Abstract
In the title compound, C19H18N4O4, the nitrophenyl and phenyl rings are twisted by 67.0 (6) and 37.4 (4)°, respectively, with respect to the pyrazole ring plane [maximum deviation = 0.0042 (16) Å]. The dihedral angle between the mean planes of the phenyl rings is 59.3 (3)°. The amide group, with a C—N—C—C torsion angle of 177.54 (13)°, is twisted away from the plane of the pyrazole ring in an antiperiplanar conformation. In the crystal, N—H⋯O hydrogen bonds involving the carbonyl group on the pyrazole ring and the amide group, together with weak C—H⋯O interactions forming R 2 2(10) graph-set motifs, link the molecules into chains along [100]. Additional weak C—H⋯O interactions involving the nitrophenyl rings further link the molecules along [001], also forming R 2 2(10) graph-set motifs, thereby generating (010) layers.
Related literature
For the structural similarity of N-substituted 2-arylacetamides to the lateral chain of natural benzylpenicillin, see: Mijin & Marinkovic (2006 ▶); Mijin et al. (2008 ▶). For the coordination abilities of amides, see: Wu et al. (2008 ▶, 2010 ▶). For the pharmaceutical, insecticidal and non-linear properties of pyrazoles, see: Chandrakantha et al. (2013 ▶); Cheng et al. (2008 ▶); Hatton et al. (1993 ▶); Liu et al. (2010 ▶). For related structures, see: Fun et al. (2012 ▶); Butcher et al. (2013a
▶,b
▶); Kaur et al. (2013 ▶); Mahan et al. (2013 ▶). For standard bond lengths, see: Allen et al. (1987 ▶).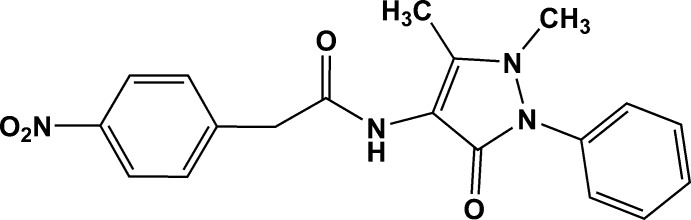
Experimental
Crystal data
C19H18N4O4
M r = 366.37
Triclinic,
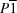
a = 6.7023 (6) Å
b = 8.6335 (8) Å
c = 15.8720 (13) Å
α = 76.305 (7)°
β = 84.399 (7)°
γ = 77.252 (7)°
V = 869.33 (13) Å3
Z = 2
Cu Kα radiation
μ = 0.84 mm−1
T = 173 K
0.28 × 0.22 × 0.12 mm
Data collection
Agilent Eos Gemini diffractometer
Absorption correction: multi-scan (CrysAlis PRO and CrysAlis RED; Agilent, 2012 ▶) T min = 0.851, T max = 1.000
5113 measured reflections
3262 independent reflections
2913 reflections with I > 2σ(I)
R int = 0.030
Refinement
R[F 2 > 2σ(F 2)] = 0.046
wR(F 2) = 0.129
S = 1.07
3262 reflections
247 parameters
H-atom parameters constrained
Δρmax = 0.27 e Å−3
Δρmin = −0.21 e Å−3
Data collection: CrysAlis PRO (Agilent, 2012 ▶); cell refinement: CrysAlis PRO; data reduction: CrysAlis RED (Agilent, 2012 ▶); program(s) used to solve structure: SUPERFLIP (Palatinus & Chapuis, 2007 ▶); program(s) used to refine structure: SHELXL2012 (Sheldrick, 2008 ▶); molecular graphics: OLEX2 (Dolomanov et al., 2009 ▶); software used to prepare material for publication: OLEX2.
Supplementary Material
Crystal structure: contains datablock(s) I. DOI: 10.1107/S1600536814009738/hg5393sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536814009738/hg5393Isup2.hkl
Supporting information file. DOI: 10.1107/S1600536814009738/hg5393Isup3.cml
CCDC reference: 1000268
Additional supporting information: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| N1—H1⋯O2i | 0.86 | 2.03 | 2.8658 (18) | 164 |
| C7—H7⋯O4ii | 0.93 | 2.54 | 3.307 (2) | 139 |
| C18—H18B⋯O2iii | 0.96 | 2.56 | 3.336 (2) | 138 |
Symmetry codes: (i) 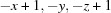 ; (ii)
; (ii) 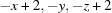 ; (iii)
; (iii) 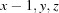 .
