Abstract
Changes in androgen signaling during prostate carcinogenesis are associated with both inhibition of cellular differentiation and promotion of malignant phenotypes. The androgen receptor (AR)-binding transcription factor (TF) RUNX2 has been linked to prostate cancer (PCa) progression but the underlying mechanisms have not been fully defined. In this study, we investigated the genome-wide influence of RUNX2 on androgen-induced gene expression and AR DNA binding in PCa cells. RUNX2 inhibited the androgen response partly by promoting the dissociation of AR from its target genes such as the tumor suppressor NKX3-1. However, AR activity persists in the presence of RUNX2 at other AR target genes, some of which are co-operatively stimulated by androgen and RUNX2 signaling. These genes are associated with putative enhancers co-occupied by AR and RUNX2. One such gene, the invasion-promoting Snail family TF SNAI2, was co-activated by AR and RUNX2. Indeed, these two TFs together, but neither alone stimulated PCa cell invasiveness, which could be abolished by SNAI2 silencing. In support of our results, an immunohistochemical analysis of SNAI2 in archived primary PCa specimens revealed a correlation with the RUNX2 histoscore; and, simultaneous strong staining for SNAI2, RUNX2 and AR (but not any pair alone) was associated with disease recurrence. Overall, our findings suggest that AR and RUNX2 cooperate to stimulate certain invasion-promoting genes like SNAI2, which might be targeted for individualized PCa therapy.
Keywords: combinatorial transcriptional control, mRNA profiling, ChIP-seq, metastasis, invasion, recurrence
INTRODUCTION
Contrasting their role in prostate epithelial cell differentiation and physiological functions, androgens acquire oncologic roles during prostate carcinogenesis, including promotion of cellular proliferation, survival and aerobic glycolysis (1–4). These changes are associated with redistribution of the androgen receptor (AR) across the prostate cancer (PCa) cell genome and alterations to its transcriptional regulatory properties (5, 6). Contributing to changes in its genomic locations and activities are AR co-activators and collaborating DNA-binding proteins such as FOXA1, NKX3-1, GATA2, RUNX2 and members of the ETS family of transcription factors (6–9).
The mammalian RUNX family consists of three transcription factors with well-established roles in both development and cancer (10–12). RUNX2, best known for its roles in skeletal development (13, 14), has also been implicated in carcinogenesis, including the promotion of breast and prostate cancer metastasis (8, 15–20). RUNX2 activity in PCa is negatively regulated by PTEN through a FOXO1-dependent mechanism (21), RUNX2 expression progressively increases during PCa development in the PTEN conditional knockout mouse model (22) and its immunoreactivity is higher in human PCa than in prostatic intraepithelial neoplasia (PIN) and normal prostate epithelium (16, 23, 24). Furthermore, manipulation of RUNX2 in tissue culture and xenograft mouse models of PCa metastasis alters invasiveness and tissue destruction (16, 17).
RUNX2 directly interacts with and influences the activity of other transcription factors, including members of the nuclear hormone receptor family. In both breast cancer cells and osteoblasts, RUNX2 and estrogen signaling modulate each other’s activity in a locus-specific manner, with implications for the regulation of both breast cancer progression and bone mass control (12, 20, 25–27). In osteoblasts, RUNX2 interacts with and augments the transcriptional activity of the vitamin D receptor at the osteocalcin gene (28). Finally, RUNX2 directly binds the AR, and this interaction is potentially important for both modulating and interpreting androgen signaling in various physiological and pathological contexts including bone metabolism and PCa progression (8, 27, 29).
In PCa and other cell types, physical interaction between AR and RUNX2’s DNA-binding domain inhibits RUNX2’s recruitment to and activation of target genes (8, 27, 29, 30). Limited investigations of the reciprocal effects, those of RUNX2 on AR led to apparently conflicting results indicating either inhibition (29, 31) or stimulation (30, 32) of AR activity. To address the hypothesis that RUNX2 influences AR activity in a locus-dependent manner, we set out to characterize genome-wide the influence of RUNX2 on AR-regulated gene expression by comprehensive mRNA profiling of C4-2B/Rx2dox PCa cells after activation of the AR with dihydrotestosterone (DHT) and/or induction of RUNX2 by doxycycline (dox). As previously described dox increases RUNX2 expression in these cells from hardly detectable to levels normally seen in other cell lines (17). The gene expression profiles, in combination with ChIP-seq analyses of RUNX2 and AR, demonstrate complex remodeling of the AR-regulated gene network: whereas RUNX2 generally attenuated recruitment of AR and stimulation of target genes, AR remained bound and active upon a specific subset of genes and even synergized with RUNX2 in some cases. Here we pursued the mechanistic basis of these diverse interactions and then investigated the significance of the synergistic activation of SNAI2 by RUNX2 and AR.
MATERIALS AND METHODS
Reagents
DHT and dox, both from Sigma-Aldrich (St. Louis, MO) were used at final concentrations of 10 nM and 0.25μg/ml, respectively. AR (N-20), RUNX2 (M70) and GAPDH (V-18) antibodies were from Santa Cruz Biotechnology (Santa Cruz, CA), Flag (M2) and SNAI2 (C19G7) antibodies were from Sigma-Aldrich and Cell Signaling Technology (Danvers, MA), respectively. RUNX2 (ab76956) and AR (F.39.4.1) antibodies for immunohistochemistry were from Abcam (Cambridge, MA) and Biogenex Laboratories (Fremont, CA) respectively. Protein-A dynabeads were from Invitrogen (Carlsbad, CA). DMEM and RPMI-1640 media were from Mediatech, Inc (Manassas, VA). Fetal bovine serum (FBS) was from Omega Scientific (Tarzana, CA). Charcoal dextran stripped serum (CSS) was from Gemini Bio Products (West Sacramento, CA).
