Abstract
Auxin plays an essential role in root development. It has been a long-held dogma that auxin required for root development is mainly transported from shoots into roots by polarly localized auxin transporters. However, it is known that auxin is also synthesized in roots. Here we demonstrate that a group of YUCCA (YUC) genes, which encode the rate-limiting enzymes for auxin biosynthesis, plays an essential role in Arabidopsis root development. Five YUC genes (YUC3, YUC5, YUC7, YUC8 and YUC9) display distinct expression patterns during root development. Simultaneous inactivation of the five YUC genes (yucQ mutants) leads to the development of very short and agravitropic primary roots. The yucQ phenotypes are rescued by either adding 5 nM of the natural auxin, IAA, in the growth media or by expressing a YUC gene in the roots of yucQ. Interestingly, overexpression of a YUC gene in shoots in yucQ causes the characteristic auxin overproduction phenotypes in shoots; however, the root defects of yucQ are not rescued. Our data demonstrate that localized auxin biosynthesis in roots is required for normal root development and that auxin transported from shoots is not sufficient for supporting root elongation and root gravitropic responses.
Keywords: Auxin, Biosynthesis, Hormone, Root development, Transport
Introduction
Auxin is known to regulate almost every aspect of root development. In Arabidopsis, auxin has been shown to play a key role in primary root elongation, lateral root initiation, root hair development and root gravitropism (Benjamins and Scheres 2008, Petricka et al. 2012). Arabidopsis seedlings grown on auxin-containing media develop shorter primary roots, more lateral roots and more and longer root hairs than plants grown on media without exogenous auxin (Hobbie and Estelle 1994).
The leading hypothesis regarding how auxin regulates root development is that the formation of an auxin gradient is a key step for several processes of root development (Petricka et al. 2012). An auxin gradient is often visualized using the auxin reporters DR5–green fluorescent protein (GFP)/β-glucosidase (GUS) or DII-VENUS (Sabatini et al. 1999, Brunoud et al. 2012). During embryogenesis, a maximum of DR5-GFP activity is observed in the uppermost suspensor cells (Friml et al. 2003). The auxin maximum as shown by DR5–GFP is proposed to play an essential role for the specification of the hypophysis, which is the precursor for the root meristem. During seedling stages, DR5–GFP is highly expressed in the quiescent center, the adjacent columella cells and root cap. It is suggested that the observed auxin gradient instructs the patterning of the distal part of the roots (Sabatini et al. 1999). Lateral root initiation is also known to depend on the formation of an auxin maximum centered at the founder cells (Benkova et al. 2003). It has long been recognized that polar auxin transport, particularly the auxin transport mediated by PIN (PINFORMED) proteins, plays an essential role in creating and maintaining an auxin gradient (Wisniewska et al. 2006). Mathematic modeling based on the polarity of PIN proteins suggests that polar auxin transport is sufficient for generating an auxin maximum/gradient that guides Arabidopsis root growth (Grieneisen et al. 2007). It is generally believed that auxin required for root development is first synthesized in shoots and then transported into roots, mainly through the action of directional transporters such as the PINs (Petricka et al. 2012). However, recent studies have clearly demonstrated that auxin is also synthesized locally in roots (Ljung et al. 2005, Stepanova et al. 2008). It is important to determine whether root-produced auxin actually plays any physiological roles. Interestingly, localized auxin biosynthesis has been recognized to play essential roles in other developmental processes including embryogenesis, seedling growth, vascular patterning, phyllotaxis and flower development (Cheng et al. 2006, Cheng et al. 2007, Pinon et al. 2013).
In Arabidopsis, auxin is mainly synthesized from the TAA (TRYPTOPHAN AMINOTRANSFERASE OF ARABIDOPSIS)/YUCCA (YUC) pathway, in which tryptophan is first converted to indole-3-pyruvate (IPA) by the TAA family of aminotransferases (Zhao 2012). Subsequently, IAA is synthesized from IPA by the YUC family of flavin-containing monooxygenases (Zhao 2012). The founding member of the TAA family of aminotransferases was identified from three different genetic screens for mutants with: (i) altered shade avoidance responses; (ii) insensitivity to the ethylene biosynthetic precursor 1-aminocyclopropane-1-carboxylic acid (ACC); and (iii) resistance to the auxin transport inhibitor 1-N-naphthylphthalamic acid (NPA) (Stepanova et al. 2008, Tao et al. 2008, Yamada et al. 2009). The latter two screens were based on root phenotypes. The taa1 mutants have longer primary roots than wild-type plants when grown on media containing ACC or NPA. Inactivation of TAA1 and its close homolog TRYPTOPHAN AMINOTRANSFERASE RELATED2 (TAR2) leads to short and agravitropic roots. Both TAA1 and TAR2 are expressed in roots and shoots, and the taa1 tar2 double mutants display pleiotropic phenotypes in both roots and shoots (Stepanova et al. 2008, Tao et al. 2008, Yamada et al. 2009). Therefore, it is difficult to determine whether the root defects in taa1 tar2 are caused by deficiency of local auxin biosynthesis in roots or decreased auxin transport from the shoots to roots, or both. The YUC genes were identified in an activation-tagging screen for mutants with long hypocotyls, and the YUCs have been suggested as a rate-limiting enzyme for auxin biosynthesis (Zhao et al. 2001). There are 11 YUC genes in Arabidopsis, and the expression of each YUC gene is temporally and spatially regulated (Cheng et al. 2006, Cheng et al. 2007). So far, YUC genes have been shown to be essential for embryogenesis, seedling growth, vascular pattern formation and flower development. The physiological roles of YUC genes correlate well with their expression patterns. For example, YUC1 and YUC4 have overlapping expression patterns during flower development. Consequently, the yuc1 yuc4 double mutants have dramatic defects in flower development, whereas the single mutants do not have obvious defects (Cheng et al. 2006, Cheng et al. 2007).
