Abstract
Leishmaniasis is a significant worldwide health problem for which no vaccine exists. Activation of CD4+ and CD8+ T cells is crucial for the generation of protective immunity against parasite. Recent trend in vaccine design has been shifted to epitope-based vaccines that are more specific, safe, and easy to produce. In the present study, four known antigenic Leishmania infantum proteins, cysteine peptidase A (CPA), histone H1, KMP-11, and Leishmania eukaryotic initiation factor (LeIF) were analyzed for the prediction of binding epitopes to H2d MHC class I and II molecules, using online available algorithms. Based on in silico analysis, eight peptides including highly scored MHC class I- and II-restricted epitopes were synthesized. Peptide immunogenicity was validated in MHC compatible BALB/c mice immunized with each synthetic peptide emulsified in complete Freund’s adjuvant/incomplete Freund’s adjuvant. CPA_p2, CPA_p3, H1_p1, and LeIF_p6 induced strong spleen cell proliferation upon in vitro peptide re-stimulation. In addition, the majority of the peptides, except of LeIF_p1 and KMP-11_p1, induced IFN-γ secretion, while KMP-11_p1 indicated a suppressive effect on IL-10 production. CPA_p2, CPA_p3, LeIF_p3, and LeIF_p6 induced IFN-γ-producing CD4+ T cells indicating a TH1-type response. In addition, CPA_p2, CPA_p3, and H1_p1 induced also the induction of CD8+ T cells. The induction of peptide-specific IgG in immunized mice designated also the existence of B cell epitopes in peptide sequences. Combining immunoinformatic tools and experimental validation, we demonstrated that CPA_p2, CPA_p3, H1_p1, H1_p3, CPA_p2, LeIF_p3, and LeIF_p6 are likely to include potential epitopes for the induction of protective cytotoxic and/or TH1-type immune responses supporting the feasibility of peptide-based vaccine development for leishmaniasis.
Keywords: in silico analysis, cysteine peptidase A, histone H1, kinetoplastid membrane protein 11, Leishmania eukaryotic initiation factor, lymphocyte proliferation, CD8+IFN-γ+ T cells, CD4+IFN-γ+ T cells
Introduction
Leishmaniasis, a vector-borne parasitic disease, is caused by dimorphic protozoan flagellates of the genus Leishmania with a worldwide distribution. The disease is characterized by diversity and complexity, presenting a wide spectrum of clinical forms in humans ranging from self-healing cutaneous leishmaniasis (CL) to fatal visceral leishmaniasis (VL). In VL, parasites colonize internal organs, primarily the spleen, liver, and bone marrow. With an estimated 0.5 million cases per year, VL has emerged as an important public-health concern with major clinical and socioeconomic impacts1. In South Europe, VL is caused almost exclusively by Leishmania (L.) infantum, which is transmitted as a zoonosis with the domestic dog serving as the main reservoir of the parasite (1). Current attempts against leishmaniasis are based on chemotherapy to alleviate disease (2, 3) and on vector control to reduce transmission (4). Toxic side effects and growing resistance to available therapeutic drugs against VL has made the global demand for an effective vaccine capable to elicit a protective immune response, a major public-health priority.
Despite the substantial knowledge regarding the various life stages of the parasite, the considerable inter-specific diversity, the extraordinary host evasive mechanisms of parasite and the heterogeneity of population (5, 6), effective vaccine development against human VL represents an unprecedented challenge. Although a great number of potent vaccine candidates has shown promising results in mice (7, 8) and dogs (9–11), none of them has entered human trials except for LeishF1 with reported phase I and II clinical trials (12, 13).
Antigen identification is considered as a significant barrier in vaccine design, as this is usually achieved through time consuming and labor-intensive in vitro and in vivo experiments. Efforts have thus focused on developing novel strategies for more rational and faster antigen identification among large numbers of pathogen proteins. Furthermore, recent reports support that epitope-based vaccines appear to be capable of inducing more potent responses than whole protein vaccines (14). Until recently, the search of immunodominant peptides relied on the direct testing of overlapping peptides or peptide libraries. T cell epitope prediction by bioinformatic analysis of protein sequences has been proposed as a promising strategy for vaccine development and an increasing number of tools have been developed, based on different algorithms and methods (15, 16). There is great possibility of missing the emergence of the sequence mutants that would potentially escape the vaccine’s protective effect. Moreover, the fact that T cells from genetically distinct populations would recognize and respond to a single peptide epitope, underline the need of identifying one or more epitope(s) that bind to multiple HLA alleles and cover close to 100% of the genetically diverse human population (17). Multi-peptide-based vaccines are designed to generate a diverse immune response to incorporate antigens and to reduce limitations due to MHC restriction into a single entity.
The effectiveness of a vaccine depends on its capacity to ensure long-lasting cell-mediated immunity. In VL, there is evidence that an interplay of T helper cytokines (TH1/TH2) is observed, while resistance or resolution of infection is associated with dominant TH1 response and CD8+ T cells (18–20). Furthermore, successful treatment of VL with sodium stibogluconate requires the presence of both CD4+ and CD8+ T cells (21) accompanied with IL-12 and IFN-γ production (22). In contrast, TH2 response with IL-4 and IL-10 production results in susceptibility to infection and development of severe disease. Murine models of leishmaniasis have been extensively used to study the pathogenesis of the disease and to test novel therapeutic agents or potent vaccine candidates in pre-clinical studies. One of the most widely studied and commonly used model of VL is the BALB/c strain of mice infected intravenously with L. infantum. Although this strain is considered to be susceptible and the infection progresses during the first month, the infection is then controlled by the host immune response. This mouse model is comparable to self-controlled oligosymptomatic cases and therefore is useful for the study of the protective immune response (23).
Several reports demonstrate that different leishmanial antigens elicit desired TH1 and CTL responses capable to sustain protection against experimental challenges (24). Among these antigens, cysteine peptidase A (CPA), histone H1, kinetoplastid membrane protein 11 (KMP-11), and Leishmania eukaryotic initiation factor (LeIF) are considered important immunogens, as supported by numerous studies. Specifically, CPA induces protection against L. major in the experimental model of CL through development of specific TH1 immune responses (25–27). Histone H1 and KMP-11, structural highly conserved proteins, are able to trigger specific immune responses (28–33), and immunization with these proteins confer protection against L. major or L. infantum infections in experimental animal models (34–38). Moreover, LeIF, originally described as a TH1-type natural adjuvant, is capable of stimulating IL-12 mediated TH1 responses in PBMCs of patients (39). Furthermore, recombinant forms of CPA, histone H1, and KMP-11 act as potent B cell immunogens since they are recognized by sera from either recovered or active cases of CL and VL, as well as by sera from asymptomatic or symptomatic dogs with leishmaniasis (40–45).
In the present study, we applied immunoinformatics using currently available online algorithms in order to identify potentially immunogenic T cell epitopes from the above mentioned L. infantum proteins, and design multi-epitope peptides containing both MHC class I and II-restricted epitopes as possible candidate peptide vaccines for VL. Immunogenicity of the synthetic multi-epitope peptides in terms of T cell activation was validated in immunized BALB/c mice by analyzing peptide-specific proliferative responses and cytokine production by CD8+ and CD4+ T cells.
Materials and Methods
Protein sequence retrieval and prediction of MHC class I and II binding epitopes
Full protein sequences of selected proteins, CPA, Histone H1, KMP-11, and LeIF, were retrieved from GenBank data2 on JPCM5 strain (MCAN/ES/98/LLm-887) and analyzed by SignalP3 for the prediction of signal peptides and transmembrane domains (Table 1). Potential MHC class I and II binding epitopes derived from the four L. infantum proteins, were predicted by in silico analysis, using three online available, binding algorithms named SYFPEITHI4, BIMAS5, and NetMHCII6. The cut-off score was adjusted to ≥18 for SYFPEITHI, ≥100 for BIMAS, and a default prediction threshold (binding affinity <500 nM) depicting accuracy >85% was used for NetMHCII (Tables 2 and 3).
Table 1.
L. infantum proteins selected as candidate antigens for epitope mapping.
Table 2.
In silico predicted MHC class I-restricted 9-mer epitopes of L. infantum proteins.
| Protein | No. of epitope | Epitope sequence | Scorea |
||||
|---|---|---|---|---|---|---|---|
| SYFPEITHI |
BIMAS |
||||||
| H2-Kd | H2-Ld | H2-Kd | H2-Ld | H2-Dd | |||
| CPA | 1 | 8-FFAIVVTIL-18 | 23 | – | 1152 | – | – |
| 2 | 84-HYDVSGKFA-92 | 23 | – | – | – | – | |
| 3 | 273-LYFGGVVTL-281 | 23 | – | 2400 | – | – | |
| 4 | 254-AYVGKNGPV-262 | 22 | – | 1200 | – | – | |
| 5 | 23-SALIAQTPL-31 | 20 | – | – | – | – | |
| 6 | 102-LYLNPNYYA-110 | 20 | – | 120 | – | – | |
| 7 | 334-NYVVTATID-343 | 20 | – | – | – | – | |
| 8 | 7-FFFAIVVTI-15 | – | – | 1152 | – | – | |
| 9 | 165-QWALKNHSL-173 | – | – | 240 | – | – | |
| 10 | 62-RFNAFKQNM-70 | – | – | 144 | – | – | |
| 11 | 319-GYIRLAMGS-327 | – | – | 120 | – | – | |
| 12 | 179-QVLVSCDNI-187 | – | – | 115.20 | – | – | |
| 13 | 68-QNMQTAYFL-76 | – | – | 115.20 | – | – | |
| 14 | 160-GNIEGQWAL-168 | – | – | 115.20 | – | – | |
| Histone H1 | 15 | 46-KKAGAKKAV-54 | 18 | – | – | – | – |
| 16 | 2-SSDSAVAAL-10 | 17 | 19 | – | – | – | |
| KMP-11 | 17 | 4-TYEEFSAKL-12 | 20 | – | 2400 | – | – |
| 18 | 47-HYEKFERMI-55 | 19 | – | 2000 | – | – | |
| 19 | 72-HFKQFAEL-79 | 18 | – | 960 | – | – | |
| 20 | 69-HSEHFKQKF-77 | – | 18 | – | – | – | |
| 21 | 7-EFSAKLDRL-15 | – | – | 960 | – | – | |
| LeIF | 22 | 385-HYHTQIDEL-393 | 24 | – | 2000 | – | – |
| 23 | 360-RYGRKGVAI-368 | 23 | – | 2000 | – | – | |
| 24 | 8-APQDQDSFL-16 | – | 22 | – | 225 | – | |
| 25 | 25-IPSFDDMPL-33 | – | 22 | – | 150 | – | |
| 26 | 199-LPKDIQVAL-207 | – | 22 | – | – | ||
| 27 | 344-LPTNKENYL-352 | – | 22 | – | 150 | – | |
| 28 | 393-LPVDFAAYL-401 | – | 22 | – | 150 | – | |
| 29 | 14-SFLDDQPGV-22 | 21 | – | 576 | – | – | |
| 30 | 197-RFLPKDIQV-205 | 21 | – | 480 | – | – | |
| 31 | 100-LSPTRELAL-108 | – | 21 | – | – | ||
| 32 | 187-GFADQIYEI-195 | 20 | – | 960 | – | – | |
| 33 | 318-SRVLVTTDL-326 | 20 | – | – | – | – | |
| 34 | 23-RPIPSFDDM-31 | – | 20 | – | 150 | – | |
| 35 | 123-NSSKFCETF-131 | – | 20 | – | – | – | |
| 36 | 265-VSIAQSVIF-273 | – | 20 | – | – | – | |
| 37 | 222-KFMRDPVRI-230 | – | – | 2304 | – | – | |
| 38 | 195-IFRFLPKDI-203 | – | – | 960 | – | – | |
| 39 | 272-IFANTRRKV-280 | – | – | 288 | – | – | |
| 40 | 312-TFRSGSSRV-320 | – | – | 288 | – | – | |
| 41 | 164-RGALRTESL-172 | – | – | – | – | 120 | |
aThe cut-off score was adjusted to ≥18 for SYFPEITHI, ≥100 for BIMAS, and <500 for NetMHCII.
