Abstract
In the title hydrated zwitterion, C11H13NO3S2·H2O, the N—C—C—C and C—C—C—S torsion angles in the side-chain are 171.06 (14) and 173.73 (12)°, respectively. In the crystal, inversion-related molecules are π-stacked with an interplanar separation of 3.3847 (2) Å. O—H⋯O hydrogen bonds link inversion-related molecules with a pair of water molecules to form R 4 2(8) rings. The closest S⋯S contact is 3.4051 (15) Å between inversion-related molecules.
Related literature
The crystal structure of a related benzothiazole derivative is described by Lynch (2002 ▶). An analysis of bond angles in the thiazole ring system has been given by Muir et al. (1987 ▶). Applications of benzothiazole derivatives have been described by Vicini et al. (2003 ▶); Bondock et al. (2010 ▶); Paramashivappa et al. (2003 ▶) and Sayama et al. (2002 ▶). 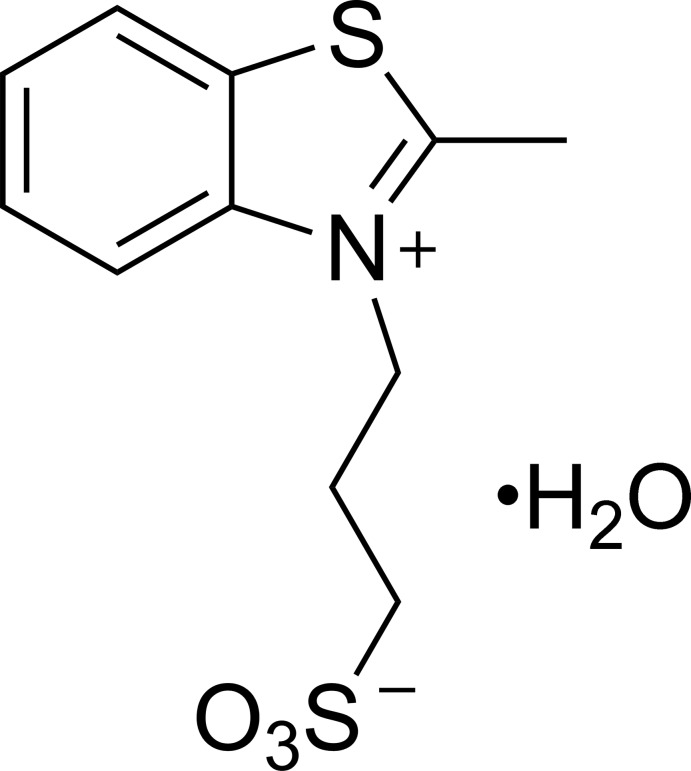
Experimental
Crystal data
C11H13NO3S2·H2O
M r = 289.36
Monoclinic,

a = 10.936 (5) Å
b = 8.708 (5) Å
c = 13.794 (5) Å
β = 109.529 (5)°
V = 1238.0 (10) Å3
Z = 4
Mo Kα radiation
μ = 0.44 mm−1
T = 296 K
0.30 × 0.20 × 0.20 mm
Data collection
Bruker SMART CCD area-detector diffractometer
Absorption correction: multi-scan (SADABS; Bruker, 2001 ▶) T min = 0.880, T max = 0.919
8500 measured reflections
2182 independent reflections
2105 reflections with I > 2sσ(I)
R int = 0.016
Refinement
R[F 2 > 2σ(F 2)] = 0.030
wR(F 2) = 0.080
S = 1.01
2182 reflections
164 parameters
H-atom parameters constrained
Δρmax = 0.31 e Å−3
Δρmin = −0.42 e Å−3
Data collection: SMART (Bruker, 2007 ▶); cell refinement: SAINT (Bruker, 2007 ▶); data reduction: SAINT; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: SHELXTL (Sheldrick, 2008 ▶); software used to prepare material for publication: SHELXTL.