Cell culture and immunofluorescence
COS7 cells and the human prostate cancer cell lines C4-2B/Rx2dox, 22Rv1/Rx2dox and LNCaP/Rx2dox were previously described (8, 17) and have been passaged for less than 6 months. PCa cells were maintained in RPMI-1640 supplemented with 10% FBS and COS7 cells were maintained in DMEM with 5% FBS. Hygromycin (50μg/ml) and puromycin (1μg/ml) were used to select cells that had incorporated the Rx2dox and the shSNAI2 lentiviral vectors, respectively. Two days before initiation of hormone treatment, 10% FBS was replaced with 5% CSS, and all experiments were performed in the absence of any selection marker. AR and RUNX2 immunofluorescence was performed using the N20 and M70 primary antibodies and fluorescein- and rhodamine-conjugated secondary antibodies respectively. Cells were mounted using Vectashield mounting medium with DAPI (Vector Laboratories Inc., Burlingame, CA) and viewed using an LSM 510 Zeiss confocal microscope (Carl Zeiss, Thornwood, NY). Fluorescence recovery after photobleaching (FRAP) was carried out as previously described (8).
ChIP, mRNA, DNA and protein assays
AR ChIP and Flag-RUNX2 ChIP were performed essentially as described previously (9, 33). Processing and quantification of mRNA and ChIP by qPCR was as described (33) using the primers listed in Supplemental Table S1. Western blot analyses were carried out essentially as described (33).
Invasion Assay
C4-2B/Rx2dox/Luc cells, expressing RUNX2 conditionally and firefly luciferase constitutively (17) were suspended in serum-free medium and seeded in 24 well plates for morphology assessment, or in Matrigel™-coated inserts (BD Bioscience, San Jose, CA) for evaluating invasiveness. The inserts were placed for 24h in wells containing 5% CSS, and non-migrating cells were removed. Results are presented as invasion indices, defined as the ratio between the luciferase activity in cells that invaded through Matrigel™-coated membranes and the respective values obtained from cells plated in control inserts with uncoated membranes. Treatment with DHT and/or dox commenced 48h prior to seeding in the inserts and lasted throughout the experiment. Silencing of SNAI2 was performed as described (20).
Bioinformatics
Gene expression profiling was performed as described previously (17, 33) and in the supplemental methods. Briefly, total RNA from C4-2B/Rx2dox cells was extracted in biological triplicates and hybridized to BeadChip HumanHT-12 v4, (Illumina Inc., San Diego, CA).
For RUNX2 and AR genomic occupancy, read coordinates (aligned to hg18) for RUNX2 and AR ChIP-seq experiments were obtained from our recent paper (33) and from Massie et al. (3), respectively. A total of 36,698 RUNX2 peaks and 10,949 AR peaks were detected using MACS (34) with a p-value threshold of p≤1E-10, Scoring profiles were constructed as described previously (35). Detailed methodologies and the combinatorial effects of AR and RUNX2 were described in the Supplemental Methods. Microarray gene expression data has been uploaded to GEO, Accession GSE52627.
Immunohistochemistry
A series of 95 patients with lymph node involvement who had undergone radical prostatectomy for locally advanced prostate cancer were selected from the institutional database at the University of Southern California. Clinical characteristics are described in Supplemental Table S2. Detailed methodologies are described in Supplemental Methods. Scoring of the SNAI2 (0,1,2,3), AR (high/low) and RUNX2 (high/low) immunoreactivity was performed under the supervision of a certified PCa pathologist, and only regions of invasive carcinoma were considered. The Institutional Review Board of USC approved the tissue procurement protocol for this study (IRB approval HS-08-00590). Appropriate written informed consent was obtained from all patients.
RESULTS
RUNX2 antagonizes AR recruitment to and stimulation of the majority (Type I) of DHT-stimulated genes
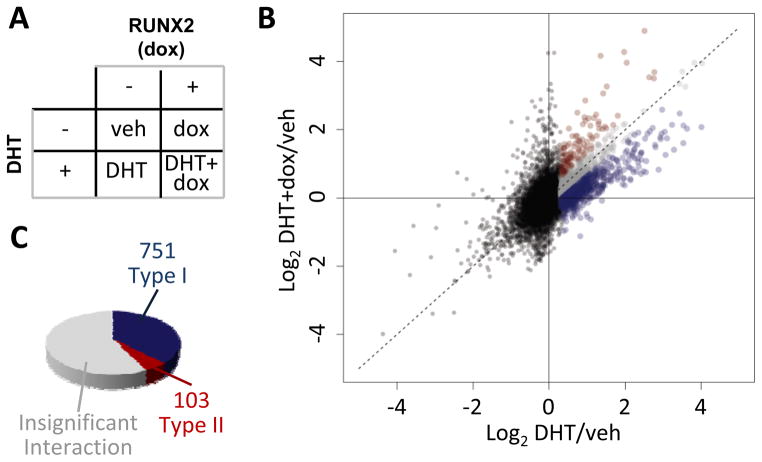
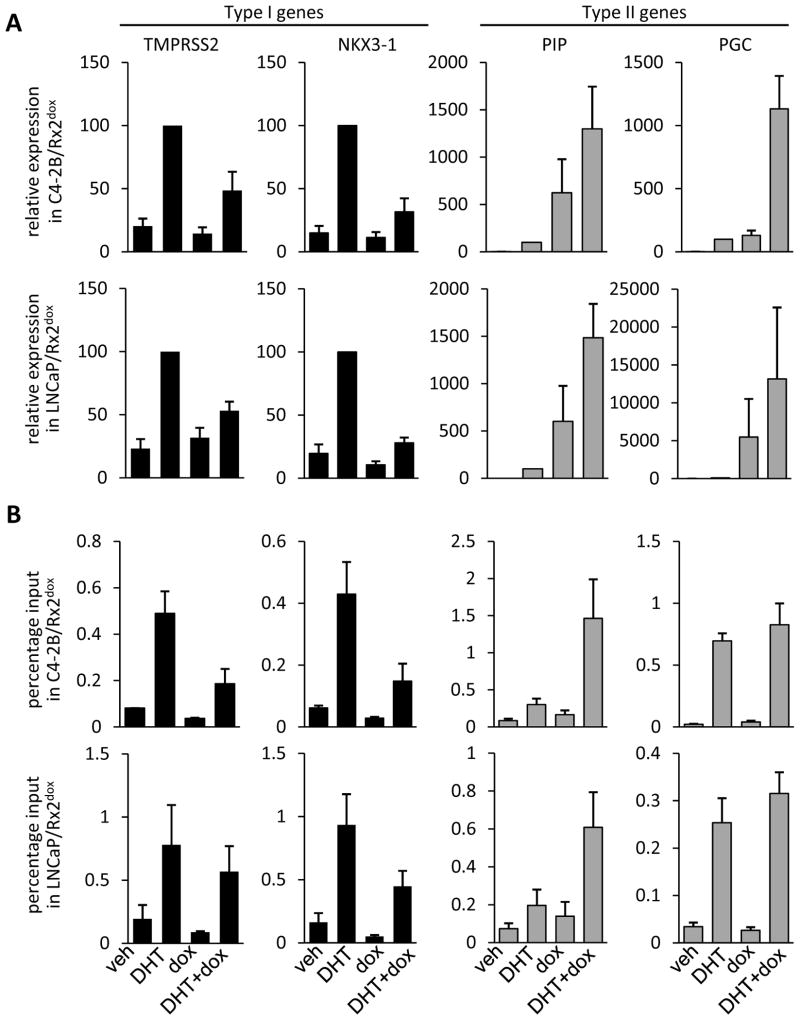