Table 3.
In silico predicted MHC class II-restricted 15-mer epitopes of L. infantum proteins.
| Protein | No. of epitope | Epitope sequence | Scorea |
||
|---|---|---|---|---|---|
| SYFPEITHI |
NetMHCII |
||||
| H2-IAd | H2-IEd | H2-IAd | |||
| CPA | 1 | 149-MCGSCWAFATTGNIE-163 | 28 | – | – |
| 2 | 246-PHDEEEIAAYVGKNG-260 | 27 | – | – | |
| 3 | 114-KDYKEHVHVDDSVRS-128 | 26 | – | – | |
| 4 | 257-GKNGPVAVAVDATTW-271 | 26 | – | – | |
| 5 | 32-GVDDFIASAHYGRFK-46 | 25 | – | – | |
| 6 | 312-GSSWGEKGYIRLAMG-326 | – | 24 | – | |
| 7 | 4-RNPFFFAIVVTILFV-18 | 23 | – | – | |
| 8 | 5-NPFFFAIVVTILFVV-19 | 23 | – | – | |
| 9 | 13-VTILFVVCYGSALIA-27 | 22 | – | – | |
| 10 | 172-SLVSLSEQVLVSCDN-186 | 22 | – | – | |
| 11 | 174-VSLSEQVLVSCDNID-188 | 22 | – | – | |
| 12 | 12-VVTILFVVCYGSALI-26 | 21 | – | – | |
| 13 | 260-GPVAVAVDATTWQLY-274 | 21 | – | – | |
| 14 | 279-VTLCFGLSLNHGVLV-293 | 21 | – | – | |
| 15 | 67-KQNMQTAYFLNAHNP-81 | 20 | – | – | |
| 16 | 273-LYFGGVVTLCFGLSL-287 | 20 | – | – | |
| 17 | 301-KPPYWIVKNSWGSSW-315 | 20 | – | – | |
| 18 | 328-NQCLLKNYVVTATID-342 | 20 | – | – | |
| 19 | 216-SYPYTSAGGTRPPCH-230 | – | 20 | – | |
| 20 | 308-KNSWGSSWGEKGYIR-322 | – | 20 | – | |
| Histone H1 | 21 | 1-MSSDSAVAALSAAMT-15 | 31 | – | 88.3 |
| 22 | 27-KTAAKKAAAKKAAAK-41 | 29 | – | 182.6 | |
| 23 | 32-KAAAKKAAAKKAGAK-46 | 29 | – | 239.9 | |
| 24 | 37-KAAAKKAGAKKAGAK-51 | 29 | – | – | |
| 25 | 42-KAGAKKAGAKKAVRK-56 | 29 | – | – | |
| 26 | 2-SSDSAVAALSAAMTS-16 | 28 | – | 123.7 | |
| 27 | 56-KVATPKKPAKKAAKK-70 | 24 | – | – | |
| 28 | 36-KKAAAKKAGAKKAGA-50 | – | 24 | – | |
| 29 | 41-KKAGAKKAGAKKAVR-55 | – | 24 | – | |
| 30 | 71-AAKKPAKKVAKKPAK-85 | – | 24 | – | |
| 31 | 16-SPQKSPRSSPKKTAA-30 | – | 22 | – | |
| 32 | 21-PRSSPKKTAAKKAAA-35 | – | 22 | – | |
| 33 | 26-KKTAAKKAAAKKAAA-40 | – | 22 | 163.5 | |
| 34 | 31-KKAAAKKAAAKKAGA-45 | – | 22 | 190.3 | |
| 35 | 45-AKKAGAKKAVRKVAT-59 | – | 22 | – | |
| 36 | 51-KKAVRKVATPKKPAK-65 | – | 22 | – | |
| 37 | 55-RKVATPKKPAKKAAK-69 | – | 22 | – | |
| 38 | 59-TPKKPAKKAAKKAAK-73 | – | 22 | – | |
| 39 | 63-PAKKAAKKAAKKPAK-77 | – | 22 | – | |
| 40 | 67-AAKKAAKKPAKKVAK-81 | – | 22 | – | |
| 41 | 75-PAKKVAKKPAKKAAK-89 | – | 22 | – | |
| 42 | 79-VAKKPAKKAAKKPAK-93 | – | 22 | – | |
| 43 | 83-PAKKAAKKPAKKPAK-97 | – | 22 | – | |
| 44 | 87-AAKKPAKKAAKKAAK-101 | – | 22 | – | |
| 45 | 91-PAKKPAKKAAKKAAK-105 | – | 22 | – | |
| 46 | 95-PAKKAAKKAAKKAAA-109 | – | 22 | – | |
| 47 | 5-SAVAALSAAMTSPQK-19 | 21 | – | 382.8 | |
| 48 | 76-AKKVAKKPAKKAAKK-90 | 21 | 20 | – | |
| 49 | 96-AKKAAKKAAKKAAAK-110 | 21 | – | – | |
| 50 | 29-AAKKAAAKKAAAKKA-43 | – | – | 107.9 | |
| 51 | 30-AKKAAAKKAAAKKAG-44 | – | – | 119.1 | |
| 52 | 25-PKKTAAKKAAAKKAA-39 | – | – | 119.6 | |
| 53 | 24-SPKKTAAKKAAAKKA-38 | – | – | 138.5 | |
| 54 | 3-SDSAVAALSAAMTSP-17 | – | – | 166.2 | |
| 55 | 28-TAAKKAAAKKAAAKK-42 | – | – | 183.3 | |
| 56 | 35-AKKAAAKKAGAKKAG-49 | – | – | 208.6 | |
| 57 | 34-AAKKAAAKKAGAKKA-48 | – | – | 232.0 | |
| 58 | 4-DSAVAALSAAMTSPQ-18 | – | – | 272.1 | |
| 59 | 48-AGAKKAVRKVATPKK-62 | – | – | 295.2 | |
| 60 | 33-AAAKKAAAKKAGAKK-47 | – | – | 317.8 | |
| 61 | 49-GAKKAVRKVATPKKP-63 | – | – | 457.6 | |
| KMP-11 | 62 | 4-TYEEFSAKLDRLDEE-18 | 23 | – | – |
| 63 | 75-QKFAELLEQQKAAQN-89 | 19 | – | – | |
| 64 | 45-KEHYEKFERMIKEHT-59 | – | 18 | – | |
| 65 | 74-KQKFAELLEQQKAAQ-88 | – | 18 | – | |
| LeIF | 66 | 100-LSPTRELALQTAEVI-114 | 28 | – | 264.2 |
| 67 | 320-VLVTTDLVARGICVH-334 | 28 | – | – | |
| 68 | 199-LPKDIQVALFSATMP-213 | 27 | – | – | |
| 69 | 387-HTQIDELPVDFAAYL-401 | 27 | – | – | |
| 70 | 138-QDDLRKLQAGVIVAV-152 | 26 | – | 205.3 | |
| 71 | 166-ALRTESLRVLVLDEA-180 | 26 | – | – | |
| 72 | 169-TESLRVLVLDEADEM-183 | 26 | – | – | |
| 73 | 62-RGGDIIAQAQSGTGK-76 | 25 | – | – | |
| 74 | 140-DLRKLQAGVIVAVGT-154 | 24 | – | 256.8 | |
| 75 | 142-RKLQAGVIVAVGTPG-156 | 24 | – | – | |
| 76 | 259-MDLYETVSIAQSVIF-273 | 24 | – | – | |
| 77 | 223-FMRDPVRILVKRESL-237 | – | 24 | – | |
| 78 | 325-DLVARGIDVHHVNIV-339 | 23 | – | – | |
| 79 | 168-RTESLRVLVLDEADE-182 | 22 | – | – | |
| 80 | 263-ETVSIAQSVIFANTR-277 | 22 | – | – | |
| 81 | 293-TVSSMHAEMPKSDRE-307 | 22 | – | – | |
| 82 | 314-RSGSSRVLVTTDLVA-328 | 22 | – | – | |
| 83 | 268-AQSVIFANTRRKVDW-282 | – | 22 | – | |
| 84 | 16-LDDQPGVRPIPSFDD-30 | 20 | – | – | |
| 85 | 71-QSGTGKTGAFSIGLL-85 | 20 | – | – | |
| 86 | 107-ALQTAEVISRIGEFL-121 | 20 | – | – | |
| 87 | 174-VLVLDEADEMLSQGF-188 | 20 | – | – | |
| 88 | 255-LDTLMDLYETVSIAQ-269 | 20 | – | – | |
| 89 | 376-VELLHEIEAHYHTQI-390 | 20 | – | – | |
| 90 | 77-TGAFSIGLLQRLDFR-91 | – | 20 | – | |
| 91 | 81-SIGLLQRLDFRHNLI-95 | – | 20 | – | |
| 92 | 158-VSDVIKRGALRTESL-172 | – | 20 | – | |
| 93 | 102-PTRELALQTAEVISR-116 | – | – | 95.3 | |
| 94 | 103-TRELALQTAEVISRI-117 | – | – | 95.4 | |
| 95 | 101-SPTRELALQTAEVIS-115 | – | – | 163.8 | |
| 96 | 139-DDLRKLQAGVIVAVG-153 | – | – | 249.0 | |
| 97 | 99-VLSPTRELALQTAEV-113 | – | – | 263.1 | |
| 98 | 98-LVLSPTRELALQTAE-112 | – | – | 287.6 | |
| 99 | 141-LRKLQAGVIVAVGTP-155 | – | – | 296.7 | |
| 100 | 49-PSSIQQRAIAPFTRG-63 | – | – | 320.9 | |
| 101 | 48-KPSSIQQRAIAPFTR-62 | – | – | 378.0 | |
| 102 | 50-SSIQQRAIAPFTRGG-64 | – | – | 434.6 | |
| 103 | 97-GLVLSPTRELALQTA-111 | – | – | 452.0 | |
| 104 | 290-SNHTVSSMHAEMPKS-304 | – | – | 489.1 | |
aThe cut-off score was adjusted to ≥18 for SYFPEITHI, ≥100 for BIMAS, and <500 for NetMHCII.
Synthetic multi-epitope peptides
Based on the prediction results of the algorithms used, 9-mer epitopes MHC class I-restricted and 15-mer epitopes MHC class II-restricted giving high score against H2d alleles were extracted and combined in order to generate multi-epitope peptides for each L. infantum protein. Thus, 8 peptides, 20–30 amino acid (aa) length, were designed in a way that each peptide included at least one MHC class I-restricted epitope scored very high, as well as adjacent or overlapping MHC class II-restricted epitopes scored also high. Sequence homology between each multi-epitope peptide and mouse proteome were analyzed on BLAST database7 and peptides with 100% identity were excluded or re-designed to avoid potential autoimmunity (Table 4). Two multi-epitope peptides of CPA (160–189 and 273–302 aa) and Histone H1 (1–20 and 43–61 aa), one peptide of KMP-11 (4–23 aa), and three peptides of LeIF (6–35, 181–210, and 371–400 aa) were synthesized by GeneCust (Labbx, Dudelange, Luxembourg) with purity ≥95%. Synthetic peptides were dissolved in DMSO, acetic acid (10% in dH2O), or dH2O according to their hydrophobicity, by vigorous pipetting and stored in aliquots, in −80°C until use. Peptides solutions were found endotoxin free, since LPS concentration was <5 EU/mg as determined by LAL Test Cartridges Portable Test System (Endosafe, Charles River Laboratories, USA). Synthetic multi-epitope peptides were also checked for the presence of 9-mer or/and 15-mer epitopes able to bind to HLA alleles (A2, A3, A24, B7, B15, DP, DQ, DR supertypes) using the above mentioned algorithms (Table 4). In addition, data on the crystal structure of HLA-A2 and HLA-DRB1 molecules were obtained from Protein Data Bank (PDB, codes 1HHG and 2SEB, respectively) and multi-epitope peptides of length 30 aa were transformed into PDB files using SWISS-MODEL, and each of them was docked with HLA-A2 or HLA-DRB1 molecule using the ClusPro program8 for structure-based analysis (46–49).
