Abstract
In Arabidopsis, trichome formation is regulated by the interplay of R3 MYBs and several others transcription factors including the WD40-repeat protein TRANSPARENT TESTA GLABRA1 (TTG1), the R2R3 MYB transcription factor GLABRA1 (GL1), the bHLH transcription factor GLABRA3 (GL3) or ENHANCER OF GLABRA3 (EGL3), and the homeodomain protein GLABRA2 (GL2). R3 MYBs including TRICHOMELESS1 (TCL1), TCL2, TRYPTICHON (TRY), CAPRICE (CPC), ENHANCER OF TRY AND CPC1 (ETC1), ETC2 and ETC3 negatively regulate trichome formation by competing with GL1 for binding GL3 or EGL3, thus blocking the formation of TTG1–GL3/EGL3–GL1, an activator complex required for the activation of the trichome positive regulator gene GL2. However, it is largely unknown if R3 MYBs in other plant species especially woody plants have similar functions. By BLASTing the Populus trichocarpa protein database using the entire amino acid sequence of TCL1, an Arabidopsis R3 MYB transcription factor, we identified a total of eight R3 MYB transcription factor genes in poplar, namely P. trichocarpa TRICHOMELESS1 through 8 (PtrTCL1–PtrTCL8). The amino acid signature required for interacting with bHLH transcription factors and the amino acids required for cell-to-cell movement of R3 MYBs are not fully conserved in all PtrTCLs. When tested in Arabidopsis protoplasts, however, all PtrTCLs interacted with GL3. Expressing each of the eight PtrTCL genes in Arabidopsis resulted in either glabrous phenotypes or plants with reduced trichome numbers, and expression levels of GL2 in all transgenic plants tested were greatly reduced. Expression of PtrTCL1 under the control of TCL1 native promoter almost completely complemented the mutant phenotype of tcl. In contrast, expression of PtrTCL1 under the control of TRY native promoter in the try mutant, or under the control of CPC native promoter in the cpc mutant resulted in glabrous phenotypes, suggesting that PtrTCL1 functions similarly to TCL1, but not TRY and CPC.
Keywords: trichome formation, R3 MYBs, transcription factors, Arabidopsis, Populus trichocarpa
INTRODUCTION
Single-repeat R3 MYB transcription factors (R3 MYBs) are small proteins that typically contain ~100 amino acids, largely consist of a single MYB DNA-binding repeat, and are best characterized for their regulatory roles in trichome and root hair development (Wang and Chen, 2014). R3 MYBs are widely distributed in the plant kingdom, and they are encoded by a small subset of MYB transcription factor genes (Dubos et al., 2010). In Arabidopsis, there are a total of seven genes encoding R3 MYBs, including TRYPTICHON (TRY; Schnittger et al., 1999), CAPRICE (CPC; Wada et al., 1997), TRICHOMELESS1 (TCL1; Wang et al., 2007), TCL2/CAPRICE-LIKE MYB4 (CPL4; Gan et al., 2011; Tominaga-Wada and Nukumizu, 2012), ENHANCER OF TRY AND CPC1 (ETC1; Esch et al., 2004; Kirik et al., 2004a), ETC2 (Kirik et al., 2004b), and ETC3/CPL3 (Simon et al., 2007; Tominaga et al., 2008). All seven R3 MYBs contain the residues [D/E]L×2[R/K]×3L×6L×3R, a conserved amino acid signature required for interaction of MYBs with R/B-like bHLH transcription factors (Zimmermann et al., 2004), and W×M, a sequence motif that has been shown to be required for cell-to-cell movement of CPC (Kurata et al., 2005).
Trichome formation in Arabidopsis is controlled by the interplay of R3 MYBs and several other transcription factors including the WD40-repeat protein TRANSPARENT TESTA GLABRA1 (TTG1; Walker et al., 1999), the R2R3 MYB-type transcription factor GLABRA1 (GL1; Oppenheimer et al., 1991), the bHLH transcription factor GLABRA3 (GL3) or ENHANCER OF GLABRA3 (EGL3; Payne et al., 2000; Zhang et al., 2003), and the homeodomain protein GLABRA2 (GL2; Rerie et al., 1994). TTG1, GL1, and GL3 or EGL3 form an activator complex to induce the expression of GL2, a positive regulatory gene of trichome formation (Rerie et al., 1994). The same TTG1–GL3/EGL3–GL1 activator complex can also induce the expression of R3 MYB genes. R3 MYBs can move from a trichome precursor cell to its neighboring cells, and compete with GL1 for binding GL3 or EGL3, thus limiting the formation of the activator complex (Hülskamp et al., 1994; Schellmann et al., 2002; Esch et al., 2003; Schiefelbein, 2003; Pesch and Hülskamp, 2004; Ishida et al., 2008), resulting in inhibition of trichome initiation.
In addition to competing with GL1 for binding GL3, some of the R3 MYBs including TCL1 and TCL2 also directly suppress the expression of GL1 (Wang et al., 2007; Gan et al., 2011). Not all the R3 MYB genes in Arabidopsis are activated by the TTG1–GL3/EGL3–GL1 activator complex (Wang et al., 2008), and microRNA156 (MIR156)-targeted SQUAMOSA PROMOTER BINDING PROTEIN LIKE (SPL) 9 has been shown to activate TCL1, TCL2, and TRY (Yu et al., 2010; Gan et al., 2011). These results indicate that R3 MYBs may use different mechanisms to regulate trichome formation in Arabidopsis.
Although single mutants of Arabidopsis R3 MYB genes have different phenotypes (Wada et al., 1997, 2002; Schnittger et al., 1999; Schellmann et al., 2002; Wang et al., 2007), over-expression of any of the R3 MYB genes in Arabidopsis resulted in glabrous phenotypes. Analysis of double, triple, and higher mutants also revealed that all the seven R3 MYBs function in a highly redundant manner to control trichome formation (Esch et al., 2004; Kirik et al., 2004a, b; Wang et al., 2007, 2008; Tominaga et al., 2008; Wester et al., 2009; Gan et al., 2011).
Functional homologs of some of these transcription factors that regulate trichome formation have been identified in other plants. For example, GaHOX1 from cotton has been identified as a functional homolog of GL2 (Guan et al., 2008) and GaMYB2 as a functional homolog of GL1 (Wang et al., 2004; Guan et al., 2011, 2014). MYB like genes from Mimulus guttatus and peach regulate trichome formation (Scoville et al., 2011; Vendramin et al., 2014), and expression of a tomato R3 MYB gene in Arabidopsis resulted in glabrous phenotypes (Tominaga-Wada et al., 2013). On the other hand, expression of Arabidopsis GL3 in Brassica napus induced ectopic trichome formation (Gruber et al., 2006). These results suggest that trichome formation in other plant species may be controlled by similar mechanisms as in Arabidopsis. However, trichome regulators in plants other than Arabidopsis remain largely unidentified.
