Abstract
Background
Recently it has been shown that Ensifer adhaerens can be used as a plant transformation technology, transferring genes into several plant genomes when equipped with a Ti plasmid. For this study, we have sequenced the genome of Ensifer adhaerens OV14 (OV14) and compared it with those of Agrobacterium tumefaciens C58 (C58) and Sinorhizobium meliloti 1021 (1021); the latter of which has also demonstrated a capacity to genetically transform crop genomes, albeit at significantly reduced frequencies.
Results
The 7.7 Mb OV14 genome comprises two chromosomes and two plasmids. All protein coding regions in the OV14 genome were functionally grouped based on an eggNOG database. No genes homologous to the A. tumefaciens Ti plasmid vir genes appeared to be present in the OV14 genome. Unexpectedly, OV14 and 1021 were found to possess homologs to chromosomal based genes cited as essential to A. tumefaciens T-DNA transfer. Of significance, genes that are non-essential but exert a positive influence on virulence and the ability to genetically transform host genomes were identified in OV14 but were absent from the 1021 genome.
Conclusions
This study reveals the presence of homologs to chromosomally based Agrobacterium genes that support T-DNA transfer within the genome of OV14 and other alphaproteobacteria. The sequencing and analysis of the OV14 genome increases our understanding of T-DNA transfer by non-Agrobacterium species and creates a platform for the continued improvement of Ensifer-mediated transformation (EMT).
Keywords: Ensifer adhaerens, Transformation, Agrobacterium tumefaciens, Genome sequencing
Background
The ability of Agrobacterium tumefaciens to transfer DNA into a plant cell via horizontal gene transfer has been instrumental in progressing the field of plant molecular biology, enabling methods such as T-DNA tagging [1,2], Agrobacterium-mediated transformation (AMT) for delivery of gene expression and silencing vectors [3,4], and the introduction of genes of interest into plant genomes [5]. In effect, these abilities have underpinned the integration of crop biotechnology into mainstream agriculture, driving the development of genetically modified crop varieties equipped with novel traits, which in 2013 were planted across 175 million hectares [6]. Indeed, based on the use of AMT, commodity crop improvement through genetic engineering has become the fastest adopted crop technology in the world with global value of the biotech/GM seed market estimated to be in excess of $13 billion [6]. However, the complexity of the Agrobacterium patent landscape remains a challenge for non-patent holders [7,8], as the execution of existing patents on crop biotechnology can restrict the widespread application of AMT technology by non-patent holders [9].
The possibility of modifying non-Agrobacterium strains to facilitate horizontal gene transfer was first described by Hooykaas et al. (1977), with work by van Veen et al. [10] showing that while Phyllobacterium myrsinacearum (harbouring the A. tumefaciens tumour inducing (Ti) plasmid) could cause tumorigenesis on plants, Rhizobium meliloti could not. It was not until 2005 though that the potential of non-Agrobacterium species’ to horizontally transfer genes into plant genomes was re-visited through CAMBIA’s Transbacter™ Project. Using the rhizobial species Sinorhizobium meliloti 1021, Rhizobium sp. NGR234 (now Sinorhizobium fredii NGR234) and Mesorhizobium loti MAFF303099, it was demonstrated that non-Agrobacterium rhizobia could indeed transfer T-DNA into plant cells [7]. However, the transformation frequency of these species was inadequate to provide a viable alternative to A. tumefaciens[11], which prompted the search for alternatives from a collection of diverse soil bacteria [11,12]. This initiative unearthed a lesser known rhizobial species, Ensifer adhaerens[13], as a rhizosphere inhabiting bacterium with the ability to successfully transform potato, tobacco and Arabidopsis. Designated Ensifer adhaerens OV14 (OV14), this strain can deliver sufficient transformation frequencies to present Ensifer-mediated transformation (EMT) as a viable alternative to existing transformation technology platforms [12].
The genetic and molecular mechanisms supporting the stable integration of A. tumefaciens T-DNA (transfer-DNA) into plant genomes have been the focus of intense research efforts since the first reports of AMT in the 1980s. A bacterial pathogen that causes ‘crown gall’ disease across a broad range of dicotyledonous and (some) monocotyledonous species [14], A. tumefaciens genetically transforms its host by transferring a single stranded DNA fragment (T-DNA) from its Ti plasmid into the host cell genome [15,16]. The T-DNA is exported from the bacterial cell into the plant cell together with several virulence effector proteins via a Type IV secretion system. By coating the T-DNA on its journey into the plant cell nucleus, this T-DNA structure appears more as a protein complex than a single strand of DNA [17]. For the purposes of genetic transformation, existing bacterial sequences within the left and right border of the T-DNA can be replaced with genes of interest (e.g. sequences coding for herbicide tolerance/disease resistance/synthesis of therapeutics), which can then be delivered into the targeted host genome using AMT. The reader is directed to a number of excellent reviews for an in-depth explanation and discussion of this process [18-20].
The genome sequences of A. tumefaciens C58 (C58) and S. meliloti 1021 (1021) were completed in 2001 [21-23]. Although these two gram-negative alphaproteobacteria are members of the same phylogenetic family (the Rhizobiaceae) and inhabit the rhizosphere, they operate very different lifestyles (pathogen vs. symbiont, respectively). The primary circular chromosomes of C58 and 1021 have been shown to share large-scale synteny, while only limited stretches of synteny can be found among additional replicons [24]. It is upon these more unique replicons that genes encoding functions leading to the different lifestyles of these organisms are found. For example, the above-mentioned T-DNA transfer mechanism of A. tumefaciens is located on the large Ti plasmid and genes key to the symbiotic interaction of 1021 with legumes are found on two megaplasmids namely pSymA and pSymB [25,26].
The application of functional genomic studies to dissect the processes of AMT have identified a number of genes located on the A. tumefaciens circular and linear chromosomes that are implicated in virulence through the processes of attachment, vir gene regulation, and resisting plant defence responses. Initial reversible attachment to plant cells involving beta-1,2-glucan and secondary irreversible attachment involving cellulose fibrils are early requirements for A. tumefaciens virulence while beta-1,2-glucan in S. meliloti plays an important role in symbiosis [27-30]. While the pAtC58 plasmid is non-essential for virulence of A. tumefaciens, it contains several att genes involved in attachment and pAtC58’s presence has been shown to have a positive effect on vir gene expression [31]. Mutations to a group of chv genes plus ros, aopB and miaA have all been shown to restrict, and in some cases halt virulence [32-37]. The ability of the bacterial cell to protect itself against plant derived reactive oxygen species (ROS) is also required for virulence by both plant pathogens and symbionts [38,39]. For example, a catalase (KatA) conferring gene has been shown to be upregulated in response to H2O2 via the peroxide sensor OxyR in both C58 and 1021 [38,40]. The rhizosphere is typically an acidic environment (~pH5.5) enriched by plant exudates including but not limited to sugars, ions, free oxygen and water [41,42]. Transcriptomic profiling of C58 and 1021 in response to a shift to acidic pH, has revealed a shared regulational change in genes involved in membrane composition and motility [42,43]. Separately, a chromosomally located two component sensor gene key to virulence in C58 and to symbiosis in 1021 termed chvG(exoS)/chvI has been cited as a global pH regulator [44]. A recently published study of 48 Sinorhizobium strains concluded that subtle differences in the presence of symbiosis associated genes involved in Nod-factor and polysaccharide biosynthesis, denitrification and Type III, IV, and VI secretion systems leads to varying compatibility among strains in legume-Sinorhizobium interactions [45]. An independent study looking at 14 rhizobia strains, including 1021, noted differences in gene content in key groups of genes, including those involved in nodulation, nitrogen fixation, production of exopolysaccharides and Type I to Type VI secretion systems with the authors concluding no simple ‘core symbiome’ exists among rhziobia [46]. In contrast to the number of comparative studies focused on symbiotic interactions that have been carried out to date, no study has yet focused on the ability of plant transformation within the rhizobia.
While a draft genome of E. adhaerens CSBa has recently been reported, to the best of our knowledge only DNA sequences for cobalamin biosynthetic (cob) genes are publicly available [47]. In this study the genome of OV14 was sequenced and functionally annotated by comparing to the already sequenced genomes of C58 and 1021 using the eggNOG database [48,49]. Subsequently, the literature was screened for all genes reported to have a positive effect on A. tumefaciens virulence and then homologs to these genes were sought for in OV14, and also in 1021 for additional comparison. The level of homology between genes was compared and, where relevant, gene copy number was considered. In addition, a phylogenetic analysis was completed on a core group of housekeeping genes and Rhizobiales chromosomal-located virulence related genes to clarify the position of OV14 within the large, diverse Rhizobiaceae family.
Results
General features of the E. adhaerens OV14 genome versus that of A. tumefaciens C58 and S. meliloti 1021
The OV14 genome is the largest of the three species at 7.71 Mb; 2.04 Mb bigger than the C58 genome and 1.01 Mb larger than the 1021 genome (Table 1). Composed of four replicons of sizes 3.96 Mb, 2.01 Mb, 1.61 Mb and 125 kb, the OV14 genome is similar to that of 1021, which is also made up of three large circular replicons minus the small accessory plasmid. In contrast the C58 genome differs dramatically with the circular chromosome being approximately 25% less than the size of OV14’s and 1021’s counterpart and the presence of a linear chromosome being a feature unique to C58. That said, OV14 (Additional file 1: Figure S1) and C58 share a similar sized 120–180 kb mobile plasmid not found in 1021. The GC content of C58’s genome is notably lower (at 58%) compared to that of OV14 (60.75%) and 1021 (62.17%) genomes (Table 1), with a total genome comparison highlighting a greater level of synteny between OV14 and 1021 (Figure 1).
Table 1.
Basic genome information for three species; Ensifer adhaerens OV14, Sinorhizobium meliloti 1021, and Agrobacterium tumefaciens C58
| OV14 | 1021 | C58 |
|---|---|---|
| CHR 1 |
CHR |
CHR Circular |
| 3956045 bp |
3654135 bp |
2841580 bp |
| 916 average gene length |
939 average gene length |
902 average gene length |
| 62.24 GC % |
62.7 GC % |
59.4 GC % |
| 9 rRNA |
3 rRNA |
2 rRNA |
| 52 tRNA |
51 tRNA |
40 tRNA |
| CHR 2 |
pSymA |
CHR Linear |
| 2012811 bp |
1354226 bp |
2075577 bp |
| 916 average gene length |
875 average gene length |
988 average gene length |
| 61.77 GC % |
60.4 GC % |
59.3 GC % |
| 3 rRNA |
0 rRNA |
2 rRNA |
| 4 tRNA |
2 tRNA |
13 tRNA |
| pOV14b |
pSymB |
pAt |
| 1614950 bp |
1683333 bp |
542868 bp |
| 860 average gene length |
949 average gene length |
849 average gene length |
| 60.65 GC % |
62.4 GC % |
57.3 GC % |
| 3 rRNA |
0 rRNA |
0 rRNA |
| 4 tRNA |
1 tRNA |
0 tRNA |
| pOV14c |
|
pTi |
| 125203 bp |
|
214233 bp |
| 815 average gene length |
|
938 average gene length |
| 58.37 GC % |
|
56.7 GC % |
| 0 rRNA |
|
0 rRNA |
| 0 tRNA | 0 tRNA |
For each entry information is as follows; Replicon id, Replicon size, average gene length, Replicons GC content as percentage, No. rRNA, and No. tRNA.
Figure 1.