Table 4.
Synthetic multi-epitope peptides including MHC class I and II-restricted epitopes.
| Peptide name | Synthetic multi-epitope peptide sequence | Included epitopes | HLA supertype |
|---|---|---|---|
| CPA_p2 | 160-GNIEGQWALKNHSLVSLSEQVLVSCDNIDD-189 | 165-QWALKNHSL-173 | HLA-A2 (A*0201), HLA-A3 (A*03), HLA-DRB1, HLA-DPA1, HLA-DQA1 |
| 179-QVLVSCDNI-187 | |||
| 160-GNIEGQWAL-168 | |||
| 172-SLVSLSEQVLVSCDN-186 | |||
| 174-VSLSEQVLVSCDNID-188 | |||
| CPA_p3 | 273-LYFGGVVTLCFGLSLNHGVLVVGFNRQAKP-302 | 273-LYFGGVVTL-281 | HLA-A2 (A*0201), HLA-A3 (A*03), HLA-A24 (A*2402), HLA-DRB1, HLA-DPA1, HLA-DQA1 |
| 279-VTLCFGLSLNHGVLV-293 | |||
| 273-LYFGGVVTLCFGLSL-287 | |||
| H1_p1 | 1-MSSDSAVAALSAAMTSPQKS-20 | 2-SSDSAVAAL-10 | HLA-A2 (A*0201), HLA-A3 (A*03), HLA-DRB1, HLA-DQA1 |
| 1-MSSDSAVAALSAAMT-15 | |||
| 2-SSDSAVAALSAAMTS-16 | |||
| 5-SAVAALSAAMTSPQK-19 | |||
| H1_p3 | 43-AGAKKAGAKKAVRKVATPKK-61 | 46-KKAGAKKAV-54 | HLA-A2 (A*0201), HLA-A3 (A*03), HLA-DRB1 |
| 42-KAGAKKAGAKKAVRK-56 | |||
| 45-AKKAGAKKAVRKVAT-59 | |||
| KMP-11_p1 | 4-TYEEFSAKLDRLDEEFNRKM-23 | 4-TYEEFSAKL-12 | HLA-A3 (A*03), HLA-A24, HLA-DRB1, HLA-DPA1, HLA-DQA1 |
| 7-EFSAKLDRL-15 | |||
| 4-TYEEFSAKLDRLDEE-18 | |||
| LeIF_p1 | 6-KIAPQDQDSFLDDQPGVRPIPSFDDMPLHQ-35 | 8-APQDQDSFL-16 | HLA-B7 (B*5101), HLA-B15 (B62), HLA-DRB1, HLA-DPA1, HLA-DQA1 |
| 25-IPSFDDMPL-33 | |||
| 14-SFLDDQPGV-22 | |||
| 23-RPIPSFDDM-31 | |||
| 16-LDDQPGVRPIPSFDD-30 | |||
| LeIF_p3 | 181-DEMLSQGFADQIYEIFRFLPKDIQVALFSA-210 | 199-LPKDIQVAL-207 | HLA-B7 (B*3501, B*5101), HLA-DRB1, HLA-DPA1, HLA-DQA1 |
| 197-RFLPKDIQV-205 | |||
| 187-GFADQIYEI-195 | |||
| 195-IFRFLPKDI-203 | |||
| 199-LPKDIQVALFSATMP-213 | |||
| LeIF_p6 | 371-VTEKDVELLHEIEAHYHTQIDELPVDFAAY-400 | 385-HYHTQIDEL-393 | HLA-A3 (A*0301), HLA-A24, HLA-DRB1, HLA-DPA1, HLA-DQA1 |
| 393-LPVDFAAYL-401 | |||
| 320-VLVTTDLVARGICVH-334 | |||
| 387-HTQIDELPVDFAAYL-401 | |||
| 376-VELLHEIEAHYHTQI-390 |
Immunization of BALB/c mice
Eight groups of female BALB/c mice (n = 8/group), 6–8 weeks old, were immunized subcutaneously at upper and lower dorsal region, with 100 μl emulsion consisting of 50 μg of each synthetic multi-epitope peptide in complete Freund’s adjuvant (CFA). Mice were also received a second immunization with 100 μl emulsion of 50 μg of the same peptide in incomplete Freund’s adjuvant (IFA), as well as a third immunization with 50 μg of peptide alone in PBS at 2 weeks intervals. Two sex and age matched groups of mice (n = 8/group) immunized similarly either with the adjuvant or with PBS alone, were served as control groups.
Animals were obtained from the breeding unit of the Hellenic Pasteur Institute (Athens, Greece) and reared in institutional facilities under specific pathogen-free conditions, receiving a diet of commercial food pellets and water ad libitum. All experimental procedures had been approved by the institutional Animal Bioethics Committee regulating according to the EU Directive 2010/63 and the National Law 2013/56.
Culture of lymphocytes and proliferation assays
Fifteen days post the third immunization, spleens from immunized and control mice (n = 3/group) were collected in aseptic conditions and used for the preparation of single cell suspensions in RPMI-1640 medium (Biochrom AG, Berlin, Germany) supplemented with 2 mM l-glutamine, 10 mM Hepes, 24 mM NaHCO3, 0.05 mM β-mercaptoethanol, 100 U/ml penicillin, 100 μg/ml streptomycin, and 10% (v/v) heat inactivated fetal bovine serum (FBS; Gibco, Paisley, UK) at a density of 1 × 106 cells/ml. Cell viability was >95% as determined by trypan blue exclusion. A volume of 200 μl/well were placed in triplicate into 96-well U-bottomed plates in the presence of various concentrations of each synthetic multi-epitope peptide ranging from 5 to 40 μg/ml and incubated for 96 h in 5% CO2 at 37°C in a humidified atmosphere. The optimal concentration for each peptide was determined at 10 μg/ml and used thereafter for recall stimulation. Cells cultured in medium alone or in the presence of Concanavalin A (6 μg/ml) were served as negative or positive control, respectively. Cells were pulsed with 1 μCi/ml of 3H-TdR (GE Healthcare, Buckinghamshire, UK) for the final 18 h of the culture period. Cells were harvested and 3H-TdR incorporation was determined on a microplate scintillation counter (Microbeta Trilux, Wallac, Turku, Finland). The results were expressed as Δcpm (cpm of cells from immunized mice stimulated with peptide – cpm of immunized mice cultured in medium alone). Proliferative response against to each synthetic peptide giving Δcpm > 2000 was considered as positive.
Cytokine detection and flow cytometry
Spleen cells from immunized and control mice (n = 5/group) were also used for cytokine detection and flow cytometry. Briefly, 1 ml/well of lymphocytes in complete RPMI-1640 medium at a density of 2 × 106 cells/ml were placed in triplicate into 24-well plates and stimulated with 10 μg/ml of each synthetic multi-epitope peptide. Cells were cultured for 72 h in 5% CO2 at 37°C in a humidified atmosphere. At the end of the incubation period, culture supernatants were collected and stored at −80°C until analyzed for their cytokine content. The concentrations of IFN-γ and IL-10 in the supernatants were determined by sandwich ELISA kits (900-K98, 900-K53; PeproTech, Rocky Hill, NJ, USA) according to the manufacturer’s instructions. The cytokine concentrations were calculated by reference to standard curves; detection threshold for IFN-γ and IL-10 was 23 and 47 pg/ml, respectively.
In parallel, at 48 h of culture period, similarly cultured cells were exposed for 4 h to 2.5 μg/ml brefeldin A (Fluka, Buchs, Germany), washed in FACS buffer (PBS-2% FBS) and stained with anti-CD4 and anti-CD8 monoclonal antibodies (mAbs) conjugated either with FITC (anti-CD4-FITC, clone RM4-5) or PE (anti-CD4-PE, clone H129.19; anti-CD8-PE, clone 53-6.7) for 30 min. For the identification of intracellular cytokine production, cells were permeabilized using FACS buffer supplemented with 0.1% (v/v) saponin (Sigma) and stained for 30 min on ice with anti-IFN-γ conjugated with FITC (clone XMG1.2) or anti-IL-4 conjugated with PE (clone BVD4-1D11) mAbs. In all cases, control cells were processed similarly using matched isotype control. All mAbs used in the study, were purchased from BD Biosciences (Erembodegem, Belgium). For each sample, 20,000 cells were analyzed on a FACSCalibur (Becton-Dickinson, San Jose, CA, USA) and the data were processed with Cell Quest Software (Becton-Dickinson). The percentage of specific cytokine-producing CD4+ or CD8+ T cells relative to total numbers of CD4+ or CD8+ T cells was determined by analysis of FACS data using the FlowJO software package (Tree Star, Inc., Ashland, OR, USA). The percentage of peptide-specific cytokine-producing cells was normalized to their respective proportion in unstimulated cells from mice immunized with CFA/IFA alone, in order to allow for comparison among all the synthetic multi-epitope peptides.
Enzyme linked immunosorbent assays
Blood collected from each group of mice (n = 8/group) at fifteenth day post the third immunization, were centrifuged at 4000 × g for 5 min and separated sera were aliquoted for the detection of specific antibodies against each synthetic multi-epitope peptide by specific ELISAs as previously described (50). In brief, 96-well microtiter plates were coated with 5 μg/ml of each individual peptide in carbonate buffer (15 mM Na2CO3, 35 mM NaHCO3), pH 9.6 and left overnight at 4°C. For the detection of total IgG antibodies, 10-fold dilutions of each serum sample in 1% BSA in PBS-T were added and incubated with HRP-labeled goat anti-mouse IgG (1/1000 dilution; GE Healthcare, Buckinghamshire, UK). For the detection of IgG1 and IgG2a isotypes, serum samples (1/100 dilution) were added and plates were similarly incubated either with biotin-labeled rat anti-mouse IgG1 (500 ng/ml; AbD Serotec, Oxford, UK) or IgG2a (250 ng/ml; AbD Serotec) followed by the addition of streptavidin–HRP (1/5000 dilution; AbD Serotec) and incubation for 1 h at 37°C. The cut-off value was determined as the mean OD value of normal mouse serum in a 1/100 dilution + 2SD.
Statistical analysis
Data were expressed as the mean value with the standard deviation (SD) indicated. Statistical significant differences of the mean values between groups of mice immunized with synthetic multi-epitope peptide emulsified in CFA/IFA and mice immunized with CFA/IFA alone were assessed by unpaired Student’s t-test. The probability (p) of <0.05 was considered to indicate statistical significance.
Results
In silico prediction of promising epitopes of L. infantum proteins bind to MHC class I and II molecules
CPA, Histone H1, KMP-11, and LeIF have already been defined as candidate antigens. CPA, a protein significantly up-regulated in mature amastigotes (40), is predicted as a secretory protein by SignalP (cleavage site between position 24 and 25 residue), while Histone H1, an also highly expressed protein in mature amastigotes (51), KMP-11, a cytoskeleton-associated protein, and LeIF constitutively expressed in both promastigotes and amastigotes (45, 52), are predicted as non-secretory. In silico analysis of proteins for the prediction of binding epitopes to H2d MHC class I and II molecules revealed, in total, 41 9-mer and 104 15-mer peptides, respectively, which scored above the cut-off value of each algorithm used for the prediction (Tables 2 and 3).
