Abstract
It is poorly understood how driver mutations in cancer genes work together to promote tumor development. Renal cell carcinoma (RCC) offers a unique opportunity to study complex relationships among cancer genes. The four most commonly mutated genes in RCC of clear-cell type (the most common type) are two-hit tumor suppressor genes and they cluster in a 43 Mb region on chromosome 3p that is deleted in ~90% of tumors: VHL (mutated in ~80%), PBRM1 (~50%), BAP1 (~15%) and SETD2 (~15%). Meta-analyses that we conducted show that mutations in PBRM1 and SETD2 co-occur in tumors at a frequency higher than expected by chance alone, indicating that these mutations may cooperate in tumorigenesis. In contrast, consistent with our previous results, mutations in PBRM1 and BAP1 tend to be mutually exclusive. Mutation exclusivity analyses (often confounded by lack of statistical power) raise the possibility of functional redundancy. However, mutation exclusivity may indicate negative genetic interactions, as proposed herein for PBRM1 and BAP1, and mutations in these genes define RCC with different pathologic features, gene expression profiles, and outcomes. Negative genetic interactions among cancer genes point toward broader context-dependencies of cancer gene action beyond tissue dependencies. Understanding cancer genes dependencies may unravel vulnerabilities that can be exploited therapeutically.
Introduction
Cancer research has been revolutionized by massively parallel sequencing. Up to 5% of protein-coding genes are potential cancer genes implicated in the development of the disease (1, 2). Many novel cancer genes have been discovered providing inroads into the molecular pathogenesis of tumors and setting a foundation for a molecular classification of cancer (3, 4).
Cancer drivers may be distinguished from passenger genes by their mutation at frequencies higher than expected by chance alone. Oncogene drivers, which are typically activated by mutation and tend to be dominant, may be recognized by the presence of recurrent missense mutations at a limited number of residues. However, multiple residues may be targeted by mutations that disrupt autoinhibitory domains (as in mTOR). In contrast, tumor suppressor genes, which are inactivated by mutation and are typically recessive, may be disrupted by a variety of alterations including insertions, deletions, nonsense, missense and splice-site mutations. Missense mutations in tumor suppressor genes are often used to identify domains important for function, but these analyses are confounded by mutations disrupting secondary or tertiary structure and causing protein instability. Typically one allele of a tumor suppressor gene is disrupted by an intragenic mutation and the other is lost as part of a large deletion, which results in loss of heterozygosity (LOH).
Further complexity arises from mutation heterogeneity in tumors (5), which results from plasticity and clonal evolution (6). According to their prevalence, somatic mutations may be divided into ubiquitous, shared, and private. Ubiquitous mutations (present in all tumor cells) encompass truncal driver events. However, not every ubiquitous mutation is a driver mutation (pre-existing mutations in the lineage giving rise to the initial tumor clone make up ubiquitous passengers) (7). Conversely, not every driver mutation may be ubiquitous, and mutations conferring invasive or metastatic potential may be found in only a subset of cells in primary tumors. This complexity can be advantageous. It can be harnessed to identify driver genes that tend to be mutated early, ubiquitous drivers. Pathways deregulated by ubiquitous drivers are optimal targets for drug therapies that seek to affect all tumor cells.
One of the challenges hampering mutation detection in solid tumors is contamination by normal stroma. DNA from stromal cells dilutes tumor DNA and reduces the sensitivity for mutation discovery. In addition, contamination makes it difficult to assess how homogeneously a given mutation is present across tumor cells in a sample. The prevalence of a mutation can be estimated by the mutant allele ratio (MAR) – the fraction of the mutant over the mutant plus wild-type sequences for a given mutation (8). A heterozygous mutation would be expected to have a MAR of 0.5. Lower MARs may indicate that the mutation is present in only a subset of tumor cells in a sample (not a ubiquitous mutation), but this assessment is precluded by stromal contamination (8). Mutation sensitivity and MAR accuracy can be improved by the analysis of tumorgrafts, human tumors implanted in mice (8). In tumorgrafts, the stroma is replaced by the host, and tumorgrafts preserve the characteristics of human tumors (9). However, studies in tumorgrafts rely on the ability to specifically query the human genome. Despite this limitation, MAR analyses in tumorgrafts can be instrumental to determine the prevalence of a mutation in a sample. Accurate MARs are helpful in a variety of other contexts. When mutations are found in areas of copy neutral LOH, MARs can show whether the mutation is homozygous (8). In addition, for mutations in areas of amplification, whether the mutant or the wild-type sequence is amplified can be determined with accurate MARs (8).
A comprehensive list of mutations in a tumor, together with an understanding of their prevalence and functional significance should pave the way for better analyses of genetic interactions among cancer genes.