.
Acknowledgments
MK is grateful to the CPEPA–UGC for the award of a JRF and thanks the University of Mysore for research facilities. JPJ acknowledges the NSF–MRI program (grant No. CHE-1039027) for funds to purchase the X-ray diffractometer.
supplementary crystallographic information
1. Comment
N-Substituted 2-arylacetamides are biologically active compounds because of their structural similarity to the lateral chain of natural benzylpenicillin (Mijin et al., 2006, 2008). Amides are also used as ligands due to their excellent coordination abilities (Wu et al., 2008, 2010). In a variety of biological heterocyclic compounds, N-pyrazole derivatives are of great interest because of their chemical and pharmaceutical properties (Cheng et al., 2008). Some of the N-pyrazole derivatives have been found to exhibit good insecticidal activities (Hatton et al., 1993), antifungal activities (Liu et al., 2010) and non-linear optical properties (Chandrakantha et al., 2013). Crystal structures of some related acetamide and pyrazole derivatives including: N-(1,5-dimethyl-3-oxo-2-phenyl-2,3-dihrdro-1H-pyrazol-4-yl)-2- [4-(methylsulfanyl)phenyl]acetamide, (Fun et al., 2012), 2-(2,4-dichlorophenyl)-N-(1,5-dimethyl-3-oxo-2- phenyl)-N-(1,5-dimethyl-3-oxo-2-phenyl-2,3-dihydro-1H-pyrazol- 4-yl)acetamide (Butcher et al., 2013a,b), 2-(3,4-Dichloro phenyl)-N-(1,5-dimethyl-3-oxo-2-phenyl-2,3-dihydro-1H-pyrazol-4-yl) acteamide (Mahan et al., 2013) and recently N-(1,5-Dimethyl- 3-oxo-2-phenyl-2,3-dihydro-1H-pyrazol-4-yl)-2-phenylacetamide (Kaur et al., 2013) have been reported. In view of the importance of amide derivatives of pyrazoles, this paper reports the crystal structure of the title compound (I), C19H18N4O4.
The title compound, (I), C19H18N4O4, crystallizes with one independent molecule in the asymmetric unit (Fig. 1). In the molecule, the pyrazole ring is nearly planar with C9 atom showing a maximum deviation of 0.0042 (16)Å. The mean planes of the 4-nitrophenyl and phenyl rings is twisted with respect to that of the pyrazole ring by 67.0 (6)° and 37.4 (4)°, respectively. The dihedral angle between the mean planes of the two phenyl rings is 59.3 (3)°. The amide group, with a torsion angle of 177.54° is twisted away from the plane of the pyrazole ring in an anti-periplanar conformation. Bond lengths are in normal ranges (Allen et al., 1987). Classical N—H···O intermolecular hydrogen bonds involving the pyrazole ring and the amide group along with weak C—H···O intermolecular interactions forming R22(10) graph set motifs link the molecules into chains along [100]. Additional weak C—H···O intermolecular interactions involving the nitrophenyl rings link the molecules further along [001] also forming R22(10) graph set motifs and further extending crystal packing into a 2-D supramolecular network (Fig. 2).
2. Experimental
4-Nitrophenylacetic acid (0.181 g, 1 mmol) and 4-aminoantipyrine (0.203 g, 1 mmol), 1-ethyl-3-(3-dimethylaminopropyl)-carbodiimide hydrochloride (1.0 g, 0.01 mol) and were dissolved in dichloromethane (20 mL). The mixture was stirred in presence of triethylamine at 273 K for about 3 h (Fig. 3). The reaction completion was confirmed by thin layer chormatography. The contents were poured into 100 ml of ice-cold aqueous hydrochloric acid with stirring, which was extracted thrice with dichloromethane. The organic layer was washed with a saturated NaHCO3 solution and brine solution, dried and concentrated under reduced pressure to give the title compound, (I). Single crystals were grown from dichloromethane and and further recrystallised from methanol solution by the slow evaporation method which were subsequently used for x-ray studies.