In particular, 14 and 20 highly scored binding peptides to H2-Kd and H2-IAd/IEd alleles, respectively, were predicted by BIMAS and SYFPEITHI for CPA. These peptides were spanning throughout the protein amino-acid sequence and they covered the 69.2% of its length. In addition, 2 and 41 binding peptides to H2-Kd/Ld and H2-IAd/IEd alleles, respectively, were predicted by SYFPEITHI and NetMHCII for Histone H1. The peptides were also spanning throughout Histone H1 amino-acid sequence, covering the entire length of the protein. Five and four binding peptides to H2-Kd/Ld and H2-IAd/IEd alleles, respectively, were predicted by BIMAS and SYFPEITHI for KMP-11. Peptides were gathered in the middle, as well as in the amino- and carboxy-terminal region of KMP-11 sequence, and covered the 55.4% of its length. In regards to LeIF, 20 and 39 binding peptides to H2-Kd/Ld/Dd and H2-IAd/IEd alleles, respectively, were predicted by BIMAS, SYFPEITHI, and NetMHCII. Peptides were spanning throughout LeIF amino-acid sequence and covered the 79.4% of the entire protein.
Synthetic multi-epitope peptides containing both MHC class I- and II-restricted epitopes
Based on the above data, eight peptides, 20–30 amino-acid length, were designed and synthesized. At least, one MHC class I-restricted epitope scored very high, as well as adjacent or overlapping MHC class II-restricted epitopes scored also high were nested in each synthetic peptide (Table 4). These multi-epitope peptides included CPA_p2 (160-GNIEGQWALKNHSLVSLSEQVLVSCDNIDD-189) and CPA_p3 (273-LYFGGVVTLCFGLSLNHGVLVVGFNRQAKP-302) from CPA, H1_p1 (1-MSSDSAVAALSAAMTSPQKS-20) and H1_p3 (43-AGAKKAGAKKAVRKVATPKK-61) from Histone H1, KMP-11_p1 (4-TYEEFSAKLDRLDEEFNRKM-23) from KMP-11, LeIF_p1 (6-KIAPQDQDSFLDDQPGVRPIPSFDDMPLHQ-35), LeIF_p3 (181-DEMLSQGFADQIYEIFRFL PKDIQVALFSA-210), and LeIF_p6 (371-VTEKDVELLHEIEAHYHTQIDELPVDFAAY-400) from LeIF. Synthetic peptide sequences showed to be retrieved from highly conserved regions of L. infantum proteins, since protein BLAST analysis revealed up to 95% residue identity to homologous sequences of corresponding proteins of strains belonging to L. major and L. donovani complexes. In addition, promiscuous 9-mer and 15-mer epitopes bound to HLA alleles (A2, A3, A24, B7, B15, DP, DQ, DR supertypes) were also nested in synthetic multi-epitope peptides as predicted by in silico analysis using the above mentioned algorithms.
Validation of synthetic multi-epitope peptides immunogenicity in mice
Immunogenicity of the eight synthetic multi-epitope peptides was validated in BALB/c mice (H2d haplotype) immunized with each synthetic peptide in combination with CFA/IFA, 15 days post third immunization. Specific proliferative T cell responses induced by synthetic multi-epitope peptides were firstly assessed. As shown in Figure 1A, CPA_p2, CPA_p3, H1_p1, and LeIF_p6 induced strong proliferation of spleen cells upon in vitro re-stimulation (Δcpm > 1128 ± 165) at the optimal dose of 10 μg/ml. Of these, CPA_p2 induced the strongest proliferation, followed by LeIF_p6, CPA_p3, and H1_p1. The results indicated that four of the eight candidate peptides could effectively induce spleen cell proliferation.
Figure 1.
Multi-epitope peptide-specific proliferative responses and cytokine secretion. (A) Proliferative responses. Spleen cells from BALB/c mice (n = 3/group) immunized either with individual peptide emulsified in CFA/IFA or PBS alone, were re-stimulated in vitro with the respective peptide (10 μg/ml) for 72 h. Cultures were pulsed for the final 18 h with 1 μCi of [3H]-TdR and results are depicted as Δcpm ± SD as described in Section “Materials and Methods.” Spleen cells derived from mice immunized with PBS alone, stimulated in vitro with ConA (Δcpm: 39743 ± 843) were used for comparison purposes. (B) IFN-γ and (C) IL-10 secretion. Cytokines were detected in culture supernatants of spleen cells from immunized BALB/c mice (n = 5/group), re-stimulated in vitro with the respective peptide (10 μg/ml) for 72 h, by ELISA. The results are expressed as pg/ml ± SD. Significant differences between groups of mice immunized with each synthetic peptide emulsified in CFA/IFA and the group of mice immunized with CFA/IFA alone are indicated by * (P < 0.05).
To validate the profile of cytokines secreted in response to the eight synthetic multi-epitope peptides, spleen cell culture supernatants from immunized mice were analyzed for their content in IFN-γ and IL-10 at 72 h post respective peptide in vitro re-stimulation. Quantitation by ELISA revealed that all peptides, except from LeIF_p1 and KMP-11_p1, induced the secretion of high amounts of IFN-γ in comparison to mice immunized with CFA/IFA alone (Figure 1B). CPA_p3, H1_p1, CPA_p2, and H1_p3 were able to induce the highest secretion of IFN-γ, followed by LeIF_p3 and LeIF_p6. In contrast, unstimulated spleen cells from immunized mice produced low levels of IFN-γ spontaneously, similar to those measured in the culture supernatants of spleen cells from mice immunized with CFA/IFA alone. The results suggest that the majority of the candidate peptides could induce IFN-γ secretion.
In addition, low levels of IL-10 were detected in the supernatants of spleen cells stimulated in vitro with each synthetic multi-epitope peptide (Figure 1C). These levels were comparable to those detected in the supernatants of unstimulated spleen cells, as well as in the supernatants of spleen cells from mice immunized with CFA/IFA alone. In particular, KMP-11_p1 indicated a rather suppressive effect on IL-10 production.
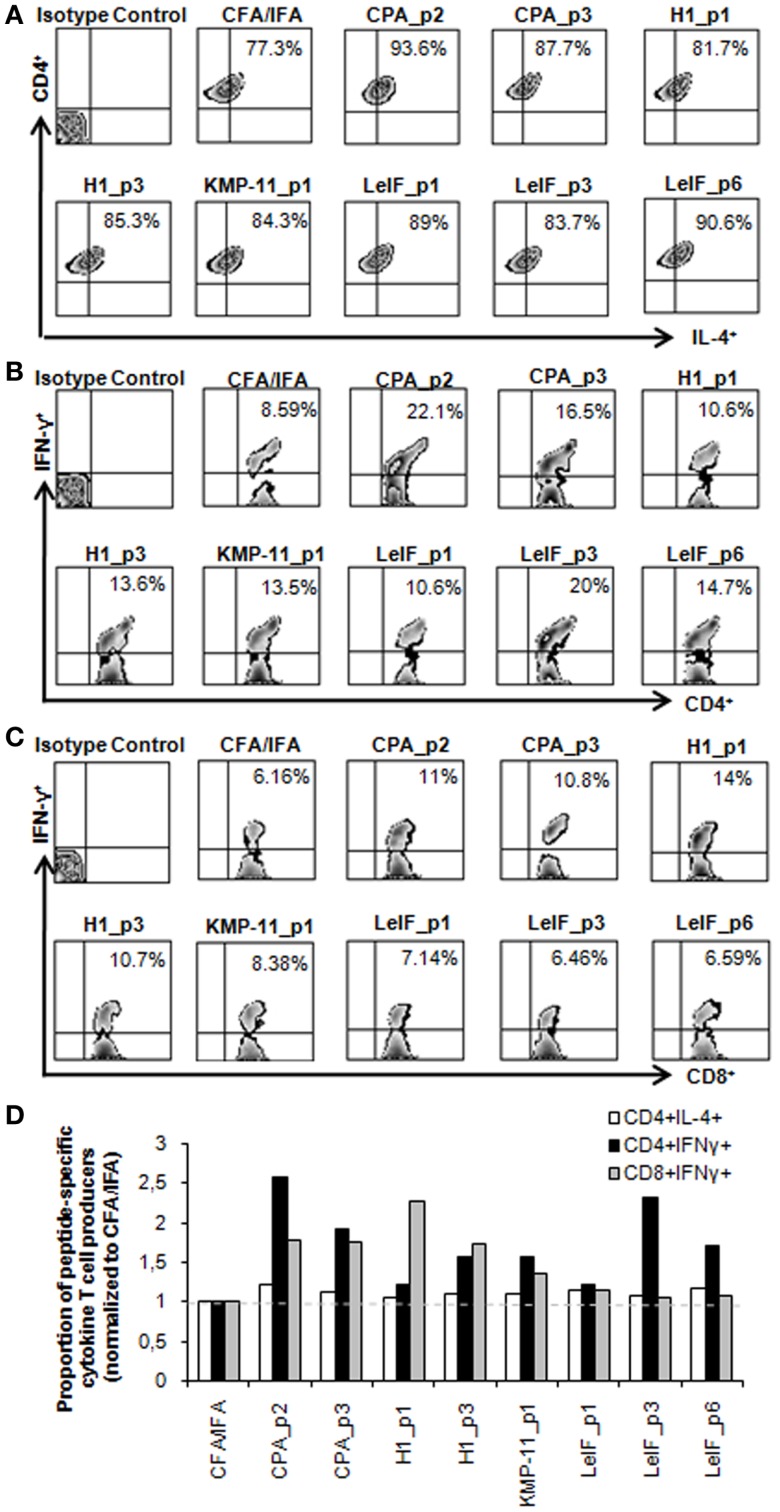
To further confirm the pattern of cytokines induced by each synthetic multi-epitope peptide, intracellular cytokine production was determined in spleen cells from immunized mice at 48 h post peptide in vitro re-stimulation using flow cytometry. As shown in Figure 2A, none of the peptides tested were able to stimulate important peptide-specific IL-4 production by CD4+ T cells, although a certain predisposition in BALB/c mice has been documented by previous studies (53). In contrast, CPA_p2, CPA_p3, H1_p3, LeIF_p3, and LeIF_p6 were able to stimulate important peptide-specific IFN-γ production by CD4+ T cells, indicating a TH cell driven toward the TH1 type.
Figure 2.
Multi-epitope peptide-specific cytokine production by CD4+ and CD8+ T cells. Spleen cells from BALB/c mice (n = 5/group) immunized either with individual peptide emulsified in CFA/IFA or PBS alone, were re-stimulated in vitro with the respective peptide (10 μg/ml) for 48 h, and analyzed for CD4+ and CD8+ T cells producing IL-4 and IFN-γ. (A–C) Representative FACS plots of intracellular staining used to define IL-4- and IFN-γ-expressing CD4+ and CD8+ T cells in spleens derived from immunized mice. Values represent the percentages of (A) IL-4+ cells among CD4+ T cells and (B,C) IFN-γ+ cells among CD4+ and CD8+ T cell populations. (D) Quantification of IL-4 and IFN-γ producing T cells in immunized mice. All data were normalized to CFA/IFA control group. Each bar represents the mean proportion of IL-4- and IFN-γ-producing CD4+ and CD8+ T cells induced by the respective peptide.
Regarding the ability of the synthetic multi-epitope peptides to induce the production of IFN-γ by CD8+ T cells, flow cytometry revealed that one of them, H1_p1 strongly induced the production of IFN-γ by splenic CD8+ T cells of immunized mice. H1_p3, CPA_p2, and CPA_p3 were also able to stimulate peptide-specific IFN-γ production by CD8+ T cells in a lower level than that detected in H1_p1 (Figure 2C). Flow cytometry overall results indicated that most of the peptides tested induced IFN-γ production from CD4+ and/or CD8+ T cells confirming the results obtained with in silico analysis (Figures 2B,D).