Herein, we identified five YUC genes (YUC3, YUC5, YUC7, YUC8 and YUC9) that had detectable expression in Arabidopsis roots at seedling stages. Simultaneous inactivation of the five YUC genes (yucQ mutants) caused auxin deficiency in roots and led to the development of short primary roots and abnormal gravitropic responses. The root defects of yucQ were rescued either by adding auxin to growth media or by expressing a YUC gene in roots. However, overproduction of auxin in shoots did not rescue yucQ. We conclude that localized auxin biosynthesis in roots plays an essential role in root development and that the shoot-produced auxin is not sufficient to support normal root development.
Results
YUC genes are expressed in root cells
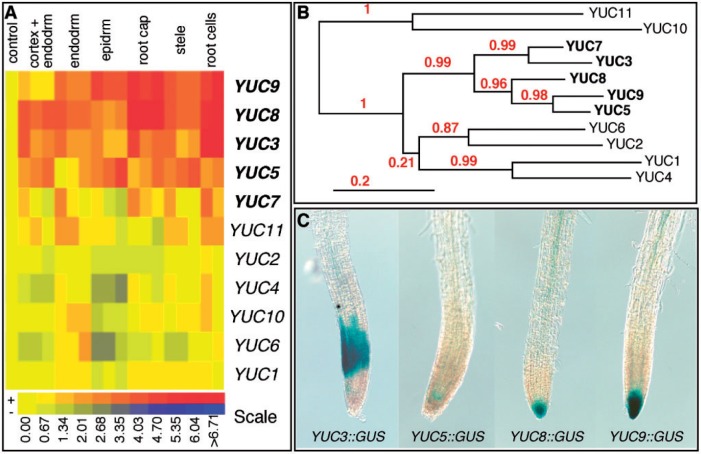
We analyzed the microarray data (Birnbaum et al. 2003, Toufighi et al. 2005) that are available to the general public to determine whether YUC genes are expressed in Arabidopsis roots. As shown in Fig. 1A, four YUC genes (YUC3, YUC5, YUC8 and YUC9) are highly expressed in root cells. YUC7 is also expressed in roots, but the expression levels are much lower compared with that of the above four YUC genes. The microarray data also revealed that the other six YUC genes had very low or no expression in the roots (Fig. 1A). Interestingly, the five YUC genes that are expressed in roots cluster together in a phylogenetic tree (Fig. 1B) (Cheng et al. 2006).
Fig. 1.
Identification of auxin biosynthesis genes in Arabidopsis roots. (A) Cluster analysis of the expression patterns of YUC genes in root cells. Microarray data from the public domain were clustered using methods previously described in (Birnbaum et al. (2003) and Toufighi et al. (2005). Note that YUC3, YUC5, YUC7, YUC8 and YUC9 are expressed in root cells whereas other YUC genes have low or no expression in roots. (B) Arabidopsis has 11 YUC flavin monooxygenases that can be divided into five clades (Cheng et al. 2006). The YUC genes with expression in roots form two close clades in the phylogenetic tree. (C) The patterns of GUS expression in roots of Arabidopsis transgenic plants that harbor YUC promoter::GUS constructs.
We further tested the expression patterns of the five YUC genes in roots by generating promoter::GUS reporter lines. Consistent with the microarray data analysis (Fig. 1A), four of the five YUC::GUS constructs showed expression in roots (Fig. 1C). We did not detect the expression of our YUC7::GUS construct. It is likely that the YUC7 promoter we used did not contain all of the necessary elements because other groups have shown that YUC7::GUS displayed expression in roots (Lee et al. 2011).
Overexpression of the root YUC genes leads to auxin overproduction phenotypes
We overexpressed the five YUC genes that are expressed in roots, using the Cauliflower mosaic virus (CaMV) 35S promoter to determine whether the YUC genes have the capacity to mediate auxin biosynthesis. We have previously shown that overexpression of YUC1, YUC2, YUC4 or YUC6 leads to the characteristic auxin overproduction phenotypes (Cheng et al. 2006, Cheng et al. 2007). The YUC1 overexpression lines have long hypocotyls and epinastic cotyledons when grown in light (Zhao et al. 2001). It was reported that overexpression of YUC5, YUC7, YUC8 and YUC9 using the CaMV 35S promoter also leads to auxin overproduction phenotypes similar to those observed in YUC1 overexpression lines (Woodward et al. 2005, Lee et al. 2011, Hentrich et al. 2013). We confirmed that overexpression of YUC5, YUC7, YUC8 and YUC9 caused auxin overproduction phenotypes (Fig. 2A). Moreover, we also observed the auxin overproduction phenotypes in the YUC3 overexpression lines (Fig. 2A). Our analyses of the YUC overexpression lines indicated that the five YUC genes in this study participate in auxin biosynthesis.
Fig. 2.