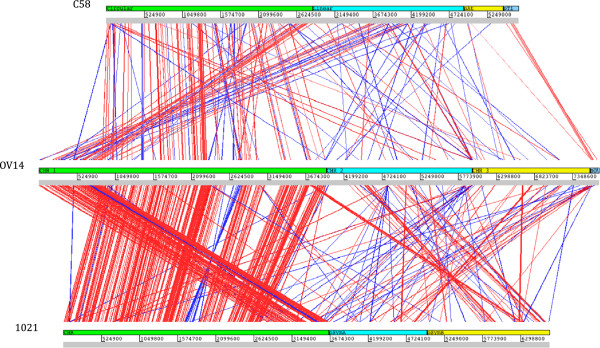
Comparative synteny plots showing total genome content of Agrobacterium tumefaciens C58 (top bar), Ensifer adhaerens OV14 (middle bar), and Sinorhizobium meliloti 1021 (bottom bar), computed using DoubleACT version2 on tBLASTx setting. Visualised in Artemis ACT with cut off set at 1000. The replicons within each genome are separated by coloured bars and labelled. Homology between the genomes is displayed via interconnecting lines; red lines representing direct homology with blue lines corresponding to inverted homologous sequence.
Structurally, chromosome 1 of OV14 shares 54% nucleotide homology with the chromosome of 1021, while the circular and linear chromosomes of C58 share 20% and 5% homology, respectively with chromosome 1 of OV14. Chromosome 2 of OV14 shows reduced homology of 20% to pSymB, 2% to pSymA, and 3% to the 1021 chromosome. In regards to C58, 3% and 7% homology to chromosome 2 of OV14 is noted for the circular and linear chromosome, respectively. The third replicon of OV14 shows 7% homology to both the pSymA and pSymB of 1021, and 4% homology to the linear chromosome of C58. Finally the small accessory plasmid pOV14c shows between 1–2% homology to each of the replicons. Homology of pOV14 to C58 replicons are found in pTi and pAt at 22% and 3%, respectively.
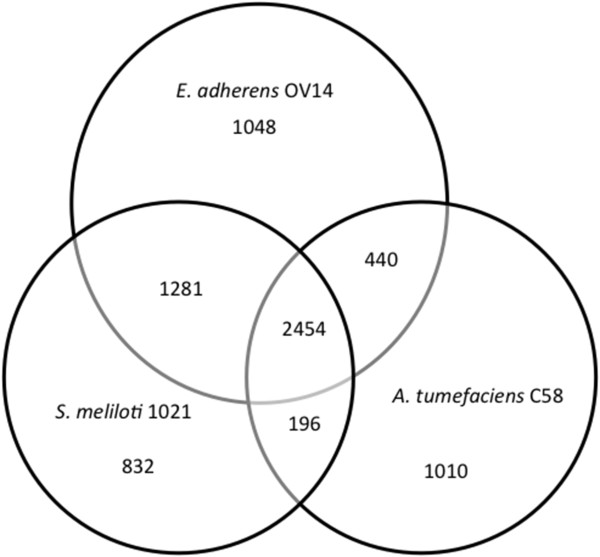
The three genomes of OV14, 1021 and C58 were compared by using evolutionary genealogy of genes: Non-supervised Orthologous Groups (eggNOG) assignments. The eggNOG database is formatted to functionally categorise genes within twenty-five groups. Twenty-one of the 25 eggNOG functional categories have representatives in the three genomes in this study (Table 2). Those categories that are not represented are RNA processing and modification [A], nuclear structure [Y], cytoskeleton [Z] and extracellular structures [W]. In total 7261 NOGs were identified in this study with 2454 (33.8%) being shared among the three species (Table 2). OV14 has the most species-specific NOGs at 1048 (14.4%), marginally ahead of C58 with 1010 (13.9%) whilst 1021 has less at 832 (11.5%). Of the NOGs that are shared between two species and not present within the third species; OV14 and 1021 share 1281 (17.6%) almost 3-fold more than that shared by OV14 and C58 at 440 (6.1%), and 6.5-fold more than that shared between C58 and 1021 at 196 (2.7%) (Figure 2).
Table 2.
Comparison of eggNOG assignments for Ensifer adhaerens OV14, Agrobacterium tumefaciens C58, and Sinorhizobium meliloti 1021
| eggNOG functional category | Shared by 3 species | OV14 & C58 only | OV14 & 1021 only | C58 & 1021 only | OV14 only | C58 only | 1021 only |
|---|---|---|---|---|---|---|---|
| Information storage and processing |
|
|
|
|
|
|
|
| [J] Translation, ribosomal structure and biogenesis |
143 |
4 |
18 |
3 |
10 |
14 |
10 |
| [K] Transcription |
160 |
58 |
135 |
19 |
140 |
116 |
83 |
| [L] Replication, recombination and repair |
99 |
16 |
40 |
7 |
42 |
27 |
37 |
| [B] Chromatin structure and dynamics |
1 |
0 |
0 |
0 |
1 |
0 |
0 |
| Cellular processes and signaling |
|
|
|
|
|
|
|
| [D] Cell cycle control, cell division, chromosome partitioning |
22 |
2 |
5 |
0 |
2 |
6 |
5 |
| [V] Defense mechanisms |
28 |
4 |
12 |
3 |
17 |
11 |
12 |
| [T] Signal transduction mechanisms |
83 |
30 |
49 |
5 |
35 |
45 |
35 |
| [M] Cell wall/membrane/envelope biogenesis |
119 |
16 |
59 |
5 |
34 |
49 |
30 |
| [N] Cell motility |
31 |
2 |
5 |
0 |
2 |
4 |
3 |
| [U] Intracellular trafficking, secretion, and vesicular transport |
34 |
13 |
13 |
10 |
16 |
13 |
9 |
| [O] Posttranslational modification, protein turnover, chaperones |
112 |
8 |
29 |
2 |
21 |
13 |
17 |
| Metabolism |
|
|
|
|
|
|
|
| [C] Energy production and conversion |
164 |
21 |
82 |
14 |
52 |
55 |
58 |
| [G] Carbohydrate transport and metabolism |
194 |
38 |
121 |
17 |
76 |
61 |
59 |
| [E] Amino acid transport and metabolism |
317 |
51 |
128 |
27 |
120 |
118 |
61 |
| [F] Nucleotide transport and metabolism |
67 |
7 |
14 |
3 |
7 |
7 |
5 |
| [H] Coenzyme transport and metabolism |
97 |
6 |
29 |
3 |
10 |
12 |
9 |
| [I] Lipid transport and metabolism |
96 |
9 |
48 |
9 |
32 |
36 |
26 |
| [P] Inorganic ion transport and metabolism |
140 |
23 |
46 |
11 |
35 |
57 |
41 |
| [Q] Secondary metabolites biosynthesis, transport and catabolism |
56 |
12 |
45 |
6 |
36 |
15 |
31 |
| Poorly characterized |
|
|
|
|
|
|
|
| [R] General function prediction only |
151 |
39 |
90 |
14 |
97 |
80 |
56 |
| [S] Function unknown | 340 | 81 | 313 | 38 | 263 | 271 | 245 |
Figure 2.
Venn diagram illustrating the number of eggNOGs found across the three species. Ensifer adhaerens OV14, Sinorhizobium meliloti 1021, and Agrobacterium tumefaciens C58.
Five of the functional categories grouped under the heading of cellular processing and signalling were of most interest to this study (Table 2). In category [V], ‘defence mechanisms’, a total of 87 NOGs were recorded with 28 (32.2%) shared across all three species. Within this category OV14 recorded the most individual NOGs at 17 (19.5%) followed by 1021 and C58 with 12 (13.8%) and 11 (12.6%), respectively. For the signal transduction mechanisms [T] category a total of 282 NOGs were found of which 83 (29.4%) were shared by all species, while within this category C58 contained 45 individual NOGs, 10 more than both OV14 and 1021. Also to be noted were the minimal number of NOGs shared by C58 and 1021 (n = 5) compared to OV14 and C58 (n = 30) and OV14 and 1021 (n = 49). Within the category cell wall/membrane/envelope biogenesis [M] 119 (38.2%) of a total of 312 were shared by all three species; OV14 and 1021 sharing 59 (19%), which was more than the 16 (5%) shared across OV14 and C58 and the 5 (1.6%) for C58 and 1021. Although C58 shares a lower number of NOGs with the other two species in category [M] it does possess the highest number of species specific NOGs at 49 (15.7%). Cell motility is category [N] representing 47 NOGs for which 31 (66%) were shared by all three species, with no NOGs shared by C58 and 1021. In category [U] (Intracellular trafficking, secretion, and vesicular transport) there were a total of 108 NOGs of which 34 (31.5%) were shared by all three species. Category [U] showed the most even distribution of any category. The final category of specific interest is post-translational modification, protein turnover, chaperones [O] with a total of 202 NOGs for which 112 (55.4%) were shared by all three species; OV14 and 1021 with 29 (14.4%), OV14 and C58 and C58 and 1021 with 8 (4%) and 2 (1%), respectively. Individually, OV14 possessed 21 (10.4%), 1021 possessed 17 (8.4%) and C58 possessed 13 (6.4%).
Attachment
The chromosomal virulence gene A (chvA) is a member of a group of orthologous genes found in the eggNOG database under the code aproNOG01094 (Table 3). Encoding a cyclic beta-1,2-glucan ABC transporter, its function is linked to chvB a member of aproNOG01088, which encodes a cyclic beta 1-2 glucan synthase. Together chvA and chvB function to synthesise and transport beta-1,2 glucan across the inner membrane, which is required for attachment of the bacterial cell to the plant cell surface. Genes with parallel function named ndvA and ndvB are found in 1021. All three species were found to possess one gene in the aproNOG01094 representative of chvA/ndvA (Table 3). Located on chromosome one of OV14 is a gene showing 89% protein sequence identity to ndvA of 1021 and 77% identity to chvA of C58. Two genes downstream of OV14’s chvA/ndvA homolog was a chvB/ndvB homolog (a member of aproNOG01088) showing 86% and 68% protein sequence identity to respective sequences in 1021 and C58, respectively. OV14 also has a second chvB/ndvB gene sharing 47% and 50% protein sequence identity to 1021’s and C58’s chvB/ndvB respectively. A third gene involved in the synthesis of beta 1-2 glucan is pscA/exoC (aproNOG01465) encoding a phosphoglucomutase, which recorded 90% and 81% to the respective target in 1021 and C58, respectively.
Table 3.
