Abstract
The title compound, C9H8N2O3, was prepared by reaction of phenol, glyoxylic acid and urea in water. The imidazolidine ring adopts an almost planar conformation (r.m.s. deviation = 0.012 Å) and is twisted by 89.3 (1)° relative to the benzene ring. In the crystal, molecules are linked by N—H⋯O and O—H⋯O hydrogen bonds into a three-dimensional framework.
Related literature
For general background to the synthesis and applications of hydantoin derivatives, see: Liu & Zhao (2001 ▶); Dhar et al. (2002 ▶); Goodnow & Kang (2003 ▶). For related compounds, see: Ji et al. (2002 ▶).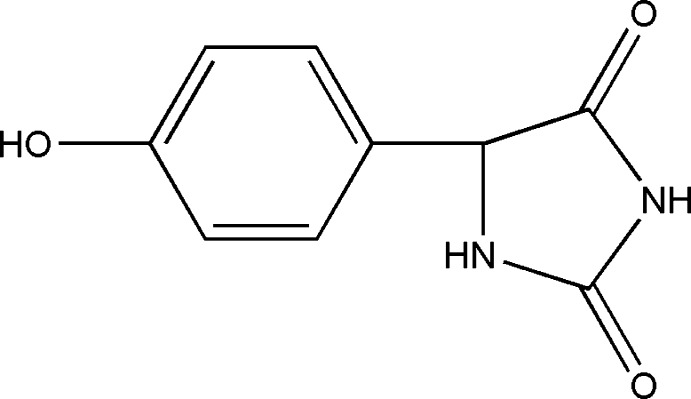
Experimental
Crystal data
C9H8N2O3
M r = 192.17
Monoclinic,
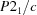
a = 10.3694 (11) Å
b = 6.9914 (8) Å
c = 12.3857 (13) Å
β = 105.619 (2)°
V = 864.76 (16) Å3
Z = 4
Mo Kα radiation
μ = 0.11 mm−1
T = 296 K
0.30 × 0.20 × 0.20 mm
Data collection
Bruker SMART CCD area-detector diffractometer
4721 measured reflections
1558 independent reflections
1100 reflections with I > 2σ(I)
R int = 0.035
Refinement
R[F 2 > 2σ(F 2)] = 0.035
wR(F 2) = 0.091
S = 1.02
1558 reflections
137 parameters
H atoms treated by a mixture of independent and constrained refinement
Δρmax = 0.15 e Å−3
Δρmin = −0.14 e Å−3
Data collection: SMART (Bruker, 1997 ▶); cell refinement: SAINT (Bruker, 1997 ▶); data reduction: SAINT; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL2013 (Sheldrick, 2008 ▶); molecular graphics: SHELXTL (Sheldrick, 2008 ▶); software used to prepare material for publication: SHELXTL.
Supplementary Material
Crystal structure: contains datablock(s) I. DOI: 10.1107/S1600536814010034/kq2013sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536814010034/kq2013Isup2.hkl
Supporting information file. DOI: 10.1107/S1600536814010034/kq2013Isup3.cml
CCDC reference: 1000728
Additional supporting information: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| N2—H2⋯O4i | 0.883 (18) | 1.952 (19) | 2.8180 (19) | 166.7 (17) |
| N1—H1⋯O4ii | 0.85 (2) | 2.535 (19) | 3.204 (2) | 136.5 (16) |
| N1—H1⋯O6iii | 0.85 (2) | 2.36 (2) | 3.067 (2) | 141.1 (17) |
| O6—H6⋯O5iv | 0.95 (2) | 1.78 (2) | 2.7223 (18) | 169.0 (18) |
Symmetry codes: (i) 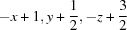 ; (ii)
; (ii) 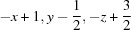 ; (iii)
; (iii) 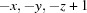 ; (iv)
; (iv) 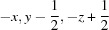 .
.