Evaluating functional relationships among cancer genes
Unraveling relationships among genes driving tumorigenesis is a challenge and represents the next frontier. A form of genetic interaction commonly reported is mutation exclusivity. Exclusivity is predicated of genes that are mutated in a particular tumor type, but not simultaneously. Often, mutation exclusivity is interpreted as evidence of functional redundancy. This is illustrated, for instance, by mutations in p16, D-type cyclins, CDK4, and retinoblastoma, which tend to be exclusive and disrupt the same cell cycle regulatory pathway.
Mutation exclusivity is frequently misinterpreted due to insufficient statistical power. As an example, when two genes are mutated at a frequency of 5%, the number of tumors required to show that a lack of mutation co-occurrence is due to a genetic interaction (as opposed to chance alone – after all, each gene is mutated in only 5% of the tumors) is 1,330. Thus, mutation exclusivity analyses may require meta-analyses of multiple studies, particularly when the interactions involve genes mutated at low frequencies.
Mutations and physical location
Another level of complexity is introduced by the physical location of cancer genes in chromosomes. In fact, the architecture of amplifications and deletions in tumors may be far more informative that previously appreciated. Traditionally, amplifications and deletions have typically been thought to be driven by a single gene, but more than one gene may be implicated in each region (see below). This has important methodologically implications as the hunt for cancer genes may need to be redirected towards genes flanking a common region of amplification or deletion, rather than those at the center.
Renal cancer, a paradigm
Renal cell carcinoma (RCC) offers a unique opportunity to study complex relationships among cancer genes. RCC is classified histologically into several types, including clear-cell (ccRCC), the most common type. Positional cloning studies of kindreds with a ccRCC predisposition syndrome, von Hippel-Lindau (VHL), led to the identification of the eponymic gene, VHL (10). Subsequently, VHL was found to be frequently mutated in sporadic ccRCC (11). VHL is mutated in ~80% of sporadic ccRCC and is inactivated by methylation in an additional 10% (12, 13). VHL is rarely mutated in other sporadic tumors (14), suggesting that the tumor suppressor function of VHL is limited to a small number of cell types. The VHL gene encodes the substrate recognition subunit of an E3 ubiquitin ligase complex that triggers the degradation of, among others, the α-subunit of hypoxia-inducible factor (HIF) transcription factors (15). The VHL gene is on 3p25.3, and for many years it was thought to explain LOH at 3p in ccRCC.
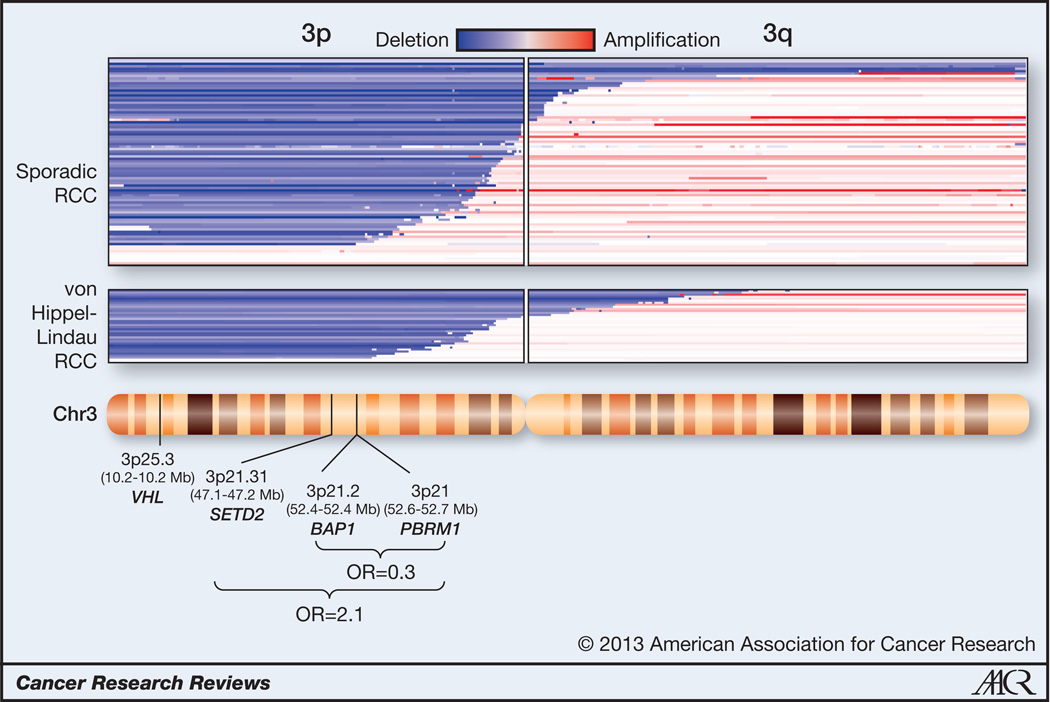
Interestingly, 4 tumor suppressor genes have been identified in a relatively small (43 Mb) region on 3p. These genes are: VHL, SET domain containing 2 (SETD2) (16), BRCA1 associated protein-1 (BAP1) (8, 17) and Polybromo 1 (PBRM1) (18). Each functions as a classical two-hit tumor suppressor gene, and an analysis we performed of previously published data (8, 19), shows that this region is lost in 90% of sporadic ccRCC (Fig. 1). PBRM1 is mutated in approximately 50% of ccRCC (18), and encodes BAF180 (herein referred to as PBRM1), the chromatin targeting subunit of a SWI/SNF nucleosome remodeling complex. Both SETD2 and BAP1 are mutated in ~15% of ccRCC. SETD2 is a histone H3K36 methyltransferase (20) specifically implicated in trimethylation (21). BAP1 is a nuclear deubiquitinase (22, 23), and while substrates have been identified in Drosophila (24) and mammals (25–28), the relevant substrate(s) in RCC remain unknown (8).