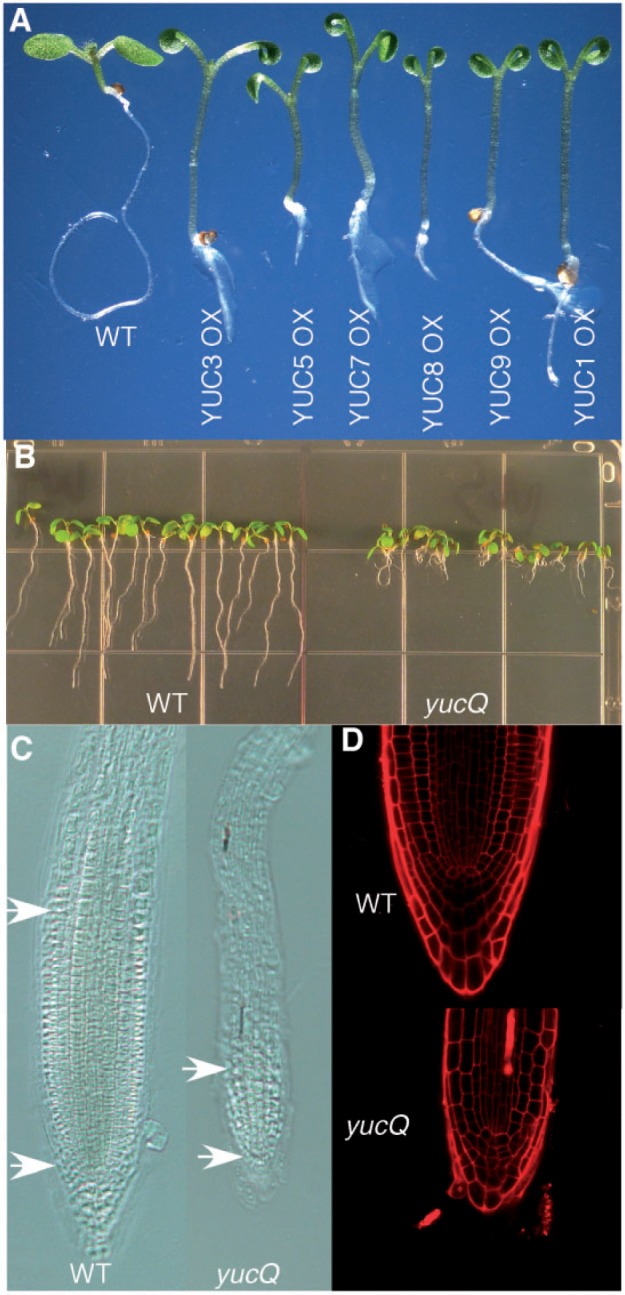
A group of YUC genes are important for auxin biosynthesis and root development. (A) Overexpression of the YUC genes causes auxin overproduction phenotypes. (B) Simultaneous inactivation of YUC3, YUC5, YUC7, YUC8 and YUC9 leads to dramatic defects in Arabidopsis root development. The yucQ mutants develop short and agravitropic primary roots. (C). Roots of yucQ are smaller in diameter and have a smaller meristem. (D) The yucQ roots have a smaller root cap and an abnormal meristem.
Inactivation of the YUC genes causes dramatic defects in root development
We obtained mutants for the YUC genes from publicly available sources (see the Materials and Methods). Single mutants of the YUC genes did not display obvious developmental defects. However, simultaneous inactivation of the five YUC genes (YUC3, YUC5, YUC7, YUC8 and YUC9) (yucQ mutants) led to severe developmental defects in roots (Fig. 2B). The yucQ mutants developed very short primary roots and had agravitropic root growth (Fig. 2B; Supplementary Fig. S1). Compared with wild-type plants, the primary roots of yucQ had much smaller meristems (Fig. 2C), and the cells in the meristem region in yucQ were enlarged and appeared to be partially differentiated (Fig. 2D), supporting the conclusion that auxin plays a key role in root meristem maintenance. Overall, the root cap of yucQ was small and less organized (Fig. 2D). We conclude that these YUC genes play critical roles in root development, including root meristem maintenance, and cell division/differentiation in the root cap.
The root defects are caused by partial auxin deficiency in roots
Because overexpression of each YUC gene led to auxin overproduction (Fig. 2A), we hypothesized that these YUC genes participate in auxin biosynthesis in roots and that the phenotypes of yucQ mutants are caused by auxin deficiency in roots. We first measured auxin levels in the yucQ roots and discovered that yucQ had about 55% less free IAA than wild-type roots (Fig. 3A). The observed low auxin levels were also consistent with the findings that the expression levels of the auxin reporter DR5–GFP were decreased in yucQ roots (Fig. 3B). More importantly, adding 5 nM free IAA into the growth media rescued the root growth phenotypes of yucQ (Fig. 3C), demonstrating that the root defects of yucQ were caused by partial auxin deficiency in roots. We noticed that the IAA-treated yucQ were still slightly agravitropic compared with wild-type plants (Supplementary Fig. S1). This could be due to the fact that the rescue conditions such as IAA concentrations were not optimum.
Fig. 3.