Acknowledgments
The authors thank the Colleges and Universities Technology Project of Shanxi Provine (20121033), and the Natural Science Fund of Lvliang University (contracts ZRXN201206 and ZRXN201210).
supplementary crystallographic information
1. Comment
Hydantoin derivatives can be used as intermediates in pharmaceutical products, pesticides and photosensitive material. It is very important to the development of hydantoins compounds. Pharmacological functions of hydantoin derivatives are mainly shown in antibacterial (Liu & Zhao, 2001), diminishing inflammation (Dhar et al., 2002), relieving cough and asthma, lowering blood sugar (Goodnow & Kang, 2003), and inhibiting agent of uremic toxin. Different substituted hydantoin and its derivatives show good application future, such as the treatment of diabetes, kidney disease, autoimmune disease and blood disease. The spectrum of hydantoin derivatives is broad as bacterial disinfectant. They are widely used in aquaculture, pest and disease control, disinfection treatment of health equipment, mildew prevention and control of crops, preservation of vegetable & Fruit, and mildew anti-corrosion of industrial products and living goods.
In the molecule of the title compound, C9H8N2O3, I (Fig. 1) bond lengths and angles are generally normal (Ji et al., 2002). The imidazolidine ring adopts a planar conformation (r.m.s. deviation is 0.012 Å) and is twisted by 89.3 (1)° relative to the benzene plane.
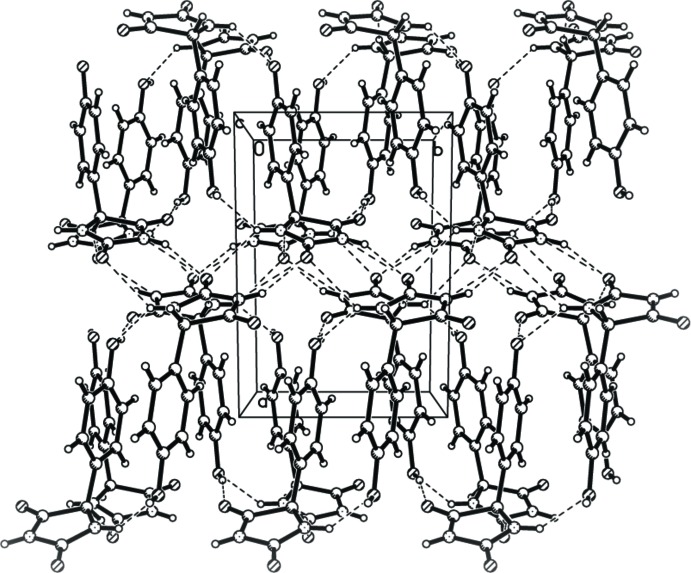
In the crystal, molecules are bound by intermolecular N—H···O and O—H···O hydrogen bonds (Table 1) into three-dimensional framework (Fig. 2).
2. Experimental
The title compound was prepared by reaction of phenol (0.05 mol), glyoxylic acid (0.06 mol) and urea (0.06 mol) in hydrochloric acid (37%, 80 ml) at 370 K for 6 h, cooling, filtering, affording the tile compound by recrystallization in water. Single crystals of the title compound suitable for X-ray measurements was obtained by recrystallization from ethanol at room temperature.
3. Refinement
The hydroxyl and amino hydrogen atoms were objectively localized in the difference-Fourier map and refined isotropically with fixed displacement parameters. The other hydrogen atoms were placed in the calculated positions with C—H distances = 0.93–0.98 Å and refined in the riding model with fixed isotropic displacement parameters: Uiso(H) = 1.2Ueq(C).
Figures
Fig. 1.
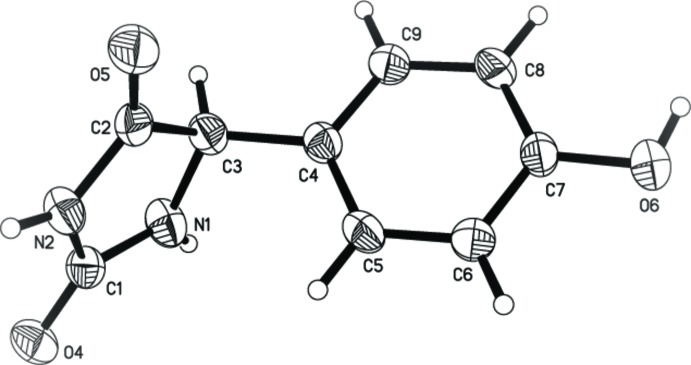
Molecular structure of I. Displacement ellipsoids are presented at the 40% probability level. H atoms are depicted as small spheres of arbitrary radius.
Fig. 2.