3. Refinement
All of the H atoms were placed in their calculated positions and then refined using the riding model with Atom—H lengths of 0.93Å (CH); 0.97Å (CH2); 0.96Å (CH3) or 0.86Å (NH). Isotropic displacement parameters for these atoms were set to 1.2 (CH, CH2, NH)and 1.5 (CH3) times Ueq of the parent atom. Idealised Me refined as rotating group.
Figures
Fig. 1.

ORTEP drawing of (I) (C19H18N4O4) showing the labeling scheme of the molecule with 30% probability displacement ellipsoids.
Fig. 2.
Molecular packing for (I) viewed along the b axis. Dashed lines indicate N—H···O intermolecular hydrogen bonds and weak C—H···O intermolecular interactions forming R22(10) graph set motifs linking the molecules into chains along [100]. Additional weak C—H···O intermolecular interactions involving the nitrophenyl rings link the molecules further along [001] also forming R22(10) graph set motifs viewed with dashed lines. H atoms not involved in hydrogen bonding have been removed for clarity.
Fig. 3.

Synthesis scheme of (I).
Crystal data
| C19H18N4O4 | Z = 2 |
| Mr = 366.37 | F(000) = 384 |
| Triclinic, P1 | Dx = 1.400 Mg m−3 |
| a = 6.7023 (6) Å | Cu Kα radiation, λ = 1.54184 Å |
| b = 8.6335 (8) Å | Cell parameters from 2476 reflections |
| c = 15.8720 (13) Å | θ = 5.4–71.4° |
| α = 76.305 (7)° | µ = 0.84 mm−1 |
| β = 84.399 (7)° | T = 173 K |
| γ = 77.252 (7)° | Block, colourless |
| V = 869.33 (13) Å3 | 0.28 × 0.22 × 0.12 mm |
Data collection
| Agilent Eos Gemini diffractometer | 3262 independent reflections |
| Radiation source: Enhance (Cu) X-ray Source | 2913 reflections with I > 2σ(I) |
| Detector resolution: 16.0416 pixels mm-1 | Rint = 0.030 |
| ω scans | θmax = 71.6°, θmin = 5.4° |
| Absorption correction: multi-scan (CrysAlis PRO and CrysAlis RED; Agilent, 2012) | h = −4→8 |
| Tmin = 0.851, Tmax = 1.000 | k = −10→10 |
| 5113 measured reflections | l = −19→19 |
Refinement
| Refinement on F2 | Hydrogen site location: inferred from neighbouring sites |
| Least-squares matrix: full | H-atom parameters constrained |
| R[F2 > 2σ(F2)] = 0.046 | w = 1/[σ2(Fo2) + (0.0731P)2 + 0.1699P] where P = (Fo2 + 2Fc2)/3 |
| wR(F2) = 0.129 | (Δ/σ)max < 0.001 |
| S = 1.07 | Δρmax = 0.27 e Å−3 |
| 3262 reflections | Δρmin = −0.21 e Å−3 |
| 247 parameters | Extinction correction: SHELXL2012 (Sheldrick, 2008), Fc*=kFc[1+0.001xFc2λ3/sin(2θ)]-1/4 |
| 0 restraints | Extinction coefficient: 0.0160 (14) |
| Primary atom site location: structure-invariant direct methods |