Populus trichocarpa is the first tree whose genome has been fully sequenced (Tuskan et al., 2006), and it is also a good model plant for studies in areas such as wood development, ecological interactions, and other aspects of perennial plants that cannot be studied in the annual model plant Arabidopsis (Groover, 2005; Whitham et al., 2006; Jansson and Douglas, 2007). By using the entire amino acid sequence of TCL1 to BLAST search the P. trichocarpa protein database, we found there are a total of eight genes in poplar encoding R3 MYB transcription factors, namely P. trichocarpa TRICHOMELESS1 through 8 (PtrTCL1–PtrTCL8). In the study described here, we examined if R3 MYBs from poplar can functionally substitute for Arabidopsis R3 MYBs to regulate trichome formation in Arabidopsis plants.
MATERIALS AND METHODS
IDENTIFICATION OF POPLAR HOMOLOGS OF Arabidopsis R3 MYB TRANSCRIPTION FACTOR TCL1
To identify poplar homologs of Arabidopsis R3 MYB transcription factors, the entire amino acid sequence of Arabidopsis R3 MYB transcription factor TCL1 was used in BLAST searches of the P. trichocarpa proteome (www.phytozome.net). The entire amino acid sequences of identified poplar R3 MYB transcription factors were then used in BLAST searches until no more poplar R3 MYBs were identifiable. Full-length amino acid sequences of Arabidopsis and poplar R3 MYBs were subjected to phylogenetic analysis using “One Click” mode of Phylogeny (www.phylogeny.fr) with default settings.
PLANT MATERIALS AND GROWTH CONDITIONS
Poplar xylem tissue from P. trichocarpa was collected as described previously (Geraldes et al., 2011; Liu et al., 2013; Wang et al., 2014), and used for RNA isolation and poplar R3 MYB gene cloning. The tcl1 and try mutants, and the 35S:HA-TCL1 transgenic plant were in the Columbia-0 (Col-0) background (Esch et al., 2003; Wang et al., 2007). The cpc mutant was in the Ws background (Wada et al., 1997).
Unless specified otherwise, Arabidopsis ecotype Col-0 was used for plant transformation. Seedlings used for RNA isolation were obtained by growing sterilized seeds on 1/2 Murashige and Skoog (MS) basal medium with vitamins (Plantmedia) and 1% (w/v) sucrose. Seedlings used for phenotypic analysis were obtained either by growing seeds on 1/2 MS medium or by directly sowing seeds into soil. All plants were grown in growth rooms at 22°C with 14/10 h photoperiod, and light density of approximately 120 μmol m-2 s-1.
RNA ISOLATION AND RT-PCR
Total RNA from poplar samples was isolated using PureLink Plant RNA Reagent (Invitrogen), and cleaned with RNeasy Plant Mini Kit (Qiagen) as described previously (Geraldes et al., 2011; Wang et al., 2014). Total RNA from Arabidopsis seedlings was isolated using EasyPureTM Plant RNA Kit (Transgene) according to the manufacturer’s instructions. All RNA samples were treated with RNase-Free DNase set (Qiagen) to eliminate possible DNA contamination.
cDNA was synthesized using 2 μg total RNA by Oligo(dT)-primed reverse transcription using Omniscript RT Kit (Qiagen). Some of the primers used for cloning or examining the expression of corresponding genes have been described previously (Wang et al., 2007, 2008, 2010; Gan et al., 2011), and poplar R3 MYB gene-specific primers are shown in Table 1.
Table 1.
Primers using for poplar R3 MYB gene cloning and expression assays.
| Primers | Sequences |
|---|---|
| PtrTCL1-Nde1F | 5′-CAACATATGGATAGACGTCGCAGG-3′ |
| PtrTCL1-Sac1R | 5′-CAAGAGCTCTTAAGAGGTATTAGGAATTAC-3′ |
| PtrTCL2-Nde1F | 5′-CAACATATGGATAGACGTCGCAAG-3′ |
| PtrTCL2-Sac1R | 5′-CAAGAGCTCTTAGGAATGACATCTC-3′ |
| PtrTCL3-Nde1F | 5′-CAACATATGGAGAGTATGAACCGC-3′ |
| PtrTCL3-Sac1R | 5′-CAAGAGCTCTCAACTAGAAGTCCTAG-3′ |
| PtrTCL4-Nde1F | 5′-CAACATATGTCCCTTCAATTTCAC-3′ |
| PtrTCL4-Sac1R | 5′-CAAGAGCTCTTACTGACTTGTAGAGTATC-3′ |
| PtrTCL5-Nde1F | 5′-CAACATATGGCTGACTTGGATCAC-3′ |
| PtrTCL5-Sac1R | 5′-CAAGAGCTCTTACTGACTTGTAGAGTATC-3′ |
| PtrTCL6-Nde1F | 5′-CAACATATGGCTGACTCTGAACATTC-3′ |
| PtrTCL6-Sac1R | 5′-CAAGAGCTCTCATTCACTTGTAGAGTATC-3′ |
| PtrTCL7-Nde1F | 5′-CAACATATGGCTGACACTGAACATTC-3′ |
| PtrTCL7-Sac1R | 5′-CAAGAGCTCTCATTCACTCGTAGAGC-3′ |
| PtrTCL8-Nde1F | 5′-CAACATATGGCTTGCTCGGGTCAC-3′ |
| PtrTCL8-Sac1R | 5′-CAAGAGCTCTCATCTTTCCTTTGATGATC-3′ |
CONSTRUCTS
To generate HA (human influenza hemagglutinin)- or GD (Gal4 DNA binding domain)-tagged constructs for poplar R3 MYB genes, the full-length, open-reading frames (ORF) of corresponding poplar R3 MYB genes were amplified by RT-PCR using RNA isolated from poplar xylem samples, and the PCR products were then cloned in-frame with an N-terminal HA or GD tag into the pUC19 vector under the control of the double 35S enhancer promoter of CaMV (Wang et al., 2005).
The 35S:PtrTCL1-GFP construct was cloned by fusing PtrTCL1 in frame with GFP (Green fluorescent protein) and then cloned into the pUC19 vector under the control of the 35S promoter. The TCL1p:HA-PtrTCL1, TRYp:HA-PtrTCL1, and CPCp:HA-PtrTCL1 constructs were cloned by replacing the 35S promoter in 35S:HA-PtrTCL1 with TCL1, TRY, and CPC promoters, respectively (Wada et al., 1997; Esch et al., 2003; Wang et al., 2007).
For plant transformation, corresponding constructs in the pUC19 vector were digested with EcoRI and subcloned into the binary vector pPZP211 or pPZP221 (Hajdukiewicz et al., 1994).
PLANT TRANSFORMATION AND TRANSGENIC PLANT SELECTION
About five-week-old plants with several mature flowers on the main inflorescence were used for plant transformation. Plants were transformed by using the floral dip method via Agrobacterium tumefaciens GV3101 (Clough and Bent, 1998). T1 seeds were geminated on plates containing antibiotics to select transgenic plants. For each construct, more than 70 transgenic lines were obtained. Phenotypes of transgenic plants were examined in the T1 generation and at least five transgenic lines with similar phenotypes were collected. The phenotypes observed were confirmed in the following two to three generations. Expression of corresponding genes in related lines was confirmed by RT-PCR. Homozygous T3 or T4 seeds were used for further experiments, and data from one representative line for each construct are presented.