Supplementary Material
Crystal structure: contains datablock(s) I, Global. DOI: 10.1107/S1600536814011660/pk2524sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536814011660/pk2524Isup2.hkl
Supporting information file. DOI: 10.1107/S1600536814011660/pk2524Isup3.cml
CCDC reference: 1004303
Additional supporting information: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| O4—H11⋯O3i | 0.76 | 2.07 | 2.831 (2) | 176 |
| O4—H12⋯O3 | 0.82 | 2.21 | 2.994 (3) | 160 |
| C3—H3A⋯O1ii | 0.97 | 2.39 | 3.269 (3) | 151 |
| C4—H4C⋯O4iii | 0.96 | 2.54 | 3.487 (3) | 169 |
Symmetry codes: (i)  ; (ii)
; (ii)  ; (iii)
; (iii)  .
.
Acknowledgments
This work was supported by the Anhui Provincial Natural Science Foundation (1308085MB24) and the Educational Commission of Anhui Province of China (KJ2012A025).
supplementary crystallographic information
1. Comment
Benzothiazole, a small and simple heterocyclic molecule, has raised considerable interest. It can be used to synthesize some Schiff bases (Vicini et al., 2003), and other derivatives that are antimicrobial (Bondock et al., 2010) and bioactive (Paramashivappa et al., 2003). They have also been used in dye-sensitized solar cells (Sayama et al., 2002). Spurred by this, we synthesized 3-(2-methylbenzo[d]thiazol-3-ium-3-yl)propane-1-sulfonate (Fig. 1), which contains a sulfonic group, with the aim of increased solubility. The single-crystal structure contains one water molecule. Comparing with C7H5N3O2S1·H2O (Lynch, 2002), both of them are in a hydrogen-bonding network with water molecules. The water H atoms are connected with O atoms of sulfonic moieties and the molecules are interconnected, via hydrogen bonds (Table 1) [O4—H11···O3i, symmetry codes: (i) -x, -y + 1, -z + 1; C3—H3A···O1ii, symmetry codes: (ii) -x, y + 1/2, -z + 3/2; C4—H4C···O4iii, symmetry codes: (iii) -x, y - 1/2, -z + 3/2]. There is a R24(8) ring formed by hydrogen-bonded water to O3—S1 interactions. Two characteristic O4—H12···O3 and O4—H11···O3i distances are 2.994 (3) Å and 2.831 (2) Å, respectively (Fig.2). In the crystal, inversion related (1-x,1-y,2-z) molecules are π-stacked with an interplanar separation of 3.3847 (2) Å. O—H···O hydrogen bonds link inversion-related (-x,1-y,1-z) molecules with a pair of water molecules to form R24(8) rings. The closest ring S···S contact is 3.4051 (15) Å between inversion-related (1-x,-y,2-z) molecules (Fig.3). The bond length between N1 and C5 [1.3216 (2) Å] indicates some double bond character and is conjugated with neighbouring bonds. The two distances of S2—C6 and S2—C5 are nearly the same [1.7327 (19) Å and 1.7024 (18) Å, respectively]. In addition, the large size of the S atom compared with N results in a reduction of the C5—S2—C6 angle [91.069 (8)°] compared with the C5—N1—C11 angle [114.001 (14)°] in thiazole ring. This reveals that the S atom might be using unhybridized p-orbitals for bonding (Muir et al., 1987).
2. Experimental
The title complex, 3-(2-methylbenzo[d]thiazol-3-ium-3-yl)propane-1-sulfonate, was prepared by mixing 2-methylbenzo[d]thiazole (1.49 g, 0.010 mol) with 1,2-oxathiolane 2,2-dioxide (1.47 g, 0.012 mol) in toluene (20 ml). The mixture was heated to reflux for 4 h. After the reaction was complete, the solution was cooled to room temperature. The mixture was filtered and washed with ethanol 3 times to give a white solid. Colorless block-shaped crystals were grown by slow evaporation an acetonitrile/ethanol mixture. 1H NMR: (400 Hz, DMSO-d6), d(p.p.m.): 8.43 (t, 2H), 7.90 (t, 1H), 7.80 (t, 1H), 4.90 (t, 2H), 3.20 (s, 3H), 2.65 (t, 2H), 2.15 (q, 2H).