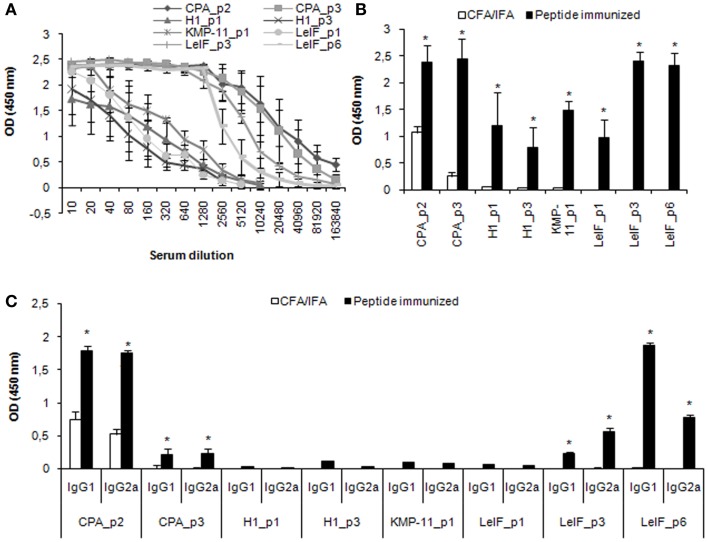
Furthermore, specific antibodies of IgG class, as well as of IgG1 and IgG2a isotypes, were detected in the serum of mice immunized with each synthetic peptide emulsified in CFA/IFA, 15 days post third immunization, in order to evaluate peptide effect on humoral response. According to the results, all the synthetic multi-epitope peptides were able to induce the secretion of specific IgG antibodies (Figures 3A,B). Of these, CPA_p2, CPA_p3, LeIF_p3, and LeIF_p6 induced the highest secretion, followed by KMP-11_p1, LeIF_p1, H1_p1, and H1_p3. Analysis of isotype pattern showed that CPA_p2 strongly induced the production of both IgG2a and IgG1 isotypes, followed by CPA_p3, while LeIF_p3 induced the production of IgG2a > IgG1 (Figure 3C). In contrast, LeIF_p6 strongly induced the production of IgG1 isotype and weakly the production of IgG2a isotype. The other four peptides, KMP-11_p1, LeIF_p1, H1_p1, and H1_p3, had insignificant effect on the production of these two IgG isotypes.
Figure 3.
Multi-epitope peptide-specific antibody production. BALB/c mice (n = 9/group) immunized either with individual peptide emulsified in CFA/IFA or PBS alone, were bled 15 days post third immunization and sera were separated. (A,B) total IgG Abs, and (C) IgG1 and IgG2a Abs against each peptide were assessed by ELISA. The results are expressed as OD450 ± SD. Significant differences between groups of mice immunized with each synthetic peptide emulsified in CFA/IFA and the group of mice immunized with CFA/IFA alone are indicated by * (P < 0.05).
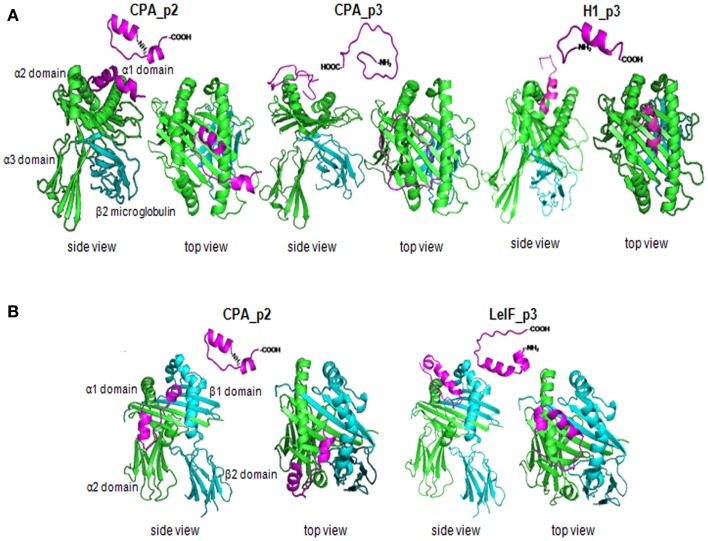
Next, we employed a structure-based method for further analysis of the tertiary structure of the most promising synthetic peptides (CPA_p2, CPA_p3, H1_p3, LeIF_p3) that bound to HLA-A2 or HLA-DRB1 molecule, since HLA-restricted epitopes were also nested in peptide sequences according to algorithms prediction (Table 4). Selection of HLA-A2 and HLA-DRB1 molecules was based on published data demonstrating high frequency of these supertypes in human population (54, 55). The ClusPro program was run to predict docked conformations presenting good surface complementarity with the two MHC molecules mentioned above. The most probable 3D models according to algorithm analysis indicating peptides located onto the peptide-binding cleft of the MHC molecules with good surface complementarity are presented in Figure 4.
Figure 4.
Synthetic multi-epitope peptides docking. Ribbon diagram of 3D structural analysis of interactions between (A) HLA-A2 molecule (PDB-code: 1HHG) and the synthetic peptides CPA_p2, CPA_p3, H1_p3, and LeIF_p3, and (B) HLA-DRB1 molecule (PDB-code: 2SEB) and the synthetic peptides CPA_p2 and LeIF_p3. Candidate peptides were predicted to locate onto the peptide-binding cleft of the HLA molecules by using ClusPro program. The side and top view are shown, the α strands were shown in green, the β strands in blue, and the multi-epitope peptides in magenta.
Discussion
In the perspective of second generation vaccines, a variety of different parasite molecules, such as secretory or transmembrane proteins, including enzymes and receptors, has been tested to date as candidate antigens for anti-Leishmania vaccine development (56). Among them, CPA, Histone H1, KMP-11, and LeIF were found to be highly immunogenic as described in murine experimental models, cured VL patients and L. infantum infected dogs and have been considered as potential vaccine candidates (39–41, 44, 45, 57). The induction of an effective T cell response against vaccine antigens requires antigen processing and peptide presentation by antigen-presenting cells (APCs), and it is well-established that T cells recognize the peptide sequence in association to appropriate MHC molecules. The discovery of MHC-binding motifs in proteins has led to the development of several algorithms predicting MHC class I- and II-restricted epitopes for presentation to CD8+ or CD4+ T cells, respectively, accelerating research related to peptide-based vaccine approach (15).
In the present study, we investigated the use of three algorithms, SYFPEITHI, BIMAS, and NetMHCII to predict sequences in CPA, Histone H1, KMP-11, and LeIF able to bind to MHC class I and II molecules of the H2d haplotype. Furthermore, combining this approach with experimental validation in MHC compatible BALB/c mice, we determined epitopes in each protein and designed multi-epitope peptides capable to induce peptide-specific T cell proliferation and cytokine production by CD4+ and/or CD8+ T cells.
The analysis of protein sequences yielded a significant number of possible epitopes from all four proteins, but only few of them were predicted by all algorithms used with binding efficiency to more than one supertypes or alleles. Interestingly, comparison of predicted peptides for each MHC class I and II alleles showed a low overlapping level between the results obtained from different algorithms used in the study, indicating the significant differences existing in the database source of building matrix motifs and different forms of scoring function of each algorithm. Also, it was observed an antigenic region clustering. Based on these findings and to the fact that prediction of MHC class I-restricted epitopes is considered more reliable (>85%) than that of MHC class II-restricted epitopes, we designed eight multi-epitope peptides for all proteins, based predominantly on highly scored MHC class I-restricted epitopes. Adjacent or overlapping MHC class II-restricted epitopes scored high were also nested in each synthetic peptide. These multi-epitope peptides contained epitopes recognized also by HLA class I and II molecules as defined by in silico analysis. Until now, very few vaccine antigens against different pathogens such as viruses, bacteria, and parasites, contain promiscuous T cell epitopes that have the ability to induce T cell-mediated protective immune responses both in mice and human by binding to several alleles of a supertype or between different supertypes (58, 59). Thus, these promiscuous epitope-driven vaccines could have the capacity of increasing the frequency of responders in genetically variable species, such as human populations (60).
The success of many vaccines is dependent on IFNγ-secreting CD4+ T cells recruitment for long term protection. This accounts for better immunologic memory leading to sustained immunity after healing of live infections (61, 62). CD4+ T cells are activated in terms of recognition of peptides-MHC class II complexes in the surface of APCs after protein processing in cells’ endocytic compartment. Activation of IFN-γ-producing CD4+ T cells plays a pivotal role in protective immune responses against leishmaniasis. Specifically, IFN-γ mediates macrophage activation against both the promastigote and amastigote forms in H2O2-dependent manner (63, 64) and nitric oxide production for parasite killing (65). According to our results, CPA_p2, CPA_p3, LeIF_p3, and LeIF_p6 induced peptide-specific IFN-γ production from CD4+ T cells in immunized mice indicating the processing and recognition of MHC class II-specific epitopes by CD4+ T cells. CPA and LeIF are considered significant candidate proteins for vaccine design against leishmaniasis. In the case of CPA, it has been shown that administration of plasmid encoding CPA induced specific TH1 immune responses resulting to partial protection against L. major in the experimental model of CL. However, protection was significantly enhanced when co-administered with CPB or as CPA/B hybrid protein (25–27), indicating the need of CPA co-administration with another protein or adjuvant. On the other hand, LeIF was originally described as a TH1-type natural adjuvant and as an antigen inducing an IL-12 mediated TH1 response in the PBMCs of leishmaniasis patients (39). LeIF is also capable of inducing the secretion of cytokines IL-12, IL-10, and TNF-α by APCs from healthy individuals (52, 66, 67). Furthermore, recombinant trifusion vaccines (leish111; leish110f) were developed by incorporating the amino-terminal region of LeIF antigen. These vaccines were shown to be efficient in experimental or clinical trials for vaccination or immunotherapy (68).
Existing data suggest that secretory and surface exposed proteins strongly induce specific CD8+ T cell responses (69, 70). A previous study applying in silico analysis revealed that a high number of peptides derived from L. major secretome could bind to H2 BALB/c molecules (71). Several studies have shown the great role played by CD8+ T cells in protective immune responses against parasite in the susceptible BALB/c strain (72–74). Specifically, CD8+ T cells either contributed in the destruction of Leishmania-infected cells by activating macrophages to oxidative burst via cytokines produced upon antigen stimulation (75, 76), or regulating CD4+ T cell-mediated immune responses (77, 78). In our study, both synthetic multi-epitope peptides of CPA, CPA_p2, and CPA_p3, except from CD4+IFN-γ+ T cells activation, induced significant IFN-γ production by CD8+ T cells. CPA_p2 and CPA_p3 belong to the secreted region of CPA as SignalP analysis showed (30, 31). Interestingly, another study applying in silico analysis in CPA sequence with MULTIPRED algorithm indicated the existence of four highly immunogenic regions recognized by the HLA-A2 supertype (79), which harbored parts from our CPA multi-epitope peptides.