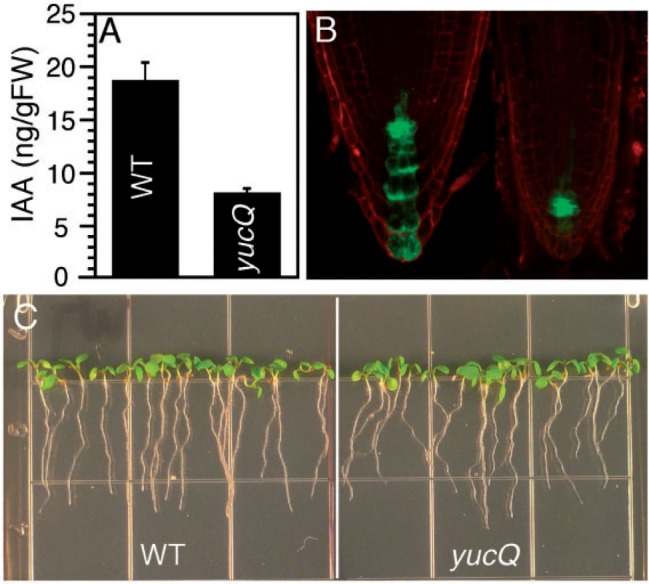
The root defects in yucQ are caused by partial auxin deficiency in roots. (A) Lower auxin levels are detected in yucQ roots than in wild-type roots. (B) The expression level of the auxin reporter DR5–GFP is dramatically decreased in yucQ. (C) The root defects of yucQ are partially rescued by 5 nM IAA.
Shoot-produced auxin is not sufficient for root development
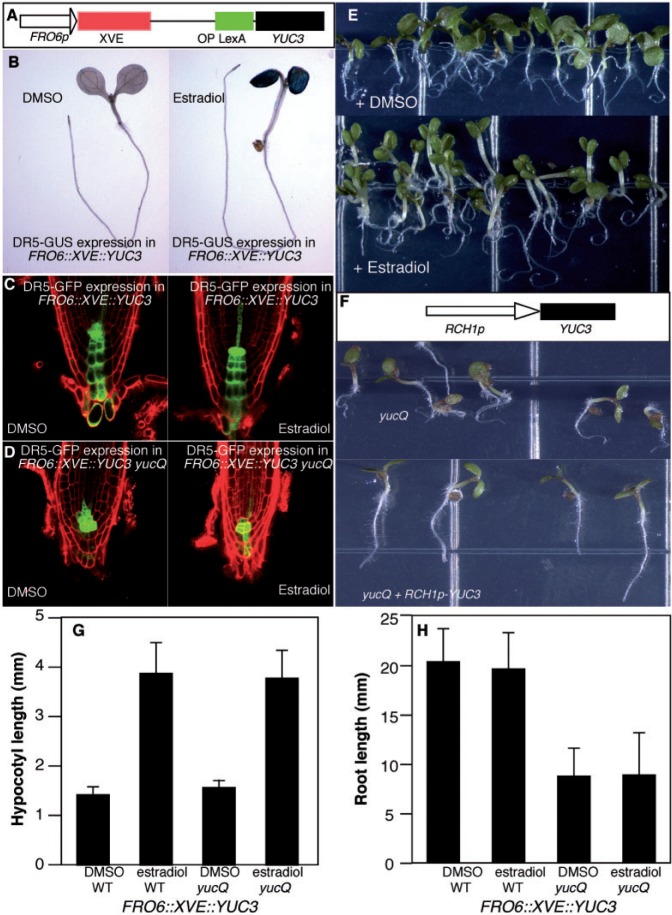
Early experiments showed that exogenously applied auxin is transported from shoots into roots (Eliasson 1972). However, the physiological functions of such a long-distance transport have not been thoroughly assessed. The yucQ mutants, which display defects in root development caused by partial auxin deficiency in roots, provide a system to test the role of shoot-produced auxin in root development. We used a shoot-specific promoter to express YUC3 in yucQ to determine whether shoot-produced auxin can rescue the root defects observed in yucQ. Because overexpression of YUC genes in shoots leads to dramatic auxin overproduction phenotypes and the resulting low seed set, we used an inducible system (Zuo et al. 2000) to overexpress YUC3 in shoots. YUC3 was under the control of a synthetic promoter and the synthetic transcription factor XVE (Zuo et al. 2000) (Fig. 4A). The shoot-specific expression was achieved by using the promoter FRO6 (Feng et al. 2006) to drive the expression of XVE. In the presence of estradiol, XVE moves into the nucleus and specifically activates the expression of YUC3 in shoots.
Fig. 4.
Shoot-produced auxin is not sufficient for rescuing the root defects of yucQ. (A) A construct for inducible expression of YUC3 in Arabidopsis shoots. The synthetic transcription factor XVE is under the control of a shoot-specific promoter FRO6p. Upon estradiol treatment, YUC3 is specifically expressed in the shoots. (B) Overexpression of YUC3 in wild-type shoots causes auxin overproduction in shoots. Note that the estradiol-treated seedling has long hypocotyls, epinastic cotyledons and elevated expression of the auxin reporter DR5–GUS in cotyledons. (C) DR5–GFP expression in roots in the mock-treated plants in the wild-type background and in the estradiol-treated plants. Although induction of YUC3 expression in shoots caused auxin overproduction in shoots, the expression of DR5–GFP was similar to that of the mock-treated plants. (D) DR5–GFP levels were reduced in yucQ, and overexpression of YUC3 in shoots did not lead to an increase in expression of DR5–GFP in roots. (E) Overproduction of auxin in shoots in yucQ did not rescue the root defects of yucQ. (F) The root phenotypes of yucQ (3 days old) were reversed when YUC3 was expressed using a root-specific promoter RCH1p. (G) Hypocotyl length of wild-type/FRO6::XVE::YUC3 and yucQ/FRO6::XVE::YUC3 plants treated with DMSO (dimethylsulfoxide) or estradiol. (H) The effects of induction of YUC3 expression in wild-type/FRO6::XVE::YUC3 and yucQ/FRO6::XVE::YUC3 plants on root length.