Fig. 1. Schematic of chromosome 3 with the estimated position of VHL, SETD2, BAP1, and PBRM1 genes and corresponding DNA copy number alterations in sporadic and familial (von Hippel-Lindau syndrome) clear cell renal cell carcinoma.
Chromosome 3 ideogram (NCBI build 37.5 [hg19]) with superimposed copy number analyses of primary ccRCC tumors from GSE14994 and GSE25540. Odds ratios (OR) for the finding of simultaneous mutations among the indicated genes are shown.
Cooperation among tumor suppressor genes on 3p
We performed meta-analyses to test for genetic interactions among 3p genes. Given, the mutation frequencies of the different genes, we used PBRM1 as a reference. PBRM1 is mutated at a high enough frequency to be able to draw conclusions and yet not uniformly as VHL. We identified studies (or datasets) reporting mutations in PBRM1 together with either SETD2 or BAP1. Adding to the challenge of uncovering genetic interactions (as determined by the frequency of VHL mutations detected), mutation sensitivity was seemingly low across all of the studies available (16, 17, 29).
The Sanger Institute sequenced 348 ccRCC for both PBRM1 and SETD2. One hundred and eleven tumors were found to be solely mutated for PBRM1 and seven were found solely mutant for SETD2 (18). Given the individual mutation rates for PBRM1 and SETD2, five tumors were expected to have mutations in both genes, but eight were found (p=0.16; Table 1). In a study by Guo et al. involving 98 ccRCC, eighteen tumors were solely mutant for PBRM1 and one was solely mutant for SETD2 (17). Again, given the mutation rates for PBRM1 and SETD2, one tumor may have been expected to have mutations in both genes, but three were found (p=0.030; Table 1). Hakimi et al. analyzed 185 ccRCC; forty-eight were found solely mutated for PBRM1 and eight solely mutated for SETD2. Given these mutation frequencies, four double mutant tumors were expected, but six were found (p=0.24; Table 1). Finally, The Cancer Genome Atlas consortium (30) released results of 293 tumors and ninety tumors were found with mutations in PBRM1 and seventeen with mutations in SETD2. Twelve tumors were expected to have mutations in both PBRM1 and SETD2, but sixteen were found (p=0.13; Table 1). Across all the studies, the number of tumors with mutations in both PBRM1 and SETD2 exceeded the number expected by chance alone. However, this difference reached statistical significance only in the study by Guo et al. (17). Nonetheless, when considered together, among 924 ccRCC, there were 267 tumors with mutations only in PBRM1, 33 with mutations only in SETD2, and 33 with mutations in both SETD2 and PBRM1 (Table 1). The number of tumors expected to have mutations in both genes by chance alone was 21, and 33 were found. While the difference in absolute numbers is small, it represents an increase by one half, and the probability that this finding occurred by chance alone is 0.003. Overall, the frequency of mutations in SETD2 was two-fold higher for PBRM1-mutant tumors than wild-type tumors (Odds ratio [OR] = 2.1; 95% confidence interval [CI] 1.3 – 3.5).
Table 1.