As for KMP-11, KMP-11_p1 belonged to the amino-terminal region of the protein and in contrast to previous observations this synthetic peptide was proved to be poorly immunogenic, indicated by the absence of peptide-specific proliferative response and cytokine secretion in immunized mice. Previous studies concerning the identification of T cell epitopes using infected macrophages or DCs as APCs, revealed the existence of potential HLA class I- and II-restricted T cell epitopes in the amino-terminal region, characterizing a dominant cluster between position 1 and 33 of KMP-11 sequence that could trigger specific cellular immune responses in L. donovani- or L. panamensis-infected volunteers (80, 81). Furthermore, hybrid-cell, DNA-based or heterologous KMP-11-DNA/rVV based vaccination exhibited immunoprotective capacity in susceptible VL murine models. Protection was accompanied with generation of antigen specific CD4+ and CD8+ T cells that produced effector cytokines such as IFN-γ, IL-2, and TNF-α (36–38, 82, 83). Also in a previous work, we demonstrated that vaccination with ex vivo pulsed bone marrow-derived dendritic cells with KMP-1112–31aa peptide and CpG as adjuvant induced strong Th1 and Th17 protective immune responses in murine model of VL (50). However, in the present study it is noteworthy that secretion of IL-10 was also abrogated. These results together suggest that KMP-11_p1 may be consisted from natural epitopes contributing in parasite host immunomodulation, allowing parasite dissemination rather than stimulate protective immune responses.
However, not only external or secreted Leishmania antigens are able to be presented in the context of MHC class I molecules but also intracellular proteins (84, 85). As such, in our study H1_p1 and H1_p3 induced a T cell response characterized mainly by CD8+ T cell priming and production of IFN-γ in immunized mice. Although, this way of cell activation in leishmaniasis remains controversial and it is not clear how non-secretory parasite antigens such as histone H1 can be presented endogenously in the context of MHC class I molecules, a number of studies supports the induction of specific CD8+ T cell responses against structural parasite proteins in animal models and VL patients (82, 84, 86, 87). Previous results from our group supported that ex vivo pulsed bone marrow-derived dendritic cells with the Leishmania histone H1 elicited significant protection in the experimental model of VL, with a pronounced enhancement of parasite-specific IFNγ-producing CD8+ T cells (88). The protective effect of Leishmania histone H1 against L. major or L. infantum infections was also shown in different experimental animal models (34, 35) suggesting that it is also a promising vaccine candidate against leishmaniasis. In contrast, none of the LeIF peptides tested could evoke specific CD8+ T cell responses. This finding was in agreement with the study of Rafati et al. showing that PBMCs from patients recovered from L. major failed to elicit HLA-A2-restricted CD8+ T cell responses against three synthetic nonamer peptides of LeIF, suggesting that these peptides are not able to induce a CD8+ T cell-induced protective immunity (86).
The relative low concentrations of IL-10 detected in the supernatants of immune lymphocytes compared to IFN-γ after peptide re-stimulation were consistent with the suggestion of a dynamic reciprocal relationship between these two cytokines. IL-10 primarily down-modulates innate as well as acquired immunity leading to parasite establishment or disease progression. In experimental model of VL, IL-10 prevents DCs migration in spleen to activate T cells (89, 90) and suppresses both TH1 and TH2 cells (91). Also, the CD4+IL-4+ T lymphocytes detected in the presence of all peptides may be attributed to BALB/c intrinsic feature to induce the production of type-2 cytokines, such as IL-4 (53), since there was not any significant difference of IL-4 levels between peptide-immunized mice and control mice receiving the adjuvant alone. Furthermore, IFA adjuvant has a propensity to induce preferentially TH2 cytokines (92, 93). Similar study for the evaluation of immunoreactivity of in silico predicted TH1 epitopes of Schistosoma japonicum showed that high levels of IL-4 were attributed to Freund’s adjuvant and BALB/c strain used (94). Therefore, in terms of proportion of intracellular cytokine production of recall CD8+ and CD4+ T cells, it is concluded that CPA_p3, H1_p1, H1_p3, CPA_p2, LeIF_p3, and LeIF_p6 synthetic multi-epitope peptides are likely to include potential epitopes for the induction of protective cytotoxic (CTL) and TH1-type immune responses. Taken into account that the sequences of these synthetic peptides are highly conserved and bind in a promiscuous manner to murine or human MHC molecules according to in silico analysis and structure-based techniques, make them candidate vaccines against leishmaniasis. Based on these results, it would be worthwhile conducting future investigations for the verification of peptides’ possible ability to induce protection in common or humanized mouse models of leishmaniasis. The incorporation of alternative and/or additional epitopes, the use of modern adjuvants and new antigen delivery systems should be combined. Conclusively, these findings give complementary data on epitope mapping for Leishmania proteins and demonstrate that combination of immunoinformatic approaches with experimental validation enables peptide identification with greater accuracy contributing to rational epitope-based vaccine development.
Author Contributions
Evita Athanasiou and Olga Koutsoni contributed equally to this work. Conceived and designed the experiments: Evdokia Karagouni. Performed computational analysis: Maria Agallou, Evita Athanasiou, Olga Koutsoni, Evdokia Karagouni. Performed the experiments: Maria Agallou, Olga Koutsoni, Evita Athanasiou. Analyzed the data: Maria Agallou, Evita Athanasiou, Olga Koutsoni, Evdokia Karagouni, Eleni Dotsika. Wrote the paper: Evdokia Karagouni, Maria Agallou.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by a grant (09SYN-14-643) from EU and the National Ministry of Education and Religion Affairs under the Operational Strategic Reference Framework (NSFR 2007–2013) awarded to Evdokia Karagouni. The authors also thank Dr. Petros Giastas (Department of Neurobiology, Hellenic Pasteur Institute) for their assistance in structure-based analysis.
Footnotes
References
- 1.Ready PD. Leishmaniasis emergence in Europe. Euro Surveill (2010) 15(10):19505. [PubMed] [Google Scholar]
- 2.Singh N, Kumar M, Singh RK. Leishmaniasis: current status of available drugs and new potential drug targets. Asian Pac J Trop Med (2012) 5(6):485–97 10.1016/S1995-7645(12)60084-4 [DOI] [PubMed] [Google Scholar]
- 3.Kobets T, Grekov I, Lipoldova M. Leishmaniasis: prevention, parasite detection and treatment. Curr Med Chem (2012) 19(10):1443–74 10.2174/092986712799828300 [DOI] [PubMed] [Google Scholar]
- 4.Picado A, Dash AP, Bhattacharya S, Boelaert M. Vector control interventions for visceral leishmaniasis elimination initiative in South Asia, 2005–2010. Indian J Med Res (2012) 136(1):22–31 [PMC free article] [PubMed] [Google Scholar]
- 5.Beverley SM, Turco SJ. Lipophosphoglycan (LPG) and the identification of virulence genes in the protozoan parasite Leishmania. Trends Microbiol (1998) 6(1):35–40 10.1016/S0966-842X(97)01180-3 [DOI] [PubMed] [Google Scholar]
- 6.Kaye PM, Aebischer T. Visceral leishmaniasis: immunology and prospects for a vaccine. Clin Microbiol Infect (2011) 17(10):1462–70 10.1111/j.1469-0691.2011.03610.x [DOI] [PubMed] [Google Scholar]
- 7.Khamesipour A, Rafati S, Davoudi N, Maboudi F, Modabber F. Leishmaniasis vaccine candidates for development: a global overview. Indian J Med Res (2006) 123(3):423–38 [PubMed] [Google Scholar]
- 8.Kedzierski L. Leishmaniasis. Hum Vaccin (2011) 7(11):1204–14 10.4161/hv.7.11.17752 [DOI] [PubMed] [Google Scholar]
- 9.Molano I, Alonso MG, Miron C, Redondo E, Requena JM, Soto M, et al. A Leishmania infantum multi-component antigenic protein mixed with live BCG confers protection to dogs experimentally infected with L. infantum. Vet Immunol Immunopathol (2003) 92(1–2):1–13 10.1016/S0165-2427(02)00315-X [DOI] [PubMed] [Google Scholar]
- 10.Rafati S, Nakhaee A, Taheri T, Taslimi Y, Darabi H, Eravani D, et al. Protective vaccination against experimental canine visceral leishmaniasis using a combination of DNA and protein immunization with cysteine proteinases type I and II of L. infantum. Vaccine (2005) 23(28):3716–25 10.1016/j.vaccine.2005.02.009 [DOI] [PubMed] [Google Scholar]
- 11.Carson C, Antoniou M, Ruiz-Arguello MB, Alcami A, Christodoulou V, Messaritakis I, et al. A prime/boost DNA/modified vaccinia virus Ankara vaccine expressing recombinant Leishmania DNA encoding TRYP is safe and immunogenic in outbred dogs, the reservoir of zoonotic visceral leishmaniasis. Vaccine (2009) 27(7):1080–6 10.1016/j.vaccine.2008.11.094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Llanos-Cuentas A, Calderon W, Cruz M, Ashman JA, Alves FP, Coler RN, et al. A clinical trial to evaluate the safety and immunogenicity of the LEISH-F1+MPL-SE vaccine when used in combination with sodium stibogluconate for the treatment of mucosal leishmaniasis. Vaccine (2010) 28(46):7427–35 10.1016/j.vaccine.2010.08.092 [DOI] [PubMed] [Google Scholar]
- 13.Chakravarty J, Kumar S, Trivedi S, Rai VK, Singh A, Ashman JA, et al. A clinical trial to evaluate the safety and immunogenicity of the LEISH-F1+MPL-SE vaccine for use in the prevention of visceral leishmaniasis. Vaccine (2011) 29(19):3531–7 10.1016/j.vaccine.2011.02.096 [DOI] [PubMed] [Google Scholar]
- 14.De Groot AS, McMurry J, Marcon L, Franco J, Rivera D, Kutzler M, et al. Developing an epitope-driven tuberculosis (TB) vaccine. Vaccine (2005) 23(17–18):2121–31 10.1016/j.vaccine.2005.01.059 [DOI] [PubMed] [Google Scholar]
- 15.Davies MN, Flower DR. Harnessing bioinformatics to discover new vaccines. Drug Discov Today (2007) 12(9–10):389–95 10.1016/j.drudis.2007.03.010 [DOI] [PubMed] [Google Scholar]
- 16.Lundegaard C, Lund O, Kesmir C, Brunak S, Nielsen M. Modeling the adaptive immune system: predictions and simulations. Bioinformatics (2007) 23(24):3265–75 10.1093/bioinformatics/btm471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cunha-Neto E. MHC-restricted antigen presentation and recognition: constraints on gene, recombinant and peptide vaccines in humans. Braz J Med Biol Res (1999) 32(2):199–205 10.1590/S0100-879X1999000200008 [DOI] [PubMed] [Google Scholar]
- 18.Stager S, Rafati S. CD8(+) T cells in Leishmania infections: friends or foes? Front Immunol (2012) 3:5. 10.3389/fimmu.2012.00005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Murray HW. Endogenous interleukin-12 regulates acquired resistance in experimental visceral leishmaniasis. J Infect Dis (1997) 175(6):1477–9 10.1086/516482 [DOI] [PubMed] [Google Scholar]