PLASMID DNA ISOLATION, PROTOPLAST TRANSFECTION, AND GUS ACTIVITY ASSAY
All reporter and effector plasmids were prepared using the GoldHi EndoFree Plasmid Maxi Kit (Kangwei) according to the manufacturer’s instructions. The procedures for protoplast isolation, transfection, and GUS activity assay have been described previously (Tiwari et al., 2003; Wang et al., 2005, 2008; Wang and Chen, 2008). Briefly, protoplasts were isolated from rosette leaves collected from ~4-week-old Arabidopsis plants. Effector and reporter plasmids were co-transfected into protoplasts and incubated at room temperature for 20–22 h under darkness. GUS activities were measured using a SynergyTM HT microplate reader (BioTEK).
MICROSCOPY
Trichomes were analyzed and photographed using a Motic K microscope equipped with a Canon digital camera. Localization of PtrTCL1-GFP proteins in transgenic plants expressing PtrTCL1–GFP under the control of the 35S promoter was examined under an Olympus FV1000 confocal microscope. Protoplast cells isolated from the PtrTCL1–GFP transgenic plants were stained with DAPI and then examined under an Olympus FV1000 microscope.
RESULTS
IDENTIFICATION OF R3 MYB TRANSCRIPTION FACTORS IN POPLAR
A total of eight poplar R3 MYB transcription factors were identified, and collectively named as PtrTCL1 to PtrTCL8. Corresponding gene names for the PtrTCLs identified are as follows: PtrTCL1, Potri.002G168900; PtrTCL2, Potri.014G096300; PtrTCL3, Potri.015G022000; PtrTCL4, Potri.007G122800; PtrTCL5, Potri.017G03700; PtrTCL6, Potri.011G026300; PtrTCL7, Potri.004G021300; and PtrTCL8, Potri.004G015100.
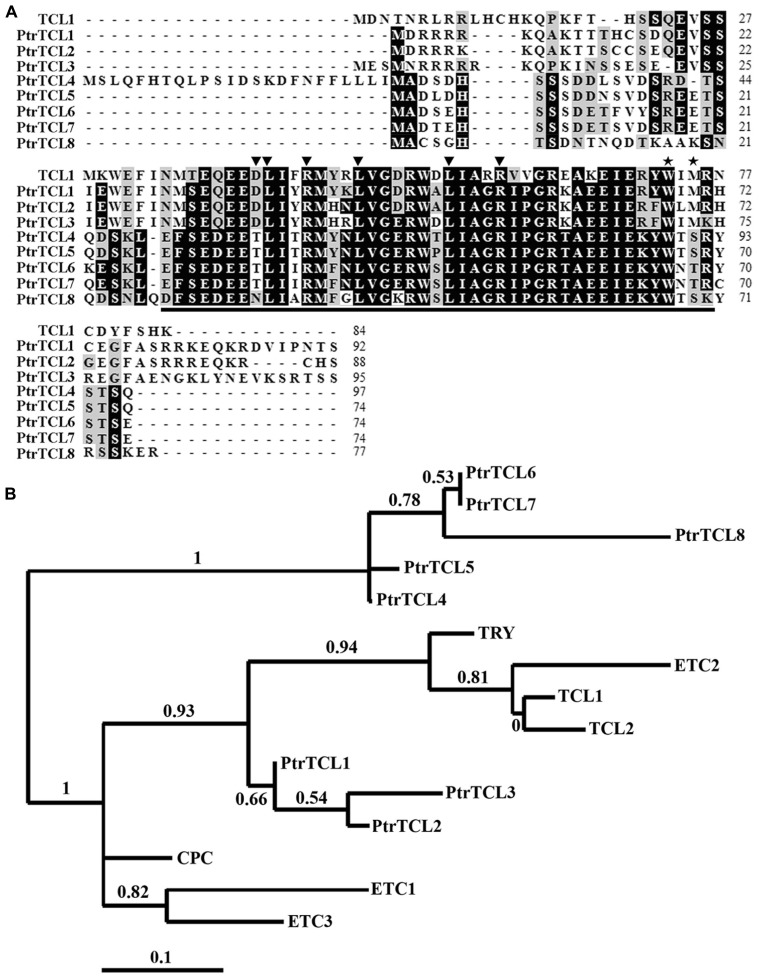
Similar to their homologs in Arabidopsis, nearly the entire protein of poplar R3 MYBs is made up of the single MYB domain (Figure 1A). The amino acid signature [D/E]L×2[R/K]×3L×6L×3R, that is required for the interaction with R/B-like bHLH transcription factors (Zimmermann et al., 2004), is fully conserved in all seven R3 MYB transcription factors in Arabidopsis (Wang et al., 2007, 2008; Gan et al., 2011), but is found in only three poplar R3 MYBs including PtrTCL1, PtrTCL2, and PtrTCL3 (Figure 1A). In the other four poplar R3 MYBs, D/E, the first amino acid in the amino acid signature is replaced by T/N (Figure 1A). Similarly, the amino acid motif W×M, that has been shown to be required for the cell-to-cell movement of CPC (Kurata et al., 2005), is conserved in all seven Arabidopsis R3 MYBs (Wang et al., 2007, 2008; Gan et al., 2011), but is found in only PtrTCL1, PtrTCL2, and PtrTCL3 (Figure 1A). In the other four poplar R3 MYBs, the M in the motif is replaced by S/T (Figure 1A).
FIGURE 1.
R3 MYB transcription factors in poplar. (A) Sequence alignment of TCL1 with eight poplar R3 MYB proteins. Identical amino acids are shaded in black and similar amino acids in gray. The R3 MYB domain is indicated by underline, the amino acid signature [D/E]L×2[R/K]×3L×6L×3R that is required for interacting with R/B-like BHLH transcription factors is indicated by arrowheads, and the amino acids within the MYB domain that are crucial for cell-to-cell movement of CPC are indicated by asterisks. (B) Phylogenetic analysis of seven Arabidopsis and the eight poplar R3 MYB transcription factors. The entire amino acid sequences were used to generate the phylogenetic tree by using “One Click” mode of Phylogeny (www.phylogeny.fr) with default settings. Branch support values are indicated above or below branches, bar indicates branch length.
Phylogenetic analysis using full-length protein sequences of poplar R3 MYBs and TCL1 showed that the clade of PtrTCL1–PtrTCL3 is most closely related to the clade of TCL1, TCL2, ETC2 and TRY (Figure 1B). Together, PtrTCL1–PtrTCL3 and seven Arabidopsis R3 MYBs formed one subgroup, and PtrTCL4–PtrTCL8 formed another subgroup.
PtrTCLs INTERACT WITH GL3
We have previously demonstrated that TRY, CPC, ETC1, ETC2, TCL1, TCL2, and ETC3 interact with GL3 in plant cells (Wang et al., 2008; Gan et al., 2011), supporting the proposal that R3 MYBs control trichome formation in Arabidopsis by competing with GL1 for binding GL3, and thus eliminating the formation of TTG1–GL3–GL1 activator complex. Considering that only three of the identified poplar R3 MYBs have the fully conserved amino acid signature that is required for the interaction with R/B-like bHLH transcription factors, we tested if PtrTCLs interact with GL3 in plant cells.