We also analyzed the effects of overexpressing YUC3 in shoots in wild-type plants. Induction of YUC3 expression in shoots resulted in the development of long hypocotyls and epinastic cotyledons, which are the characteristic auxin overproduction phenotypes (Fig. 4B). Consistent with auxin overproduction in the estradiol-treated plants, we observed elevated and ectopic expression of the auxin reporter DR5–GUS in the shoots (Fig. 4B). However, the DR5–GUS expression levels in root tips in estradiol-treated and mock-treated plants were very similar (Fig. 4B). We further obtained high-resolution images of DR5–GFP in both estradiol-treated and mock-treated plants (Fig. 4C). Consistent with our DR5–GUS data, induction of YUC3 expression in shoots in wild-type plants did not dramatically alter the expression of DR5–GFP expression in the root tip (Fig. 4C).
We then introduced the FRO6::XVE::YUC3 construct into yucQ to investigate whether shoot-produced auxin can rescue yucQ (Fig. 4E). As expected, estradiol treatments induced YUC3 expression and led to the development of long hypocotyls and epinastic cotyledons in yucQ (Fig. 4E). However, the roots of estradiol-treated plants still displayed the same agravitropic growth as yucQ plants (Fig. 4E). Furthermore, the expression levels of DR5–GFP in the estradiol-treated plants were not restored to the wild-type level (Fig. 4D). The root morphology of yucQ was not rescued either (Fig. 4D). We further measured the hypocotyl and the root length of wild-type/FRO6::XVE::YUC3 and yucQ/FRO6::XVE::YUC3 in the presence and absence of estradiol treatment (Fig. 4G, H). Induction of YUC3 expression in shoots by estradiol led to long hypocotyls in both wild-type and yucQ plants (Fig. 4G). Overexpression of YUC3 in shoots did not affect root length in either wild-type or yucQ plants (Fig. 4H). Moreover, auxin measurements in shoot and root tissues further demonstrated that overexpression of YUC3 in shoots led to auxin overproduction in shoots, but did not change the IAA concentrations in roots (Supplementary Fig. S2). Our results demonstrated that shoot-produced auxin is not sufficient to support normal root development.
We expressed YUC3 under the root-specific promoter RCH1p (Casamitjana-Martinez et al. 2003) in the yucQ background (Fig. 4F). As shown in Fig. 4F, the root defects of yucQ were rescued when YUC3 was expressed in roots. The root length of the rescued yucQ was very similar to that of the wild type.
Discussion
Spatial and temporal regulation of auxin levels is believed to be essential for normal root development. The predominant dogma regarding auxin in root development is that polar auxin transport, particularly auxin transport mediated by the PIN auxin efflux carriers, is necessary and sufficient for controlling auxin dynamics during root development (Petricka et al. 2012). The main auxin source for root development has been hypothesized to come from shoots. In this study, we clearly demonstrate that a group of YUC genes, which encode the rate-limiting enzymes for tryptophan-dependent auxin biosynthesis, were expressed in Arabidopsis roots. We show that auxin synthesized in roots by the YUC flavin-containing monooxygenases is required for normal root development and root gravitropic responses. Furthermore, we provide evidence that the shoot-produced auxin is not sufficient to support the development of roots.
The Arabidopsis genome contains 11 YUC genes, which have distinct and overlapping expression patterns (Fig. 1). Interestingly, the physiological functions of the YUC genes appear to correlate well with their expression patterns. We have previously shown that YUC1, YUC2, YUC4 and YUC6, which form two close subclades in a phylogenetic tree (Fig. 1B), are expressed in discrete groups of cells in seedlings, leaves and flowers (Cheng et al. 2006, Cheng et al. 2007). Consequently, YUC1, YUC2, YUC4 and YUC6 had overlapping functions in vegetative growth, the formation of veins and flower development (Cheng et al. 2006, Cheng et al. 2007). During flower development, the YUC2 and YUC6 subclade is involved in stamen development, whereas YUC1 and YUC4 play a more dominant role in the development of the carpel and other floral organs (Cheng et al. 2006, Cheng et al. 2007). We also showed previously that YUC1, YUC4, YUC10 and YUC11 are essential for embryogenesis (Cheng et al. 2006, Cheng et al. 2007). We have shown that YUC1, YUC2, YUC4, YUC6, YUC10 and YUC11 are expressed during embryogenesis and in shoots, but have very low or no expression in roots (Fig. 1A). The yuc1 yuc2 yuc4 yuc6 quadruple mutants have very severe defects in vascular patterning and flower development, but the quadruple mutants did not have obvious root defects, which is consistent with the expression patterns of the YUC genes (Fig. 1A). The five YUC genes that are expressed in roots are clustered together in a phylogenetic tree (Fig. 1B), and are essential for root development. The existence of multiple YUC genes in the Arabidopsis genome provides an effective way to control auxin levels temporally and spatially during development.
It is clear that Arabidopsis uses different sets of YUC genes to synthesize auxin for root and shoot development. Inactivation of the root YUC genes caused dramatic defects in root development (Fig. 2B), but did not cause obvious defects in shoots, indicating that local auxin production by the YUC proteins is required for normal root development. On the other hand, mutations in the shoot YUC genes led to defects in shoot development without severely affecting root development, suggesting that shoot-produced auxin may not be necessary for normal root development. Our studies on various yuc combinations suggest that auxin transport between shoots and roots cannot compensate local auxin deficiency in shoots or in roots. We further showed clearly that overproduction of auxin in shoots is not sufficient to rescue the root defects observed in yucQ mutants (Fig. 4), which are defective in auxin biosynthesis in roots.