Comparative analysis for the presence/absence of genes identified to have a positive effect on Agrobacterium virulence that are located in the genomic background (not on Ti plasmid) of Ensifer adhaerens OV14, Agrobacterium tumefaciens C58, and Sinorhizobium meliloti 1021
|
Gene id |
eggNOG id |
OV14 gene |
C58 gene |
1021 gene |
No. species |
No. proteins |
Virulence function |
Product |
|---|---|---|---|---|---|---|---|---|
| In C58 | Copy number | Copy number | Copy number | In NOG | In NOG | In C58 | ||
|
chvA |
aproNOG01094 |
1 |
1 |
1 |
52 |
52 |
Attachment |
Cyclic beta-1,2-glucan ABC transporter |
|
chvB |
aproNOG01088 |
2 |
1 |
1 |
39 |
40 |
Attachment |
Cyclic beta 1-2 glucan synthase |
|
pscA(exoC) |
aproNOG01465 |
1 |
1 |
1 |
65 |
65 |
Attachment |
Phosphoglucomutase |
|
pcs |
aproNOG02893 |
1 |
1 |
1 |
37 |
37 |
Attachment |
Phosphatidylcholine synthase |
|
pmtA |
aproNOG06650 |
1 |
1 |
1 |
58 |
58 |
Attachment |
Phospholipid N-methyltransferase |
|
choX |
aproNOG00993 |
2 |
2 |
2 |
35 |
41 |
Attachment |
Choline SBP |
|
choW |
aproNOG00971 |
1 |
1 |
1 |
35 |
35 |
Attachment |
Choline ABC ATPase |
|
choV |
aproNOG02245 |
2 |
3 |
1 |
45 |
78 |
Attachment |
Choline ABC permease |
|
chvD |
aproNOG00260 |
1 |
1 |
1 |
102 |
104 |
vir gene regulation |
Uracil phosphoribosyltransferase |
|
chvE |
aproNOG03985 |
1 |
1 |
1 |
29 |
20 |
vir gene regulation |
Multiple sugar-binding periplasmic receptor |
|
chvG |
aproNOG00593 |
1 |
1 |
1 |
86 |
89 |
vir gene regulation |
DNA-binding/iron metalloprotein/AP endonuclease |
|
chvI |
aproNOG03091 |
1 |
1 |
1 |
85 |
87 |
vir gene regulation |
Transcriptional regulator |
|
chvH |
aproNOG03687 |
1 |
1 |
1 |
50 |
50 |
vir gene regulation |
Elongation factor P |
|
miaA |
aproNOG00010 |
1 |
1 |
1 |
116 |
116 |
vir gene regulation |
tRNA delta (2)-isopentenylpyrophosphate transferase |
|
ros |
aproNOG09171 |
1 |
1 |
1 |
16 |
17 |
vir gene regulation |
Ros/MucR family transcriptional regulator |
|
picA |
aproNOG09265 |
1 |
2 |
0 |
13 |
15 |
Host cell wall degradation |
Polygalacturonase |
|
kdgF |
aproNOG11632 |
1 |
1 |
1 |
15 |
15 |
Host cell wall degradation |
Pectin degradation protein |
|
ligE |
aproNOG03997 |
1 |
1 |
1 |
31 |
31 |
Host cell wall degradation |
Lignin degradation protein |
|
acvB |
aproNOG05730 |
1 |
1 |
2 |
28 |
31 |
Forms complex with T-strand |
Acid tolerance and virulence protein |
|
aopB |
aproNOG08879 |
1 |
1 |
1 |
17 |
17 |
Defence |
Porin-like membrane protien |
|
katA |
aproNOG00015 |
2 |
1 |
1 |
51 |
54 |
Defence |
Catalase-Peroxidase |
|
dps |
aproNOG08385 |
0 |
1 |
0 |
19 |
19 |
Defence |
DNA starvation/stationary phase protection protein |
|
catE |
aproNOG02507 |
0 |
1 |
1 |
40 |
40 |
Defence |
Catalase |
|
oxyR |
aproNOG01190 |
1 |
1 |
0 |
34 |
48 |
Defence |
Oxidative stress transcription regulator |
|
oxyR (1021) |
aproNOG01330 |
0 |
0 |
1 |
33 |
33 |
Defence |
Oxidative stress transcription regulator |
| sodB | aproNOG00877 | 2 | 3 | 1 | 115 | 122 | Defence | Super oxide dismutase |
The OV14 genome was also screened for the presence/absence of genes linked to the C58 pAt att locus, which contains up to 24 genes for which some have been implicated in the early stages of attachment and virulence. The attR gene is part of aproNOG01724 and encodes an acetyltransferase but no copies were found across the OV14 genome, compared to a single copy in 1021 and two attR copies in C58. The genes attB and attD are implicated in bacteria-plant signalling during root colonisation and at the wound site during pathogenesis, with attB part of aproNOG06835 annotated as part of a binding-protein-dependent transport system, which is predicted to transport mannopine in C58 [50]. While 1021 and C58 both possess a single copy, a member of the aproNOG06835 was not found in OV14. The attD gene of C58 appears to be unique, unassigned to any aproNOG and having no similar sequence in OV14 or 1021. Mutations to genes attC and attG can render Agrobacterium avirulent on tomato and carrot by preventing attachment to the host cell [51]. Both genes are annotated as ABC transporters for which no orthologs exist in OV14: an attC ortholog in aproNOG06683 is present in 1021 and C58 (Table 4). The attKLM (renamed blcABC) operon within this locus has been linked to quorum sensing and found to be up regulated in response to salicylic acid (SA) in C58 [52]. All three species contain multiple gene entries (four copies in OV14, three in C58 and five in 1021) in aproNOG00713, which houses the C58 blcA (a NAD-dependent succinyl dehydrogenase). No genes homologous to C58 blcB (aproNOG04363) or blcC (aproNOG02812) were found in OV14. While the role of the remaining genes of the att locus in virulence remains unclear, the genes attE, attF, attG, attH, attO, attT, attV, attY and attZ were not found within the OV14 genome, or that of 1021 either (Table 4). However, they are represented by aproNOGs highlighting their presence among other alphaproteobacteria. Genes found in the C58 att locus not represented by aproNOGs include attD, attP, attS, attU, attW, and attX.
Table 4.
Comparative analysis for the presence/absence of genes involved in attachment to plant cell that are located in the genomic background (not on Ti plasmid) of Ensifer adhaerens OV14, Agrobacterium tumefaciens C58, and Sinorhizobium meliloti 1021
|
Gene id |
eggNOG |
OV14 gene |
C58 gene |
1021 gene |
No. species |
No. protein |
Virulence function |
Product |
|---|---|---|---|---|---|---|---|---|
| In C58 | Copy number | Copy number | Copy number | In NOG | In NOG | In C58 | ||
|
celD |
aproNOG11524 |
0 |
1 |
0 |
7 |
7 |
Attachment |
Cellulose biosynthesis protein |
|
celE |
aproNOG04598 |
1 |
1 |
0 |
7 |
7 |
Attachment |
Cellulose synthesis protein |
|
celG |
aproNOG12411 |
1 |
1 |
0 |
23 |
26 |
Attachment |
Cellulose synthesis protein |
|
celC |
aproNOG07630 |
1 |
1 |
0 |
25 |
28 |
Attachment |
Endoglucanase |
|
celB |
aproNOG09454 |
1 |
1 |
0 |
23 |
26 |
Attachment |
Cellulose synthase |
|
celA |
aproNOG05761 |
1 |
1 |
0 |
27 |
30 |
Attachment |
Cellulose synthase |
|
attK2 |
aproNOG02908 |
0 |
1 |
1 |
17 |
18 |
Attachment |
Semialdehyde dehydrogenase |
|
attA1 |
aproNOG00407 |
3 |
2 |
4 |
47 |
73 |
Attachment |
ABC transporter, nucleotide binding/ATPase protein [putrescine] |
|
attA2 |
aproNOG05011 |
0 |
1 |
1 |
15 |
15 |
Attachment |
ABC transporter, membrane spanning protein [mannopine] |
|
attB |
aproNOG06835 |
0 |
1 |
1 |
16 |
16 |
Attachment |
ABC transporter, membrane spanning protein [mannopine] |
|
attC |
aproNOG06683 |
0 |
1 |
1 |
12 |
12 |
Attachment |
ABC transporter, substrate binding protein [mannopine] |
|
attE |
aproNOG05033 |
0 |
2 |
0 |
24 |
25 |
Attachment |
ABC transporter nucleotide binding/ATPase protein |
|
attF |
aproNOG00433 |
0 |
3 |
0 |
24 |
26 |
Attachment |
ABC transporter, membrane spanning protein |
|
attG |
aproNOG00433 |
0 |
3 |
0 |
24 |
26 |
Attachment |
ABC transporter, membrane spanning protein |
|
attH |
aproNOG04763 |
0 |
2 |
0 |
22 |
23 |
Attachment |
Hypothetical protein |
|
attJ/blcR |
aproNOG06067 |
0 |
1 |
0 |
17 |
17 |
Attachment |
Transcriptional repressor of the blcABC operon |
|
attK/blcA |
aproNOG00713 |
4 |
3 |
5 |
88 |
141 |
Attachment |
NAD-dependent succinyl-semialdehyde dehydrogenase |
|
attL/blcB |
aproNOG04363 |
0 |
1 |
1 |
15 |
19 |
Attachment |
Gamma hydroxybutyrate dehydrogenase |
|
attM/blcC |
aproNOG02812 |
0 |
2 |
0 |
19 |
23 |
Attachment |
Zn-dependent gamma butyryl lactone lactonase |
|
attO |
aproNOG09383 |
0 |
1 |
0 |
5 |
5 |
Attachment |
Transcriptional regulator, AraC family |
|
attR |
aproNOG01724 |
0 |
2 |
1 |
24 |
27 |
Attachment |
Transacetylase |
|
attT |
aproNOG12782 |
0 |
1 |
0 |
3 |
3 |
Attachment |
GNAT family acetyltransferase |
|
attV |
aproNOG09482 |
0 |
1 |
0 |
12 |
12 |
Attachment |
Mg (2+) transport ATPase |
|
attY |
aproNOG07710 |
0 |
1 |
0 |
45 |
48 |
Attachment |
Glutathione S-transferase |
| attZ | aproNOG08980 | 0 | 1 | 0 | 21 | 26 | Attachment | Transcriptional regulator |
The cel locus is comprised of six genes celABCDEG and encodes a synthase for cellulose fibrils implicated in the second stage of attachment referred to as tight binding, which is irreversible [53] and critical for the virulence of Agrobacterium cells [53]. The genome of OV14 had genes orthologous to celABCEG, but not celD, which are thought to be cytoplasmic lipid carriers (Table 4). The aproNOGs representing celABCG genes were found in 23–27 separate alphaproteobacteria while aproNOGs including celDE were located in only 7 alphaproteobacteria. Of interest, 1021 did not contain any orthologs to the cel genes of C58 (Table 4).
The presence of phosphatidylcholine in prokaryotic membranes is generally confined to species that intimately interact with eukaryotic cells [54]. Two pathways present in C58 can lead to phosphatidylcholine production; the methylation pathway that requires pmtA (aproNOG06650) and the pcs pathway that requires pcs (aproNOG02893) [54]. In C58 phosphatidylcholine is found in the inner and outer membrane constituting around 23% of total membrane lipids. The pcs gene of OV14 shares 92% and 85% protein sequence identity with 1021 and C58 respectively, while the pmtA gene of OV14 shares 83% and 67% protein sequence identity with 1021’s and C58’s respecitvely. The pcs pathway is dependent on the uptake of choline from the environment [55]. Screening OV14 for the choline ABC transporter genes choXWV (that have been identified in both C58 and 1021), revealed that the choX solute binding protein component (aproNOG00993) was represented by two orthologs in all three species (Table 3). The ABC ATPase choW (aproNOG00971) has one member in each species and the choline permease (aproNOG02245) was noted to have three members in C58, two in OV14 and one in 1021.
Host cell wall degradation
The C58 genome contains two copies of the picA gene (aproNOG09265), which encodes a polygalacturonase to degrade the pectin network in targeted cell walls and aid the secretion of bacterial proteins into the plant cell [56]. The genome of OV14 was equipped with one copy showing 78% identity to picA of C58, while 1021 has no recorded picA homolog (Table 3). A complementary gene involved in pectin degradation is kdgF (aproNOG11632), with all three species possessing a kdgF homolog; OV14 sharing 72% and 47% protein sequence identity with C58 and 1021 respectively. Finally all three species possessed a member of the aproNOG03997, a beta-etherase linked to a lignin degradation protein annotated as ligE in 1021 and C58, with OV14’s ligE homolog sharing 77% and 65% protein sequence identity to 1021 and C58, respectively.