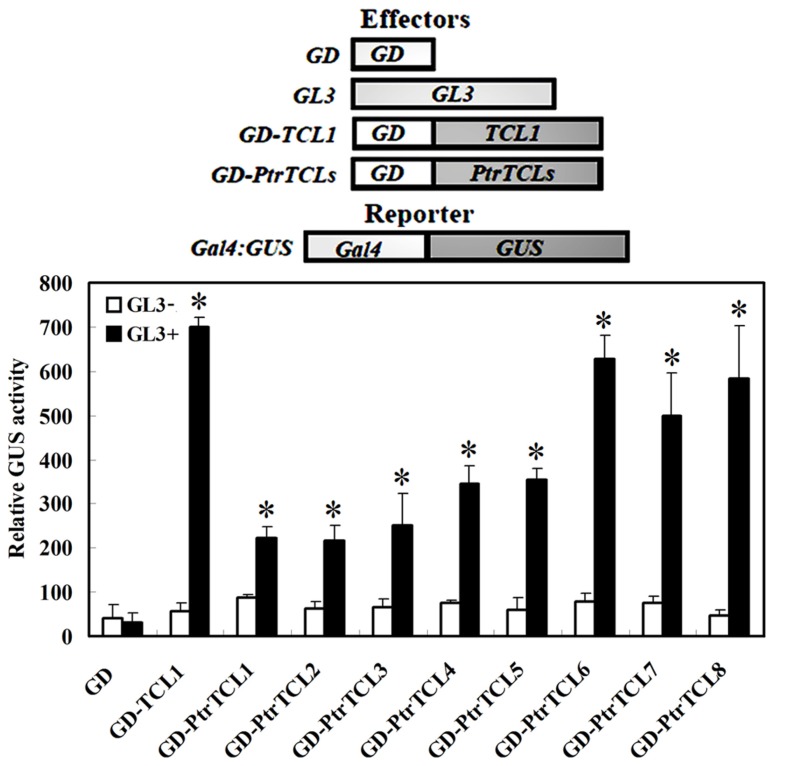
Arabidopsis protoplast transfection assays were used to test the interaction between PtrTCLs and GL3. Plasmids of the reporter gene Gal4-GUS, together with the effector genes GL3 and GD fused PtrTCLs (GD–PtrTCLs; Figure 2), were co-transfected into Arabidopsis protoplasts. GD and GD–TCL1 were used as negative and positive controls, respectively. As expected, neither GD nor GD–TCL1 activated the reporter gene in the absence of GL3. In the presence of GL3, GD–TCL1 but not GD activated the reporter gene. Similarly, none of the eight poplar R3 MYBs activated the reporter gene in the absence of GL3; however, all of them activated the reporter gene in the presence of GL3 (Figure 2).
FIGURE 2.
Poplar R3 MYBs interact with GL3 in plant cells. Reporter gene and effector gene plasmids were co-transfected into protoplasts isolated from Arabidopsis rosette leaves. Protoplasts were incubated in darkness for 20–22 h after transfection, and then GUS activity was measured. Data represent the mean±SD of three replicates. Reporter and effector constructs are diagrammed at the top of the figure. *significantly different from the absence of GL3 (GL3-; P < 0.001).
PtrTCLs NEGATIVELY REGULATE TRICHOME FORMATION WHEN EXPRESSED IN Arabidopsis
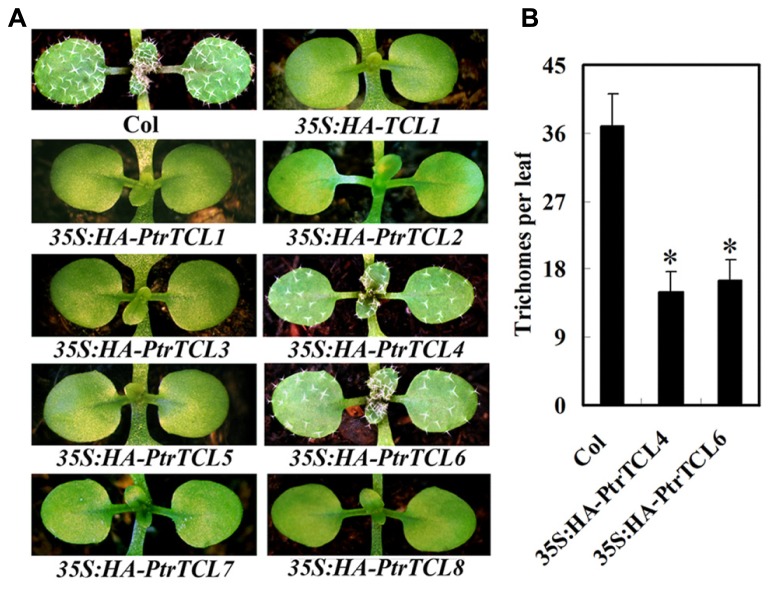
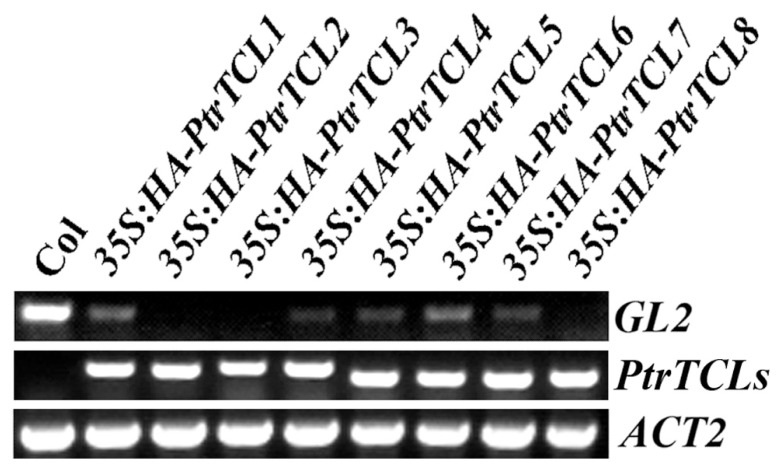
Having shown that all the eight poplar R3 MYBs interact with GL3 in plant cells, we further analyzed if PtrTCLs regulate trichome formation by generating transgenic Arabidopsis plants expressing HA-tagged PtrTCLs under the control of 35S promoter (35S:HA-PtrTCLs). As shown in Figure 3A, expression of PtrTCL1, PtrTCL2, PtrTCL3, PtrTCL5, PtrTCL7 and PtrTCL8 in Arabidopsis resulted in glabrous phenotypes, similar to the phenotype observed in plants over-expressing TCL1 (Wang et al., 2007). In contrast, transgenic plants expressing PtrTCL4 and PtrTCL6 had greatly reduced number of trichomes on rosette leaves, but none of the transgenic plants obtained showed glabrous phenotypes (Figure 3). RT-PCR analysis showed that poplar R3 MYB genes were highly expressed in their corresponding transgenic lines (Figure 4), indicating that the phenotypes observed in PtrTCL4 and PtrTCL6 transgenic plants were not due to relative lower expression levels of the corresponding genes.
FIGURE 3.
Poplar R3 MYBs negatively regulate trichome formation in Arabidopsis. (A) Trichome phenotypes in Arabidopsis plants expressing poplar R3 MYB genes. All transgenic lines are in Col background. Pictures were taken from 2-week-old, soil-grown seedlings. (B) Trichome density on the first two rosettle leaves of Arabidopsis plants expressing PtrTCL4 and PtrTCL6. Data represent the mean±SD of 18 plants. *significantly different from Col wild-type plants (P < 0.001).