A portion of the crystal structure of I viewed along [001]. The intermolecular hydrogen bonding interactions are depicted by dashed lines.
Crystal data
| C9H8N2O3 | F(000) = 400 |
| Mr = 192.17 | Dx = 1.476 Mg m−3 |
| Monoclinic, P21/c | Mo Kα radiation, λ = 0.71073 Å |
| a = 10.3694 (11) Å | Cell parameters from 933 reflections |
| b = 6.9914 (8) Å | θ = 2.0–25.0° |
| c = 12.3857 (13) Å | µ = 0.11 mm−1 |
| β = 105.619 (2)° | T = 296 K |
| V = 864.76 (16) Å3 | Rectangle, colourless |
| Z = 4 | 0.30 × 0.20 × 0.20 mm |
Data collection
| Bruker SMART CCD area-detector diffractometer | 1100 reflections with I > 2σ(I) |
| Radiation source: fine-focus sealed tube | Rint = 0.035 |
| Graphite monochromator | θmax = 25.2°, θmin = 2.0° |
| phi and ω scans | h = −12→12 |
| 4721 measured reflections | k = −8→8 |
| 1558 independent reflections | l = −7→14 |
Refinement
| Refinement on F2 | Secondary atom site location: difference Fourier map |
| Least-squares matrix: full | Hydrogen site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.035 | H atoms treated by a mixture of independent and constrained refinement |
| wR(F2) = 0.091 | w = 1/[σ2(Fo2) + (0.0426P)2 + 0.0206P] where P = (Fo2 + 2Fc2)/3 |
| S = 1.02 | (Δ/σ)max < 0.001 |
| 1558 reflections | Δρmax = 0.15 e Å−3 |
| 137 parameters | Δρmin = −0.14 e Å−3 |
| 0 restraints | Extinction correction: SHELXL2013 (Sheldrick, 2008), Fc*=kFc[1+0.001xFc2λ3/sin(2θ)]-1/4 |
| Primary atom site location: structure-invariant direct methods | Extinction coefficient: 0.024 (3) |
Special details
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| N1 | 0.36582 (16) | 0.1852 (2) | 0.60788 (13) | 0.0438 (4) | |
| H1 | 0.3651 (18) | 0.070 (3) | 0.6298 (15) | 0.053* | |
| N2 | 0.39496 (14) | 0.4952 (2) | 0.61795 (12) | 0.0394 (4) | |
| H2 | 0.4272 (18) | 0.605 (3) | 0.6494 (15) | 0.047* | |
| O4 | 0.46179 (12) | 0.32043 (18) | 0.78108 (10) | 0.0464 (4) | |
| O5 | 0.32237 (12) | 0.58433 (19) | 0.43383 (10) | 0.0508 (4) | |
| O6 | −0.23743 (12) | 0.1408 (2) | 0.29172 (11) | 0.0512 (4) | |
| H6 | −0.2595 (19) | 0.108 (3) | 0.2145 (17) | 0.061* | |
| C1 | 0.41256 (16) | 0.3268 (3) | 0.67946 (15) | 0.0360 (4) | |
| C2 | 0.34366 (16) | 0.4631 (3) | 0.50733 (15) | 0.0362 (4) | |
| C3 | 0.31532 (17) | 0.2501 (2) | 0.49270 (14) | 0.0385 (5) | |
| H3 | 0.3691 | 0.1943 | 0.4466 | 0.046* | |
| C4 | 0.16868 (17) | 0.2098 (2) | 0.43983 (14) | 0.0353 (4) | |
| C5 | 0.07338 (17) | 0.2567 (3) | 0.49525 (15) | 0.0410 (5) | |
| H5 | 0.1009 | 0.3060 | 0.5676 | 0.049* | |
| C6 | −0.06081 (18) | 0.2319 (3) | 0.44558 (15) | 0.0417 (5) | |