Chromosomal regulation of Ti based virulence genes
Key to the regulation of vir genes in Agrobacterium is the chvG/chvI two-component sensor, with a mutation to either chvG or chvI halting virulence [34]. Responsive to acidic pH, the chvG/chvI sensor regulates aopB and katA, two genes involved indirectly in virulence by promoting homeostasis in acidic conditions. A homologous system in 1021 is also responsive to acidic pH. This two component sensor encoded by exoS/chvI regulates the production of succinoglycan and is vital for symbiosis with alfalfa [57]. The OV14 genome has genes homologous to both chvG (exoS) (aproNOG00593) and chvI (aproNOG03091) situated in an operon as expected for two-component sensors (Table 3). The OV14 exoS (chvG) homolog shared 91% and 79% protein sequence identity to 1021’s exoS and C58’s chvG, whereas the OV14 chvI homolog shared 94% and 91% protein sequence identity to 1021 and C58 chvI homologs, respectively. The chvD from C58 interacts with virB8 and has a positive effect on virulence, with both virulence and vir gene expression greatly reduced when the function of chvD is disrupted in C58 [32]. A member of aproNOG00260, all three species contained a homolog with the OV14 homolog recording 89% and 87% protein sequence identity, corresponding to that of 1021 and C58.
Chromosomal virulence gene E (chvE) codes for a multiple sugar binding periplasmic sensor, which interacts with the periplasmic domain of virA aiding in the regulation of the vir operon through the virA/G two-component sensor. OV14 possesses chvE as a member of the aproNOG03985, sharing 93% and 77% protein sequence identity to its 1021 and C58 counterparts. The chvE homologs of 1021 and OV14 are ~100 bp shorter than the version located in C58, with the difference found towards the N terminus of the gene where a putative ligand binding site is positioned. On the C58 circular chromosome, chvE is located adjacent to gguABC components of an ABC sugar transporter. The same operon arrangement is found in all three species chvE-gguABC with all species’ gguABC genes present in the same aproNOGs (A = aproNOG01497, B = aproNOG03238, and C = aproNOG05875). In regards to the C58 gene chvH (encoding elongation P factor, member of aproNOG03687), virulence of A. tumefaciens is decreased in the chvH mutant, which has been linked to reduced levels of virE2 [33]. The OV14 chvH homolog was found to share 95% and 84% protein sequence identity to 1021’s and C58’s respective copy.
The A. tumefaciens ros regulator (aproNOG09171) has been shown to repress the virC and virD operons by binding to a ros box within promoter regions of both genes, but the binding activity of virG is able to overcome this repression although the exact mechanism is unclear. The 1021 ros homolog named mucR (aproNOG09171) is involved in the regulation of both motility and exopolysaccharide production [58]. The OV14 genome has one gene homologous to the ros gene sharing 92% and 79% to 1021 (mucR) and C58 counterparts, correspondingly (Table 3). A homolog of the C58 miaA gene was found in OV14, with mutations of the miaA gene in A. tumefaciens reported to marginally decrease virB, virD and virE gene expression [36]. The miaA gene (aproNOG00010) encodes a tRNA delta (2)-isopentenylpyrophosphate transferase which is involved in protein translation; a homolog was also identified in 1021.
Chromosomal based acvB
C58’s acvB (aproNOG05730) contains multiple annotations, the most common being an acid induced virulence protein and the virJ-like protein. The acvB protein has been reported to bind to the T-strand in the periplasm increasing transport efficiency to the plant cell compared to an acvB - strain [59]. In this regard, OV14 contained one entry in aproNOG05730 as did C58, while 1021 possessed two. The OV14 acvB homolog shares 53% protein identity to C58 acvB and the two 1021 acvB orthologs SMc00612 and SMc00613 were found to share disrupted homology to the C58 and OV14 acvB genes (Table 3).
Protecting against plant defences
The C58 gene katA (aproNOG00015) encodes a catalase-peroxidase implicated in virulence through detoxification of hydrogen peroxide encountered during bacteria-plant interactions [39]. Three catalase genes have been previously identified in 1021; katA, katB and katC[38], with the 1021 katB being a member homologous to and a member of the same aproNOG as the C58 katA. OV14 had two gene members in aproNOG00015; one sharing 89% protein sequence identity with C58’s katA and 61% identity to katB of 1021 and a second more putative gene with 64% similarity to the C58 katB gene. The oxyR peroxide sensor regulates transcription of katA in C58 with hydrogen peroxide and superoxide anions indirectly/directly oxidizing oxyR leading to katA activation [40]. Although the oxyR gene of C58 and 1021 are found separately in aproNOG01190 and aproNOG01330 respectively, OV14 was found to only contain a homolog of the C58 oxyR. While OV14 does possess a dps family protein (aproNOG06937); not found in C58 or 1021. No homologs to the C58 dps (aproNOG08385), which functions to protect DNA from hydroxyl radicals produced during oxidation of Fe (II) by hydrogen peroxide [60] were detected in OV14, or 1021 (Table 3).
C58 and 1021 both have a single catalase gene in aproNOG02507 that functions in protecting cells from the toxic effects of hydrogen peroxide, annotated as catE in C58 and catC in 1021. No homolog was detected in OV14. Superoxide dismutases help to protect the cell via dismutation of superoxide into oxygen and hydrogen peroxide and three copies of the sodB gene (aproNOG00877) were found in C58, two copies in OV14 and one copy in 1021 (Table 3). Knockout of all three sodB genes in A. tumefaciens results in avirulence, while only the sodBI mutant shows reduced virulence when targeted individually [61].
Ti based virulence
The vir operon found on the C58 Ti plasmid encodes the core machinery for the production and transport of T-DNA from the bacterial cell with the two-component regulator virA/G switching on expression of ancillary vir genes upon detection of plant phenolics. No homologs of this system were found to exist in OV14 (Additional file 1: Figure S2) or 1021. A combination of virB genes and virD4 form the Type IV secretion system of C58. Part of aproNOG03383 (Table 5), the virD4 aproNOG is shared by seventy-two species. The aproNOG03383 has two entries in C58 (Atu4858 and Atu6184), named virD4-like and virD4, respectively. Four virD4-like genes were identified in OV14. Upon inspection only one was found to share a protein sequence identity exceeding 50% with any known alphaproteobacteria gene, sharing 71% protein sequence identity to virD4 (Arad_15020) of A. rhizogenes K84.
Table 5.
Comparative analysis for the presence/absence of Ti-based virulence genes in Ensifer adhaerens OV14, Agrobacterium tumefaciens C58, and Sinorhizobium meliloti 1021
|
Gene id |
eggNOG |
OV14 gene |
C58 gene |
1021 gene |
No. species |
No. protein |
Virulence function |
Product |
|---|---|---|---|---|---|---|---|---|
| In C58 | Copy number | Copy number | Copy number | In NOG | In NOG | In C58 | ||
|
virA |
aproNOG05576 |
0 |
1 |
0 |
7 |
7 |
vir operon regulation |
Sensor kinase |
|
virG |
aproNOG04872 |
0 |
1 |
0 |
9 |
10 |
vir operon regulation |
Regulatory protein |
|
virB1 |
aproNOG10673 |
0 |
1 |
0 |
21 |
22 |
Type IV secretion |
Type IV secretion system lytic transglycosylase |
|
virB2 |
aproNOG12925 |
0 |
1 |
0 |
8 |
10 |
Type IV secretion |
Type IV secretion system Pilin subunit protein |
|
virB3 |
aproNOG08388 |
0 |
2 |
1 |
20 |
27 |
Type IV secretion |
Type IV secretion system Pilin-like protein |
|
virB4 |
aproNOG02013 |
2 |
2 |
1 |
71 |
117 |
Type IV secretion |
Type IV secretion system ATPase |
|
virB5 |
N/A |
0 |
1 |
0 |
N/A |
N/A |
Type IV secretion |
Type IV secretion system protein |
|
virB6 |
N/A |
0 |
1 |
0 |
N/A |
N/A |
Type IV secretion |
Type IV secretion system protein |
|
virB7 |
N/A |
0 |
1 |
0 |
N/A |
N/A |
Type IV secretion |
Type IV secretion system protein |
|
virB8 |
N/A |
0 |
1 |
0 |
N/A |
N/A |
Type IV secretion |
Type IV secretion system protein |
|
virB9 |
aproNOG01070 |
0 |
1 |
1 |
20 |
29 |
Type IV secretion |
Type IV secretion system protein |
|
virB10 |
aproNOG01880 |
0 |
2 |
1 |
25 |
32 |
Type IV secretion |
Type IV secretion system ATP energy sensor |
|
virB11 |
aproNOG02544 |
1 |
2 |
1 |
70 |
87 |
Type IV secretion |
Type IV secretion system ATPase |
|
virC1 |
aproNOG17216 |
0 |
1 |
0 |
6 |
6 |
Generation of the T-strand |
DNA-binding protein |
|
virC2 |
N/A |
0 |
1 |
0 |
N/A |
N/A |
Generation of the T-strand |
Hypothetical protein |
|
virD1 |
aproNOG18795 |
0 |
1 |
0 |
5 |
5 |
T-DNA processing |
Endonuclease accessory protein |
|
virD2 |
aproNOG06745 |
0 |
1 |
0 |
13 |
16 |
T-DNA processing |
Endonuclease |
|
virD3 |
aproNOG10158 |
0 |
1 |
0 |
14 |
14 |
T-DNA processing |
Hypothetical protein |
|
virD4 |
aproNOG03383 |
1 |
2 |
0 |
72 |
121 |
Type IV secretion |
Coupling protein |
|
virD5 |
N/A |
0 |
1 |
0 |
N/A |
N/A |
T-DNA processing |
Hypothetical protein |
|
virE0 |
N/A |
0 |
1 |
0 |
N/A |
N/A |
Generation of the T-strand |
Hypothetical protein |
|
virE1 |
N/A |
0 |
1 |
0 |
N/A |
N/A |
Generation of the T-strand |
Chaperone protein |
|
virE2 |
N/A |
0 |
1 |
0 |
N/A |
N/A |
Generation of the T-strand |
ss-DNA binding protein |
|
virE3 |
N/A |
0 |
1 |
0 |
N/A |
N/A |
Generation of the T-strand |
Hypothetical protein |
|
virF |
N/A |
0 |
1 |
0 |
N/A |
N/A |
Effector |
Hypothetical protein |
|
virH1 |
aproNOG14518 |
0 |
1 |
0 |
3 |
3 |
Non-essential |
Hypothetical protein |
|
virH2 |
aproNOG15187 |
0 |
1 |
0 |
3 |
3 |
Non-essential |
Hypothetical protein |
| virK | aproNOG20065 | 0 | 1 | 0 | 4 | 4 | Non-essential | Hypothetical protein |
The virB operon encodes for eleven proteins (numbered 1–11), which form the T-DNA transporting type IV secretion system. C58 has three similar Type IV secretion systems, a Ti-plasmid based virB, a Ti plasmid trb operon and a linear chromosome based avh. VirB3, virB4, virB10, and virB11 form part of the same aproNOGs as their avh counterparts, with virB1, virB2, and virB9 found in different aproNOGs from counterpart’s avhB1, avhB2, avhB11. While avhB5, avhB6, avhB7, and avhB8 are all found in aproNOGs, virB5, virB6, virB7, and virB8 are not associated with any aproNOG. In this analysis, OV14 shares aproNOG02013 (housing avhB3), aproNOG04596 (housing avhB6) and aproNOG02544 (housing avhB11) only. The genes present from OV14 identified as part of aproNOG02013 showed less than 50% protein sequence identity to the closest matches from Phenylobacterium zucineum HLK1 and Erythrobacter litoralis HTCC2594, with OV14 containing 4 genes for aproNOG04596.