FIGURE 4.
Expression of GL2 and poplar R3 MYB genes in transgenic Arabidopsis plants. RNA was isolated from Col wild-type and transgenic Arabidopsis seedlings, RT-PCR was used to examine the expression of GL2 and polar R3 MYB genes. The expression of ACTIN2 (ACT2) was used as a control.
GL2 is one of the target genes of the activator complex TTG1–GL3/EGL3–GL1, and it positively regulates trichome formation in Arabidopsis. Because binding of PtrTCLs to GL3 indicated that expression of PtrTCLs in Arabidopsis resulted in the inhibition of the formation of TTG1–GL3/EGL3–GL1 activator complex, we examined the expression of GL2 in Arabidopsis transgenic plants expressing poplar R3 MYB genes. As shown in Figure 4, expression of GL2 was dramatically reduced in transgenic plants expressing any of the poplar R3 MYB genes.
SUBCELLULAR LOCALIZATION OF PtrTCL1
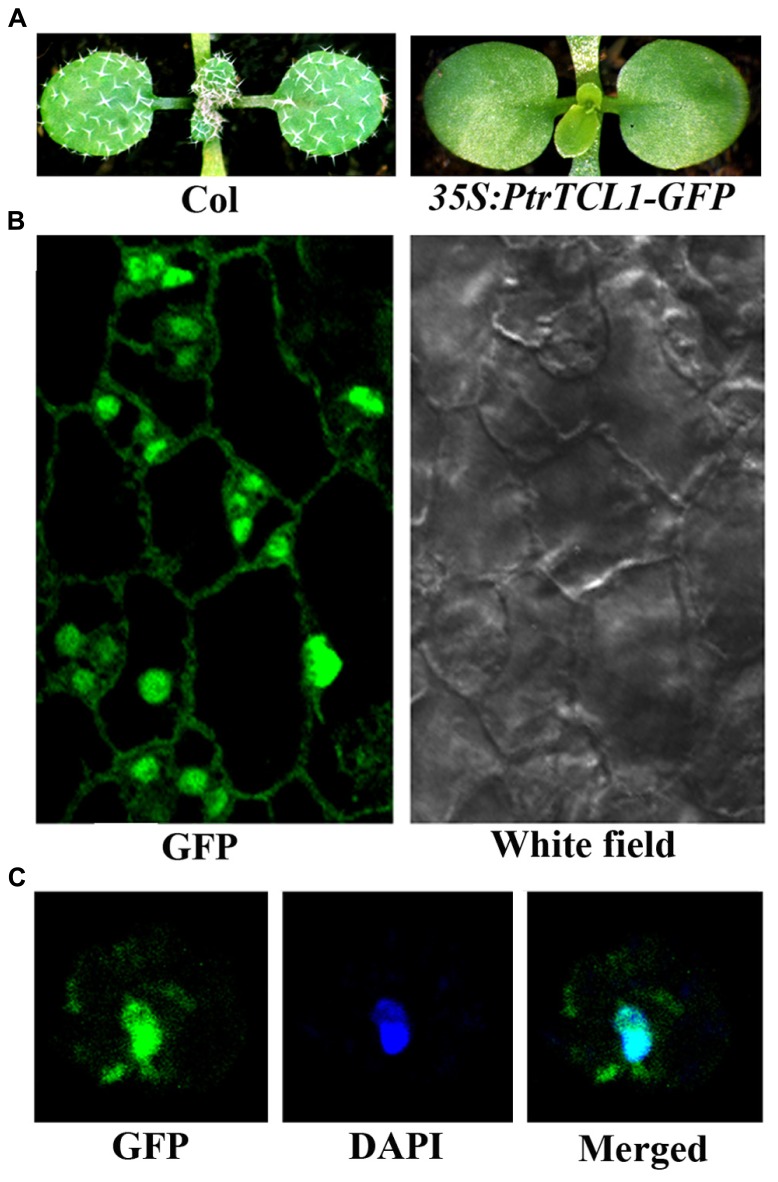
Among the eight poplar R3 MYB transcription factors, PtrTCL1 has highest amino acid similarity with TCL1, and phylogenetic analysis also showed that the clade of PtrTCL1–PtrTCL3 is most closely related to TCL1 (Figure 1). We wanted to further explore the functions of poplar R3 MYB transcription factors in the regulation of trichome formation in Arabidopsis by taking PtrTCL1 as an example. We first examined the subcellular localization of the PtrTCL1 protein by generating transgenic plants expressing PtrTCL1–GFP under the control of the 35S promoter. As shown in Figure 5A, expression of PtrTCL1–GFP in Arabidopsis resulted in a glabrous phenotype, a phenotype similar to transgenic plants expressing PtrTCL1 (Figure 3). This indicated that the PtrTCL1–GFP fusion protein is likely functional. By examining the transgenic plants obtained, we found that GFP florescence was mainly observed in the nucleus of epidermal cells (Figure 5B); examination of DAPI stained protoplast cells isolated from the PtrTCL1–GFP transgenic plants confirmed that the GFP was mainly observed in the nucleus (Figure 5C), indicating that PtrTCL1 is a nuclear localized protein.
FIGURE 5.
Subcellular localization of PtrTCL1. (A) Expression of PtrTCL1-GFP under the control of 35S promoter in Arabidopsis resulted in a glabrous phenotype. Pictures were taken from 2-week-old, soil-grown seedlings. (B) GFP fluorescence in the epidermal cells of the rosettle leaves in a 2-week-old transgenic plant expressing PtrTCL1-GFP under the control of the 35S promoter. Left panel: GFP channel, right panel: white field image. (C) GFP fluorescence in a protoplast cell isolated from transgenic plant expressing PtrTCL1-GFP under the control of the 35S promoter. Left panel: GFP channel; Middle panel: DAPI channel; right panel: merged.
PtrTCL1 ALMOST COMPLETELY RESCUED tcl1 MUTANT TRICHOME PHENOTYPES
Previously, we identified TCL1 as a major R3 MYB transcription factor that regulates trichome formation on the inflorescences and pedicels, and we showed that knock-out of TCL1 resulted in ectopic trichome formation on the inflorescence stems and pedicels. In addition, we showed that over-expression of TCL1 in Arabidopsis resulted in a glabrous phenotype (Wang et al., 2007). Here, we showed that PtrTCL1, that is in the most closely related clade to TCL1 (Figure 1), resulted in a glabrous phenotype when over-expressed in Arabidopsis (Figure 3). Therefore, we wanted to further examine if PtrTCL1 is the functional equivalent of TCL1 by testing if PtrTCL1 could rescue the tcl1 mutant phenotype when expressed under the control of TCL1 native promoter.
Transgenic plants were generated in a tcl1 background to express PtrTCL1 under the control of the TCL1 native promoter (TCL1p:HA-PtrTCL1/tcl1). Previously, we showed that expression of TCL1–GFP under the control of the TCL1 promoter fully rescued the tcl1 trichome phenotype (Wang et al., 2007), indicating that the TCL1 promoter used is functional in Arabidopsis. As shown in Figure 6, expression of PtrTCL1 under the control of TCL1 native promoter almost completely rescued tcl1 mutant phenotype, i.e., tcl1 mutant forms ectopic trichomes in both stem internodes above the first flower and pedicels, while TCL1p:HA-PtrTCL1/tcl1 plants do not have trichomes on stem internodes after the first flowers, but have a few trichomes on first pedicel only. These results indicate that PtrTCL1 is likely the functional equivalent of TCL1 in controlling trichome formation on the inflorescence stems and pedicels.