Three cell lines were used in this study to investigate the influence of RUNX2 on AR-driven gene expression in PCa cells. All three lines are essentially RUNX2 negative and each was engineered with the Rx2dox lentiviral system, which facilitates RUNX2 induction upon dox treatment (8). The LNCaP and the C4-2B cell lines require presence of androgens for AR activation, whereas the 22Rv1 cell line also expresses AR variants that are active independent of ligand (36). We first analyzed global mRNA profiles of C4-2B/Rx2dox cells treated with DHT to activate the AR and/or with dox to induce RUNX2 expression (Figure 1A). DHT significantly upregulated 2002 genes (FDR-adjusted p<0.01). To illustrate the global influence of RUNX2 on the DHT response, we plotted the normalized gene expression values from cells co-treated with DHT plus dox against the respective values from cells treated with DHT alone (Figure 1B). Approximately half (1148) of the genes responded in a similar manner to DHT alone and to DHT plus dox (Figure 1B–C, grey). The remaining 854 genes responded differently to DHT plus dox compared to DHT alone, and of these, 751 (88%, henceforth Type I) were less strongly stimulated in the presence of RUNX2 (Figure 1B–C, blue). Similarly, in the reciprocal orientation, the predominant influence of DHT was attenuation of RUNX2-mediated stimulation of gene expression (Supplemental Figure S1). Thus, in PCa cells, AR and RUNX2 are generally antagonistic, consistent with the expression patterns of hand-picked genes previously investigated in this and other cell types (29, 30). Interestingly, however, 12% of the DHT-stimulated genes whose expression was modified by RUNX2 were further stimulated, rather than inhibited when RUNX2 was induced. These 103 genes were designated Type II (Figure 1B–C, red). Supplemental Tables S3 and S4 list the DHT-stimulated genes, whereby the response to DHT is attenuated (Type I) or augmented (Type II) in the presence versus absence of RUNX2. RT-qPCR analysis essentially confirmed the expression pattern of several Type I and Type II genes (Figure 2A, and Supplemental Tables S3, S4). RT-qPCR analysis of these genes in another PCa cell line, LNCaP/Rx2dox, demonstrated similar locus-dependent effects of RUNX2 on DHT-stimulated genes (Figure 2A, lower panels and Supplemental Tables S3, S4). We also investigated the effects of RUNX2 on DHT-stimulated genes by RT-qPCR in the 22Rv1/Rx2dox cell line, a model of castration resistant PCa (CRPC). Interestingly, the results from 22Rv1 cells were unlike those in LNCaP and C4-2B cells, with type II behavior representing the most common mode of interaction in this CRPC model (Supplemental Figure S2). These results suggest that the interaction between RUNX2 and AR signaling is not only locus-dependent, but may also be modified during the transition from ADPC to CRPC. This speculation is the focus of an ongoing investigation, which is outside the scope of the present study.
Figure 1. RUNX2 modulates AR activity in a locus-specific manner.
A. C4-2B/Rx2dox cells were treated as indicated with dox and/or DHT, and mRNA expression was profiled using Illumina’s Bead-chip arrays. B. Scatter plot describing the response to DHT+dox versus the response to DHT alone. Black dots represent genes not significantly up-regulated by DHT and grey dots represent genes with DHT response not significantly influenced by RUNX2. Blue and red dots represent Type I and type II genes, defined, respectively, based on attenuated or augmented response to DHT+dox versus DHT alone. C. Pie chart illustrating the frequency of genes whose stimulation by DHT is attenuated (blue), augmented (red), or not significantly changed (grey) in the presence versus absence of RUNX2. Data represent a combined analysis of three biological replicates.
Figure 2. Androgen receptor activity and occupancy at type I and type II genes.
A. C4-2B/Rx2dox (upper four panels) or LNCaP/Rx2dox cells (lower four panels) were treated with dox and/or DHT and expression of the indicated type I (black) and type II (grey) genes was assessed by RT-qPCR. B. C4-2B/Rx2dox (upper four panels) or LNCaP/Rx2dox cells (lower four panels) were treated as in A and AR recruitment to known AREs associated with the indicated genes was measured by ChIP-qPCR. Data in A are a combined analysis of four independent experiments and are normalized to the values measured in the presence of DHT (defined as 100%). Data in B are a combined analysis of three independent experiments. (Mean±SEM).