Cooperativity between SETD2 and PBRM1 in ccRCC.
| Study | n | PBRM1 | SETD2 |
SETD2/ PBRM1 |
Expected Double mutants |
p value | Odds Ratio (95% CI) |
|---|---|---|---|---|---|---|---|
| Sanger Institute (16, 18) | 348 | 111 | 7 | 8 | 5 (2–8) | 0.16 | 2.3 (0.8 – 6.5) |
| Guo et al. (17) | 98 | 18 | 1 | 3 | 1 (0–2) | 0.03 | 12.7 (1.2 – 129) |
| Hakimiet al. (29) | 185 | 48 | 8 | 6 | 4 (2–7) | 0.24 | 1.9 (0.6 – 5.8) |
| TCGA | 293 | 90 | 17 | 16 | 12 (7–14) | 0.13 | 1.8 (0.9 – 3.7) |
| Total | 924 | 267 | 33 | 33 | 21 (17–25) | 0.003 | 2.1 (1.3 – 3.5) |
Number of tumors with specific mutations and expected frequencies. The range of expected double mutants was calculated based on a hypergeometric distribution. Differences between actual and expected values were evaluated with a Fisher’s exact test. The Mantel-Haenszel test was used to integrate odds ratios from the different studies. KIRC TCGA data obtained from January, 2013 release. Data from the combination of these studies (the total) are highlighted in bold. Significant P values are also in bold.
These results suggest that mutations in PBRM1 and SETD2 cooperate in renal tumorigenesis. They assume that mutations occur independently and in the same tumor cells, and functional studies will be required for confirmation. The biological basis for this cooperation remains to be determined, but plausible models may be proposed based on the function of these proteins, in particular, because both proteins converge on histones, one as writer of a histone mark (SETD2) and the other one as a reader (PBRM1). Despite this cooperation, however, no differences in overall survival were found between patients with PBRM1-mutated tumors and those with tumors mutated in both SETD2 and PBRM1 (Suppl. Fig. 1).
Antagonism among tumor suppressor genes on 3p
We reported previously that mutations in PBRM1 and BAP1 are largely mutually exclusive (8), which stands in contrast to the findings from meta-analyses reported herein of PBRM1 and SETD2. Among 176 ccRCC we analyzed, we found 89 tumors with mutations solely in PBRM1 and 21 with mutations solely in BAP1. By chance, 13 tumors would have been expected to have mutations in both genes, but only 3 tumors were found. The probability that this observation was by chance alone was very low (p=0.00003) (8).
Guo et al. and Hakimi et al. have also reported BAP1 and PBRM1 mutations in ccRCC (17, 29). In both studies, the sensitivity for mutation detection was seemingly low, and consequently statistical power was insufficient. However, in both instances, fewer tumors were found with simultaneous mutations in both genes than were expected by chance alone (Table 2). Similarly, in data from the TCGA, there was an under-representation of tumors with mutations in both BAP1 and PBRM1 (Table 2). In the TCGA study, which is the largest, among 293 tumors, there were 101 with mutations solely in PBRM1 (independently of SETD2) and 22 with mutations solely in BAP1. Given the relative frequencies of tumors individually mutated for BAP1 and PBRM1, 10 tumors would have been expected to have mutations in both genes, but only 5 were found (p=0.058; Table 2). When combined, these 3 studies evaluated 576 tumors and among them there were 175 tumors with mutations solely in PBRM1 and 40 with mutations solely in BAP1. Considered together, 14 tumors would have been expected with mutations in both BAP1 and PBRM1, but only 6 were found, and the p value was significant (p=0.004). Thus, the odds of having a BAP1 mutation in PBRM1-mutant tumors are one third of those for wild-type tumors (OR=0.29; 95% CI, 0.12 – 0.70). While we cannot exclude that mutation co-occurrence rates may be affected by epigenetic changes (31) (or other factors), these data suggest that simultaneous mutations in BAP1 and PBRM1 are negatively selected for in ccRCC.
Table 2.
Antagonism between BAP1 and PBRM1 in ccRCC.
| Study | n | PBRM1 | BAP1 |
BAP1/ PBRM1 |
Expected double mutants |
p value | Odds Ratio (95% CI) |
|---|---|---|---|---|---|---|---|
| Peña-Llopis et al. (8) | 176 | 89 | 21 | 3 | 13 (9–16) | 0.00003 | 0.10 (0.03 – 0.35) |
| Guo et al. (17) | 98 | 21 | 8 | 0 | 2 (0–4) | 0.20 | 0.19 (0.01 – 3.43) |
| Hakimiet al. (29) | 185 | 53 | 10 | 1 | 3 (1–5) | 0.18 | 0.23 (0.03 – 1.83) |
| TCGA | 293 | 101 | 22 | 5 | 10 (7–13) | 0.058 | 0.37 (0.14 – 1.01) |
| Total | 576 | 175 | 40 | 6 | 14 (11–18) | 0.004 | 0.29 (0.12 – 0.70) |
Number of tumors with specific mutations and expected frequencies. The range of expected double mutants was calculated based on a hypergeometric distribution. Differences between actual and expected values were evaluated with a Fisher’s exact test. The Mantel-Haenszel test was used to integrate odds ratios from the different studies. A fixed 0.5 correction was used to find the odds ratio when there was a frequency of zero. KIRC TCGA data obtained from January, 2013 release. Note that differences in the number of tumors with mutations in PBRM1 with Table 1 reflect how tumors are divided according to SETD2 or BAP1 status. Data from the combination of these studies (the total) are highlighted in bold. Significant P values are also in bold.