FIGURE 6.
Rescue of tcl1 phenotype by PtrTCL1. Trichome formation on inflorescences in Col, tcl1 and TCL1p:HA-PtrTCL1/tcl1-1 plants. Pictures were taken from 5-week-old, soil-grown plants.
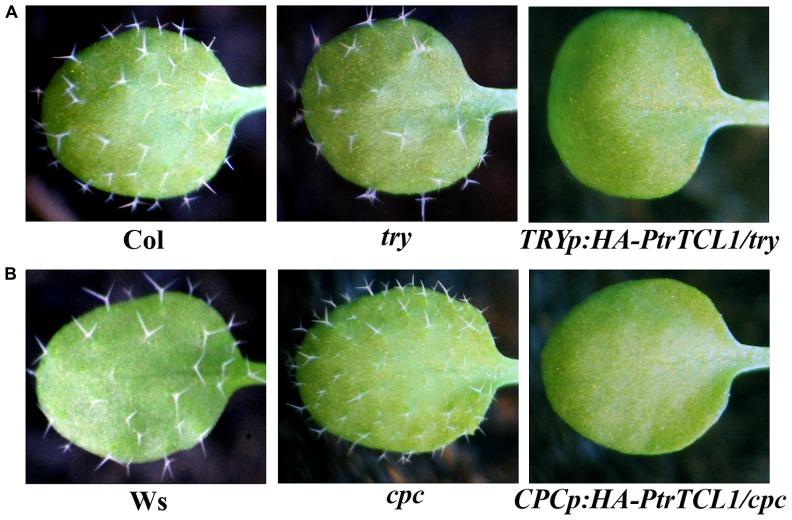
In addition to tcl1, single mutants try and cpc showed trichome phenotypes; try mutants have trichome clusters, and cpc mutants have increased numbers of trichome on leaves (Wada et al., 1997; Schnittger et al., 1999), so we also examined if PtrTCL1 is functionally equivalent to TRY or CPC. Transgenic plants were generated to express PtrTCL1 under the control of TRY native promoter (TRYp:HA-PtrTCL1/try) in the try background, and under the control of CPC native promoter (CPCp:HA-PtrTCL1/cpc) in the cpc background. As shown in Figure 7, expression of PtrTCL1 under the control of TRY native promoter in a try mutant background, or under the control of CPC native promoter in a cpc mutant background resulted in glabrous phenotypes.
FIGURE 7.
Expression of PtrTCL1 in try and cpc resulted in glabrous phenotypes. (A) Expression of PtrTCL1 under the control of TRY native promoter in the try mutant background resulted in glabrous phenotype. (B) Expression of PtrTCL1 under the control of CPC native promoter in the cpc mutant background resulted in glabrous phenotype. Pictures were taken from 2-week-old, soil-grown seedlings.
DISCUSSION
In this study we identified poplar homologs of Arabidopsis R3 MYB transcription factors, and analyzed their function in trichome formation in Arabidopsis. We showed that expression of any of the eight poplar R3 MYB genes under the control of the 35S promoter in Arabidopsis resulted in either a glabrous phenotype or a great reduction in trichome numbers (Figure 3). These results suggest that poplar R3 MYBs act as negative regulators for trichome formation and may have overlapping functions, similar to their Arabidopsis R3 MYB homologs.
Among the eight poplar R3 MYB transcription factors, PtrTCL1 is in the most closely related clade to TCL1 (Figure 1B), is localized in the nucleus (Figure 5), and expression of PtrTCL1 under the control of the TCL1 promoter in the tcl1 mutant background (TCL1p:HA-PtrTCL1/tcl1) almost fully restored the trichome phenotype of the tcl1 mutant (Figure 6). Expression of PtrTCL1 under the control of the TRY native promoter in the try background (TRYp:HA-PtrTCL1/try), or under the control of the CPC native promoter in the cpc background (CPCp:HA-PtrTCL1/cpc) resulted in glabrous phenotypes (Figure 7), suggesting that PtrTCL1 may be the functional equivalent of TCL1, rather than TRY and CPC. It should be noted that ETC2 and TCL2 are also in the clade that is sister to the clade of PtrTCL1–PtrTCL3; however, because no mutant available for TCL2 and etc2 mutant does not have any trichome phenotype, we could not test whether PtrTCL1 might be functionally equivalent to TCL2 or ETC2.
In Arabidopsis, R3 MYBs inhibit trichome formation by competing with GL1 for binding of GL3 or EGL3, thus inhibiting the formation of the TTG1–GL3/EGL3–GL1 activator complex (Schellmann et al., 2002; Esch et al., 2003; Schiefelbein, 2003; Pesch and Hülskamp, 2004; Ishida et al., 2008). In accordance with these results, all seven Arabidopsis R3 MYBs have the conserved [D/E]L×2[R/K]×3L×6L×3R amino acid signature that is required for interaction with R/B-like bHLH transcription factors (Zimmermann et al., 2004), and protoplast transfection assays showed that they all interact with GL3 in plant cells (Wang et al., 2008; Gan et al., 2011). Sequence alignment results showed that only three of the poplar R3 MYBs, namely PtrTCL1, PtrTCL2, and PtrTCL3, have the fully conserved amino acid signature required for interacting with bHLH transcription factors (Figure 1A). When tested in protoplasts, however, all the eight poplar R3 MYBs interacted with GL3 (Figure 2), and the interaction with GL3 may be stronger for those R3 MYBs without the fully conserved amino acid signature as judged by the GUS activities (Figure 2). Our results indicate that either the conserved amino acid signature is not required for the interaction of poplar R3 MYBs with GL3, or a single-amino-acid substitution (D/E > T/N) in poplar R3 MYBs does not affect their interaction with GL3. It is also possible that previous designations of conserved residues may be biased due to a smaller set of proteins analyzed. Furthermore, interaction of poplar R3 MYBs with GL3 in plant cells suggests that poplar R3 MYBs can block the formation of the activator complex required for trichome formation. In accordance with this, RT-PCR results showed that expression of GL2 is dramatically reduced in the transgenic plants expressing poplar R3 MYB genes (Figure 4). Since our previous results showed that R3 MYBs in Arabidopsis may regulate trichome formation in a GL2 independent manner (Wang et al., 2010), we cannot rule out the possibility that poplar R3 MYBs may also regulate trichome formation through other mechanisms.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors thank Drs. Tom Guilfoyle and Gretchen Hagen (University of Missouri-Columbia) for providing vectors for protoplast transfection assays, and Dr. Tom Guilfoyle for critical reading of the manuscript. This work was supported by the National Natural Science Foundation of China (grant no. 31170262), the Programme for Introducing Talents to Universities (B07017), and the startup funds from Northeast Normal University (www.nenu.edu.cn) to Shucai Wang.