YUC genes have been shown to function non-cell autonomously, suggesting that auxin is transported from the source cells to target cells (Tobena-Santamaria et al. 2002). The five YUC genes studied in this work were expressed in roots, but their expression patterns are distinct and overlapping (Fig. 1). The fact that single yuc mutants did not display obvious root phenotypes indicates that defects in auxin biosynthesis in some cells can be compensated by auxin synthesized in other cells. However, it is not clear how far auxin can travel to compensate auxin deficiency in cells. We have previously shown that auxin from both localized auxin biosynthesis and polar auxin transport contributes to the initiation of true leaves (Cheng et al. 2007). Development of Arabidopsis roots has been studied extensively, and many tools for root research are available. Further studies using cell type-specific promoters to drive auxin biosynthesis in roots of yucQ and in auxin transport mutants such as aux1 and pin2 will provide insights into the distance and directions of auxin transport required for root development.
Materials and Methods
The yuc mutants
The yuc mutants were genotyped using the following genotyping primer sets. Primers for yuc3 (GABI_376G12) are: yuc3LP, TTATAGCCCCTAAAACAGAGCATC; yuc3RP, GAAATGTGAAATACAACGACG; and the T-DNA primer CCCATTTGGACGTGAATGTAGACAC. The yuc5 (CSHL_GT6160) mutant was genotyped with primers: yuc5LP, CGGACTCTAATCAAAGTCCC; yuc5RP, GGAGATTTCAAAACTAGATTTG; and the T-DNA primer Ds3_1, ACCCGACCGGATCGTATCGGT. Primers for the yuc7 (SALK_059832) mutant are: yuc7LP, CATGGAGTGGGCTTATCTCTTTG; yuc7RP, ACGAAAAACAGAGCACCCTG; and the T-DNA primer JM-LB1, GGCAATCAGCTGTTGCCCGTCTCACTGGTG. Primers for yuc8 (SM_3.23299) are: the T-DNA primer spm32, TACGAATAAGAGCGTCCATTTTAGAGTGA; Yuc8LP, CATCCTCTCCACGTGGCTTCC; and yuc8RP, GAACTGACGCTTCGTCGGGTAC. Genotyping primers for yuc9 (SAIL_762_D07) are: yuc9-LP, GCTCGGTAAGCAAAACAAAACTG; yuc9RP, GAAGGAAATGCCCAATGAGAC; and the T-DNA primer SAIL-LB1, GCCTTTTCAGAAATGGATAAATAGCCTTGCTTCC. The genotyping results are shown in Supplementary Fig. S3.
The YUC::GUS reporter lines
The lines were constructed by amplifying the YUC promoters using the primers specified below. The YUC promoters then were cloned into the pBI101.3 vector with SalI at the 5′ end and SmaI at the 3′ end to drive the expression of the GUS reporter. Primers for YUC3 promoters are: Y3pro5p, TCAATCAACGAGAATGGAACACGCC; and Y3pro3p, CTTCCACAGAGTTTAGAGTTTGC. The YUC5 promoter was amplified using Y5pro5p, CCAAACTCAAGTGCCTATGG; and Y5pro3p, CTTTAGGGGTGAGTTTGATCG. Y7pro5p, GCCCGATTCAACTCCACTACG; and Y7pro3p, CTTATTGGGAGCTTTTTAGTG were used to amplify the YUC7 promoter. Primers for the YUC9 promoter are: Y9pro3p, TTTCTTGAGTGAGTTTTTGAATG; and Y9pro5p, CACAAAGATCAATACAAAAGCC. We cloned the YUC8 promoter using primers: Y8pro5p, TCTTTTTTTATAAGTTTCTTTAATAAG; and Y8pro3p, CGCATGACTCAGTTGTTGCGATC.
Plant growth conditions
Seeds were surface-sterilized and sown on vertical square plates with half-strength Murashige and Skoog (MS) medium (1/2 MS salts, 1.5% sucrose, 0.65% agar), stratified in the dark at 4°C for 2 d, germinated, and grown at 22°C under a 16 h light/8 h dark cycle for 5 d or for times specifically mentioned.
Microscopic analysis
Propidium iodide staining was performed as previously described (Helariutta et al. 2000). Fluorescence was visualized under a Zeiss LSM 710 confocal microscope. DIC (differential interference contrast) pictures were taken with a Leica DM5000 microscope. GUS staining and measurements of the gravitropic response were performed as previously described (Li et al. 2010).
Auxin measurements
Endogenous levels of IAA in Arabidopsis were analyzed by a previously reported method (Okumura et al. 2013).
Tissue-specific auxin production
Root-specific auxin production was achieved by expressing YUC3 under the control of the RCH1 promoter (Casamitjana-Martinez et al. 2003). The RCH1 (At5g48940) promoter and YUC3 genomic DNA were fused together and cloned into the XhoI and SpeI sites of the pER8 plasmid (Zuo et al. 2000). The RCH1 promoter was amplified using the primers: RCH1pF, AGCTAGTCGACTCTAGCCTGTGTGAGAAGCGAGGAGGGGT; and RCH1pR, TGATAGACTTCTTGTTATTATTTCCATACATAAGAGTTTTTTTCTTTGCATTTGGCTCT. The primers for amplifying YUC3 were: YUC3F1, AGAGCCAAATGCAAAGAAAAAAACTCTTATGTATGGAAATAATAACAAGAAGTCTATCA; and YUC3R1, CTGGGAGGCCTGGATCGATTAGAAATGTGAAATACAACGACGATGA.