Mutation exclusivity is often interpreted to indicate functional redundancy. However, differences in pathological features, gene expression and outcomes between tumors with BAP1 and PBRM1 mutations suggest that BAP1 and PBRM1 are not functionally redundant. BAP1-mutant tumors tend to be of high grade, whereas tumors exclusively mutated for PBRM1 are typically of low grade (8). BAP1-mutant tumors, but not PBRM1-mutant tumors, are associated with activation of the mTORC1 pathway (8, 32), a critical pathway in ccRCC (33). BAP1- and PBRM1-mutant tumors are associated with different gene expression signatures. We analyzed 308 ccRCC from the TCGA that had RNA-Seq data available and found that when compared to the rest, 3,250 genes distinguished the BAP1-mutant group (n=20) and 2,235 genes distinguished the PBRM1-mutant group (n=66) (32). In contrast, when groups of tumors were assembled arbitrarily, the number of genes that distinguished these random groups from the rest was less than 200. The difference in the number of genes associated with the BAP1- and PBRM1-mutant groups vs. the random groups was highly statistically significant (p<0.0001) (32). These data indicate that the signatures identified are highly specific. The BAP1- and PBRM1-mutant signatures did not overlap beyond what was expected by chance alone, indicating that they were different (32). BAP1-mutant tumors and PBRM1-mutant tumors are also associated with different outcomes in patients (32). Whereas the median overall survival for patients with BAP1-mutant tumors is 4.6 years (95% CI, 2.1–7.2), for patients with PBRM1-mutant tumors, median survival is 10.6 years (95% CI, 9.8–11.5; hazard ratio, 2.7 [95%CI, 0.99–7.6]; p=0.044) (32).
Taken together differences in pathology, gene expression and outcomes strongly suggest that the BAP1 and PBRM1 proteins regulate different processes. Thus, the observation that BAP1 and PBRM1 mutations co-occur in tumors at a frequency lower than expected suggests that even within a tumor type, a context-dependency of tumor suppressor function may exist. Whereas in some contexts, mutations may be tolerated and be advantageous, the same mutations may not be tolerated in other contexts.
The context-dependency of tumor suppressor function fits well with the empiric observation that genes exert their tumorigenic properties in a contextual, tissue-dependent manner. This is illustrated in familial cancer syndromes, in which a germline mutation (typically in a tumor suppressor gene), predisposes to a limited spectrum of tumors. Thus, despite the presence of the mutation in all diploid cells, tumors arise in a limited number of tissues. Other factors could contribute to explain the limited tissue repertoire, including differences in rates of mutation of the remaining allele across cell types. Nevertheless, a limited tumor spectrum is also observed in familial cancer syndromes resulting from germline mutations in oncogenes, such as RET, which are not associated with a mutation of the second allele. Other examples of contextual effects are provided by the overexpression of certain oncogenes, which depending on the cellular context, may induce senescence or proliferation (34). We conjecture that contextual differences in cancer gene action extend beyond tissue boundaries so that even within a specific tumor type there may be permissive and non-permissive contexts (dictated perhaps by other mutations). Thus, tumors may be viewed as an evolving set of conditional dependencies, which if understood, may uncover vulnerabilities that could be exploited therapeutically.
A model for ccRCC development
We propose that ccRCC development evolves along two different paths. Following a VHL mutation, which is an early event (35, 36), and the loss of 3p, which is frequently observed (Fig. 1), mutations in the remaining PBRM1 or BAP1 allele may lead to tumors of different characteristics. Thus, tumor aggressiveness may be programmed early during ccRCC development. This model may explain why despite the discovery of the VHL gene in 1993 (10), a mouse model of ccRCC does not exist today. Interestingly, whereas the VHL gene is linked to PBRM1 and BAP1 on the same 3p arm in humans, Vhl is on a different chromosome than Pbrm1 and Bap1 in the mouse. Thus, loss of heterozygosity of the Vhl region in the mouse would not simultaneously inactivate one copy of Pbrm1 and Bap1. If this model is correct, ccRCC should develop in mice with simultaneous inactivation of Vhl and either Pbrm1 or Bap1 genes, a testable hypothesis presently under evaluation.