Some proteins encoded by the vir regulon are non-essential for transformation but are known to increase transformation efficiency. The respective proteins of virC1 and virC2 may enhance nicking at the right border of T-DNA and virE2 is exported to the plant cell along with the T-strand potentially protecting the ss-DNA from degradation or detection. The virE1 protein binds to virE2 within the bacterial cell blocking interaction with the T-strand until within the plant cell. No homologs for either were found in OV14 or 1021. The genes of the virE operon and virC2 were not found in any aproNOG but the virC1 gene was a member of aproNOG17216 that was identified to be present in 6 alphaproteobacteria species. The virulence genes virF, virH1 and virH2 have been implicated in the expansion of the host range during Agrobacterium-mediated transformation [22], with virF involved in the stripping of virE2 proteins off the T-DNA and virH1 and virH2 involved in the detoxification of anti-bacterial phenolics. With gene virF not part of an aproNOG and virH1 and virH2 of C58 sharing aproNOGs with only 2 other species, Agrobacterium rhizogenes K84 and Chelativorans sp. BNC1, no homologs were found in OV14 or 1021.
Type IV secretion systems
Compared to the three T4SS found in C58 (based on virB, avhB, and trb), only one T4SS was identified in OV14 (based on trb); equivalent to a single system also present in 1021 (based on avhB). The virB T4SS, which is known to export T-DNA from the bacterial cell into the plant cell is only found on the Ti-plasmid of A. tumefaciens. C58’s trb is known to be responsible for conjugation of the Ti-plasmid between bacterial cells. A homologous trb was found on pOV14c (Additional file 1: Figure S2), with the trb operon sharing the same gene arrangement and located immediately upstream of repABC in both the OV14 and C58 genome. The protein sequence identities of the 11 genes comprising the trb operon ranged from 71% to 90%.
pH responsive gene networks
Key to vir gene regulation is chvI, which functions with chvG (known as exoS in S. meliloti 1021) and is up regulated in both C58 and 1021 [42,43]. Nine exo genes (exoF, exoH, exoK, exoL, exoN, exoQ, exoT, exoW, and exoY) involved in the synthesis and metabolism of succinoglycan are shared and up regulated in both organisms and all nine genes were found to be present in OV14. Two additional exo genes (exoM and exoU) were also noted to be in all three species. The acid inducible membrane protein aopB (aproNOG08879), which is involved in C58 virulence was also present as a single copy in OV14 and 1021 (ropB1) (Table 3). All three species are represented in this NOG with the OV14 homolog sharing 55% and 82% protein sequence homology with the C58 aopB and 1021 ropB1, respectively. Finally a recently identified imp type VI (T6SS) secretion system which is up regulated in C58 in response to low pH (5.5) [42] was not found in OV14 nor 1021. The function of this T6SS in C58 has yet to be determined.
Phylogenetic positioning of E. adhaerens OV14 in the Rhizobiales
By concatenating the full length sequence of 12 housekeeping (16S rRNA, 23S rRNA, atpD, dnaK, exoC, gap, gyrB, infB, nusA, pnp, recA, rpoB, thrC) and 8 rhizobial virulence genes (chvA(ndvA), chvB, chvD, chvG(exoS), miaA, and pcs) OV14 grouped with the Sinorhizobium of the Ensifer/Sinorhizobium group forming a clade which is a sister group to the Rhizobium/Agrobacterium clade. Within the Ensifer/Sinorhizobium group, E. adhaerens formed a sister group to the Sinorhizobium species (Figure 3). BLASTn searches of each replicon of OV14 revealed the highest synteny to S. fredii for the chromosomes and to A. vitis S4 pTiS4 and A. tumefaciens pTiC58 for the E. adhaerens pOV14 (Additional file 2). If considering the main chromosome, for which large scale synteny was observed, a gradient of sequence identity can be observed in OV14’s chromosome one; S. fredii strains shows 58% query coverage, S. meliloti strains shows 54% query coverage, S. medicae 49%, Rhizobium species including A. rhizogenes from 34–23%, and A. tumefaciens and Mesorhizobium species show 20% query coverage. Upon a BLASTn of the A. tumefaciens circular chromosome (Additional file 3) the query coverage to the closet matched rhizobia species was 34% dropping to 27%. Interestingly S. fredii species show higher coverage of the C58 circular chromosome at 26% than A. vitis does at 23%. Finally S. meliloti and E. adhaerens share 22% and 20% coverage with the A. tumefaciens C58 circular chromosome, respectively. Further support for this is available from the comparative NOG analysis, which reported OV14 as sharing more genes with 1021 than with C58, whilst also showing that OV14 shares more genes with C58 than 1021 and C58 share with each other (Figure 2).
Figure 3.
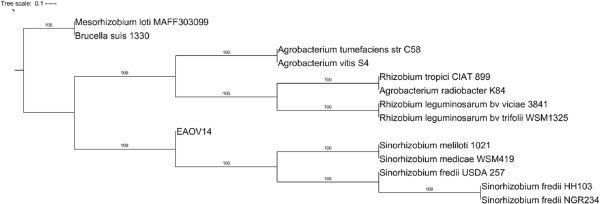
Phylogenetic reconstruction based on the concatenated 16S rRNA, 23S rRNA, atpD, chvA (ndvA), chvB, chvD, chvG (exoS), dnaK, exoC, gap, gyrB, infB, miaA, nusA pcs, pnp, recA, rpoB, and thrC gene sequences. Analyses were conducted using the consensus method (majority rule extended) with 100 bootstrap replicates. Bootstrap scores are represented numerically above branches. EAOV14 represents E. adhaerens.
Discussion
While the ability of OV14 to genetically transform plant genomes has previously been demonstrated [12], OV14 has not acquired a known suite of symbiosis or pathogenesis genes but instead appears to effectively utilise supplementary vir or sym genes for virulence and symbiosis. For example; E. adhaerens strain ATCC 33499 was found to form nitrogen-fixing nodules on Phaseolus vulgaris (bean) and Leucaena leucocephala when equipped with the two symbiotic plasmids from Rhizobium tropici CFN299 [62], strain OV14 used in this study has shown the ability to deliver DNA into plants when equipped with the unitary pCambia5105 vector and strain 5D19 has been found to inhabit alfalfa nodules during a screen for diversity among the S. meliloti population [63]. Of interest, R. etli CE3 has been shown to possess vir gene homologs on a self-transmissible vector indicating a potential source of novel vir genes in soil bacteria [64].
For this study, the genome of OV14 was compared against the genomes of C58 and 1021 using eggNOG assignments to gain an understanding of their relatedness, with a focus being on an in/ability to transform plant cells. The potential of OV14 and 1021 to transfer T-DNA has only been achieved when equipped with a Ti-plasmid [7,12]. In this study we confirmed that there are no Agrobacterium vir gene homologs present in the OV14 genome. In the cases where homologs appeared in NOGs alongside virB and virD4 it was likely due to parallel functions within type IV secretion systems. For the virB type IV secretion system virB5, virB6, virB7 and virB8 were not found as part of any NOG suggesting no similar genes in fully sequenced alphaproteobacteria to date. However, Sugawara et al. [45] detected five clades of type IV secretion systems within 48 sequenced Sinorhizobium species. Phylogenetic analysis found A. tumefaciens virB genes to be present in clade I with 1021 virB genes found in clade II along with A. tumefaciens avh genes. Interestingly, seven Sinorhizobium strains were found to possess type IV secretion systems in clade I [45].
Homologs to all chromosomal-based genes cited to be essential for T-DNA transfer in C58 were found to be present in OV14 and 1021, as indeed they are also present in several other alphaproteobacteria. Additionally all genes shown to be beneficial for Agrobacterium virulence were found in OV14, however not all of these genes were identified in 1021. At first it may appear that these genes are advantageous to life in the rhizosphere, however their presence in bacteria such as the animal pathogen Brucella broadens this hypothesis. Transgression into the rhizosphere or an animal host represents a dramatic change in environmental conditions. While nutrient availability may increase, changes in pH and eukaryotic cells defending themselves against invasion becomes a new challenge for the cell to overcome. Transcriptomic profiles in response to acidic pH (5.5) in both C58 and 1021 have shown the expression of genes involved in succionoglycan biogenesis and the regulation of acid inducible genes [42,65]. Down regulation of genes involved in motility via flagellum is also a shared response. Separately, electron microscopy analysis of OV14 completed by this research group has already confirmed the presence of functional flagella (Rathore et al. unpublished). The production of flagellum requires a large amount of energy and down regulation may free up energy for rearrangement of the cell envelope. Consequently, down regulation of flagellum may make the bacterial cell difficult to detect for the plant cell.
While many exo genes have been shown to be upregulated in C58 in response to acidic pH, the chvG/chvI sensor in C58 has not been shown to control their expression. Exerting control over cell envelope composition is most likely critical to the type of interaction that occurs with the plant cell directly via cell-to-cell contact or indirectly via the ability of the membrane to incorporate influential proteins and protein complexes. Indeed, differences in polysaccharide biosynthesis among Sinorhizobium species has been predicted as a host determination factor allowing for varying strategies for legume-Sinorhizobium interactions [45]. The C58 aopB has been shown to have a positive effect on virulence and was upregulated by acidic pH while the 1021 aopB homolog ropB1 was not [42,66]. Of interest, a ropB1 R. leguminosarum bv. viciae VF39SM mutant was shown to have increased sensitivity to detergents, hydrophobic antibiotics, and weak organic acids, with a suggestion that ropB1 plays a role in membrane stability [66]. For OV14, it would be of interest to complete a transcriptomic analysis to monitor the expression of these key genes under varying induction conditions.
Another cell wall component present in OV14, C58 and 1021 is phosphatidylcholine, a phospholipid and major component of eukaryotic membranes. Phosphatidylcholine has been shown to be essential for virulence in C58 and for symbiosis and normal growth in 1021 [67]. Interestingly an A. tumefaciens mutant deficient in phosphatidylcholine production was unable to support a type IV secretion system in the bacterial cell wall and subsequently lost its virulence potential [54]. The role of the virB type IV secretion system is known to be critical for T-DNA transfer to plants and is potentially the only system capable of this feat. Differences in phosphatidylcholine content in the cell wall of OV14, C58 and 1021 could affect the cells ability to support the complete virB type IV secretion system and greatly affect the transfer of T-DNA. The phosphatidylcholine synthase (pcs) pathway is choline-dependent and requires the uptake of choline into the cell. An Agrobacterium pcs/pmtA double mutant has been shown to be attenuated in expression of the virB operon [54] while a high-affinity choline ABC transport choXWV has been identified in C58 and 1021 [68,69]; OV14 was found to contain homologs to this system. The choV gene encodes the ATPase component of the ABC transporter. The fact that C58 has an additional copy of choV compared to OV14 and two additional copies of choV compared to 1021 suggests C58 may have an increased ability to actively acquire choline for phosphatidylcholine synthesis and ultimately complete T-DNA transfer into targeted host cells.
One of the most notable differences across the studied genomes was that OV14 and C58 possess cel homologs that are absent in 1021. The cel locus has been implicated in the attachment of Agrobacterium to the plant cell but is not required for tumour formation [70]. The presence of cel homologs in OV14 may suggest a role in attachment to plant surfaces or potentially other bacteria, which may explain its discovery within nodules alongside S. meliloti and the recorded ability of E. adhaerens to phagocytose other bacteria. Another observed difference was the lack of a type VI (T6SS) secretion system in OV14 and 1021 compared to C58. The lack of a type VI secretion system in 1021 was also noted by Sugawara et al. [45]. Type VI secretion systems are a relatively new discovery and their application is not well understood. However studies in Vibrio cholerae and Pseudomonas aeruginosa have shown the T6SS system to be involved in aggressive bacteria-to-bacteria cell interactions [71].