To express YUC3 in Arabidopsis shoots, the FRO6 promoter was used. The FRO6 (At5g49730) promoter (Feng et al. 2006) was amplified using the following two primers: FRO6pF, 5′-CAATATATCCTGTCAAACCGATGCTCTCAAGGCCAAAGAACA-3′; and FRO6pR, 5′-GCCGTTAACGCTTTCATCTTTATTTGAATTTCCACTTCTCAGTG-3′. The G10-90 promoter in pER8 was removed and an Mfel site was introduced before XVE. The FRO6 promoter was inserted at the MfeI site. YUC3 genomic DNA was cloned into XhoI and SpeI sites using the primers YUC3F2, AGCTAGTCGACTCTAGCCATGTATGGAAATAATAACAAGAAGTCTATCA, and YUC3R1.
Supplementary data
Supplementary data are available at PCP online.
Funding
This work was supported by the National Institutes of Health [R01GM68631 to Y.Z.]; Japan Science and Technology Agency (JST) [PRESTO (to H.K.)].
Disclosures
The authors have no conflicts of interest to declare.
Supplementary Material
Glossary
Abbreviations
- ACC
1-aminocyclopropane-1-carboxylic acid
- GFP
green fluorescent protein
- GUS
β-glucuronidase
- IPA
indole-3-pyruvate
- NPA
1-N-naphthylphthalamic acid
- TAA
TRYPTOPHAN AMINOTRANSFERASE OF ARABIDOPSIS
- TAR
TRYPTOPHAN AMINOTRANSFERASE RELATED
- YUC
YUCCA flavin monooxygenase
References
- Benjamins R, Scheres B. Auxin: the looping star in plant development. Annu. Rev. Plant Biol. 2008;59:443–465. doi: 10.1146/annurev.arplant.58.032806.103805. [DOI] [PubMed] [Google Scholar]
- Benkova E, Michniewicz M, Sauer M, Teichmann T, Seifertova D, Jurgens G, et al. Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell. 2003;115:591–602. doi: 10.1016/s0092-8674(03)00924-3. [DOI] [PubMed] [Google Scholar]
- Birnbaum K, Shasha DE, Wang JY, Jung JW, Lambert GM, Galbraith DW, et al. A gene expression map of the Arabidopsis root. Science. 2003;302:1956–1960. doi: 10.1126/science.1090022. [DOI] [PubMed] [Google Scholar]
- Brunoud G, Wells DM, Oliva M, Larrieu A, Mirabet V, Burrow AH, et al. A novel sensor to map auxin response and distribution at high spatio-temporal resolution. Nature. 2012;482:103–106. doi: 10.1038/nature10791. [DOI] [PubMed] [Google Scholar]
- Casamitjana-Martinez E, Hofhuis HF, Xu J, Liu CM, Heidstra R, Scheres B. Root-specific CLE19 overexpression and the sol1/2 suppressors implicate a CLV-like pathway in the control of Arabidopsis root meristem maintenance. Curr. Biol. 2003;13:1435–1441. doi: 10.1016/s0960-9822(03)00533-5. [DOI] [PubMed] [Google Scholar]
- Cheng Y, Dai X, Zhao Y. Auxin biosynthesis by the YUCCA flavin monooxygenases controls the formation of floral organs and vascular tissues in Arabidopsis. Genes Dev. 2006;20:1790–1799. doi: 10.1101/gad.1415106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y, Dai X, Zhao Y. Auxin synthesized by the YUCCA flavin monooxygenases is essential for embryogenesis and leaf formation in Arabidopsis. Plant Cell. 2007;19:2430–2439. doi: 10.1105/tpc.107.053009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eliasson L. Translocation of shoot-applied indolylacetic acid into the roots of Populus tremula. Physiol. Plant. 1972;27:412–416. [Google Scholar]
- Feng H, An F, Zhang S, Ji Z, Ling HQ, Zuo J. Light-regulated, tissue-specific, and cell differentiation-specific expression of the Arabidopsis Fe(III)-chelate reductase gene AtFRO6. Plant Physiol. 2006;140:1345–1354. doi: 10.1104/pp.105.074138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friml J, Vieten A, Sauer M, Weijers D, Schwarz H, Hamann T, et al. Efflux-dependent auxin gradients establish the apical–basal axis of Arabidopsis. Nature. 2003;426:147–153. doi: 10.1038/nature02085. [DOI] [PubMed] [Google Scholar]
- Grieneisen VA, Xu J, Maree AF, Hogeweg P, Scheres B. Auxin transport is sufficient to generate a maximum and gradient guiding root growth. Nature. 2007;449:1008–1013. doi: 10.1038/nature06215. [DOI] [PubMed] [Google Scholar]
- Helariutta Y, Fukaki H, Wysocka-Diller J, Nakajima K, Jung J, Sena G, et al. The SHORT-ROOT gene controls radial patterning of the Arabidopsis root through radial signaling. Cell. 2000;101:555–567. doi: 10.1016/s0092-8674(00)80865-x. [DOI] [PubMed] [Google Scholar]
- Hentrich M, Bottcher C, Duchting P, Cheng Y, Zhao Y, Berkowitz O, et al. The jasmonic acid signaling pathway is linked to auxin homeostasis through the modulation of YUCCA8 and YUCCA9 gene expression. Plant J. 2013;74:626–637. doi: 10.1111/tpj.12152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobbie L, Estelle M. Genetic approaches to auxin action. Plant Cell Environ. 1994;17:525–540. doi: 10.1111/j.1365-3040.1994.tb00147.x. [DOI] [PubMed] [Google Scholar]