3. Refinement
The water H atoms were located in a difference map and refined isotropically with Uiso(H) = 1.5 Ueq(O). Other hydrogens were placed in geometrically idealized positions (C—H = 0.93–0.97 Å) and allowed to ride on their parent atoms with Uiso(H) = 1.2 Ueq(C) or 1.5Ueq(CMe).
Figures
Fig. 1.
: The molecular structure of the title compound showing 30% probability displacement ellipsoids.
Fig. 2.
: View of the R24(8) ring formed by O4—H12···O3 and O4—H11···O3i intermolecular interactions, showing O—H···O hydrogen-bonding interactions as dashed lines.
Fig. 3.
: Packing diagram of the title compound.
Crystal data
| C11H13NO3S2·H2O | F(000) = 608 |
| Mr = 289.36 | Dx = 1.552 Mg m−3 |
| Monoclinic, P21/c | Mo Kα radiation, λ = 0.71069 Å |
| Hall symbol: -P 2ybc | Cell parameters from 7517 reflections |
| a = 10.936 (5) Å | θ = 2.8–27.1° |
| b = 8.708 (5) Å | µ = 0.44 mm−1 |
| c = 13.794 (5) Å | T = 296 K |
| β = 109.529 (5)° | Block, white |
| V = 1238.0 (10) Å3 | 0.30 × 0.20 × 0.20 mm |
| Z = 4 |
Data collection
| Bruker SMART CCD area-detector diffractometer | 2182 independent reflections |
| Radiation source: fine-focus sealed tube | 2105 reflections with I > 2sσ(I) |
| Graphite monochromator | Rint = 0.016 |
| φ and ω scans | θmax = 25.0°, θmin = 2.0° |
| Absorption correction: multi-scan (SADABS; Bruker, 2001) | h = −13→12 |
| Tmin = 0.880, Tmax = 0.919 | k = −10→10 |
| 8500 measured reflections | l = −16→16 |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.030 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.080 | H-atom parameters constrained |
| S = 1.01 | w = 1/[σ2(Fo2) + (0.0449P)2 + 0.799P] where P = (Fo2 + 2Fc2)/3 |
| 2182 reflections | (Δ/σ)max < 0.001 |
| 164 parameters | Δρmax = 0.31 e Å−3 |
| 0 restraints | Δρmin = −0.42 e Å−3 |
Special details
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| C1 | −0.04082 (15) | 0.49554 (19) | 0.80688 (12) | 0.0271 (3) | |
| H1A | −0.0210 | 0.6044 | 0.8110 | 0.033* | |
| H1B | −0.0771 | 0.4720 | 0.8604 | 0.033* | |
| C2 | 0.08437 (15) | 0.4060 (2) | 0.82760 (12) | 0.0294 (4) | |
| H2A | 0.1180 | 0.4207 | 0.7715 | 0.035* | |
| H2B | 0.0682 | 0.2972 | 0.8327 | 0.035* | |
| C3 | 0.18249 (16) | 0.46262 (19) | 0.92775 (13) | 0.0301 (4) | |
| H3A | 0.2076 | 0.5670 | 0.9186 | 0.036* | |
| H3B | 0.1426 | 0.4636 | 0.9809 | 0.036* | |
| C4 | 0.23671 (18) | 0.2245 (2) | 1.09534 (14) | 0.0388 (4) | |
| H4A | 0.2531 | 0.2983 | 1.1499 | 0.058* | |
| H4B | 0.2549 | 0.1232 | 1.1240 | 0.058* | |