The chromosomal-based acvB gene has been cited as essential for Agrobacterium to achieve T-DNA transfer in the absence of virJ and has been found to localise to the periplasmic space and associate with the T-stand in Agrobacterium[72]. Homologs to the acvB gene were found in all three species in this study and another 25 alphaproteobacteria upon inspection of aproNOG05730. The homology of the two 1021 copies to the C58 acvB and OV14 homologs was found to be low, with it appearing that the 1021 acvB has split into two genes, which may be why acvB was not initially detected in 1021 [59]. The C58 and OV14 acvB shares 50% homology with virJ a gene found on octopine-type Ti plasmids which can complement a acvB mutant [73]. While acvB appears to play a role in export maybe as a chaperone to the T-strand [59,73], the OV14 acvB homolog shares 50% identity to its C58 counterpart and could be a key target for future studies focussed on the improvement of EMT.
The ability to defend against oxidative stress leads to increased virulence as the bacterial cell survives plant cell defences and acidic pH allowing expression of virulence genes and delivery of T-DNA to plant cell. All three species in this study were found to possess genes involved in protection against reactive oxygen species. One such gene that was absent in OV14 and 1021 was dps, (DNA-binding proteins from starved cells), which in A. tumefaciens protects the cell by acting as an hydroxyl radical scavenger and could well function with a catalase such as katA to increase the cell’s tolerance to the toxic effect of hydrogen peroxide [60].
In 1982, E. adhaerens was first described as a gram-negative predatory bacteria [13] but more recently, a request to rename E. adhaerens to S. adhaerens initiated a debate as to the appropriate nomenclature [74]. Following on from this the International Committee on Systematics of Prokaryotes ruled all Sinorhizobium species were to be transferred to Ensifer based on Ensifer being an early synonym of Sinorhizobium[13,75,76]. The current standing appears to be that Ensifer is the correct name for the amended genus, but the Judicial Commission also acknowledges the later synonym Sinorhizobium[76]. The work detailed here describes the genome sequencing of OV14 and based on a phylogenetic analysis of 20 housekeeping genes shows OV14 to form a branch separated from the Sinorhizobia. While the primary chromosome of OV14 shows a high level of synteny with the Sinorhizobia (highest to S. fredii) the remaining replicons share minimal synteny to any known species and are a potential resource of novel alpha-proteobacterial genes.
Conclusions
This study has confirmed the presence of genes in OV14 that are confirmed homologs of chromosomal-based C58 virulence genes. As to how much their sequence diversity affects their function during T-DNA transfer remains unknown. Whereas the reported transformation efficiency of OV14 was achieved with environmental conditions optimal for A. tumefaciens, it is possible that Ti virulence induction conditions for non-Agrobacterium species may be different to A. tumefaciens and this is therefore an area that requires further attention. Re-engineering these non-Agrobacterium species with improved virulence functions offers the opportunity to increase the range of bacterial species that can be used for the genetic transformation of plant cells. Considering the limitations to the host range of A. tumefaciens have already been described [16,18,77], the use of non-pathogenic bacterial species may increase the range of plant species amenable to agronomic enhancement via genetic transformation.
Methods
OV14 was originally isolated from the rhizosphere of Brassica napus at Oak Park in Carlow, Ireland. The strain was selected for sequencing based on its ability to transform plant cells [12].
DNA isolation
Strain OV14 was grown to midlogarithmic phase in TY medium at 28°C, 200 rpm. DNA was isolated from 20 ml of cells using a modified CTAB (Cetyl thrimethylammonium bromide) genomic DNA isolation method [78]. RNase (20 mg/ml) was added to lysis buffer in step 3, and centrifugal spins were extended to 20 mins to allow separation to lysate and supernatant.
Genome sequencing
The OV14 genome was sequenced and constructed by BaseClear B. V. Leiden, Netherlands. A hybrid approach using the Illumina HiSeq and PacBio RS platforms was selected. The genome was constructed from 1GB Illumina paired-end reads, 500 MB Illumina mate paired end reads, and 100 MB PacBio RS reads. The assembly was built in the following manner. First Illumina raw reads filtered using CASAVA version 1.8.2 and subsequently trimmed based on the Phred quality scores using the CLC Genomics workbench 1.8.3. Filtering of PacBio CLR reads was performed using the PacBio SMRT analysis suite. The quality-trimmed sequence reads were puzzled into a number of contig sequences with the CLCbio de novo assembler. This set defines the draft assembly. Subsequently the contigs were linked and placed into super-scaffolds based on the alignment against the long PacBio CLR reads. Alignment of the contigs was performed with BLASR [79]. From the alignment the orientation, order and distance between the contigs was estimated. As a result contigs were placed in super-scaffolds. This analysis was performed using a modified version of the SSPACE Premium scaffolder version 2.3 [80]. Finally gapped regions within the super-scaffolds were (partially) closed in an automated manner using GapFiller version 1.10 [81]. The method takes advantage of the insert size between the Illumina paired-end reads. The resulting scaffolds define the draft genome and plasmids, with the genome sequence available in the NCBI database under accession numbers CP007236.1, CP007237.1, CP007238.1 and CP007239.1.
eggNOG analysis
Glimmer-predicted coding regions in the OV14 genome were BLASTp searched against an alphaproteobacteria database downloaded from eggnog.embl.de and assigned to NOGs based on similarity with a cut-off of 60 bits used to filter data. A reciprocal blast analysis (genome to EGGNOG and EGGNOG to genome) was also completed to ensure that recorded hits were evident in both directions, regardless of obtained low r values, which may have been due to evolutionary distinctness of the species. For comparative analysis all alphaproteobacterial gene families and their corresponding functional classifications were retrieved from eggNOG. The Functional categories used are based on: A Genomic Perspective on Protein Families [82]. The literature was screened for all genes known and predicted to be involved in T-DNA transfer and genes induced by the rhizosphere/rhizoplane environment across all NOG categories.
Phylogenetic analysis
FASTA files for individual genes were obtained from NCBI and aligned using Clustal Omega. Clustal files were converted to Phylip format using an online tool found at http://insilico.ehu.es/tophylip/. Phylip files were concatenated using Seaview 4. The 40,470 base pair concatenated file was run using raxmlGUI producing a consensus tree with 100 bootstrap replicates. The tree was rooted by treating Brucella suis 1330 and Mesorhizobium loti MAFF303099 as the outgroup.
Availability of supporting data
The following additional data are available with the online version of this paper. Additional file 1 includes Figures S1 and S2. Additional file 2 includes BLAST search scores for OV14 replicons. Additional file 3 includes BLAST search scores for C58 replicons.
Abbreviations
EMT: Ensifer-mediated transformation; AMT: Agrobacterium-mediated transformation; Ti: Tumor inducing; T-DNA: Transfer-DNA; ROS: Reactive oxygen species; SA: Salicylic acid; T6SS: Type VI secretion system.
Competing interests
The authors declare no competing interests.
Authors’ contributions
SR, EM, FD, TW and CC developed the concepts and designed the research. SR performed the research and analysed the data with assistance from CC, EM and FD. EM and FD supervised the project. SR and EM prepared the paper. All authors edited read and approved the submitted manuscript.
Supplementary Material
Circular representation of the four replicons of E. adhaerens OV14. Circles, from the inside out, show: (1) GC skew; (2) Coding regions; light blue blocks represent genes with predicted function, red blocks show transposable elements, dark blue and grey blocks show genes of hypothetical and unknown function, respectively. Figure S2. Synteny plots showing total sequence of Ensifer adhaerens OV14 pOV14c (top bar) vs Agrobacterium tumefaciens C58 pTi (bottom bar), computed using DoubleACT version2 on tBLASTx setting with cut off set at 100. Visualised in Artemis ACT. Homology between the genomes is displayed via interconnecting lines; red lines representing direct homology while blue lines represent homologues but inverted sequence.
BLAST of Ensifer adhaerens OV14 replicons.xlsx. Excel file includes tables of BLAST search of individual Ensifer adhaerens OV14 replicons.
BLAST of Agrobacterium tumefaciens C58 replicons.xlsx. Excel file includes tables of BLAST searches of individual Agrobacterium tumefaciens C58.
Contributor Information
Steven Rudder, Email: steven.rudder@ucdconnect.ie.
Fiona Doohan, Email: fiona.doohan@ucdconnect.ie.
Christopher J Creevey, Email: chris.creevy@gmail.com.
Toni Wendt, Email: toni.wendt@agrsci.dk.
Ewen Mullins, Email: ewen.mullins@teagasc.ie.
Acknowledgements
This publication has emanated from research conducted with the financial support of Science Foundation Ireland under grant number SFI 11/RFP.1/GEN/3420.