Thus, the physical location of cancer genes in the genome may dictate the spectrum of tumors that a particular species may be predisposed to. Thus, in some species, a deletion may eliminate a combination of tumor suppressor genes conducive to tumor development, but if the genes are not collinear in another species, the species may be protected from the corresponding tumor type.
The physical location of tumor suppressor genes may also have implications within a species. For example, the type of second hit mutation observed may depend on whether there are neighboring tumor suppressor genes that function as such in the specific tissue. For instance, BAP1 is mutated in mesothelioma (37, 38), ccRCC (8, 17), and uveal melanoma (39–41). However, the “second-hit” mutation may be different in the three tissues. In mesothelioma, focal mutations may inactivate the second allele (38), whereas in ccRCC, the second allele is typically inactivated by loss of 3p (Fig. 1), and in metastatic uveal melanoma through whole chromosome 3 loss (39). These data are consistent with the notion that VHL (as well as SETD2 and PBRM1) may not function as a tumor suppressor gene in mesothelioma, in contrast with ccRCC. The data also suggest the existence of other tumor suppressor genes in metastatic uveal melanoma in 3q. We speculate that differences in the type of second hit mutations across tissues illustrate tissue-specific differences in tumor suppressor gene activities and tumor suppressor gene cooperativity.
The type of “second hit” mutation may also depend on non-cancer genes. Non-cancer genes may be subject to dosage effects and these effects may be context or tissue specific. Thus, a large deletion may be poorly tolerated in some tumor types as it may uncover tissue-specific haploinsufficient genes, diminishing thereby the fitness of the tumor cell.
Conclusion
Improved functional annotation of mutations in cancer genes and an understanding of mutant allele ratios and mutation prevalence in tumors should facilitate the development of genetic interaction maps. To uncover the full spectrum of genetic interactions among cancer genes, adequate statistical power is necessary and meta-analyses may be required. Large deletions in tumors may be driven by the loss of more than one tumor suppressor gene, and syntenic differences may explain differential tumor predisposition across species. Together with the notion that tumor suppressor genes function as such in a tissue-restricted manner, the physical location of a gene may explain the type of second hit mutation and the architecture of deletions across different tumor types. Understanding genetic interactions among driver genes and context dependencies of oncogenic (or tumor suppressor) action, which extend beyond tissue boundaries, may expose vulnerabilities that could be exploited therapeutically.
Supplementary Material
Acknowledgments
We appreciate TCGA for making their data readily available to the scientific community.
Grant Support
Cancer Prevention and Research Institution of Texas (CPRIT) RP101075 and NCI R01CA129387.
Footnotes
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Author Contributions
S.P-L. obtained data, performed the bioinformatic and statistical analyses, and prepared the figures. A.C. reviewed statistics and performed the Mantel-Haenszel test under the supervision of X-J.X. J.B. conceived the study and wrote the manuscript.
References
- 1.Lander ES. Initial impact of the sequencing of the human genome. Nature. 2011;470:187–197. doi: 10.1038/nature09792. [DOI] [PubMed] [Google Scholar]
- 2.Stratton MR, Campbell PJ, Futreal PA. The cancer genome. Nature. 2009;458:719–724. doi: 10.1038/nature07943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Haber DA, Gray NS, Baselga J. The evolving war on cancer. Cell. 2011;145:19–24. doi: 10.1016/j.cell.2011.03.026. [DOI] [PubMed] [Google Scholar]
- 4.Sellers WR. A blueprint for advancing genetics-based cancer therapy. Cell. 2011;147:26–31. doi: 10.1016/j.cell.2011.09.016. [DOI] [PubMed] [Google Scholar]
- 5.Gerlinger M, Rowan AJ, Horswell S, Larkin J, Endesfelder D, Gronroos E, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. The New England journal of medicine. 2012;366:883–892. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Greaves M, Maley CC. Clonal evolution in cancer. Nature. 2012;481:306–313. doi: 10.1038/nature10762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tomasetti C, Vogelstein B, Parmigiani G. Half or more of the somatic mutations in cancers of self-renewing tissues originate prior to tumor initiation. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:1999–2004. doi: 10.1073/pnas.1221068110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Peña-Llopis S, Vega-Rubín-de-Celis S, Liao A, Leng N, Pavía-Jiménez A, Wang S, et al. BAP1 loss defines a new class of renal cell carcinoma. Nature Genetics. 2012;44:751–759. doi: 10.1038/ng.2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sivanand S, Peña-Llopis S, Zhao H, Kucejova B, Spence P, Pavia-Jimenez A, et al. A validated tumorgraft model reveals activity of dovitinib against renal cell carcinoma. Sci Transl Med. 2012;4:137ra75. doi: 10.1126/scitranslmed.3003643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Latif F, Tory K, Gnarra J, Yao M, Duh FM, Orcutt ML, et al. Identification of the von Hippel-Lindau disease tumor suppressor gene. Science. 1993;260:1317–1320. doi: 10.1126/science.8493574. [DOI] [PubMed] [Google Scholar]
- 11.Gnarra JR, Tory K, Weng Y, Schmidt L, Wei MH, Li H, et al. Mutations of the VHL tumour suppressor gene in renal carcinoma. Nat Genet. 1994;7:85–90. doi: 10.1038/ng0594-85. [DOI] [PubMed] [Google Scholar]
- 12.Herman JG, Latif F, Weng Y, Lerman MI, Zbar B, Liu S, et al. Silencing of the VHL tumor-suppressor gene by DNA methylation in renal carcinoma. Proc Natl Acad Sci U S A. 1994;91:9700–9704. doi: 10.1073/pnas.91.21.9700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nickerson ML, Jaeger E, Shi Y, Durocher JA, Mahurkar S, Zaridze D, et al. Improved identification of von Hippel-Lindau gene alterations in clear cell renal tumors. Clin Cancer Res. 2008;14:4726–4734. doi: 10.1158/1078-0432.CCR-07-4921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Catalogue of Somatic Mutations in Cancer. [cited 2013 Jan 26] Available from: http://www.sanger.ac.uk/genetics/CGP/cosmic.