Special details
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| O1 | 0.1021 (2) | 0.22514 (15) | 0.69436 (8) | 0.0438 (3) | |
| O2 | 0.42534 (16) | 0.19900 (14) | 0.42650 (7) | 0.0318 (3) | |
| O3 | 0.5894 (2) | 0.31428 (19) | 1.03446 (9) | 0.0505 (4) | |
| O4 | 0.8856 (2) | 0.18161 (18) | 1.00103 (10) | 0.0521 (4) | |
| N1 | 0.2446 (2) | 0.05926 (16) | 0.60349 (8) | 0.0288 (3) | |
| H1 | 0.3271 | −0.0300 | 0.5983 | 0.035* | |
| N2 | −0.10661 (19) | 0.30173 (16) | 0.44159 (8) | 0.0285 (3) | |
| N3 | 0.08379 (19) | 0.31651 (16) | 0.39847 (8) | 0.0273 (3) | |
| N4 | 0.6997 (2) | 0.22341 (17) | 0.99332 (9) | 0.0342 (3) | |
| C1 | 0.2184 (2) | 0.10192 (19) | 0.68181 (10) | 0.0295 (3) | |
| C2 | 0.3372 (3) | −0.02284 (19) | 0.75387 (10) | 0.0333 (4) | |
| H2A | 0.2456 | −0.0893 | 0.7878 | 0.040* | |
| H2B | 0.4453 | −0.0940 | 0.7277 | 0.040* | |
| C3 | 0.4314 (2) | 0.04908 (18) | 0.81419 (9) | 0.0290 (3) | |
| C4 | 0.3122 (3) | 0.1581 (2) | 0.86039 (11) | 0.0345 (4) | |
| H4 | 0.1727 | 0.1924 | 0.8516 | 0.041* | |
| C5 | 0.3993 (3) | 0.2157 (2) | 0.91904 (11) | 0.0347 (4) | |
| H5 | 0.3196 | 0.2879 | 0.9502 | 0.042* | |
| C6 | 0.6072 (2) | 0.16397 (18) | 0.93061 (9) | 0.0292 (3) | |
| C7 | 0.7300 (3) | 0.0574 (2) | 0.88551 (11) | 0.0348 (4) | |
| H7 | 0.8698 | 0.0247 | 0.8940 | 0.042* | |
| C8 | 0.6403 (3) | 0.0001 (2) | 0.82728 (10) | 0.0336 (4) | |
| H8 | 0.7208 | −0.0721 | 0.7964 | 0.040* | |
| C9 | 0.1396 (2) | 0.15810 (18) | 0.53105 (9) | 0.0273 (3) | |
| C10 | −0.0656 (2) | 0.20985 (19) | 0.52365 (10) | 0.0288 (3) | |
| C11 | 0.2410 (2) | 0.22118 (18) | 0.45045 (9) | 0.0259 (3) | |
| C12 | 0.0955 (2) | 0.35883 (18) | 0.30627 (10) | 0.0270 (3) | |
| C13 | −0.0571 (3) | 0.33649 (19) | 0.25914 (10) | 0.0321 (4) | |
| H13 | −0.1676 | 0.2941 | 0.2878 | 0.039* | |
| C14 | −0.0426 (3) | 0.3779 (2) | 0.16951 (11) | 0.0377 (4) | |
| H14 | −0.1457 | 0.3660 | 0.1378 | 0.045* | |
| C15 | 0.1246 (3) | 0.4370 (2) | 0.12657 (11) | 0.0402 (4) | |
| H15 | 0.1350 | 0.4629 | 0.0662 | 0.048* | |
| C16 | 0.2765 (3) | 0.4574 (2) | 0.17385 (11) | 0.0377 (4) | |
| H16 | 0.3896 | 0.4955 | 0.1449 | 0.045* | |
| C17 | 0.2615 (2) | 0.42151 (19) | 0.26367 (10) | 0.0319 (4) | |
| H17 | 0.3612 | 0.4391 | 0.2951 | 0.038* | |
| C18 | −0.2656 (2) | 0.4522 (2) | 0.42912 (11) | 0.0346 (4) | |
| H18A | −0.2460 | 0.5174 | 0.4677 | 0.052* | |
| H18B | −0.3983 | 0.4248 | 0.4414 | 0.052* | |
| H18C | −0.2561 | 0.5122 | 0.3702 | 0.052* | |