References
- AzpirozLeehan R, Feldmann KA. T-DNA insertion mutagenesis in Arabidopsis: going back and forth. Trends Genet. 1997;15(4):152–156. doi: 10.1016/s0168-9525(97)01094-9. [DOI] [PubMed] [Google Scholar]
- Jeon JS, Lee S, Jung KH, Jun SH, Jeong DH, Lee J, Kim C, Jang S, Yang K, Nam J, An K, Han MJ, Sung RJ, Choi HS, Yu JH, Choi JH, Cho SY, Cha SS, Kim SI, An G. T-DNA insertional mutagenesis for functional genomics in rice. Plant J. 2000;15(6):561–570. doi: 10.1046/j.1365-313x.2000.00767.x. [DOI] [PubMed] [Google Scholar]
- Thomas CM, Tang SJ, Hammond-Kosack K, Jones JDG. Comparison of the hypersensitive response induced by the tomato Cf-4 and Cf-9 genes in Nicotiana spp. Mol Plant Microbe Interact. 2000;15(4):465–469. doi: 10.1094/MPMI.2000.13.4.465. [DOI] [PubMed] [Google Scholar]
- Johansen LK, Carrington JC. Silencing on the spot. Induction and suppression of RNA silencing in the Agrobacterium-mediated transient expression system. Plant Physiol. 2001;15(3):930–938. doi: 10.1104/pp.126.3.930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An G, Watson BD, Chiang CC. Transformation of tobacco, tomato, potato, and Arabidopsis-thaliana using a binary Ti vector system. Plant Physiol. 1986;15(1):301–305. doi: 10.1104/pp.81.1.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James C. ISAAA Brief 46, Global Status of Commercialized Biotech/GM Crops. Ithaca, New York: ISAAA Briefs; 2011. International Service for the Acquisition of Agri-biotech Applications (ISAAA) Retrieved February 2014. [Google Scholar]
- Broothaerts W, Mitchell HJ, Weir B, Kaines S, Smith LMA, Yang W, Mayer JE, Roa-Rodriguez C, Jefferson RA. Gene transfer to plants by diverse species of bacteria. Nature. 2005;15(7026):629–633. doi: 10.1038/nature03309. [DOI] [PubMed] [Google Scholar]
- Chi-Ham CL, Boettiger S, Figueroa-Balderas R, Bird S, Geoola JN, Zamora P, Alandete-Saez M, Bennett AB. An intellectual property sharing initiative in agricultural biotechnology: development of broadly accessible technologies for plant transformation. Plant Biotechnol J. 2012;15(5):501–510. doi: 10.1111/j.1467-7652.2011.00674.x. [DOI] [PubMed] [Google Scholar]
- Nottenburg C, Rodríguez CR. In: Agrobacterium: From Biology to Biotechnology. Tzfira T, Citovsky V, editor. New York: Springer; 2008. Agrobacterium-mediated gene transfer: a lawyer’s perspective; pp. 699–735. [Google Scholar]
- Van Veen R, den Dulk-Ras H, Schilperoort R, Hooykaas P. Ti plasmid containing Rhizobium meliloti are non-tumorigenic on plants, despite proper virulence gene induction and T-strand formation. Arch Microbiol. 1989;15(1):85–89. doi: 10.1007/BF00277546. [DOI] [Google Scholar]
- Wendt T, Doohan F, Winckelmann D, Mullins E. Gene transfer into Solanum tuberosum via Rhizobium spp. Transgenic Res. 2011;15(2):377–386. doi: 10.1007/s11248-010-9423-4. [DOI] [PubMed] [Google Scholar]
- Wendt T, Doohan F, Mullins E. Production of Phytophthora infestans-resistant potato (Solanum tuberosum) utilising Ensifer adhaerens OV14. Transgenic Res. 2012;15(3):567–578. doi: 10.1007/s11248-011-9553-3. [DOI] [PubMed] [Google Scholar]
- Casida LE. Ensifer-adhaerens gen-nov, sp-nov - a bacterial predator of bacteria in soil. Int J Syst Bacteriol. 1982;15(3):339–345. doi: 10.1099/00207713-32-3-339. [DOI] [Google Scholar]
- Pitzschke A, Hirt H. New insights into an old story: Agrobacterium-induced tumour formation in plants by plant transformation. EMBO J. 2010;15(6):1021–1032. doi: 10.1038/emboj.2010.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelvin SB. The introduction and expression of transgenes in plants. Curr Opin Biotechnol. 1998;15(2):227–232. doi: 10.1016/S0958-1669(98)80120-1. [DOI] [PubMed] [Google Scholar]
- Gelvin SB. Agrobacterium and plant genes involved in T-DNA transfer and integration. Annu Rev Plant Biol. 2000;15(1):223–256. doi: 10.1146/annurev.arplant.51.1.223. [DOI] [PubMed] [Google Scholar]
- Gelvin SB. Traversing the cell: Agrobacterium T-DNA’s journey to the host genome. Front Plant Sci. 2012;15:52. doi: 10.3389/fpls.2012.00052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzfira T, Citovsky V. Agrobacterium-mediated genetic transformation of plants: biology and biotechnology. Curr Opin Biotechnol. 2006;15(2):147–154. doi: 10.1016/j.copbio.2006.01.009. [DOI] [PubMed] [Google Scholar]
- McCullen CA, Binns AN. Agrobacterium tumefaciens and plant cell interactions and activities required for interkingdom macromolecular transfer. Annu Rev Cell Dev Biol. 2006;15:101–127. doi: 10.1146/annurev.cellbio.22.011105.102022. [DOI] [PubMed] [Google Scholar]
- Gelvin SB. Agrobacterium-mediated plant transformation: the biology behind the “gene-Jockeying” tool. Microbiol Mol Biol Rev. 2003;15(1):16. doi: 10.1128/MMBR.67.1.16-37.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galibert F, Finan TM, Long SR, Pühler A, Abola P, Ampe F, Barloy-Hubler F, Barnett MJ, Becker A, Boistard P. The composite genome of the legume symbiont Sinorhizobium meliloti. Science. 2001;15(5530):668–672. doi: 10.1126/science.1060966. [DOI] [PubMed] [Google Scholar]
- Goodner B, Hinkle G, Gattung S, Miller N, Blanchard M, Qurollo B, Goldman BS, Cao Y, Askenazi M, Halling C. Genome sequence of the plant pathogen and biotechnology agent Agrobacterium tumefaciens C58. Science. 2001;15(5550):2323–2328. doi: 10.1126/science.1066803. [DOI] [PubMed] [Google Scholar]
- Wood DW, Setubal JC, Kaul R, Monks DE, Kitajima JP, Okura VK, Zhou Y, Chen L, Wood GE, Almeida NF. The genome of the natural genetic engineer Agrobacterium tumefaciens C58. Science. 2001;15(5550):2317–2323. doi: 10.1126/science.1066804. [DOI] [PubMed] [Google Scholar]
- Slater SC, Goldman BS, Goodner B, Setubal JC, Farrand SK, Nester EW, Burr TJ, Banta L, Dickerman AW, Paulsen I. Genome sequences of three Agrobacterium biovars help elucidate the evolution of multichromosome genomes in bacteria. J Bacteriol. 2009;15(8):2501–2511. doi: 10.1128/JB.01779-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnett MJ, Fisher RF, Jones T, Komp C, Abola AP, Barloy-Hubler F, Bowser L, Capela D, Galibert F, Gouzy J. Nucleotide sequence and predicted functions of the entire Sinorhizobium meliloti pSymA megaplasmid. Proc Natl Acad Sci. 2001;15(17):9883–9888. doi: 10.1073/pnas.161294798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finan TM, Weidner S, Wong K, Buhrmester J, Chain P, Vorhölter FJ, Hernandez-Lucas I, Becker A, Cowie A, Gouzy J. The complete sequence of the 1,683-kb pSymB megaplasmid from the N2-fixing endosymbiont Sinorhizobium meliloti. Proc Natl Acad Sci. 2001;15(17):9889–9894. doi: 10.1073/pnas.161294698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomlinson AD, Fuqua C. Mechanisms and regulation of polar surface attachment in Agrobacterium tumefaciens. Curr Opin Microbiol. 2009;15(6):708–714. doi: 10.1016/j.mib.2009.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cangelosi GA, Martinetti G, Leigh JA, Lee CC, Thienes C, Nester EW. Role for [corrected] Agrobacterium tumefaciens ChvA protein in export of beta-1,2-glucan. J Bacteriol. 1989;15(3):1609–1615. doi: 10.1128/jb.171.3.1609-1615.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ielpi L, Dylan T, Ditta GS, Helinski DR, Stanfield SW. The ndvB locus of Rhizobium meliloti encodes a 319-kda protein involved in the production of beta-(1-2)-glucan. J Biol Chem. 1990;15(5):2843–2851. [PubMed] [Google Scholar]
- Stanfield SW, Ielpi L, Obrochta D, Helinski DR, Ditta GS. The ndvA gene-product of Rhizobium meliloti is required for beta-(1-2) glucan production and has homology to the Atp-binding export protein HlyB. J Bacteriol. 1988;15(8):3523–3530. doi: 10.1128/jb.170.8.3523-3530.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair GR, Liu Z, Binns AN. Re-examining the role of the accessory plasmid pAtC58 in the virulence of Agrobacterium tumefaciens strain C58. Plant Physiol. 2003;15(3):989–999. doi: 10.1104/pp.103.030262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu ZY, Jacobs M, Schaff DA, McCullen CA, Binns AN. ChvD, a chromosomally encoded ATP-binding cassette transporter-homologous protein involved in regulation of virulence gene expression in Agrobacterium tumefaciens. J Bacteriol. 2001;15(11):3310–3317. doi: 10.1128/JB.183.11.3310-3317.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng WT, Banta LM, Charles TC, Nester EW. The chvH locus of Agrobacterium encodes a homologue of an elongation factor involved in protein synthesis. J Bacteriol. 2001;15(1):36–45. doi: 10.1128/JB.183.1.36-45.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charles TC, Nester EW. A chromosomally encoded two-component sensory transduction system is required for virulence of Agrobacterium tumefaciens. J Bacteriol. 1993;15(20):6614–6625. doi: 10.1128/jb.175.20.6614-6625.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Close TJ, Rogowsky PM, Kado CI, Winans SC, Yanofsky MF, Nester EW. Dual control of Agrobacterium tumefaciens Ti plasmid virulence genes. J Bacteriol. 1987;15(11):5113–5118. doi: 10.1128/jb.169.11.5113-5118.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray J, Wang J, Gelvin SB. Mutation of the miaA gene of Agrobacterium tumefaciens results in reduced vir gene expression. J Bacteriol. 1992;15(4):1086–1098. doi: 10.1128/jb.174.4.1086-1098.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemner JM, Liang XY, Nester EW. The Agrobacterium tumefaciens virulence gene chvE is part of a putative ABC-type sugar transport operon. J Bacteriol. 1997;15(7):2452–2458. doi: 10.1128/jb.179.7.2452-2458.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo L, Qi MS, Yao SY, Cheng HP, Zhu JB, Yu GQ. Role of oxyR from Sinorhizobium meliloti in regulating the expression of catalases. Acta Biochim Biophys Sin. 2005;15(6):421–428. doi: 10.1111/j.1745-7270.2005.00055.x. [DOI] [PubMed] [Google Scholar]
- Xu XQ, Li LP, Pan SQ. Feedback regulation of an Agrobacterium catalase gene katA involved in Agrobacterium-plant interaction. Mol Microbiol. 2001;15(3):645–657. doi: 10.1046/j.1365-2958.2001.02653.x. [DOI] [PubMed] [Google Scholar]
- Nakjarung K, Mongkolsuk S, Vattanaviboon P. The oxyR from Agrobacterium tumefaciens: evaluation of its role in the regulation of catalase and peroxide responses. Biochem Biophys Res Commun. 2003;15(1):41–47. doi: 10.1016/S0006-291X(03)00535-7. [DOI] [PubMed] [Google Scholar]
- Heckman J, Strick J. Teaching plant-soil relationships with color images of rhizosphere pH. J Nat Resour Life Sci Educ. 1996;15(1):13–16. [Google Scholar]