- 15.Kaelin WG., Jr The von Hippel-Lindau tumour suppressor protein: O2 sensing and cancer. Nat Rev Cancer. 2008;8:865–873. doi: 10.1038/nrc2502. [DOI] [PubMed] [Google Scholar]
- 16.Dalgliesh GL, Furge K, Greenman C, Chen L, Bignell G, Butler A, et al. Systematic sequencing of renal carcinoma reveals inactivation of histone modifying genes. Nature. 2010;463:360–363. doi: 10.1038/nature08672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guo G, Gui Y, Gao S, Tang A, Hu X, Huang Y, et al. Frequent mutations of genes encoding ubiquitin-mediated proteolysis pathway components in clear cell renal cell carcinoma. Nat Genet. 2012;44:17–19. doi: 10.1038/ng.1014. [DOI] [PubMed] [Google Scholar]
- 18.Varela I, Tarpey P, Raine K, Huang D, Ong CK, Stephens P, et al. Exome sequencing identifies frequent mutation of the SWI/SNF complex gene PBRM1 in renal carcinoma. Nature. 2011;469:539–542. doi: 10.1038/nature09639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Beroukhim R, Brunet JP, Di Napoli A, Mertz KD, Seeley A, Pires MM, et al. Patterns of gene expression and copy-number alterations in von-hippel lindau disease-associated and sporadic clear cell carcinoma of the kidney. Cancer Res. 2009;69:4674–4681. doi: 10.1158/0008-5472.CAN-09-0146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sun XJ, Wei J, Wu XY, Hu M, Wang L, Wang HH, et al. Identification and characterization of a novel human histone H3 lysine 36-specific methyltransferase. J Biol Chem. 2005;280:35261–35271. doi: 10.1074/jbc.M504012200. [DOI] [PubMed] [Google Scholar]
- 21.Edmunds JW, Mahadevan LC, Clayton AL. Dynamic histone H3 methylation during gene induction: HYPB/Setd2 mediates all H3K36 trimethylation. EMBO J. 2008;27:406–420. doi: 10.1038/sj.emboj.7601967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jensen DE, Proctor M, Marquis ST, Gardner HP, Ha SI, Chodosh LA, et al. BAP1: a novel ubiquitin hydrolase which binds to the BRCA1 RING finger and enhances BRCA1-mediated cell growth suppression. Oncogene. 1998;16:1097–1112. doi: 10.1038/sj.onc.1201861. [DOI] [PubMed] [Google Scholar]
- 23.Ventii KH, Devi NS, Friedrich KL, Chernova TA, Tighiouart M, Van Meir EG, et al. BRCA1-associated protein-1 is a tumor suppressor that requires deubiquitinating activity and nuclear localization. Cancer Res. 2008;68:6953–6962. doi: 10.1158/0008-5472.CAN-08-0365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Scheuermann JC, de Ayala Alonso AG, Oktaba K, Ly-Hartig N, McGinty RK, Fraterman S, et al. Histone H2A deubiquitinase activity of the Polycomb repressive complex PR-DUB. Nature. 2010;465:243–247. doi: 10.1038/nature08966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Machida YJ, Machida Y, Vashisht AA, Wohlschlegel JA, Dutta A. The deubiquitinating enzyme BAP1 regulates cell growth via interaction with HCF-1. J Biol Chem. 2009;284:34179–34188. doi: 10.1074/jbc.M109.046755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Misaghi S, Ottosen S, Izrael-Tomasevic A, Arnott D, Lamkanfi M, Lee J, et al. Association of C-terminal ubiquitin hydrolase BRCA1-associated protein 1 with cell cycle regulator host cell factor 1. Mol Cell Biol. 2009;29:2181–2192. doi: 10.1128/MCB.01517-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dey A, Seshasayee D, Noubade R, French DM, Liu J, Chaurushiya MS, et al. Loss of the tumor suppressor BAP1 causes myeloid transformation. Science. 2012;337:1541–1546. doi: 10.1126/science.1221711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ruan HB, Han X, Li MD, Singh JP, Qian K, Azarhoush S, et al. O-GlcNAc transferase/host cell factor C1 complex regulates gluconeogenesis by modulating PGC-1alpha stability. Cell Metab. 