| C19 | −0.2353 (3) | 0.1816 (2) | 0.58962 (11) | 0.0402 (4) | |
| H19A | −0.2664 | 0.2676 | 0.6206 | 0.060* | |
| H19B | −0.1943 | 0.0792 | 0.6296 | 0.060* | |
| H19C | −0.3545 | 0.1799 | 0.5612 | 0.060* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| O1 | 0.0579 (8) | 0.0399 (7) | 0.0327 (6) | 0.0048 (6) | −0.0132 (5) | −0.0154 (5) |
| O2 | 0.0265 (6) | 0.0381 (6) | 0.0309 (6) | −0.0061 (4) | −0.0018 (4) | −0.0081 (5) |
| O3 | 0.0527 (8) | 0.0643 (9) | 0.0441 (8) | −0.0095 (7) | −0.0063 (6) | −0.0315 (7) |
| O4 | 0.0444 (8) | 0.0567 (8) | 0.0606 (9) | −0.0034 (6) | −0.0248 (6) | −0.0207 (7) |
| N1 | 0.0320 (7) | 0.0311 (7) | 0.0243 (6) | −0.0055 (5) | −0.0044 (5) | −0.0077 (5) |
| N2 | 0.0241 (6) | 0.0364 (7) | 0.0266 (7) | −0.0075 (5) | −0.0009 (5) | −0.0088 (5) |
| N3 | 0.0253 (6) | 0.0338 (7) | 0.0244 (6) | −0.0067 (5) | −0.0013 (5) | −0.0088 (5) |
| N4 | 0.0423 (8) | 0.0332 (7) | 0.0280 (7) | −0.0087 (6) | −0.0102 (6) | −0.0044 (6) |
| C1 | 0.0351 (8) | 0.0311 (8) | 0.0252 (7) | −0.0097 (6) | −0.0051 (6) | −0.0075 (6) |
| C2 | 0.0468 (9) | 0.0282 (8) | 0.0269 (8) | −0.0086 (7) | −0.0076 (7) | −0.0068 (6) |
| C3 | 0.0401 (9) | 0.0276 (7) | 0.0197 (7) | −0.0090 (6) | −0.0050 (6) | −0.0023 (6) |
| C4 | 0.0303 (8) | 0.0438 (9) | 0.0318 (8) | −0.0058 (7) | −0.0037 (6) | −0.0135 (7) |
| C5 | 0.0375 (9) | 0.0380 (9) | 0.0307 (8) | −0.0035 (7) | −0.0029 (6) | −0.0147 (7) |
| C6 | 0.0371 (8) | 0.0291 (8) | 0.0226 (7) | −0.0099 (6) | −0.0064 (6) | −0.0032 (6) |
| C7 | 0.0322 (8) | 0.0377 (9) | 0.0328 (8) | −0.0025 (6) | −0.0080 (6) | −0.0065 (7) |
| C8 | 0.0402 (9) | 0.0307 (8) | 0.0284 (8) | −0.0003 (6) | −0.0045 (6) | −0.0088 (6) |
| C9 | 0.0318 (8) | 0.0304 (8) | 0.0237 (7) | −0.0098 (6) | −0.0041 (6) | −0.0095 (6) |
| C10 | 0.0322 (8) | 0.0337 (8) | 0.0252 (7) | −0.0118 (6) | −0.0017 (6) | −0.0107 (6) |
| C11 | 0.0284 (7) | 0.0275 (7) | 0.0254 (7) | −0.0068 (6) | −0.0041 (6) | −0.0106 (6) |
| C12 | 0.0324 (8) | 0.0251 (7) | 0.0242 (7) | −0.0042 (6) | −0.0034 (6) | −0.0078 (5) |
| C13 | 0.0356 (8) | 0.0319 (8) | 0.0310 (8) | −0.0077 (6) | −0.0064 (6) | −0.0085 (6) |
| C14 | 0.0478 (10) | 0.0357 (9) | 0.0313 (9) | −0.0048 (7) | −0.0133 (7) | −0.0098 (7) |
| C15 | 0.0567 (11) | 0.0367 (9) | 0.0234 (8) | −0.0037 (8) | −0.0041 (7) | −0.0041 (6) |
| C16 | 0.0453 (10) | 0.0329 (8) | 0.0311 (9) | −0.0073 (7) | 0.0037 (7) | −0.0023 (7) |
| C17 | 0.0345 (8) | 0.0309 (8) | 0.0306 (8) | −0.0071 (6) | −0.0029 (6) | −0.0064 (6) |