| H4C | 0.1474 | 0.2306 | 1.0522 | 0.058* | |
| C5 | 0.32123 (15) | 0.25761 (19) | 1.03338 (12) | 0.0272 (3) | |
| C6 | 0.49490 (15) | 0.26655 (18) | 0.95809 (12) | 0.0264 (3) | |
| C7 | 0.60073 (16) | 0.2548 (2) | 0.92365 (13) | 0.0333 (4) | |
| H7 | 0.6661 | 0.1832 | 0.9517 | 0.040* | |
| C8 | 0.60501 (18) | 0.3533 (2) | 0.84639 (14) | 0.0390 (4) | |
| H8 | 0.6733 | 0.3465 | 0.8207 | 0.047* | |
| C9 | 0.50797 (19) | 0.4631 (2) | 0.80639 (13) | 0.0383 (4) | |
| H9 | 0.5145 | 0.5296 | 0.7556 | 0.046* | |
| C10 | 0.40270 (17) | 0.4758 (2) | 0.83992 (13) | 0.0328 (4) | |
| H10 | 0.3387 | 0.5494 | 0.8131 | 0.039* | |
| C11 | 0.39668 (15) | 0.37330 (18) | 0.91581 (12) | 0.0258 (3) | |
| N1 | 0.29980 (13) | 0.36398 (15) | 0.96126 (10) | 0.0257 (3) | |
| O1 | −0.18774 (14) | 0.29348 (16) | 0.68281 (11) | 0.0490 (4) | |
| O2 | −0.26970 (12) | 0.55228 (16) | 0.68129 (11) | 0.0447 (3) | |
| O3 | −0.10098 (14) | 0.50116 (19) | 0.60890 (10) | 0.0487 (4) | |
| O4 | 0.09084 (19) | 0.6948 (3) | 0.55198 (17) | 0.0951 (8) | |
| H11 | 0.0932 | 0.6459 | 0.5068 | 0.143* | |
| H12 | 0.0523 | 0.6424 | 0.5819 | 0.143* | |
| S1 | −0.15982 (4) | 0.45636 (5) | 0.68522 (3) | 0.02836 (14) | |
| S2 | 0.46338 (4) | 0.16183 (5) | 1.05392 (3) | 0.02926 (14) |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| C1 | 0.0260 (8) | 0.0278 (8) | 0.0252 (8) | 0.0026 (7) | 0.0053 (6) | −0.0020 (6) |
| C2 | 0.0262 (8) | 0.0304 (8) | 0.0289 (8) | 0.0044 (7) | 0.0058 (7) | −0.0041 (7) |
| C3 | 0.0282 (8) | 0.0283 (8) | 0.0295 (8) | 0.0087 (7) | 0.0039 (7) | −0.0034 (6) |
| C4 | 0.0365 (9) | 0.0464 (11) | 0.0353 (9) | 0.0047 (8) | 0.0144 (8) | 0.0022 (8) |
| C5 | 0.0269 (8) | 0.0271 (8) | 0.0238 (7) | 0.0020 (6) | 0.0035 (6) | −0.0043 (6) |
| C6 | 0.0265 (8) | 0.0243 (8) | 0.0254 (8) | −0.0021 (6) | 0.0049 (6) | −0.0043 (6) |
| C7 | 0.0266 (8) | 0.0352 (9) | 0.0368 (9) | −0.0014 (7) | 0.0089 (7) | −0.0075 (7) |
| C8 | 0.0353 (10) | 0.0461 (11) | 0.0382 (10) | −0.0124 (8) | 0.0156 (8) | −0.0107 (8) |
| C9 | 0.0457 (11) | 0.0394 (10) | 0.0283 (9) | −0.0146 (8) | 0.0102 (8) | −0.0020 (7) |
| C10 | 0.0351 (9) | 0.0292 (9) | 0.0269 (8) | −0.0042 (7) | 0.0010 (7) | 0.0001 (7) |
| C11 | 0.0253 (8) | 0.0245 (8) | 0.0244 (7) | −0.0025 (6) | 0.0039 (6) | −0.0052 (6) |
| N1 | 0.0247 (7) | 0.0250 (7) | 0.0243 (6) | 0.0036 (5) | 0.0038 (5) | −0.0029 (5) |
| O1 | 0.0500 (8) | 0.0333 (7) | 0.0562 (9) | −0.0105 (6) | 0.0079 (7) | −0.0086 (6) |
| O2 | 0.0295 (7) | 0.0515 (8) | 0.0460 (8) | 0.0109 (6) | 0.0032 (6) | 0.0044 (6) |