| H6A | −0.1233 | 0.2633 | 0.4842 | 0.050* | |
| C7 | −0.10245 (17) | 0.1601 (2) | 0.33802 (14) | 0.0366 (4) | |
| C8 | −0.00956 (17) | 0.1091 (3) | 0.28186 (14) | 0.0413 (5) | |
| H8 | −0.0374 | 0.0583 | 0.2099 | 0.050* | |
| C9 | 0.12569 (18) | 0.1341 (3) | 0.33336 (14) | 0.0414 (5) | |
| H9 | 0.1883 | 0.0992 | 0.2955 | 0.050* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| N1 | 0.0489 (10) | 0.0338 (9) | 0.0412 (10) | −0.0001 (8) | −0.0011 (7) | 0.0047 (7) |
| N2 | 0.0423 (9) | 0.0360 (9) | 0.0336 (9) | −0.0046 (7) | −0.0009 (7) | 0.0005 (7) |
| O4 | 0.0472 (8) | 0.0544 (9) | 0.0312 (8) | 0.0075 (6) | −0.0002 (6) | 0.0051 (6) |
| O5 | 0.0534 (9) | 0.0547 (9) | 0.0394 (8) | −0.0061 (7) | 0.0038 (6) | 0.0111 (7) |
| O6 | 0.0360 (8) | 0.0701 (10) | 0.0427 (8) | −0.0081 (6) | 0.0023 (6) | −0.0054 (7) |
| C1 | 0.0277 (9) | 0.0427 (11) | 0.0348 (11) | 0.0036 (8) | 0.0035 (8) | 0.0016 (8) |
| C2 | 0.0281 (9) | 0.0437 (11) | 0.0340 (11) | −0.0025 (8) | 0.0036 (8) | 0.0027 (9) |
| C3 | 0.0357 (10) | 0.0431 (11) | 0.0342 (10) | 0.0011 (8) | 0.0054 (8) | −0.0028 (8) |
| C4 | 0.0374 (10) | 0.0326 (10) | 0.0338 (10) | −0.0025 (8) | 0.0063 (8) | −0.0021 (8) |
| C5 | 0.0433 (12) | 0.0468 (11) | 0.0310 (10) | −0.0035 (9) | 0.0068 (8) | −0.0100 (8) |
| C6 | 0.0394 (11) | 0.0483 (12) | 0.0385 (11) | −0.0017 (9) | 0.0126 (8) | −0.0076 (9) |
| C7 | 0.0330 (10) | 0.0377 (11) | 0.0359 (10) | −0.0036 (8) | 0.0040 (8) | 0.0007 (8) |
| C8 | 0.0460 (11) | 0.0463 (11) | 0.0297 (10) | −0.0078 (9) | 0.0071 (8) | −0.0090 (8) |
| C9 | 0.0399 (11) | 0.0489 (12) | 0.0368 (11) | −0.0028 (9) | 0.0124 (8) | −0.0078 (9) |
Geometric parameters (Å, º)
| N1—C1 | 1.330 (2) | C3—H3 | 0.9800 |
| N1—C3 | 1.454 (2) | C4—C9 | 1.379 (2) |
| N1—H1 | 0.85 (2) | C4—C5 | 1.386 (3) |
| N2—C2 | 1.348 (2) | C5—C6 | 1.373 (2) |
| N2—C1 | 1.387 (2) | C5—H5 | 0.9300 |
| N2—H2 | 0.883 (18) | C6—C7 | 1.380 (2) |
| O4—C1 | 1.2251 (18) | C6—H6A | 0.9300 |
| O5—C2 | 1.220 (2) | C7—C8 | 1.378 (3) |
| O6—C7 | 1.3690 (19) | C8—C9 | 1.387 (2) |
| O6—H6 | 0.95 (2) | C8—H8 | 0.9300 |
| C2—C3 | 1.519 (2) | C9—H9 | 0.9300 |
| C3—C4 | 1.511 (2) | ||
| C1—N1—C3 | 113.08 (15) | C9—C4—C5 | 118.33 (16) |
| C1—N1—H1 | 121.6 (13) | C9—C4—C3 | 120.95 (17) |
| C3—N1—H1 | 125.3 (13) | C5—C4—C3 | 120.65 (16) |
| C2—N2—C1 | 111.98 (15) | C6—C5—C4 | 121.34 (17) |
| C2—N2—H2 | 126.3 (12) | C6—C5—H5 | 119.3 |
| C1—N2—H2 | 121.0 (12) | C4—C5—H5 | 119.3 |
| C7—O6—H6 | 112.8 (12) | C5—C6—C7 | 119.65 (17) |
| O4—C1—N1 | 129.33 (17) | C5—C6—H6A | 120.2 |
| O4—C1—N2 | 123.50 (17) | C7—C6—H6A | 120.2 |