- 20.Engwerda CR, Murphy ML, Cotterell SE, Smelt SC, Kaye PM. Neutralization of IL-12 demonstrates the existence of discrete organ-specific phases in the control of Leishmania donovani. Eur J Immunol (1998) 28(2):669–80 [DOI] [PubMed] [Google Scholar]
- 21.Murray HW, Oca MJ, Granger AM, Schreiber RD. Requirement for T cells and effect of lymphokines in successful chemotherapy for an intracellular infection. Experimental visceral leishmaniasis. J Clin Invest (1989) 83(4):1253–7 10.1172/JCI114009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Murray HW, Montelibano C, Peterson R, Sypek JP. Interleukin-12 regulates the response to chemotherapy in experimental visceral Leishmaniasis. J Infect Dis (2000) 182(5):1497–502 10.1086/315890 [DOI] [PubMed] [Google Scholar]
- 23.Goto H, Lindoso JA. Immunity and immunosuppression in experimental visceral leishmaniasis. Braz J Med Biol Res (2004) 37(4):615–23 10.1590/S0100-879X2004000400020 [DOI] [PubMed] [Google Scholar]
- 24.Singh B, Sundar S. Leishmaniasis: vaccine candidates and perspectives. Vaccine (2012) 30(26):3834–42 10.1016/j.vaccine.2012.03.068 [DOI] [PubMed] [Google Scholar]
- 25.Ahmed SB, Touihri L, Chtourou Y, Dellagi K, Bahloul C. DNA based vaccination with a cocktail of plasmids encoding immunodominant Leishmania (Leishmania) major antigens confers full protection in BALB/c mice. Vaccine (2009) 27(1):99–106 10.1016/j.vaccine.2008.10.013 [DOI] [PubMed] [Google Scholar]
- 26.Rafati S, Salmanian AH, Taheri T, Vafa M, Fasel N. A protective cocktail vaccine against murine cutaneous leishmaniasis with DNA encoding cysteine proteinases of Leishmania major. Vaccine (2001) 19(25–26):3369–75 10.1016/S0264-410X(01)00081-0 [DOI] [PubMed] [Google Scholar]
- 27.Zadeh-Vakili A, Taheri T, Taslimi Y, Doustdari F, Salmanian AH, Rafati S. Immunization with the hybrid protein vaccine, consisting of Leishmania major cysteine proteinases Type I (CPB) and Type II (CPA), partially protects against leishmaniasis. Vaccine (2004) 22(15–16):1930–40 10.1016/j.vaccine.2003.11.014 [DOI] [PubMed] [Google Scholar]
- 28.Requena JM, Alonso C, Soto M. Evolutionarily conserved proteins as prominent immunogens during Leishmania infections. Parasitol Today (2000) 16(6):246–50 10.1016/S0169-4758(00)01651-3 [DOI] [PubMed] [Google Scholar]
- 29.Galanti N, Galindo M, Sabaj V, Espinoza I, Toro GC. Histone genes in trypanosomatids. Parasitol Today (1998) 14(2):64–70 10.1016/S0169-4758(97)01162-9 [DOI] [PubMed] [Google Scholar]
- 30.Stebeck CE, Beecroft RP, Singh BN, Jardim A, Olafson RW, Tuckey C, et al. Kinetoplastid membrane protein-11 (KMP-11) is differentially expressed during the life cycle of African trypanosomes and is found in a wide variety of kinetoplastid parasites. Mol Biochem Parasitol (1995) 71(1):1–13 10.1016/0166-6851(95)00022-S [DOI] [PubMed] [Google Scholar]
- 31.Matos DC, Faccioli LA, Cysne-Finkelstein L, Luca PM, Corte-Real S, Armoa GR, et al. Kinetoplastid membrane protein-11 is present in promastigotes and amastigotes of Leishmania amazonensis and its surface expression increases during metacyclogenesis. Mem Inst Oswaldo Cruz (2010) 105(3):341–7 10.1590/S0074-02762010000300018 [DOI] [PubMed] [Google Scholar]
- 32.Tolson DL, Jardim A, Schnur LF, Stebeck C, Tuckey C, Beecroft RP, et al. The kinetoplastid membrane protein 11 of Leishmania donovani and African trypanosomes is a potent stimulator of T-lymphocyte proliferation. Infect Immun (1994) 62(11):4893–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jardim A, Tolson DL, Turco SJ, Pearson TW, Olafson RW. The Leishmania donovani lipophosphoglycan T lymphocyte-reactive component is a tightly associated protein complex. J Immunol (1991) 147(10):3538–44 [PubMed] [Google Scholar]
- 34.Solioz N, Blum-Tirouvanziam U, Jacquet R, Rafati S, Corradin G, Mauel J, et al. The protective capacities of histone H1 against experimental murine cutaneous leishmaniasis. Vaccine (1999) 18(9–10):850–9 10.1016/S0264-410X(99)00340-0 [DOI] [PubMed] [Google Scholar]
- 35.Masina S, M Gicheru M, Demotz SO, Fasel NJ. Protection against cutaneous leishmaniasis in outbred vervet monkeys using a recombinant histone H1 antigen. J Infect Dis (2003) 188(8):1250–7 10.1086/378677 [DOI] [PubMed] [Google Scholar]
- 36.Basu R, Bhaumik S, Basu JM, Naskar K, De T, Roy S. Kinetoplastid membrane protein-11 DNA vaccination induces complete protection against both pentavalent antimonial-sensitive and -resistant strains of Leishmania donovani that correlates with inducible nitric oxide synthase activity and IL-4 generation: evidence for mixed Th1- and Th2-like responses in visceral leishmaniasis. J Immunol (2005) 174(11):7160–71 10.4049/jimmunol.174.11.7160 [DOI] [PubMed] [Google Scholar]
- 37.Guha R, Das S, Ghosh J, Naskar K, Mandala A, Sundar S, et al. Heterologous priming-boosting with DNA and vaccinia virus expressing kinetoplastid membrane protein-11 induces potent cellular immune response and confers protection against infection with antimony resistant and sensitive strains of Leishmania (Leishmania) donovani. Vaccine (2013) 31(15):1905–15 10.1016/j.vaccine.2013.02.025 [DOI] [PubMed] [Google Scholar]
- 38.Bhaumik S, Basu R, Sen S, Naskar K, Roy S. KMP-11 DNA immunization significantly protects against L. donovani infection but requires exogenous IL-12 as an adjuvant for comparable protection against L. major. Vaccine (2009) 27(9):1306–16 10.1016/j.vaccine.2008.12.053 [DOI] [PubMed] [Google Scholar]
- 39.Skeiky YA, Guderian JA, Benson DR, Bacelar O, Carvalho EM, Kubin M, et al. A recombinant Leishmania antigen that stimulates human peripheral blood mononuclear cells to express a Th1-type cytokine profile and to produce interleukin 12. J Exp Med (1995) 181(4):1527–37 10.1084/jem.181.4.1527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rafati S, Salmanian AH, Hashemi K, Schaff C, Belli S, Fasel N. Identification of Leishmania major cysteine proteinases as targets of the immune response in humans. Mol Biochem Parasitol (2001) 113(1):35–43 10.1016/S0166-6851(00)00377-7 [DOI] [PubMed] [Google Scholar]
- 41.Rafati S, Nakhaee A, Taheri T, Ghashghaii A, Salmanian AH, Jimenez M, et al. Expression of cysteine proteinase type I and II of Leishmania infantum and their recognition by sera during canine and human visceral leishmaniasis. Exp Parasitol (2003) 103(3–4):143–51 10.1016/S0014-4894(03)00097-3 [DOI] [PubMed] [Google Scholar]
- 42.Nakhaee A, Taheri T, Taghikhani M, Mohebali M, Salmanian AH, Fasel N, et al. Humoral and cellular immune responses against Type I cysteine proteinase of Leishmania infantum are higher in asymptomatic than symptomatic dogs selected from a naturally infected population. Vet Parasitol (2004) 119(2–3):107–23 10.1016/j.vetpar.2003.11.013 [DOI] [PubMed] [Google Scholar]
- 43.Pascalis H, Lavergne A, Bourreau E, Prevot-Linguet G, Kariminia A, Pradinaud R, et al. Th1 cell development induced by cysteine proteinases A and B in localized cutaneous leishmaniasis due to Leishmania guyanensis. Infect Immun (2003) 71(5):2924–6 10.1128/IAI.71.5.2924-2926.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Carmelo E, Martinez E, Gonzalez AC, Pinero JE, Patarroyo ME, Del Castillo A, et al. Antigenicity of Leishmania braziliensis histone H1 during cutaneous leishmaniasis: localization of antigenic determinants. Clin Vaccine Immunol (2002) 9(4):808–11 10.1128/CDLI.9.4.808-811.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Berberich C, Requena JM, Alonso C. Cloning of genes and expression and antigenicity analysis of the Leishmania infantum KMP-11 protein. Exp Parasitol (1997) 85(1):105–8 10.1006/expr.1996.4120 [DOI] [PubMed] [Google Scholar]
- 46.Kozakov D, Brenke R, Comeau SR, Vajda S. PIPER: an FFT-based protein docking program with pairwise potentials. Proteins (2006) 65(2):392–406 10.1002/prot.21117 [DOI] [PubMed] [Google Scholar]
- 47.Kozakov D, Beglov D, Bohnuud T, Mottarella SE, Xia B, Hall DR, et al. How good is automated protein docking? Proteins (2013) 81(12):2159–66 10.1002/prot.24403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Comeau SR, Gatchell DW, Vajda S, Camacho CJ. ClusPro: a fully automated algorithm for protein-protein docking. Nucleic Acids Res (2004) 32:W96–9 10.1093/nar/gkh354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Comeau SR, Gatchell DW, Vajda S, Camacho CJ. ClusPro: an automated docking and discrimination method for the prediction of protein complexes. Bioinformatics (2004) 20(1):45–50 10.1093/bioinformatics/btg371 [DOI] [PubMed] [Google Scholar]
- 50.Agallou M, Margaroni M, Karagouni E. Cellular vaccination with bone marrow-derived dendritic cells pulsed with a peptide of Leishmania infantum KMP-11 and CpG oligonucleotides induces protection in a murine model of visceral leishmaniasis. Vaccine (2011) 29(31):5053–64 10.1016/j.vaccine.2011.04.089 [DOI] [PubMed] [Google Scholar]
- 51.Noll TM, Desponds C, Belli SI, Glaser TA, Fasel NJ. Histone H1 expression varies during the Leishmania major life cycle. Mol Biochem Parasitol (1997) 84(2):215–27 10.1016/S0166-6851(96)02801-0 [DOI] [PubMed] [Google Scholar]
- 52.Probst P, Skeiky YA, Steeves M, Gervassi A, Grabstein KH, Reed SG. A Leishmania protein that modulates interleukin (IL)-12, IL-10 and tumor necrosis factor-alpha production and expression of B7-1 in human monocyte-derived antigen-presenting cells. Eur J Immunol (1997) 27(10):2634–42 10.1002/eji.1830271024 [DOI] [PubMed] [Google Scholar]
- 53.Mills CD, Kincaid K, Alt JM, Heilman MJ, Hill AM. M-1/M-2 macrophages and the Th1/Th2 paradigm. J Immunol (2000) 164(12):6166–73 10.4049/jimmunol.164.12.6166 [DOI] [PubMed] [Google Scholar]
- 54.Cao K, Hollenbach J, Shi X, Shi W, Chopek M, Fernandez-Vina MA. Analysis of the frequencies of HLA-A, B, and C alleles and haplotypes in the five major ethnic groups of the United States reveals high levels of diversity in these loci and contrasting distribution patterns in these populations. Hum Immunol (2001) 62(9):1009–30 10.1016/S0198-8859(01)00298-1 [DOI] [PubMed] [Google Scholar]
- 55.Mack SJ, Tu B, Yang R, Masaberg C, Ng J, Hurley CK. Human leukocyte antigen-A, -B, -C, -DRB1 allele and haplotype frequencies in Americans originating from southern Europe: contrasting patterns of population differentiation between Italian and Spanish Americans. Hum Immunol (2011) 72(2):144–9 10.1016/j.humimm.2010.10.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kedzierski L. Leishmaniasis vaccine: where are we today? J Glob Infect Dis (2010) 2(2):177–85 10.4103/0974-777X.62881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jensen AT, Gasim S, Ismail A, Gaafar A, Kurtzhals JA, Kemp M, et al. Humoral and cellular immune responses to synthetic peptides of the Leishmania donovani kinetoplastid membrane protein-11. Scand J Immunol (1998) 48(1):103–9 10.1046/j.1365-3083.1998.00370.x [DOI] [PubMed] [Google Scholar]