- Lee M, Jung JH, Han DY, Seo PJ, Park WJ, Park CM. Activation of a flavin monooxygenase gene YUCCA7 enhances drought resistance in Arabidopsis. Planta. 2011;235:923–938. doi: 10.1007/s00425-011-1552-3. [DOI] [PubMed] [Google Scholar]
- Li Y, Dai X, Cheng Y, Zhao Y. NPY genes play an essential role in root gravitropic responses in Arabidopsis. Mol. Plant. 2010;4:171–179. doi: 10.1093/mp/ssq052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ljung K, Hull AK, Celenza J, Yamada M, Estelle M, Normanly J, et al. Sites and regulation of auxin biosynthesis in Arabidopsis roots. Plant Cell. 2005;17:1090–1104. doi: 10.1105/tpc.104.029272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okumura K, Goh T, Toyokura K, Kasahara H, Takebayashi Y, Mimura T, et al. GNOM/FEWER ROOTS is required for the establishment of an auxin response maximum for Arabidopsis lateral root initiation. Plant Cell Physiol. 2013;54:406–417. doi: 10.1093/pcp/pct018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petricka JJ, Winter CM, Benfey PN. Control of Arabidopsis root development. Annu. Rev. Plant Biol. 2012;63:563–590. doi: 10.1146/annurev-arplant-042811-105501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinon V, Prasad K, Grigg SP, Sanchez-Perez GF, Scheres B. Local auxin biosynthesis regulation by PLETHORA transcription factors controls phyllotaxis in Arabidopsis. Proc. Natl Acad. Sci. USA. 2013;110:1107–1112. doi: 10.1073/pnas.1213497110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabatini S, Beis D, Wolkenfelt H, Murfett J, Guilfoyle T, Malamy J, et al. An auxin-dependent distal organizer of pattern and polarity in the Arabidopsis root. Cell. 1999;99:463–472. doi: 10.1016/s0092-8674(00)81535-4. [DOI] [PubMed] [Google Scholar]
- Stepanova AN, Robertson-Hoyt J, Yun J, Benavente LM, Xie DY, Dolezal K, et al. TAA1-mediated auxin biosynthesis is essential for hormone crosstalk and plant development. Cell. 2008;133:177–191. doi: 10.1016/j.cell.2008.01.047. [DOI] [PubMed] [Google Scholar]
- Tao Y, Ferrer JL, Ljung K, Pojer F, Hong F, Long JA, et al. Rapid synthesis of auxin via a new tryptophan-dependent pathway is required for shade avoidance in plants. Cell. 2008;133:164–176. doi: 10.1016/j.cell.2008.01.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobena-Santamaria R, Bliek M, Ljung K, Sandberg G, Mol JN, Souer E, et al. FLOOZY of petunia is a flavin mono-oxygenase-like protein required for the specification of leaf and flower architecture. Genes Dev. 2002;16:753–763. doi: 10.1101/gad.219502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toufighi K, Brady SM, Austin R, Ly E, Provart NJ. The Botany Array Resource: e-Northerns, Expression Angling, and promoter analyses. Plant J. 2005;43:153–163. doi: 10.1111/j.1365-313X.2005.02437.x. [DOI] [PubMed] [Google Scholar]
- Wisniewska J, Xu J, Seifertova D, Brewer PB, Ruzicka K, Blilou I, et al. Polar PIN localization directs auxin flow in plants. Science. 2006;312:883. doi: 10.1126/science.1121356. [DOI] [PubMed] [Google Scholar]
- Woodward C, Bemis SM, Hill EJ, Sawa S, Koshiba T, Torii KU. Interaction of auxin and ERECTA in elaborating Arabidopsis inflorescence architecture revealed by the activation tagging of a new member of the YUCCA family putative flavin monooxygenases. Plant Physiol. 2005;139:192–203. doi: 10.1104/pp.105.063495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada M, Greenham K, Prigge MJ, Jensen PJ, Estelle M. The TRANSPORT INHIBITOR RESPONSE2 gene is required for auxin synthesis and diverse aspects of plant development. Plant Physiol. 2009;151:168–179. doi: 10.1104/pp.109.138859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y. Auxin biosynthesis: a simple two-step pathway converts tryptophan to indole-3-acetic acid in plants. Mol. Plant. 2012;5:334–338. doi: 10.1093/mp/ssr104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Christensen SK, Fankhauser C, Cashman JR, Cohen JD, Weigel D, et al. A role for flavin monooxygenase-like enzymes in auxin biosynthesis. Science. 2001;291:306–309. doi: 10.1126/science.291.5502.306. [DOI] [PubMed] [Google Scholar]
- Zuo J, Niu QW, Chua NH. Technical advance: an estrogen receptor-based transactivator XVE mediates highly inducible gene expression in transgenic plants. Plant J. 2000;24:265–273. doi: 10.1046/j.1365-313x.2000.00868.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.