| C18 | 0.0310 (8) | 0.0354 (8) | 0.0390 (9) | −0.0041 (6) | −0.0019 (6) | −0.0137 (7) |
| C19 | 0.0346 (9) | 0.0571 (11) | 0.0315 (9) | −0.0166 (8) | 0.0025 (7) | −0.0098 (8) |
Geometric parameters (Å, º)
| O1—C1 | 1.217 (2) | C7—H7 | 0.9300 |
| O2—C11 | 1.2436 (19) | C7—C8 | 1.384 (2) |
| O3—N4 | 1.2168 (19) | C8—H8 | 0.9300 |
| O4—N4 | 1.2278 (19) | C9—C10 | 1.357 (2) |
| N1—H1 | 0.8600 | C9—C11 | 1.435 (2) |
| N1—C1 | 1.3630 (19) | C10—C19 | 1.489 (2) |
| N1—C9 | 1.405 (2) | C12—C13 | 1.396 (2) |
| N2—N3 | 1.4047 (17) | C12—C17 | 1.390 (2) |
| N2—C10 | 1.373 (2) | C13—H13 | 0.9300 |
| N2—C18 | 1.473 (2) | C13—C14 | 1.382 (2) |
| N3—C11 | 1.3905 (19) | C14—H14 | 0.9300 |
| N3—C12 | 1.4206 (19) | C14—C15 | 1.386 (3) |
| N4—C6 | 1.463 (2) | C15—H15 | 0.9300 |
| C1—C2 | 1.523 (2) | C15—C16 | 1.386 (3) |
| C2—H2A | 0.9700 | C16—H16 | 0.9300 |
| C2—H2B | 0.9700 | C16—C17 | 1.384 (2) |
| C2—C3 | 1.508 (2) | C17—H17 | 0.9300 |
| C3—C4 | 1.394 (2) | C18—H18A | 0.9600 |
| C3—C8 | 1.391 (2) | C18—H18B | 0.9600 |
| C4—H4 | 0.9300 | C18—H18C | 0.9600 |
| C4—C5 | 1.381 (2) | C19—H19A | 0.9600 |
| C5—H5 | 0.9300 | C19—H19B | 0.9600 |
| C5—C6 | 1.382 (2) | C19—H19C | 0.9600 |
| C6—C7 | 1.378 (2) | ||
| C1—N1—H1 | 119.3 | N1—C9—C11 | 123.26 (13) |
| C1—N1—C9 | 121.36 (13) | C10—C9—N1 | 127.98 (14) |
| C9—N1—H1 | 119.3 | C10—C9—C11 | 108.77 (13) |
| N3—N2—C18 | 115.56 (12) | N2—C10—C19 | 120.59 (14) |
| C10—N2—N3 | 106.45 (12) | C9—C10—N2 | 109.94 (13) |
| C10—N2—C18 | 120.24 (13) | C9—C10—C19 | 129.46 (15) |
| N2—N3—C12 | 118.84 (12) | O2—C11—N3 | 124.28 (14) |
| C11—N3—N2 | 109.85 (12) | O2—C11—C9 | 131.03 (14) |
| C11—N3—C12 | 125.33 (12) | N3—C11—C9 | 104.68 (13) |
| O3—N4—O4 | 122.92 (14) | C13—C12—N3 | 120.30 (14) |
| O3—N4—C6 | 118.50 (14) | C17—C12—N3 | 119.20 (14) |
| O4—N4—C6 | 118.56 (14) | C17—C12—C13 | 120.49 (15) |
| O1—C1—N1 | 123.02 (14) | C12—C13—H13 | 120.3 |
| O1—C1—C2 | 122.67 (14) | C14—C13—C12 | 119.43 (16) |
| N1—C1—C2 | 114.22 (13) | C14—C13—H13 | 120.3 |
| C1—C2—H2A | 108.6 | C13—C14—H14 | 119.8 |
| C1—C2—H2B | 108.6 | C13—C14—C15 | 120.40 (16) |
| H2A—C2—H2B | 107.6 | C15—C14—H14 | 119.8 |
| C3—C2—C1 | 114.64 (13) | C14—C15—H15 | 120.1 |
| C3—C2—H2A | 108.6 | C14—C15—C16 | 119.79 (15) |
| C3—C2—H2B | 108.6 | C16—C15—H15 | 120.1 |
| C4—C3—C2 | 121.41 (15) | C15—C16—H16 | 119.7 |
| C8—C3—C2 | 119.46 (14) | C17—C16—C15 | 120.67 (16) |
| C8—C3—C4 | 119.05 (14) | C17—C16—H16 | 119.7 |
| C3—C4—H4 | 119.7 | C12—C17—H17 | 120.4 |
| C5—C4—C3 | 120.64 (15) | C16—C17—C12 | 119.16 (15) |