REFERENCES
- Clough S. J., Bent A. F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16 735–743 10.1046/j.1365-313x.1998.00343.x [DOI] [PubMed] [Google Scholar]
- Dubos C., Stracke R., Grotewold E., Weisshaar B., Martin C., Lepiniec L. (2010). MYB transcription factors in Arabidopsis Trends Plant Sci. 15 573–581 10.1016/j.tplants.2010.06.005 [DOI] [PubMed] [Google Scholar]
- Esch J. J., Chen M. A., Hillestad M., Marks M. D. (2004). Comparison of TRY and the closely related At1g01380 gene in controlling Arabidopsis trichome patterning. Plant J. 40 860–869 10.1111/j.1365-313X.2004.02259.x [DOI] [PubMed] [Google Scholar]
- Esch J. J., Chen M. A., Sanders M., Hillestad M., Ndkium S., Idelkope B., et al. (2003). A contradictory GLABRA3 allele helps define gene interactions controlling trichome development in Arabidopsis. Development 130 5885–5894 10.1242/dev.00812 [DOI] [PubMed] [Google Scholar]
- Gan L., Xia K., Chen J. G., Wang S. (2011). Functional characterization of TRICHOMELESS2, a new single repeat R3 MYB transcription factor in the regulation of trichome patterning in Arabidopsis. BMC Plant Biol. 11:176 10.1186/1471-2229-11-176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geraldes A., Pang J., Thiessen N., Cezard T., Moore R., Zhao Y., et al. (2011). SNP discovery in black cottonwood (Populus trichocarpa) by population transcriptome resequencing. Mol. Ecol. Resour. (Suppl. 1) 81–92 10.1111/j.1755-0998.2010.02960.x [DOI] [PubMed] [Google Scholar]
- Groover A. T. (2005). What genes make a tree a tree? Trends Plant Sci. 10 210–214 10.1016/j.tplants.2005.03.001 [DOI] [PubMed] [Google Scholar]
- Gruber M. Y., Wang S., Ethier S., Holowachuk J., Bonham-Smith P. C., Soroka J., et al. (2006). “HAIRY CANOLA” – Arabidopsis GL3 induces a dense covering of trichomes on Brassica napus seedlings. Plant Mol. Biol. 60 679–698 10.1007/s11103-005-5472-0 [DOI] [PubMed] [Google Scholar]
- Guan X., Lee J. J., Pang M., Shi X., Stelly D. M., Chen Z. J. (2011). Activation of Arabidopsis seed hair development by cotton fiber-related genes. PLoS ONE 6:e21301 10.1371/journal.pone.0021301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan X., Pang M., Nah G., Shi X., Ye W., Stelly D. M., et al. (2014). miR828 and miR858 regulate homoeologous MYB2 gene functions in Arabidopsis trichome and cotton fibre development. Nat. Commun. 5 3050 10.1038/ncomms4050 [DOI] [PubMed] [Google Scholar]
- Guan X. Y., Li Q. J., Shan C. M., Wang S., Mao Y. B., Wang L. J., et al. (2008). The HD-Zip IV gene GaHOX1 from cotton is a functional homologue of the Arabidopsis GLABRA2. Physiol. Plant. 134 174–182 10.1111/j.1399-3054.2008.01115.x [DOI] [PubMed] [Google Scholar]
- Hajdukiewicz P., Svab Z., Maliga P. (1994). The small, versatile pPZP family of Agrobacterium binary vectors for plant transformation. Plant Mol. Biol. 25 989–994 10.1007/BF00014672 [DOI] [PubMed] [Google Scholar]
- Hülskamp M., Misra S, Jürgens G. (1994). Genetic dissection of trichome cell development in Arabidopsis. Cell 76 555–566 10.1016/0092-8674(94)90118-X [DOI] [PubMed] [Google Scholar]
- Ishida T., Kurata T., Okada K., Wada T. (2008). A genetic regulatory network in the development of trichomes and root hairs. Annu. Rev. Plant Biol. 59 364–386 10.1146/annurev.arplant.59.032607.092949 [DOI] [PubMed] [Google Scholar]
- Jansson S., Douglas C. J. (2007). Populus: a model system for plant biology. Annu. Rev. Plant Biol. 58 435–458 10.1146/annurev.arplant.58.032806.103956 [DOI] [PubMed] [Google Scholar]
- Kirik V., Simon M., Hülskamp M., Schiefelbein J. (2004a). The ENHANCER OF TRY AND CPC1 gene acts redundantly with TRIPTYCHON and CAPRICE in trichome and root hair cell patterning in Arabidopsis. Dev. Biol. 268 506–513 10.1016/j.ydbio.2003.12.037 [DOI] [PubMed] [Google Scholar]
- Kirik V., Simon M., Wester K., Schiefelbein J, Hülskamp M. (2004b). ENHANCER of TRY and CPC 2(ETC2) reveals redundancy in the region-specific control of trichome development of Arabidopsis. Plant Mol. Biol. 55 389–398 10.1007/s11103-004-0893-8 [DOI] [PubMed] [Google Scholar]
- Kurata T., Ishida T., Kawabata-Awai C., Noguchi M., Hattori S., Sano R., et al. (2005). Cell-to-cell movement of the CAPRICE protein in Arabidopsis root epidermal cell differentiation. Development 132 5387–5398 10.1242/dev.02139 [DOI] [PubMed] [Google Scholar]
- Liu S., Wang X., Li E., Douglas C. J., Chen J. G., Wang S. (2013). R2R3 MYB transcription factor PtrMYB192 regulates flowering time in Arabidopsis by activating flowering locus C. J. Plant Biol. 56 243–250 10.1007/s12374-013-0135-1 [DOI] [Google Scholar]
- Oppenheimer D. G., Herman P. L., Sivakumaran S., Esch J., Marks M. D. (1991). A myb gene required for leaf trichome differentiation in Arabidopsis is expressed in stipules. Cell 67 483–493 10.1016/0092-8674(91)90523-2 [DOI] [PubMed] [Google Scholar]
- Payne C. T., Zhang F., Lloyd A. M. (2000). GL3 encodes a bHLH protein that regulates trichome development in Arabidopsis through interaction with GL1 and TTG1. Genetics 156 1349–1362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesch M, Hülskamp M. (2004). Creating a two-dimensional pattern de novo during Arabidopsis trichome and root hair initiation. Curr. Opin. Genet. Dev. 14 422–427 10.1016/j.gde.2004.06.007 [DOI] [PubMed] [Google Scholar]
- Rerie W. G., Feldmann K. A., Marks M. D. (1994). The GLABRA2 gene encodes a homeo domain protein required for normal trichome development in Arabidopsis. Genes Dev. 8 1388–1399 10.1101/gad.8.12.1388 [DOI] [PubMed] [Google Scholar]
- Schellmann S., Schnittger A., Kirik V., Wada T., Okada K., Beermann A., et al. (2002). TRIPTYCHON and CAPRICE mediate lateral inhibition during trichome and root hair patterning in Arabidopsis. EMBO J. 21 5036–5046 10.1093/emboj/cdf524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiefelbein J. (2003). Cell-fate specification in the epidermis: a common patterning mechanism in the root and shoot. Curr. Opin. Plant Biol. 6 74–78 10.1016/S136952660200002X [DOI] [PubMed] [Google Scholar]