- 58.Khan AM, Miotto O, Heiny AT, Salmon J, Srinivasan KN, Nascimento EJ, et al. A systematic bioinformatics approach for selection of epitope-based vaccine targets. Cell Immunol (2006) 244(2):141–7 10.1016/j.cellimm.2007.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Reche PA, Reinherz EL. Definition of MHC supertypes through clustering of MHC peptide-binding repertoires. Methods Mol Biol (2007) 409:163–73 10.1007/978-1-60327-118-9_11 [DOI] [PubMed] [Google Scholar]
- 60.Lima-Junior JC, Banic DM, Tran TM, Meyer VS, De-Simone SG, Santos F, et al. Promiscuous T-cell epitopes of Plasmodium merozoite surface protein 9 (PvMSP9) induces IFN-gamma and IL-4 responses in individuals naturally exposed to malaria in the Brazilian Amazon. Vaccine (2010) 28(18):3185–91 10.1016/j.vaccine.2010.02.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Okwor I, Uzonna J. Persistent parasites and immunologic memory in cutaneous leishmaniasis: implications for vaccine designs and vaccination strategies. Immunol Res (2008) 41(2):123–36 10.1007/s12026-008-8016-2 [DOI] [PubMed] [Google Scholar]
- 62.Nateghi Rostami M, Keshavarz H, Edalat R, Sarrafnejad A, Shahrestani T, Mahboudi F, et al. CD8+ T cells as a source of IFN-gamma production in human cutaneous leishmaniasis. PLoS Negl Trop Dis (2010) 4(10):e845. 10.1371/journal.pntd.0000845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Murray HW, Byrne GI, Rothermel CD, Cartelli DM. Lymphokine enhances oxygen-independent activity against intracellular pathogens. J Exp Med (1983) 158(1):234–9 10.1084/jem.158.1.234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Murray HW, Cartelli DM. Killing of intracellular Leishmania donovani by human mononuclear phagocytes. Evidence for oxygen-dependent and -independent leishmanicidal activity. J Clin Invest (1983) 72(1):32–44 10.1172/JCI110972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cunningham AC. Parasitic adaptive mechanisms in infection by Leishmania. Exp Mol Pathol (2002) 72(2):132–41 10.1006/exmp.2002.2418 [DOI] [PubMed] [Google Scholar]
- 66.Barhoumi M, Meddeb-Garnaoui A, Tanner NK, Banroques J, Kaabi B, Guizani I. DEAD-box proteins, like Leishmania eIF4A, modulate interleukin (IL)-12, IL-10 and tumour necrosis factor-alpha production by human monocytes. Parasite Immunol (2013) 35(5–6):194–9 10.1111/pim.12026 [DOI] [PubMed] [Google Scholar]
- 67.Barhoumi M, Garnaoui A, Kaabi B, Tanner NK, Guizani I. Leishmania infantum LeIF and its recombinant polypeptides modulate interleukin IL-12p70, IL-10 and tumour necrosis factor-alpha production by human monocytes. Parasite Immunol (2011) 33(10):583–8 10.1111/j.1365-3024.2011.01320.x [DOI] [PubMed] [Google Scholar]
- 68.Fujiwara RT, Vale AM, Franca da Silva JC, da Costa RT, Quetz Jda S, Martins Filho OA, et al. Immunogenicity in dogs of three recombinant antigens (TSA, LeIF and LmSTI1) potential vaccine candidates for canine visceral leishmaniasis. Vet Res (2005) 36(5–6):827–38 10.1051/vetres:2005033 [DOI] [PubMed] [Google Scholar]
- 69.Bertholet S, Goldszmid R, Morrot A, Debrabant A, Afrin F, Collazo-Custodio C, et al. Leishmania antigens are presented to CD8+ T cells by a transporter associated with antigen processing-independent pathway in vitro and in vivo. J Immunol (2006) 177(6):3525–33 10.4049/jimmunol.177.6.3525 [DOI] [PubMed] [Google Scholar]
- 70.Kima PE, Ruddle NH, McMahon-Pratt D. Presentation via the class I pathway by Leishmania amazonensis-infected macrophages of an endogenous leishmanial antigen to CD8+ T cells. J Immunol (1997) 159(4):1828–34 [PubMed] [Google Scholar]
- 71.Guerfali FZ, Ben-Abdallah H, Sghaier RM, Ben-Aissa K, Mkannez G, Attia H, et al. An in silico immunological approach for prediction of CD8+ T cell epitopes of Leishmania major proteins in susceptible BALB/c and resistant C57BL/6 murine models of infection. Infect Genet Evol (2009) 9(3):344–50 10.1016/j.meegid.2008.02.011 [DOI] [PubMed] [Google Scholar]
- 72.Huber M, Timms E, Mak TW, Rollinghoff M, Lohoff M. Effective and long-lasting immunity against the parasite Leishmania major in CD8-deficient mice. Infect Immun (1998) 66(8):3968–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hill JO, Awwad M, North RJ. Elimination of CD4+ suppressor T cells from susceptible BALB/c mice releases CD8+ T lymphocytes to mediate protective immunity against Leishmania. J Exp Med (1989) 169(5):1819–27 10.1084/jem.169.5.1819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Stefani MM, Muller I, Louis JA. Leishmania major-specific CD8+ T cells are inducers and targets of nitric oxide produced by parasitized macrophages. Eur J Immunol (1994) 24(3):746–52 10.1002/eji.1830240338 [DOI] [PubMed] [Google Scholar]
- 75.Diez H, Lopez MC, Del Carmen Thomas M, Guzman F, Rosas F, Velazco V, et al. Evaluation of IFN-gamma production by CD8 T lymphocytes in response to the K1 peptide from KMP-11 protein in patients infected with Trypanosoma cruzi. Parasite Immunol (2006) 28(3):101–5 10.1111/j.1365-3024.2005.00815.x [DOI] [PubMed] [Google Scholar]
- 76.Tsagozis P, Karagouni E, Dotsika E. CD8(+) T cells with parasite-specific cytotoxic activity and a Tc1 profile of cytokine and chemokine secretion develop in experimental visceral leishmaniasis. Parasite Immunol (2003) 25(11–12):569–79 10.1111/j.0141-9838.2004.00672.x [DOI] [PubMed] [Google Scholar]
- 77.Uzonna JE, Joyce KL, Scott P. Low dose Leishmania major promotes a transient T helper cell type 2 response that is down-regulated by interferon gamma-producing CD8+ T cells. J Exp Med (2004) 199(11):1559–66 10.1084/jem.20040172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mary C, Auriault V, Faugere B, Dessein AJ. Control of Leishmania infantum infection is associated with CD8(+) and gamma interferon- and interleukin-5-producing CD4(+) antigen-specific T cells. Infect Immun (1999) 67(11):5559–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Saffari B, Mohabatkar H. Computational analysis of cysteine proteases (Clan CA, Family Cl) of Leishmania major to find potential epitopic regions. Genomics Proteomics Bioinformatics (2009) 7(3):87–95 10.1016/S1672-0229(08)60037-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Basu R, Roy S, Walden P. HLA class I-restricted T cell epitopes of the kinetoplastid membrane protein-11 presented by Leishmania donovani-infected human macrophages. J Infect Dis (2007) 195(9):1373–80 10.1086/513439 [DOI] [PubMed] [Google Scholar]
- 81.Delgado G, Parra-Lopez CA, Vargas LE, Hoya R, Estupinan M, Guzman F, et al. Characterizing cellular immune response to kinetoplastid membrane protein-11 (KMP-11) during Leishmania (Viannia) panamensis infection using dendritic cells (DCs) as antigen presenting cells (APCs). Parasite Immunol (2003) 25(4):199–209 10.1046/j.1365-3024.2003.00626.x [DOI] [PubMed] [Google Scholar]
- 82.Basu R, Bhaumik S, Haldar AK, Naskar K, De T, Dana SK, et al. Hybrid cell vaccination resolves Leishmania donovani infection by eliciting a strong CD8+ cytotoxic T-lymphocyte response with concomitant suppression of interleukin-10 (IL-10) but not IL-4 or IL-13. Infect Immun (2007) 75(12):5956–66 10.1128/IAI.00944-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mukhopadhyay S, Sen P, Bhattacharyya S, Majumdar S, Roy S. Immunoprophylaxis and immunotherapy against experimental visceral leishmaniasis. Vaccine (1999) 17(3):291–300 10.1016/S0264-410X(98)90017-2 [DOI] [PubMed] [Google Scholar]
- 84.Iborra S, Soto M, Carrion J, Alonso C, Requena JM. Vaccination with a plasmid DNA cocktail encoding the nucleosomal histones of Leishmania confers protection against murine cutaneous leishmaniosis. Vaccine (2004) 22(29–30):3865–76 10.1016/j.vaccine.2004.04.015 [DOI] [PubMed] [Google Scholar]
- 85.Carneiro MW, Santos DM, Fukutani KF, Clarencio J, Miranda JC, Brodskyn C, et al. Vaccination with L. infantum chagasi nucleosomal histones confers protection against new world cutaneous leishmaniasis caused by Leishmania braziliensis. PLoS One (2012) 7(12):e52296. 10.1371/journal.pone.0052296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Seyed N, Zahedifard F, Safaiyan S, Gholami E, Doustdari F, Azadmanesh K, et al. In silico analysis of six known Leishmania major antigens and in vitro evaluation of specific epitopes eliciting HLA-A2 restricted CD8 T cell response. PLoS Negl Trop Dis (2011) 5(9):e1295. 10.1371/journal.pntd.0001295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Resende DM, Caetano BC, Dutra MS, Penido ML, Abrantes CF, Verly RM, et al. Epitope mapping and protective immunity elicited by adenovirus expressing the Leishmania amastigote specific A2 antigen: correlation with IFN-gamma and cytolytic activity by CD8+ T cells. Vaccine (2008) 26(35):4585–93 10.1016/j.vaccine.2008.05.091 [DOI] [PubMed] [Google Scholar]
- 88.Agallou M, Smirlis D, Soteriadou KP, Karagouni E. Vaccination with Leishmania histone H1-pulsed dendritic cells confers protection in murine visceral leishmaniasis. Vaccine (2012) 30(34):5086–93 10.1016/j.vaccine.2012.05.075 [DOI] [PubMed] [Google Scholar]
- 89.Basu A, Chakrabarti G, Saha A, Bandyopadhyay S. Modulation of CD11C+ splenic dendritic cell functions in murine visceral leishmaniasis: correlation with parasite replication in the spleen. Immunology (2000) 99(2):305–13 10.1046/j.1365-2567.2000.00939.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Engwerda CR, Kaye PM. Organ-specific immune responses associated with infectious disease. Immunol Today (2000) 21(2):73–8 10.1016/S0167-5699(99)01549-2 [DOI] [PubMed] [Google Scholar]
- 91.Bodas M, Jain N, Awasthi A, Martin S, Penke Loka RK, Dandekar D, et al. Inhibition of IL-2 induced IL-10 production as a principle of phase-specific immunotherapy. J Immunol (2006) 177(7):4636–43 10.4049/jimmunol.177.7.4636 [DOI] [PubMed] [Google Scholar]
- 92.Yip HC, Karulin AY, Tary-Lehmann M, Hesse MD, Radeke H, Heeger PS, et al. Adjuvant-guided type-1 and type-2 immunity: infectious/noninfectious dichotomy defines the class of response. J Immunol (1999) 162(7):3942–9 [PubMed] [Google Scholar]
- 93.Shibaki A, Katz SI. Induction of skewed Th1/Th2 T-cell differentiation via subcutaneous immunization with Freund’s adjuvant. Exp Dermatol (2002) 11(2):126–34 10.1034/j.1600-0625.2002.110204.x [DOI] [PubMed] [Google Scholar]
- 94.Zhang YL, Jia K, Zhao BP, Li Y, Yuan CX, Yang JM, et al. Identification of Th1 epitopes within molecules from the lung-stage schistosomulum of Schistosoma japonicum by combining prediction analysis of the transcriptome with experimental validation. Parasitol Int (2012) 61(4):586–93 10.1016/j.parint.2012.05.010 [DOI] [PubMed] [Google Scholar]