| C5—C4—H4 | 119.7 | C16—C17—H17 | 120.4 |
| C4—C5—H5 | 120.6 | N2—C18—H18A | 109.5 |
| C4—C5—C6 | 118.71 (15) | N2—C18—H18B | 109.5 |
| C6—C5—H5 | 120.6 | N2—C18—H18C | 109.5 |
| C5—C6—N4 | 118.90 (14) | H18A—C18—H18B | 109.5 |
| C7—C6—N4 | 118.88 (14) | H18A—C18—H18C | 109.5 |
| C7—C6—C5 | 122.21 (14) | H18B—C18—H18C | 109.5 |
| C6—C7—H7 | 120.8 | C10—C19—H19A | 109.5 |
| C6—C7—C8 | 118.36 (15) | C10—C19—H19B | 109.5 |
| C8—C7—H7 | 120.8 | C10—C19—H19C | 109.5 |
| C3—C8—H8 | 119.5 | H19A—C19—H19B | 109.5 |
| C7—C8—C3 | 121.01 (15) | H19A—C19—H19C | 109.5 |
| C7—C8—H8 | 119.5 | H19B—C19—H19C | 109.5 |
| O1—C1—C2—C3 | −43.2 (2) | C4—C5—C6—N4 | −179.40 (14) |
| O3—N4—C6—C5 | 1.4 (2) | C4—C5—C6—C7 | 0.1 (3) |
| O3—N4—C6—C7 | −178.19 (15) | C5—C6—C7—C8 | −0.5 (2) |
| O4—N4—C6—C5 | −177.43 (15) | C6—C7—C8—C3 | 0.2 (2) |
| O4—N4—C6—C7 | 3.0 (2) | C8—C3—C4—C5 | −0.7 (2) |
| N1—C1—C2—C3 | 140.27 (14) | C9—N1—C1—O1 | 1.0 (2) |
| N1—C9—C10—N2 | −178.78 (14) | C9—N1—C1—C2 | 177.54 (13) |
| N1—C9—C10—C19 | 2.0 (3) | C10—N2—N3—C11 | 5.88 (16) |
| N1—C9—C11—O2 | 3.9 (2) | C10—N2—N3—C12 | 160.12 (13) |
| N1—C9—C11—N3 | −177.63 (13) | C10—C9—C11—O2 | −176.07 (15) |
| N2—N3—C11—O2 | 173.55 (13) | C10—C9—C11—N3 | 2.41 (16) |
| N2—N3—C11—C9 | −5.07 (15) | C11—N3—C12—C13 | 130.08 (16) |
| N2—N3—C12—C13 | −19.9 (2) | C11—N3—C12—C17 | −49.4 (2) |
| N2—N3—C12—C17 | 160.63 (13) | C11—C9—C10—N2 | 1.18 (17) |
| N3—N2—C10—C9 | −4.27 (16) | C11—C9—C10—C19 | −178.02 (16) |
| N3—N2—C10—C19 | 175.01 (13) | C12—N3—C11—O2 | 21.4 (2) |
| N3—C12—C13—C14 | −179.79 (14) | C12—N3—C11—C9 | −157.26 (14) |
| N3—C12—C17—C16 | 177.76 (14) | C12—C13—C14—C15 | 1.7 (2) |
| N4—C6—C7—C8 | 179.08 (14) | C13—C12—C17—C16 | −1.8 (2) |
| C1—N1—C9—C10 | −56.9 (2) | C13—C14—C15—C16 | −1.1 (3) |
| C1—N1—C9—C11 | 123.16 (16) | C14—C15—C16—C17 | −0.9 (3) |
| C1—C2—C3—C4 | 57.6 (2) | C15—C16—C17—C12 | 2.4 (2) |
| C1—C2—C3—C8 | −125.41 (16) | C17—C12—C13—C14 | −0.3 (2) |
| C2—C3—C4—C5 | 176.33 (15) | C18—N2—N3—C11 | 142.19 (13) |
| C2—C3—C8—C7 | −176.71 (15) | C18—N2—N3—C12 | −63.56 (17) |
| C3—C4—C5—C6 | 0.4 (3) | C18—N2—C10—C9 | −138.12 (14) |
| C4—C3—C8—C7 | 0.3 (2) | C18—N2—C10—C19 | 41.2 (2) |
Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| N1—H1···O2i | 0.86 | 2.03 | 2.8658 (18) | 164 |
| C7—H7···O4ii | 0.93 | 2.54 | 3.307 (2) | 139 |
| C18—H18B···O2iii | 0.96 | 2.56 | 3.336 (2) | 138 |
Symmetry codes: (i) −x+1, −y, −z+1; (ii) −x+2, −y, −z+2; (iii) x−1, y, z.