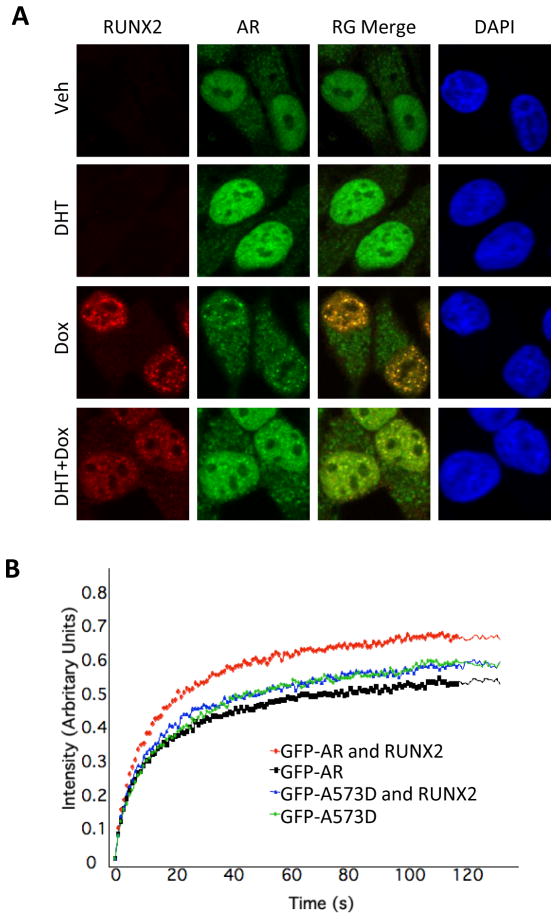
The functional interrelationships between AR and RUNX2 could be related to the physical interaction between these two transcription factors. Indeed, similar to PC3-AR, COS7, SaOS-2 and MC3T3E-1 cells (8, 29), the two transcription factors appear to physically interact in the C4-2B/Rx2dox culture model as well. This is suggested by co-immunofluorescence imaging of dox-treated C4-2B/Rx2dox cells, which demonstrated co-localization of AR and RUNX2 within distinct nuclear domains (Figure 3A) as well as alteration to AR’s cellular distribution, from a relatively uniform nuclear staining in the absence of RUNX2 to textural staining that includes nuclear speckles co-occupied by the two proteins once RUNX2 is expressed (Figure 3A). Furthermore, RUNX2 modified the FRAP of GFP-AR (Figure 3B), indicating that RUNX2 influenced AR’s intranuclear mobility, consistent with physical interaction between the two proteins in living cells. As control, mobility of the AR-A573D mutant, which binds neither DNA nor RUNX2 (8), was not influenced by RUNX2 (Figure 3B). Although binding of RUNX2 to AR within distinct subnuclear domains may underlie the modification of the androgen response by RUNX2, it cannot explain the locus-dependent interaction observed in type I versus type II genes (Figures 1 and 2A).
Figure 3. RUNX2 modifies AR localization and mobility in living cells.
A. C4-2B/Rx2dox cells were treated with dox and/or DHT, then immunostained and subjected to confocal microscopy to visualize the AR (green) and RUNX2 (red). DAPI (blue) demarcates the cell nucleus. B. GFP-AR or GFP-AR-A573D fusion proteins were expressed in COS7 cells either alone or together with RUNX2. The cells were treated with DHT and a portion of their nuclei was subjected to FRAP analysis. Curves represent fluorescence intensity relative to the respective pre-photobleaching levels.
Because recruitment of AR is central to androgen-mediated stimulation of target genes, we measured AR occupancy by ChIP-qPCR at known androgen response elements (AREs) associated with the Type I genes NKX3-1 and TMPRSS2 and the Type II genes PIP and PGC (6, 30, 37, 38). As expected, treatment of either C4-2B/Rx2dox or LNCaP/Rx2dox cells with DHT alone resulted in AR recruitment to AREs of both Type I and Type II genes (Figure 2B). When RUNX2 was induced along with DHT treatment we observed differing behaviors of the AR in both these cell lines. Whereas RUNX2 attenuated AR recruitment to the Type I genes (Figure 2B, black), likely contributing to their blunted DHT response, RUNX2 did not attenuate (PGC) and even enhanced (PIP) the recruitment of AR to AREs near Type II genes (Figure 2B, grey). Thus, RUNX2 influences DHT-mediated AR recruitment to and activation of target genes in a locus-dependent manner.
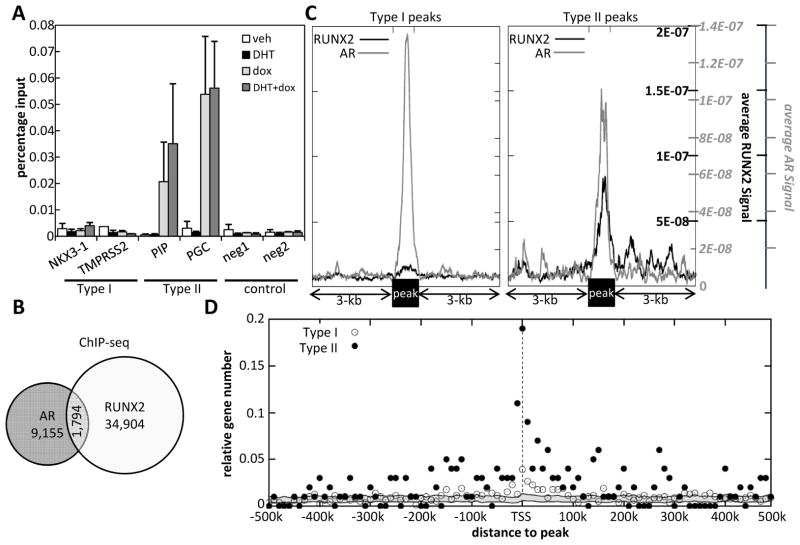
Regions doubly occupied by AR and RUNX2 are found near Type II genes
Whereas RUNX2-mediated attenuation of DHT responsiveness in Type I genes was attributable in part to lesser AR recruitment, the uninhibited DHT response of Type II genes, and in particular the enhanced response of PGC, could not be explained simply based on AR recruitment (Figure 2). We tested, initially by RUNX2 ChIP-qPCR the alternative and non-mutually exclusive hypothesis that RUNX2 itself is recruited along with AR to Type II genes. Indeed, RUNX2 was readily detectable at the AREs of the Type II genes PIP and PGC, but not at the AREs of the Type I genes NKX3-1 and TMPRSS2 (Figure 4A). We further tested this hypothesis at the whole genome level by reanalyzing our RUNX2 ChIP-seq dataset (33) along with an AR ChIP-seq dataset obtained in LNCaP cells (3). We initially determined the frequency of ARORs that also recruited RUNX2. As shown in Figure 4B, 1,794 (16%) of the 10,949 AR ChIP-seq peaks overlapped with RUNX2 peaks. Using the same two datasets, we then plotted the average RUNX2 ChIP-seq signal and the average AR signal across AR peaks adjacent to (within 10kb of) the TSS of Type I and Type II genes (Figure 4C). These genome-wide aggregate profiles clearly demonstrated the presence of a strong