| O3 | 0.0504 (8) | 0.0678 (10) | 0.0303 (7) | −0.0090 (7) | 0.0166 (6) | 0.0000 (6) |
| O4 | 0.0798 (13) | 0.1204 (18) | 0.1083 (16) | −0.0428 (13) | 0.0622 (12) | −0.0642 (14) |
| S1 | 0.0254 (2) | 0.0311 (2) | 0.0257 (2) | −0.00197 (15) | 0.00465 (17) | −0.00040 (15) |
| S2 | 0.0295 (2) | 0.0272 (2) | 0.0296 (2) | 0.00697 (16) | 0.00785 (17) | 0.00262 (16) |
Geometric parameters (Å, º)
| C1—C2 | 1.518 (2) | C6—C7 | 1.394 (2) |
| C1—S1 | 1.7793 (16) | C6—S2 | 1.7329 (17) |
| C1—H1A | 0.9700 | C7—C8 | 1.381 (3) |
| C1—H1B | 0.9700 | C7—H7 | 0.9300 |
| C2—C3 | 1.521 (2) | C8—C9 | 1.397 (3) |
| C2—H2A | 0.9700 | C8—H8 | 0.9300 |
| C2—H2B | 0.9700 | C9—C10 | 1.381 (3) |
| C3—N1 | 1.484 (2) | C9—H9 | 0.9300 |
| C3—H3A | 0.9700 | C10—C11 | 1.394 (2) |
| C3—H3B | 0.9700 | C10—H10 | 0.9300 |
| C4—C5 | 1.482 (2) | C11—N1 | 1.402 (2) |
| C4—H4A | 0.9600 | O1—S1 | 1.4489 (16) |
| C4—H4B | 0.9600 | O2—S1 | 1.4495 (14) |
| C4—H4C | 0.9600 | O3—S1 | 1.4587 (14) |
| C5—N1 | 1.322 (2) | O4—H11 | 0.7630 |
| C5—S2 | 1.7024 (17) | O4—H12 | 0.8203 |
| C6—C11 | 1.393 (2) | ||
| C2—C1—S1 | 114.11 (11) | C11—C6—S2 | 110.39 (12) |
| C2—C1—H1A | 108.7 | C7—C6—S2 | 128.39 (13) |
| S1—C1—H1A | 108.7 | C8—C7—C6 | 117.63 (17) |
| C2—C1—H1B | 108.7 | C8—C7—H7 | 121.2 |
| S1—C1—H1B | 108.7 | C6—C7—H7 | 121.2 |
| H1A—C1—H1B | 107.6 | C7—C8—C9 | 120.82 (17) |
| C1—C2—C3 | 108.81 (13) | C7—C8—H8 | 119.6 |
| C1—C2—H2A | 109.9 | C9—C8—H8 | 119.6 |
| C3—C2—H2A | 109.9 | C10—C9—C8 | 122.04 (17) |
| C1—C2—H2B | 109.9 | C10—C9—H9 | 119.0 |
| C3—C2—H2B | 109.9 | C8—C9—H9 | 119.0 |
| H2A—C2—H2B | 108.3 | C9—C10—C11 | 117.02 (16) |
| N1—C3—C2 | 111.67 (13) | C9—C10—H10 | 121.5 |
| N1—C3—H3A | 109.3 | C11—C10—H10 | 121.5 |
| C2—C3—H3A | 109.3 | C6—C11—C10 | 121.23 (16) |
| N1—C3—H3B | 109.3 | C6—C11—N1 | 111.50 (14) |
| C2—C3—H3B | 109.3 | C10—C11—N1 | 127.25 (15) |
| H3A—C3—H3B | 107.9 | C5—N1—C11 | 114.00 (13) |
| C5—C4—H4A | 109.5 | C5—N1—C3 | 123.89 (14) |
| C5—C4—H4B | 109.5 | C11—N1—C3 | 122.08 (13) |
| H4A—C4—H4B | 109.5 | H11—O4—H12 | 105.3 |
| C5—C4—H4C | 109.5 | O1—S1—O2 | 113.43 (9) |
| H4A—C4—H4C | 109.5 | O1—S1—O3 | 112.69 (9) |
| H4B—C4—H4C | 109.5 | O2—S1—O3 | 112.28 (9) |
| N1—C5—C4 | 125.63 (15) | O1—S1—C1 | 106.94 (8) |
| N1—C5—S2 | 113.01 (12) | O2—S1—C1 | 105.11 (8) |
| C4—C5—S2 | 121.31 (13) | O3—S1—C1 | 105.63 (9) |
| C11—C6—C7 | 121.22 (16) | C5—S2—C6 | 91.07 (8) |
| S1—C1—C2—C3 | 173.73 (12) | C4—C5—N1—C3 | −3.1 (2) |