2012;16:226–237. doi: 10.1016/j.cmet.2012.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hakimi AA, Chen YB, Wren J, Gonen M, Abdel-Wahab O, Heguy A, et al. Clinical and Pathologic Impact of Select Chromatin-modulating Tumor Suppressors in Clear Cell Renal Cell Carcinoma. Eur Urol. 2012;63(527):848–854. doi: 10.1016/j.eururo.2012.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.The Cancer Genome Atlas. [Cited 2013 Jan 26] Available at: https://tcga-data.nci.nih.gov/tcga.
- 31.Schuster-Bockler B, Lehner B. Chromatin organization is a major influence on regional mutation rates in human cancer cells. Nature. 2012;488:504–507. doi: 10.1038/nature11273. [DOI] [PubMed] [Google Scholar]
- 32.Kapur P, Peña-Llopis S, Christie A, Zhrebker L, Pavía-Jiménez A, Kimryn Rathmell W, et al. Effects on survival of BAP1 and PBRM1 mutations in sporadic clear cell renal cell carcinoma: a retrospective analysis with independent validation. Lancet Oncol. 2013;14:159–167. doi: 10.1016/S1470-2045(12)70584-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brugarolas J. Research Translation and Personalized Medicine. In: Figlin RA, Rathmell WK, Rini BI, editors. Renal Cell Carcinoma. New York: Springer; 2012. pp. 161–191. [Google Scholar]
- 34.Collado M, Serrano M. Senescence in tumours: evidence from mice and humans. Nat Rev Cancer. 2010;10:51–57. doi: 10.1038/nrc2772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mandriota SJ, Turner KJ, Davies DR, Murray PG, Morgan NV, Sowter HM, et al. HIF activation identifies early lesions in VHL kidneys: evidence for site-specific tumor suppressor function in the nephron. Cancer Cell. 2002;1:459–468. doi: 10.1016/s1535-6108(02)00071-5. [DOI] [PubMed] [Google Scholar]
- 36.Montani M, Heinimann K, von Teichman A, Rudolph T, Perren A, Moch H. VHL-gene deletion in single renal tubular epithelial cells and renal tubular cysts: further evidence for a cyst-dependent progression pathway of clear cell renal carcinoma in von Hippel-Lindau disease. Am J Surg Pathol. 2010;34:806–815. doi: 10.1097/PAS.0b013e3181ddf54d. [DOI] [PubMed] [Google Scholar]
- 37.Bott M, Brevet M, Taylor BS, Shimizu S, Ito T, Wang L, et al. The nuclear deubiquitinase BAP1 is commonly inactivated by somatic mutations and 3p21.1 losses in malignant pleural mesothelioma. Nat Genet. 2011;43:668–672. doi: 10.1038/ng.855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Testa JR, Cheung M, Pei J, Below JE, Tan Y, Sementino E, et al. Germline BAP1 mutations predispose to malignant mesothelioma. Nat Genet. 2011;43:1022–1025. doi: 10.1038/ng.912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Harbour JW, Onken MD, Roberson ED, Duan S, Cao L, Worley LA, et al. Frequent mutation of BAP1 in metastasizing uveal melanomas. Science. 2010;330:1410–1413. doi: 10.1126/science.1194472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wiesner T, Obenauf AC, Murali R, Fried I, Griewank KG, Ulz P, et al. Germline mutations in BAP1 predispose to melanocytic tumors. Nat Genet. 2011;43:1018–1021. doi: 10.1038/ng.910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Abdel-Rahman MH, Pilarski R, Cebulla CM, Massengill JB, Christopher BN, Boru G, et al. Germline BAP1 mutation predisposes to uveal melanoma, lung adenocarcinoma, meningioma, and other cancers. J Med Genet. 2011;48:856–859. doi: 10.1136/jmedgenet-2011-100156. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.