RUNX2 signal at AR peaks associated with Type II (Figure 4C, right) but not Type I (Figure 4C, left) genes. These results suggest that the increased expression of Type II genes in cells treated with DHT plus dox as compared to DHT alone is attributable to regulation by enhancers capable of recruiting both AR and RUNX2, and that the binding of RUNX2 to these enhancers allows them to escape RUNX2-mediated attenuation of the androgen response. Seeking further support for this view, we mapped the doubly-occupied presumptive enhancers with respect to the TSSs of Type II versus Type I genes. Enumeration of the doubly-occupied enhancers as a function of distance from their respective nearest TSSs revealed many more doubly-occupied enhancers near Type II as compared to Type I TSSs (Figure 4D). Remarkably, 27 (27%) of the 100 Type II genes with mapped Refseq co-ordinates had doubly-occupied enhancers between positions −30-kb and +30-kb, compared to only 7.6% (60/792) of Type I genes having corresponding doubly-occupied enhancers (Figure 4D). Although the enrichment for doubly-occupied enhancers near Type II compared to Type I genes dramatically dropped as a function of distance from the respective TSSs, it remained significantly higher at distances exceeding 200-kb (Figure 4D), likely reflecting looping of doubly-occupied enhancers onto Type II target genes located many kilobases away.
Figure 4. Type II genes are characterized by regions co-occupied by AR and RUNX2.
A. C4-2B/Rx2dox cells were treated with dox and/or DHT, and RUNX2 occupancy at known AREs of the indicated type I and II genes was measured by ChIP-qPCR (Mean±SEM; n=3). B. Venn diagram showing the overlap between AR and RUNX2 occupancy based on ChIP-seq datasets publically available for these two transcription factors (33, 3). These datasets were also used in Panels C and D. C. AR ChIP-seq peaks adjacent to Type I (left) or Type II genes (right) were centered and the average local AR (grey) and RUNX2 (black) ChIP-seq signals were normalized by the total number of AR peaks in each class and the total number of reads in each library. D. AR/RUNX2-doubly occupied ChIP-seq peaks within 500-kb of the TSSs of Type I (open dots) and Type II (filled dots) genes were enumerated in 10 kb windows and expressed as a fraction of the total number of Type I or Type II genes. Randomly selected genes were used to compute the background (grey ribbon) TSS-to-peak distances as described in Supplemental Materials and Methods.
RUNX2 and AR synergistically stimulate a subset of Type II genes that includes SNAI2
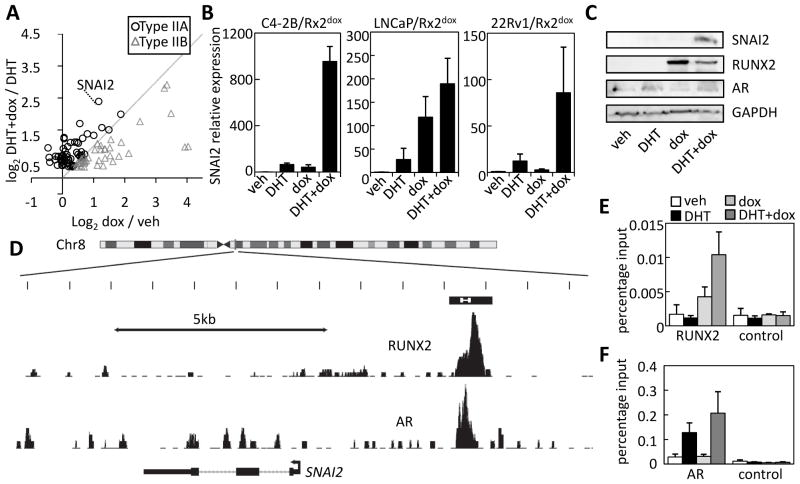
We had initially defined Type II genes based on stronger stimulation by DHT plus dox as compared to DHT alone (Figure 1). Because we observed more RUNX2 binding near Type II as compared to Type I genes (Figure 4), the high expression of Type II genes in cells treated with DHT plus dox compared to DHT alone could simply reflect the summed stimulatory effects of AR and RUNX2. Close examination of the expression profiles of Type II genes, however, revealed cases of synergistic, rather than additive stimulation by DHT and RUNX2. Indeed, a scatter plot of the RUNX2 response of Type II genes in the presence versus absence of DHT (Figure 5A) demonstrates that many (61%) of the Type II genes, hereafter Type IIA, were synergistically stimulated by DHT and dox. One of the clearest examples of synergism was SNAI2 (see Figure 5A and Supplemental Table S5). Consistent with previous investigations (17, 39), RT-qPCR analysis shows that each of DHT and RUNX2 increases SNAI2 mRNA levels in PCa cells (Figure 5B). More importantly, and consistent with the microarray analysis, the simultaneous induction of RUNX2 (by dox) and activation of AR (by DHT) results in cooperative stimulation of SNAI2 transcription in three different PCa cell lines, with particularly strong synergism in C4-2B cells (Figure 5B). Western blot analysis confirmed the synergism between AR and RUNX2 in stimulating SNAI2 expression at the protein level (Figure 5C and Supplemental Figure S3).
Figure 5. SNAI2 is synergistically stimulated by AR and RUNX2.