Footnotes
Supporting information for this paper is available from the IUCr electronic archives (Reference: HG5393).
References
- Agilent (2012). CrysAlis PRO and CrysAlis RED Agilent Technologies, Yarnton, England.
- Allen, F. H., Kennard, O., Watson, D. G., Brammer, L., Orpen, A. G. & Taylor, R. (1987). J. Chem. Soc. Perkin Trans. 2, pp. S1–19.
- Butcher, R. J., Mahan, A., Nayak, P. S., Narayana, B. & Yathirajan, H. S. (2013a). Acta Cryst. E69, o46–o47. [DOI] [PMC free article] [PubMed]
- Butcher, R. J., Mahan, A., Nayak, P. S., Narayana, B. & Yathirajan, H. S. (2013b). Acta Cryst. E69, o39. [DOI] [PMC free article] [PubMed]
- Chandrakantha, B., Isloor, A. M., Sridharan, K., Philip, R., Shetty, P. & Padaki, M. (2013). Arab. J Chem. 6, 97–102.
- Cheng, J. L., Wei, F. L., Zhu, L., Zhao, J. H. & Zhu, G. N. (2008). Chin. J. Org. Chem. 28, 622–627.
- Dolomanov, O. V., Bourhis, L. J., Gildea, R. J., Howard, J. A. K. & Puschmann, H. (2009). J. Appl. Cryst. 42, 339–341.
- Fun, H.-K., Quah, C. K., Nayak, P. S., Narayana, B. & Sarojini, B. K. (2012). Acta Cryst. E68, o2677. [DOI] [PMC free article] [PubMed]
- Hatton, L. R., Buntain, I. G., Hawkins, D. W., Parnell, E. W. & Pearson, C. J. (1993). US Patent 5232940.
- Kaur, M., Jasinski, J. P., Anderson, B. J., Yathirajan, H. S. & Narayana, B. (2013). Acta Cryst. E69, o1726–o1727. [DOI] [PMC free article] [PubMed]
- Liu, Y. Y., Shi, H., Li, Y. F. & Zhu, H. J. (2010). J. Heterocycl. Chem. 47, 897–902.
- Mahan, A., Butcher, R. J., Nayak, P. S., Narayana, B. & Yathirajan, H. S. (2013). Acta Cryst. E69, o402–o403. [DOI] [PMC free article] [PubMed]
- Mijin, D. & Marinkovic, A. (2006). Synth. Commun. 36, 193–198.
- Mijin, D. Z., Prascevic, M. & Petrovic, S. D. (2008). J. Serb. Chem. Soc. 73, 945–950.
- Palatinus, L. & Chapuis, G. (2007). J. Appl. Cryst. 40, 786–790.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Wu, W.-N., Cheng, F.-X., Yan, L. & Tang, N. (2008). J. Coord. Chem. 61, 2207–2215.
- Wu, W.-N., Wang, Y., Zhang, A.-Y., Zhao, R.-Q. & Wang, Q.-F. (2010). Acta Cryst. E66, m288. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) I. DOI: 10.1107/S1600536814009738/hg5393sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536814009738/hg5393Isup2.hkl
Supporting information file. DOI: 10.1107/S1600536814009738/hg5393Isup3.cml
CCDC reference: 1000268
Additional supporting information: crystallographic information; 3D view; checkCIF report



