |
| O11—C10—C9 | 107.40 (14) | N24—C25—C26 | 107.0 (4) |
| C12—C10—C9 | 112.49 (17) | N24—C25—H25A | 110.3 |
| O11—C10—C13 | 103.00 (16) | C26—C25—H25A | 110.3 |
| C12—C10—C13 | 112.04 (18) | N24—C25—H25B | 110.3 |
| C9—C10—C13 | 113.52 (14) | C26—C25—H25B | 110.3 |
| C10—O11—H11A | 101.6 (16) | H25A—C25—H25B | 108.6 |
| C10—C12—H12A | 109.5 | C28—C26—C25 | 118.7 (7) |
| C10—C12—H12B | 109.5 | C28—C26—C27 | 58.4 (5) |
| H12A—C12—H12B | 109.5 | C25—C26—C27 | 112.8 (6) |
| C10—C12—H12C | 109.5 | C28—C26—H26 | 117.7 |
| H12A—C12—H12C | 109.5 | C25—C26—H26 | 117.7 |
| H12B—C12—H12C | 109.5 | C27—C26—H26 | 117.7 |
| C14—C13—C16 | 108.8 (2) | C28—C27—C26 | 58.0 (5) |
| C14—C13—C15 | 106.9 (2) | C28—C27—H27A | 118.0 |
| C16—C13—C15 | 107.1 (2) | C26—C27—H27A | 118.0 |
| C14—C13—C10 | 111.42 (17) | C28—C27—H27B | 118.0 |
| C16—C13—C10 | 113.5 (2) | C26—C27—H27B | 118.0 |
| C15—C13—C10 | 108.93 (18) | H27A—C27—H27B | 115.1 |
| C13—C14—H14A | 109.5 | C26—C28—C27 | 63.6 (5) |
| C13—C14—H14B | 109.5 | C26—C28—H28A | 117.4 |
| H14A—C14—H14B | 109.5 | C27—C28—H28A | 117.4 |
| C13—C14—H14C | 109.5 | C26—C28—H28B | 117.4 |
| H14A—C14—H14C | 109.5 | C27—C28—H28B | 117.4 |
| H14B—C14—H14C | 109.5 | H28A—C28—H28B | 114.5 |
| C13—C15—H15A | 109.5 | N24—C29—C18 | 108.7 (2) |
| C13—C15—H15B | 109.5 | N24—C29—C30 | 113.6 (2) |
| H15A—C15—H15B | 109.5 | C18—C29—C30 | 112.59 (18) |
| C13—C15—H15C | 109.5 | N24—C29—H29A | 107.2 |
| H15A—C15—H15C | 109.5 | C18—C29—H29A | 107.2 |
| H15B—C15—H15C | 109.5 | C30—C29—H29A | 107.2 |
| C13—C16—H16A | 109.5 | C31—C30—C29 | 115.01 (17) |
| C13—C16—H16B | 109.5 | C31—C30—H30A | 108.5 |
| H16A—C16—H16B | 109.5 | C29—C30—H30A | 108.5 |
| C13—C16—H16C | 109.5 | C31—C30—H30B | 108.5 |
| H16A—C16—H16C | 109.5 | C29—C30—H30B | 108.5 |
| H16B—C16—H16C | 109.5 | H30A—C30—H30B | 107.5 |
| C18—C17—C9 | 109.96 (14) | C32—C31—C33 | 115.8 (2) |
| C18—C17—H17A | 109.7 | C32—C31—C30 | 118.4 (2) |
| C9—C17—H17A | 109.7 | C33—C31—C30 | 124.6 (2) |
| C18—C17—H17B | 109.7 | C3—C32—C31 | 122.89 (18) |
| C9—C17—H17B | 109.7 | C3—C32—C21 | 109.98 (15) |
| H17A—C17—H17B | 108.2 | C31—C32—C21 | 125.23 (17) |
| C19—C18—C17 | 105.59 (17) | C34—C33—C31 | 120.7 (2) |
| C19—C18—C21 | 110.22 (16) | C34—C33—H33A | 119.6 |
| C17—C18—C21 | 109.11 (14) | C31—C33—H33A | 119.6 |
| C19—C18—C29 | 111.45 (18) | C33—C34—C2 | 122.5 (2) |
| C17—C18—C29 | 115.04 (16) | C33—C34—H34A | 118.8 |
| C21—C18—C29 | 105.44 (16) | C2—C34—H34A | 118.8 |
| C18—C19—C20 | 109.58 (16) | ||
| C29—N24—C25—C26 | −155.4 (4) | C29—N24—C25A—C26A | −72.2 (7) |
Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| O1—H1A···O11i | 0.88 (4) | 1.93 (4) | 2.798 (3) | 166 (3) |
| O11—H11A···O7 | 0.92 (3) | 1.81 (3) | 2.574 (2) | 139 (3) |
Symmetry code: (i) −x+1, y+1/2, −z.
Footnotes
Supporting information for this paper is available from the IUCr electronic archives (Reference: GK2610).
References
- Allen, F. H., Johnson, O., Shields, G. P., Smith, B. R. & Towler, M. (2004). J. Appl. Cryst. 37, 335–338.
- Flippen-Anderson, J. L., George, C., Bertha, C. M. & Rice, K. C. (1994). Heterocycles, 39, 751–766.
- Hooft, R. W. W. (1998). COLLECT Nonius BV, Delft, The Netherlands.
- Huang, P., Kehner, G. B., Cowan, A. & Liu-Chen, L. Y. (2001). J. Pharmacol. Exp. Ther. 297, 688–695. [PubMed]
- Kitajgorodskij, A. I. (1973). In Molecular Crystals and Molecules New York: Academic Press.
- Kratochvil, B., Husak, M., Bulej, P. & Jegorov, A. (1994). Collect. Czech. Chem. Commun. 59, 2472–2480.
- Macrae, C. F., Bruno, I. J., Chisholm, J. A., Edgington, P. R., McCabe, P., Pidcock, E., Rodriguez-Monge, L., Taylor, R., van de Streek, J. & Wood, P. A. (2008). J. Appl. Cryst. 41, 466–470.
- Otwinowski, Z. & Minor, W. (1997). Methods in Enzymology, Vol. 276, Macromolecular Crystallography, Part A, edited by C. W. Carter Jr & R. M. Sweet, pp. 307–326. New York: Academic Press.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Weinberg, D. S., Inturrisi, C. E., Reidenberg, B., Moulin, D. E., Nip, T. J., Wallenstein, S., Houde, R. W. & Foley, K. M. (1988). Clin. Pharmacol. Ther. 44, 335–342. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) New_Global_Publ_Block, I. DOI: 10.1107/S1600536814009672/gk2610sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536814009672/gk2610Isup2.hkl
CCDC reference: 1000182
Additional supporting information: crystallographic information; 3D view; checkCIF report