A. The microarray expression data for type II genes was used to plot the stimulation by dox in the presence versus absence of DHT. Data points significantly to the left of the diagonal (circles, type IIA) represent synergism between DHT and dox, whereas those to the right (triangles, type IIB) are strongly driven by RUNX2 regardless of DHT. B. C4-2B/Rx2dox, LNCaP/Rx2dox and 22Rv1/RX2dox cells were treated as indicated and SNAI2 mRNA expression was analyzed by RT-qPCR. C. Western blot analysis of SNAI2, RUNX2 (FLAG) and AR in C4-2B/Rx2dox cells treated as indicated. D. ChIP-seq data describing RUNX2 (33) and AR (3) occupancy over a 13-kb region at the SNAI2 locus. The putative SNAI2 composite enhancer is shown as a black box with the region amplified in Panels E and F marked in white. E–F. RUNX2 (E) and AR (F) occupancy at the SNAI2 enhancer was measured by ChIP-qPCR. Controls include amplification of a remote genomic region (E) or the same region after ChIP with non-specific IgG (F). Data are Mean±SEM; n=3.
The landscape of AR and RUNX2 occupancy at the SNAI2 locus, derived from the aforementioned ChIP-seq datasets (3, 33) suggested recruitment of both RUNX2 and AR to a putative composite enhancer approximately 4-kb upstream of the SNAI2 TSS (Figure 5D). ChIP-qPCR analysis of C4-2B/Rx2dox cells treated with DHT and/or dox confirmed occupancy as well as mutual enhancement of the AR and RUNX2 recruitment (Figure 5E–F).
RUNX2 and AR signaling cooperatively induce invasiveness in a SNAI2-dependent manner
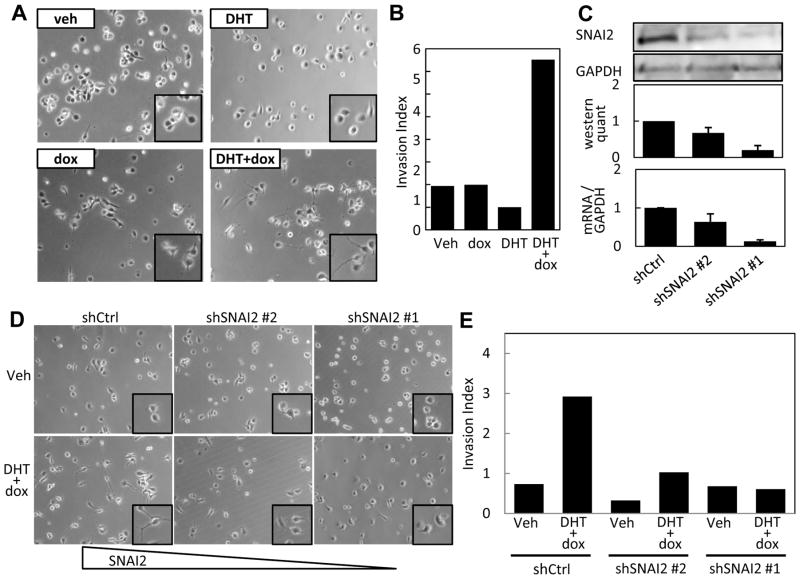
SNAI2 promotes invasiveness and other metastatic properties in various cancers (40). We therefore asked whether the synergistic stimulation of SNAI2 by AR and RUNX2 in C4-2B/Rx2dox cells might influence invasiveness. Co-activation of RUNX2 and AR induced an elongated cell morphology and dendrite-like processes (Figure 6A) often associated with invasiveness and metastasis (41). Matrigel™ invasion assays showed that combined AR activation and RUNX2 induction, but neither alone, led to a remarkable increase in cell invasiveness (Figure 6B), and western analysis confirmed the synergistic stimulation of SNAI2 by AR and RUNX2 under the conditions employed during the invasion assay (Supplemental Figure S4). Finally, to test the role of SNAI2 in this increased invasiveness, we knocked down its expression using each of two shRNAs (Figure 6C). Both the morphological changes (Figure 6D) and the synergistic stimulation of cellular invasiveness (Figure 6E) in response to DHT and dox were diminished with shRNA#1, which robustly knocked down SNAI2 expression. Somewhat weaker diminution of the invasiveness was observed with shRNA#2, which decreased SNAI2 expression to a lesser extent. These results indicate that synergistic stimulation of SNAI2 expression by RUNX2 and androgen signaling is required for the increased invasiveness observed when the two pathways are simultaneously activated.
Figure 6. RUNX2 and Androgen signaling co-operatively induce invasiveness of PCa cells via SNAI2.
A. Phase contrast images of C4-2B/Rx2dox cells treated with DHT and/or dox as indicated. B. C4-2B/Rx2dox cells constitutively expressing luciferase were treated with dox and/or DHT and invasion index was assessed based on luciferase activity in cells that had invaded through Matrigel™ coated versus non-coated membranes as described in Materials and Methods. C. C4-2B/Rx2dox cells were transduced with control (shCtrl) or SNAI2-targeting shRNA lentiviruses (shSNAI2 #1, #2) and SNAI2 silencing was assessed by Western blotting (upper and middle panels) and by RT-qPCR (lower panel). D–E. Effects of DHT and dox on cell morphology (D) and invasiveness (E) were determined as in A and B, respectively, after transduction of C4-2B/Rx2dox cells with shSNAI2#1, shSNAI2#2 or shCtrl. A, B, D and E are representative of three independent experiments. Bars in C are Mean±SEM of three experiments).