- Schnittger A., Folkers U., Schwab B., Jürgens G, Hülskamp M. (1999). Generation of a spacing pattern: the role of TRIPTYCHON in trichome patterning in Arabidopsis. Plant Cell 11 1105–1116 10.1105/tpc.11.6.1105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scoville A. G., Barnett L. L., Bodbyl-Roels S., Kelly J. K., Hileman L. C. (2011). Differential regulation of a MYB transcription factor is correlated with transgenerational epigenetic inheritance of trichome density in Mimulus guttatus. New Phytol. 191 251–263 10.1111/j.1469-8137.2011.03656.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon M., Lee M. M., Lin Y., Gish L., Schiefelbein J. (2007). Distinct and overlapping roles of single-repeat MYB genes in root epidermal patterning. Dev. Biol. 311 566–578 10.1016/j.ydbio.2007.09.001 [DOI] [PubMed] [Google Scholar]
- Tiwari S. B., Hagen G., Guilfoyle T. (2003). The roles of auxin response factor domains in auxin-responsive transcription. Plant Cell 15 533–543 10.1105/tpc.008417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tominaga R., Iwata M., Sano R., Inoue K., Okada K., Wada T. (2008). Arabidopsis CAPRICE-LIKE MYB 3 (CPL3) controls endoreduplication and flowering development in addition to trichome and root hair formation. Development 135 1335–1345 10.1242/dev.017947 [DOI] [PubMed] [Google Scholar]
- Tominaga-Wada R., Nukumizu Y. (2012). Expression analysis of an R3-type MYB transcription factor CPC-LIKE MYB4 (TRICHOMELESS2) and CPL4-related transcripts in Arabidopsis. Int. J. Mol. Sci. 13 3478–3491 10.3390/ijms13033478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tominaga-Wada R., Nukumizu Y., Sato S., Wada T. (2013). Control of plant trichome and root-hair development by a tomato (Solanum lycopersicum) R3 MYB transcription factor. PLoS ONE 8:e54019 10.1371/journal.pone.0054019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuskan G. A., Difazio S., Jansson S., Bohlmann J., Grigoriev I., Hellsten U., et al. (2006). The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science 313 1596–1604 10.1126/science.1128691 [DOI] [PubMed] [Google Scholar]
- Vendramin E., Pea G., Dondini L., Pacheco I., Dettori M. T., Gazza L., et al. (2014). A unique mutation in a MYB gene cosegregates with the nectarine phenotype in peach. PLoS ONE 9:e90574 10.1371/journal.pone.0090574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada T., Kurata T., Tominaga R., Koshino-Kimura Y., Tachibana T., Goto K., et al. (2002). Role of a positive regulator of root hair development, CAPRICE, in Arabidopsis root epidermal cell differentiation. Development 129 5409–5419 10.1242/dev.00111 [DOI] [PubMed] [Google Scholar]
- Wada T., Tachibana T., Shimura Y., Okada K. (1997). Epidermal cell differentiation in Arabidopsis determined by a Myb homolog, CPC. Science 277 1113–1116 10.1126/science.277.5329.1113 [DOI] [PubMed] [Google Scholar]
- Walker A. R., Davison P. A., Bolognesi-Winfield A. C., James C. M., Srinivasan N., Blundell T. L., et al. (1999). The TRANSPARENT TESTA GLABRA1 locus, which regulates trichome differentiation and anthocyanin biosynthesis in Arabidopsis, encodes a WD40 repeat protein. Plant Cell 11 1337–1350 10.1105/tpc.11.7.1337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Barron C., Schiefelbein J., Chen J. G. (2010). Distinct relationships between GLABRA2 and single-repeat R3 MYB transcription factors in the regulation of trichome and root hair patterning in Arabidopsis. New Phytol. 185 387–400 10.1111/j.1469-8137.2009.03067.x [DOI] [PubMed] [Google Scholar]
- Wang S., Chen J. G. (2008). Arabidopsis transient expression analysis reveals that activation of GLABRA2 may require concurrent bindings of GLABRA1 and GLABRA3 to the promoter of GLABRA2. Plant Cell Physiol. 49 1792–1804 10.1093/pcp/pcn159 [DOI] [PubMed] [Google Scholar]
- Wang S., Chen J. G. (2014). Regulation of cell fate determination by single-repeat R3 MYB transcription factors in Arabidopsis. Front. Plant Sci. 5:133 10.3389/fpls.2014.00133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Hubbard L., Chang Y., Guo J., Schiefelbein J., Chen J. G. (2008). Comprehensive analysis of single-repeat R3 MYB proteins in epidermal cell patterning and their transcriptional regulation in Arabidopsis. BMC Plant Biol. 8:81 10.1186/1471-2229-8-81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Kwak S. H., Zeng Q., Ellis B. E., Chen X. Y., Schiefelbein J., et al. (2007). TRICHOMELESS1 regulates trichome patterning by suppressing GLABRA1 in Arabidopsis. Development 134 3873–3882 10.1242/dev.009597 [DOI] [PubMed] [Google Scholar]
- Wang S., Li E., Porth I., Chen J. G., Mansfield S. D., Douglas C. J. (2014). Regulation of secondary cell wall biosynthesis by poplar R2R3 MYB transcription factor PtrMYB152 in Arabidopsis. Sci. Rep. 4 5054 10.1038/srep05054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Tiwari S. B., Hagen G., Guilfoyle T. J. (2005). AUXIN RESPONSE FACTOR7 restores the expression of auxin-responsive genes in mutant Arabidopsis leaf mesophyll protoplasts. Plant Cell 17 1979–1993 10.1105/tpc.105.031096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Wang J. W., Yu N., Li C. H., Luo B., Gou J. Y., et al. (2004). Control of plant trichome development by a cotton fiber MYB gene. Plant Cell 16 2323–2334 10.1105/tpc.104.024844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wester K., Digiuni S., Geier F., Timmer J., Fleck C, Hülskamp M. (2009). Functional diversity of R3 single-repeat genes in trichome development. Development 136 1487–1496 10.1242/dev.021733 [DOI] [PubMed] [Google Scholar]
- Whitham T. G., Bailey J. K., Schweitzer J. A., Shuster S. M., Bangert R. K., LeRoy C. J., et al. (2006). A framework for community and ecosystem genetics: from genes to ecosystems. Nat. Rev. Genet. 7 510–523 10.1038/nrg1877 [DOI] [PubMed] [Google Scholar]
- Yu N., Cai W. J., Wang S., Shan C. M., Wang L. J., Chen X. Y. (2010). Temporal control of trichome distribution by MicroRNA156-targeted SPL genes in Arabidopsis thaliana. Plant Cell 22 2322–2335 10.1105/tpc.109.072579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F., Gonzalez A., Zhao M., Payne C. T., Lloyd A. (2003). A network of redundant bHLH proteins functions in all TTG1-dependent pathways of Arabidopsis. Development 130 4859–4869 10.1242/dev.00681 [DOI] [PubMed] [Google Scholar]
- Zimmermann I. M., Heim M. A., Weisshaar B., Uhrig J. F. (2004). Comprehensive identification of Arabidopsis thaliana MYB transcription factors interacting with R/B-like BHLH proteins. Plant J. 40 22–34 10.1111/j.1365-313X.2004.02183.x [DOI] [PubMed] [Google Scholar]