| N1—C1—N2 | 107.17 (15) | O6—C7—C8 | 122.53 (16) |
| O5—C2—N2 | 125.82 (17) | O6—C7—C6 | 117.33 (16) |
| O5—C2—C3 | 127.01 (16) | C8—C7—C6 | 120.13 (15) |
| N2—C2—C3 | 107.16 (15) | C7—C8—C9 | 119.54 (16) |
| N1—C3—C4 | 114.94 (15) | C7—C8—H8 | 120.2 |
| N1—C3—C2 | 100.48 (13) | C9—C8—H8 | 120.2 |
| C4—C3—C2 | 111.97 (14) | C4—C9—C8 | 120.98 (17) |
| N1—C3—H3 | 109.7 | C4—C9—H9 | 119.5 |
| C4—C3—H3 | 109.7 | C8—C9—H9 | 119.5 |
| C2—C3—H3 | 109.7 | ||
| C3—N1—C1—O4 | −179.77 (17) | C2—C3—C4—C9 | 112.19 (19) |
| C3—N1—C1—N2 | 0.8 (2) | N1—C3—C4—C5 | 49.1 (2) |
| C2—N2—C1—O4 | 177.58 (16) | C2—C3—C4—C5 | −64.7 (2) |
| C2—N2—C1—N1 | −3.0 (2) | C9—C4—C5—C6 | −1.0 (3) |
| C1—N2—C2—O5 | −177.36 (17) | C3—C4—C5—C6 | 175.96 (17) |
| C1—N2—C2—C3 | 3.78 (19) | C4—C5—C6—C7 | −0.5 (3) |
| C1—N1—C3—C4 | −119.10 (17) | C5—C6—C7—O6 | −179.04 (16) |
| C1—N1—C3—C2 | 1.27 (19) | C5—C6—C7—C8 | 1.6 (3) |
| O5—C2—C3—N1 | 178.20 (17) | O6—C7—C8—C9 | 179.43 (16) |
| N2—C2—C3—N1 | −2.96 (17) | C6—C7—C8—C9 | −1.3 (3) |
| O5—C2—C3—C4 | −59.3 (2) | C5—C4—C9—C8 | 1.4 (3) |
| N2—C2—C3—C4 | 119.51 (16) | C3—C4—C9—C8 | −175.60 (17) |
| N1—C3—C4—C9 | −133.99 (17) | C7—C8—C9—C4 | −0.2 (3) |
Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| N2—H2···O4i | 0.883 (18) | 1.952 (19) | 2.8180 (19) | 166.7 (17) |
| N1—H1···O4ii | 0.85 (2) | 2.535 (19) | 3.204 (2) | 136.5 (16) |
| N1—H1···O6iii | 0.85 (2) | 2.36 (2) | 3.067 (2) | 141.1 (17) |
| O6—H6···O5iv | 0.95 (2) | 1.78 (2) | 2.7223 (18) | 169.0 (18) |
Symmetry codes: (i) −x+1, y+1/2, −z+3/2; (ii) −x+1, y−1/2, −z+3/2; (iii) −x, −y, −z+1; (iv) −x, y−1/2, −z+1/2.
Footnotes
Supporting information for this paper is available from the IUCr electronic archives (Reference: KQ2013).
References
- Bruker (1997). SMART and SAINT Bruker AXS Inc., Madison, Wisconsin, USA.
- Dhar, T. G., Iwanowicz, E., Launay, M., Maillet, M., Nicolai, E. & Potin, D. (2002). Hydantoin compounds useful as anti-inflammatory agents US Patent US2002/0143035A1[P].
- Goodnow, J. R. & Kang, L. (2003). Hydantoin-containing glucokinase activators US Patent US2003/0225286A1[P].
- Ji, B. M., Du, C. X., Zhu, Y. & Wang, Y. (2002). Chin. J. Struct. Chem. 21, 252–255.
- Liu, J. & Zhao, Y. (2001). Chin. J. Disinfect. 18, 218–222.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) I. DOI: 10.1107/S1600536814010034/kq2013sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536814010034/kq2013Isup2.hkl
Supporting information file. DOI: 10.1107/S1600536814010034/kq2013Isup3.cml
CCDC reference: 1000728
Additional supporting information: crystallographic information; 3D view; checkCIF report