- Yuan ZC, Liu P, Saenkham P, Kerr K, Nester EW. Transcriptome profiling and functional analysis of Agrobacterium tumefaciens reveals a general conserved response to acidic conditions (pH 5.5) and a complex acid-mediated signaling involved in Agrobacterium-plant interactions. J Bacteriol. 2008;15(2):494–507. doi: 10.1128/JB.01387-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellweg C, Puhler A, Weidner S. The time course of the transcriptomic response of Sinorhizobium meliloti 1021 following a shift to acidic pH. BMC Microbiol. 2009;15:37. doi: 10.1186/1471-2180-9-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li LP, Jia YH, Hou QM, Charles TC, Nester EW, Pan SQ. A global pH sensor: Agrobacterium sensor protein ChvG regulates acid-inducible genes on its two chromosomes and Ti plasmid. Proc Natl Acad Sci. 2002;15(19):12369–12374. doi: 10.1073/pnas.192439499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugawara M, Epstein B, Badgley B, Unno T, Xu L, Reese J, Gyaneshwar P, Denny R, Mudge J, Bharti AK, Farmer AD, May GD, Woodward JE, Médigue C, Vallenet D, Lajus A, Rouy Z, Martinez-Vaz B, Tiffin P, Young ND, Sadowsky MJ. Comparative genomics of the core and accessory genomes of 48 Sinorhizobium strains comprising five genospecies. Genome Biol. 2013;15(2):R17. doi: 10.1186/gb-2013-14-2-r17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black M, Moolhuijzen P, Chapman B, Barrero R, Howieson J, Hungria M, Bellgard M. The genetics of symbiotic nitrogen fixation: comparative genomics of 14 rhizobia strains by resolution of protein clusters. Genes. 2012;15(1):138–166. doi: 10.3390/genes3010138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thi Vu H, Itoh H, Ishii S, Senoo K, Otsuka S. Identification and phylogenetic characterization of cobalamin biosynthetic genes of Ensifer adhaerens. Microbes Environ. 2013;15(1):153–155. doi: 10.1264/jsme2.ME12069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen LJ, Julien P, Kuhn M, von Mering C, Muller J, Doerks T, Bork P. eggNOG: automated construction and annotation of orthologous groups of genes. Nucleic Acids Res. 2008;15(Database issue):D250–D254. doi: 10.1093/nar/gkm796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell S, Szklarczyk D, Trachana K, Roth A, Kuhn M, Muller J, Arnold R, Rattei T, Letunic I, Doerks T, Jensen LJ, von Mering C, Bork P. eggNOG v3.0: orthologous groups covering 1133 organisms at 41 different taxonomic ranges. Nucleic Acids Res. 2012;15(Database issue):D284–D289. doi: 10.1093/nar/gkr1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthysse AG, Yarnall H, Boles SB, McMahan S. A region of the Agrobacterium tumefaciens chromosome containing genes required for virulence and attachment to host cells. Biochim Biophys Acta. 2000;15(1–2):208–212. doi: 10.1016/s0167-4781(99)00250-x. [DOI] [PubMed] [Google Scholar]
- Matthysse AG, Jaeckel P, Jeter C. attG and attC mutations of Agrobacterium tumefaciens are dominant negative mutations that block attachment and virulence. Can J Microbiol. 2008;15(4):241–247. doi: 10.1139/W08-005. [DOI] [PubMed] [Google Scholar]
- Yuan ZC, Edlind MP, Liu P, Saenkham P, Banta LM, Wise AA, Ronzone E, Binns AN, Kerr K, Nester EW. The plant signal salicylic acid shuts down expression of the vir regulon and activates quormone-quenching genes in Agrobacterium. Proc Natl Acad Sci U S A. 2007;15(28):11790–11795. doi: 10.1073/pnas.0704866104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthysse AG, McMahan S. Root colonization by Agrobacterium tumefaciens is reduced in cel, attB, attD, and attR mutants. Appl Environ Microbiol. 1998;15(7):2341–2345. doi: 10.1128/aem.64.7.2341-2345.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessel M, Klusener S, Godeke J, Fritz C, Hacker S, Narberhaus F. Virulence of Agrobacterium tumefaciens requires phosphatidylcholine in the bacterial membrane. Mol Microbiol. 2006;15(3):906–915. doi: 10.1111/j.1365-2958.2006.05425.x. [DOI] [PubMed] [Google Scholar]
- Klusener S, Aktas M, Thormann KM, Wessel M, Narberhaus F. Expression and physiological relevance of Agrobacterium tumefaciens phosphatidylcholine biosynthesis genes. J Bacteriol. 2009;15(1):365–374. doi: 10.1128/JB.01183-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rong L, Karcher S, O’Neal K, Hawes M, Yerkes C, Jayaswal R, Hallberg C, Gelvin S. picA, a novel plant-inducible locus on the Agrobacterium tumefaciens chromosome. J Bacteriol. 1990;15(10):5828–5836. doi: 10.1128/jb.172.10.5828-5836.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng HP, Walker GC. Succinoglycan production by Rhizobium meliloti is regulated through the ExoS-ChvI two-component regulatory system. J Bacteriol. 1998;15(1):20–26. doi: 10.1128/jb.180.1.20-26.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahlawane C, McIntosh M, Krol E, Becker A. Sinorhizobium meliloti regulator MucR couples exopolysaccharide synthesis and motility. Mol Plant Microbe Interact. 2008;15(11):1498–1509. doi: 10.1094/MPMI-21-11-1498. [DOI] [PubMed] [Google Scholar]
- Wirawan IGP, Kojima M. Distribution of a chromosomal virulence gene, acvB, of Agrobacterium tumefaciens among various bacteria. Biosci Biotechnol Biochem. 1996;15(1):50–53. doi: 10.1271/bbb.60.50. [DOI] [Google Scholar]
- Ceci P, Ilari A, Falvo E, Chiancone E. The Dps protein of Agrobacterium tumefaciens does not bind to DNA but protects it toward oxidative cleavage x-ray crystal structure, iron binding and hydroxyl radical scavenging properties. J Biol Chem. 2003;15(22):20319–20326. doi: 10.1074/jbc.M302114200. [DOI] [PubMed] [Google Scholar]
- Saenkham P, Eiarnphungporn W, Farrand SK, Vattanaviboon P, Mongkolsuk S. Multiple superoxide dismutases in Agrobacterium tumefaciens: functional analysis, gene regulation, and influence on tumorigenesis. J Bacteriol. 2007;15(24):8807–8817. doi: 10.1128/JB.00960-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogel MA, Hernandez-Lucas I, Kuykendall LD, Balkwill DL, Martinez-Romero E. Nitrogen-fixing nodules with Ensifer adhaerens harboring Rhizobium tropici symbiotic plasmids. Appl Environ Microbiol. 2001;15(7):3264–3268. doi: 10.1128/AEM.67.7.3264-3268.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willems A, Fernandez-Lopez M, Munoz-Adelantado E, Goris J, De Vos P, Martinez-Romero E, Toro N, Gillis M. Description of new Ensifer strains from nodules and proposal to transfer Ensifer adhaerens Casida 1982 to Sinorhizobium as Sinorhizobium adhaerens comb. nov. Request for an opinion. Int J Syst Evol Microbiol. 2003;15(Pt 4):1207–1217. doi: 10.1099/ijs.0.02264-0. [DOI] [PubMed] [Google Scholar]
- Bittinger MA, Gross JA, Widom J, Clardy J, Handelsman J. Rhizobium etli CE3 carries vir gene homologs on a self-transmissible plasmid. Mol Plant Microbe Interact. 2000;15(9):1019–1021. doi: 10.1094/MPMI.2000.13.9.1019. [DOI] [PubMed] [Google Scholar]
- Jia YH, Li LP, Hou QM, Pan SQ. An Agrobacterium gene involved in tumorigenesis encodes an outer membrane protein exposed on the bacterial cell surface. Gene. 2002;15(1–2):113–124. doi: 10.1016/s0378-1119(02)00385-2. [DOI] [PubMed] [Google Scholar]
- Foreman DL, Vanderlinde EM, Bay DC, Yost CK. Characterization of a gene family of outer membrane proteins (ropB) in Rhizobium leguminosarum bv. viciae VF39SM and the role of the sensor kinase ChvG in their regulation. J Bacteriol. 2010;15(4):975–983. doi: 10.1128/JB.01140-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Rudder KEE, Lopez-Lara IM, Geiger O. Inactivation of the gene for phospholipid N-methyltransferase in Sinorhizobium meliloti: phosphatidylcholine is required for normal growth. Mol Microbiol. 2000;15(4):763–772. doi: 10.1046/j.1365-2958.2000.02032.x. [DOI] [PubMed] [Google Scholar]
- Aktas M, Jost KA, Fritz C, Narberhaus F. Choline uptake in Agrobacterium tumefaciens by the high-affinity ChoXWV transporter. J Bacteriol. 2011;15(19):5119–5129. doi: 10.1128/JB.05421-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupont L, Garcia I, Poggi MC, Alloing G, Mandon K, Le Rudulier D. The Sinorhizobium meliloti ABC transporter cho is highly specific for choline and expressed in bacteroids from Medicago sativa nodules. J Bacteriol. 2004;15(18):5988–5996. doi: 10.1128/JB.186.18.5988-5996.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthysse AG. Role of bacterial cellulose fibrils in Agrobacterium tumefaciens infection. J Bacteriol. 1983;15(2):906–915. doi: 10.1128/jb.154.2.906-915.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basler M, Ho BT, Mekalanos JJ. Tit-for-tat: type VI secretion system counterattack during bacterial cell-cell interactions. Cell. 2013;15(4):884–894. doi: 10.1016/j.cell.2013.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang HW, Wirawan IGP, Kojima M. Cellular-localization and functional-analysis of the protein encoded by the chromosomal virulence gene (acvb) of Agrobacterium tumefaciens. Biosci Biotechnol Biochem. 1994;15(11):2024–2032. doi: 10.1271/bbb.58.2024. [DOI] [PubMed] [Google Scholar]
- Kalogeraki VS, Winans SC. The octopine-type Ti plasmid pTia6 of Agrobacterium tumefaciens contains a gene homologous to the chromosomal virulence gene acvB. J Bacteriol. 1995;15(4):892–897. doi: 10.1128/jb.177.4.892-897.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young JM. Sinorhizobium versus Ensifer: may a taxonomy subcommittee of the ICSP contradict the Judicial Commission? Int J Syst Evol Microbiol. 2010;15(Pt 7):1711–1713. doi: 10.1099/ijs.0.025163-0. [DOI] [PubMed] [Google Scholar]
- Chen WX, Yan GH, Li JL. Numerical taxonomic study of fast-growing soybean rhizobia and a proposal that Rhizobium fredii be assigned to Sinorhizobium gen-nov. Int J Syst Bacteriol. 1988;15(4):392–397. doi: 10.1099/00207713-38-4-392. [DOI] [Google Scholar]
- Young JM. The genus name Ensifer Casida 1982 takes priority over Sinorhizobium Chen et al. 1988, and Sinorhizobium morelense Wang et al. 2002 is a later synonym of Ensifer adhaerens Casida 1982. Is the combination ‘Sinorhizobium adhaerens’ (Casida 1982) Willems et al. 2003 legitimate? Request for an opinion. Int J Syst Evol Microbiol. 2003;15(6):2107–2110. doi: 10.1099/ijs.0.02665-0. [DOI] [PubMed] [Google Scholar]
- Hood EE, Gelvin SB, Melchers LS, Hoekema A. New agrobacterium helper plasmids for gene-transfer to plants. Transgenic Res. 1993;15(4):208–218. doi: 10.1007/BF01977351. [DOI] [Google Scholar]
- Wilson K. Preparation of genomic DNA from bacteria. Curr Protoc Mol Biol. 1987;15:2.4. doi: 10.1002/0471142727.mb0204s56. 1-2.4. 5. [DOI] [PubMed] [Google Scholar]
- Chaisson MJ, Tesler G. Mapping single molecule sequencing reads using basic local alignment with successive refinement (BLASR): application and theory. BMC Bioinformatics. 2012;15(1):238. doi: 10.1186/1471-2105-13-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boetzer M, Henkel CV, Jansen HJ, Butler D, Pirovano W. Scaffolding pre-assembled contigs using SSPACE. Bioinformatics. 2011;15(4):578–579. doi: 10.1093/bioinformatics/btq683. [DOI] [PubMed] [Google Scholar]
- Boetzer M, Pirovano W. Toward almost closed genomes with GapFiller. Genome Biol. 2012;15(6):R56. doi: 10.1186/gb-2012-13-6-r56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatusov RL, Koonin EV, Lipman DJ. A genomic perspective on protein families. Science. 1997;15(5338):631–637. doi: 10.1126/science.278.5338.631. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Circular representation of the four replicons of E. adhaerens OV14. Circles, from the inside out, show: (1) GC skew; (2) Coding regions; light blue blocks represent genes with predicted function, red blocks show transposable elements, dark blue and grey blocks show genes of hypothetical and unknown function, respectively. Figure S2. Synteny plots showing total sequence of Ensifer adhaerens OV14 pOV14c (top bar) vs Agrobacterium tumefaciens C58 pTi (bottom bar), computed using DoubleACT version2 on tBLASTx setting with cut off set at 100. Visualised in Artemis ACT. Homology between the genomes is displayed via interconnecting lines; red lines representing direct homology while blue lines represent homologues but inverted sequence.
BLAST of Ensifer adhaerens OV14 replicons.xlsx. Excel file includes tables of BLAST search of individual Ensifer adhaerens OV14 replicons.
BLAST of Agrobacterium tumefaciens C58 replicons.xlsx. Excel file includes tables of BLAST searches of individual Agrobacterium tumefaciens C58.