Strong SNAI2 expression in PCa biopsies with high nuclear levels of both AR and RUNX2 predicts disease recurrence
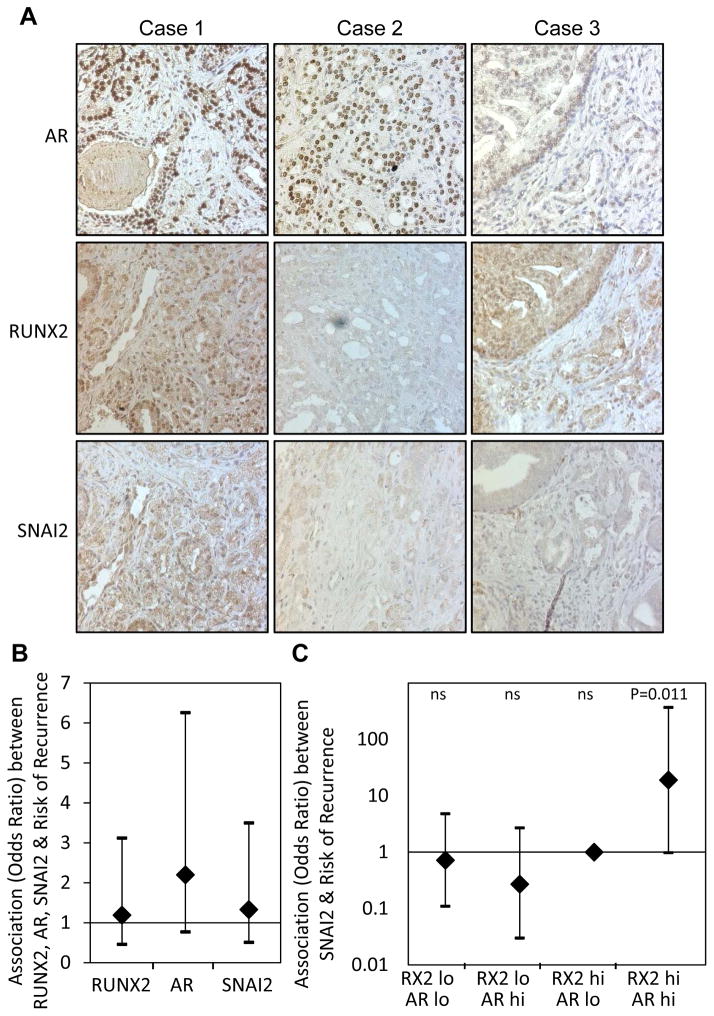
In pursuit of evidence for potential co-stimulation of SNAI2 by AR and RUNX2 in a clinical setting, we assessed by immunohistochemical staining expression of the respective proteins in 95 primary PCa tumors using a tissue microarray (TMA) containing tumors from 73 patients who remained free from clinical recurrence and 22 who relapsed. Consistent with published data (39), most of the tissue samples were stained for SNAI2 only weakly, but four sections were assigned the highest SNAI2 histoscore of 3 (Supplemental Table S6). Each of these four sections, e.g., Case 1 in Figure 7A, was also assigned high histoscores for both RUNX2 and AR (Supplemental Table S6). Reciprocally, absence or low expression of either nuclear RUNX2 or nuclear AR was most commonly associated with low or lack of detectable SNAI2 (e.g., Figure 7A, Cases 2 and 3, respectively). Overall, there was a strong correlation between the SNAI2 histocore and the sum histoscores for AR and RUNX2 (r=0.26, p=0.003, based on Kendall’s tau measure of correlation), with RUNX2 making the major contribution to the correlation (Supplemental Table S7). However, a minor yet sizable proportion of the SNAI2-negative tumors stained strongly for both nuclear RUNX2 and nuclear AR (Supplemental Table S6), possibly reflecting conditions in these cases that limit the transcriptional activity of RUNX2, AR, or the co-operation between them. Taken together, the TMA data suggest that, similar to our in vitro results, cooperation between AR and RUNX2 in stimulating SNAI2 expression exists in the majority of human PCa tumors in vivo. In our cohort, however, none of the AR, RUNX2 or SNAI2 histoscores in isolation correlated with disease recurrence (Figure 7B).
Figure 7. Evidence that RUNX2 and AR co-operate to induce SNAI2 in PCa tumors and the potential clinical significance.
A PCa tissue microarray was subjected to IHC staining of AR, RUNX2 and SNAI2. A. The relationships between SNAI2 staining and nuclear AR and RUNX2 staining, summarized in Supplemental Table S6, are represented here by three cases. Case 1 is strongly stained for SNAI2 (Score=3) as well as for nuclear RUNX2 and AR. Case 2, with high nuclear AR, but no nuclear RUNX2 staining lacks SNAI2 staining, Case 3, with a low level of nuclear AR and a high level of nuclear RUNX2 also lacks SNAI2 staining. B. Odds ratios and 95% confidence intervals for association of each of RUNX2, AR and SNAI2 with recurrence C. Odds ratios and 95% confidence intervals for association of SNAI2 with recurrence for each combination of high (hi) or low (lo) RUNX2 and AR.
Because a minority, of the tumors did not exhibit evidence for cooperation between AR and RUNX2 in stimulating SNAI2, we asked whether they differed from the majority of tumors (with evidence of cooperation) in terms of disease recurrence. Indeed, tumors with evidence of cooperation (RUNX2high/ARhigh/SNAI2high) recurred more frequently than those expressing high SNAI2, but low AR or RUNX2. Association between SNAI2 and recurrence risk was significant when RUNX2 and AR were both high (p=0.011) but not when either was low (Figure 7C and Supplemental Table S8). These results suggest that tumors in which AR and RUNX2 can interact to stimulate SNAI2 expression are more likely to recur after resection.
DISCUSSION
Expression of the osteoblast master regulator RUNX2 in PCa cells was originally investigated in the context of the osteomimetic properties displayed by these bone-seeking tumors (42). More recent studies demonstrate that RUNX2 stimulates various pro-metastatic genes and phenotypes that include, but are not limited to such related to the high predilection of PCa for bone (16, 17). Here we further demonstrate that RUNX2 modulates activity of the AR. This modulation primarily entails inhibition of androgen-stimulated expression of genes, including such that mediate cellular differentiation and tumor suppression. Examples include inhibition of the NKX3-1 and PDEF tumor suppressor genes (43, 44) and the epithelial marker KRT19 (Supplemental Table S3). On the other hand, a small subset of the AR transcriptome was resistant to attenuation by RUNX2, and in some cases RUNX2 even augmented the expression of androgen-stimulated genes. Examples for these so-called Type II genes include the anti-apoptotic genes EGFR, ITSN1 and CRYAB, the pro-proliferative gene PRKCD, the pro-metastatic gene SNAI2 and additional genes implicated in various aspects of PCa progression such as HIPK2, SOX9 and RAB3B (Supplemental Table S4). Thus, the ectopic expression of RUNX2 during PCa progression may reshape the androgen response by attenuating expression of AR-regulated tumor suppressor genes while sparing and even augmenting expression of AR-regulated oncogenes.