| C1—C2—C3—N1 | 171.06 (14) | S2—C5—N1—C3 | 179.33 (11) |
| C11—C6—C7—C8 | 0.3 (2) | C6—C11—N1—C5 | −0.11 (19) |
| S2—C6—C7—C8 | −179.16 (13) | C10—C11—N1—C5 | −178.36 (15) |
| C6—C7—C8—C9 | 1.6 (3) | C6—C11—N1—C3 | −178.26 (13) |
| C7—C8—C9—C10 | −1.6 (3) | C10—C11—N1—C3 | 3.5 (2) |
| C8—C9—C10—C11 | −0.2 (2) | C2—C3—N1—C5 | −100.77 (18) |
| C7—C6—C11—C10 | −2.2 (2) | C2—C3—N1—C11 | 77.20 (19) |
| S2—C6—C11—C10 | 177.37 (12) | C2—C1—S1—O1 | 59.13 (15) |
| C7—C6—C11—N1 | 179.47 (14) | C2—C1—S1—O2 | 179.98 (13) |
| S2—C6—C11—N1 | −1.01 (16) | C2—C1—S1—O3 | −61.12 (15) |
| C9—C10—C11—C6 | 2.1 (2) | N1—C5—S2—C6 | −1.51 (12) |
| C9—C10—C11—N1 | −179.81 (15) | C4—C5—S2—C6 | −179.18 (14) |
| C4—C5—N1—C11 | 178.76 (15) | C11—C6—S2—C5 | 1.41 (12) |
| S2—C5—N1—C11 | 1.21 (17) | C7—C6—S2—C5 | −179.11 (16) |
Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| O4—H11···O3i | 0.76 | 2.07 | 2.831 (2) | 176 |
| O4—H12···O3 | 0.82 | 2.21 | 2.994 (3) | 160 |
| C3—H3A···O1ii | 0.97 | 2.39 | 3.269 (3) | 151 |
| C4—H4C···O4iii | 0.96 | 2.54 | 3.487 (3) | 169 |
Symmetry codes: (i) −x, −y+1, −z+1; (ii) −x, y+1/2, −z+3/2; (iii) −x, y−1/2, −z+3/2.
Footnotes
Supporting information for this paper is available from the IUCr electronic archives (Reference: PK2524).
References
- Bondock, S., Fadaly, W. & Metwally, M. A. (2010). Eur. J. Med. Chem. 45, 3692–3701. [DOI] [PubMed]
- Bruker (2001). SADABS Bruker AXS Inc., Madison, Wisconsin, USA.
- Bruker (2007). SMART and SAINT Bruker AXS Inc., Madison. Wisconsin. USA.
- Lynch, D. E. (2002). Acta Cryst. E58, o1139–o1141.
- Muir, J. A., Gomez, G. M., Muir, M. M., Cox, O. & Cadiz, M. E. (1987). Acta Cryst. C43, 1258–1261.
- Paramashivappa, R., Kumar, P. P., Rao, P. V. S. & Rao, A. S. (2003). Bioorg. Med. Chem. Lett. 13, 657–660. [DOI] [PubMed]
- Sayama, K., Tsukagoshi, S., Hara, K., Ohga, Y., Shinpou, A., Abe, Y., Suga, S. & Arakawa, H. (2002). J. Phys. Chem. B, 106, 1363–1371.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Vicini, P., Geronikaki, A., Incerti, M., Busonera, B., Poni, G., Cabras, C. A. & Colla, P. L. (2003). Bioorg. Med. Chem. 11, 4785–4789. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) I, Global. DOI: 10.1107/S1600536814011660/pk2524sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536814011660/pk2524Isup2.hkl
Supporting information file. DOI: 10.1107/S1600536814011660/pk2524Isup3.cml
CCDC reference: 1004303
Additional supporting information: crystallographic information; 3D view; checkCIF report