Attenuation of the androgen response by RUNX2 at most androgen-stimulated (Type I) genes, as well as the reciprocal attenuation of the RUNX2 response by androgens (Supplemental Figure S1), are attributable to the direct interaction between the two transcription factors, demonstrated previously by co-immunoprecipitation and GST pull-down assays (8, 29) and reiterated herein based on co-localization in C4-2B/Rx2dox cells and alteration to AR intranuclear mobility in response to RUNX2. Consistent with the involvement of the respective DNA-binding domains in their physical interaction (8, 29), attenuation of the androgen response after RUNX2 induction was associated with decreased recruitment of AR to Type I genes (this study); and, attenuation of the RUNX2 response by androgens was associated with compromised recruitment to its targets (8). In breast cancer cells, a similar relationship of reciprocal attenuation has been documented for most RUNX2- and most estrogen-responsive genes (25, 26). Interestingly, however, RUNX2-stimulated SNAI2 expression in breast cancer cells followed the global trend and was attenuated by estradiol, potentially contributing to the anti-RUNX2 and protective effects that estradiol had with regard to breast cancer cell invasiveness (20). Unlike in breast cancer cells, the present work with PCa cells demonstrates that SNAI2 in this cancer type is subject to an unusual mechanism whereby androgens and RUNX2 signaling cooperate to stimulate gene expression.
How a minority of AR-stimulated genes, e.g., PIP, PGC (Figure 2) and SNAI2 (Figure 5) escape RUNX2-mediated attenuation remains to be fully elucidated. We observed retention of AR and recruitment of RUNX2 itself to AR-occupied regions (ARORs) near these so-called Type II genes (Figure 4). At first glance, the recruitment of RUNX2 could be interpreted as tethering to these ARORs via contacting DNA-bound AR. Arguing against such a tethering mechanism, RUNX2 was recruited to the doubly occupied regions even in cells not stimulated by DHT (Figure 4A and 5E). Furthermore, the doubly occupied regions near Type II genes are enriched for sequence elements resembling the RUNX consensus motif TGTGGT (91% contain such a motif, compared to 43% of AR-only peaks). Our working model therefore suggests that AR and RUNX2 bind individual elements at composite enhancers of Type II genes, and that proximity between these elements permits each transcription factors to remain bound in the presence of the other. We do not know, however, why some Type II genes merely escape attenuation of the androgen response by RUNX2 (e.g., SGK1; see Supplemental Figure S5), while others are further stimulated by AR and RUNX2 in a synergistic manner (e.g., SNAI2). We speculate that certain spatial configurations of AR- and RUNX2-binding elements render composite enhancers of the so-called Type IIA genes exceptionally attractive to co-activators, which promote the observed transcriptional synergism.
SNAI2 is a major player in cancer metastasis (20, 40). Knockdown of endogenous SNAI2 in PCa cells results in reduced expression of mesenchymal markers, corresponding morphological changes, and decreased cell invasiveness (45, 46). In frozen sections of PCa biopsies, SNAI2 mRNA was higher in microdissected metastatic lesions compared to primary PCa (39, 47). Recent studies also demonstrated positive correlation between SNAI2 immunohistochemical staining in primary tumors and disease progression (16, 39, 48). The regulation of SNAI2 by each of AR and RUNX2 has been independently reported (17, 20, 39), and here we show that the two regulatory pathways intersect to cooperatively promote SNAI2 expression and PCa cell invasiveness in vitro. Clinically, we observe that the minority of primary PCa tumor sections that are strongly immunostained for SNAI2 are typically highly positive for both AR and RUNX2 nuclear immunostaining; low or no nuclear staining of either AR or RUNX2 is usually associated with lack of SNAI2 staining. Perhaps most significantly, high SNAI2 expression in our series of primary tumor biopsies correlated with disease recurrence, but only when it was associated with strong AR and strong RUNX2 immunohistochemical staining. These ARhigh/RUNX2high/SNAI2high tumors may represent an aggressive PCa subtype with a high recurrence rate. In contrast, many tumors where high SNAI2 expression was associated with low AR or low RUNX2 had low recurrence rates. If ARhigh/RUNX2high/SNAI2high primary tumors were reproducibly found aggressive in additional patient cohorts, efforts would be warranted to screen for such patients and develop drugs, e.g., AR/RUNX2 disruptors, which may spare them the dire consequences of disease recurrence.
In conclusion, RUNX2 remodels androgen signaling in PCa cells in a locus-dependent manner. It usually attenuates AR-driven transcription, but a minority of genes remain androgen-responsive in the presence of RUNX2. Some of them, e.g., SNAI2, exhibit synergistic stimulation and recruitment of both AR and RUNX2 to composite enhancers. Targeting the AR-RUNX2 interaction presents an opportunity for the development of novel therapeutic approaches that would retain expression of androgen-stimulated tumor suppressors while preventing synergistic interaction between AR and RUNX2 at PCa-driving genes. Such novel therapeutic approaches would be particularly suited to prevent disease recurrence in patients whose primary tumor biopsies exhibit high expression of AR, RUNX2 and SNAI2
Supplementary Material
Acknowledgments
Financial Support
This work was supported by NIH grants RO1 DK07112 and RO1 DK07112S from the National Institute of Diabetes and Digestive and Kidney Diseases to BF, NIH grant P30 CA014089 from the National Cancer Institute to SG (Biostatistics Core), as well as grants from the Wright Foundation to GHL and from the Zumberge Foundation to SKB.
We thank Ms. Michaela MacVeigh Aloni at the USC Confocal Microscopy Core Facility for excellent technical assistance, and Tommy Tong and Lillian Young for help with the TMA processing and analysis. The microarray analysis was performed by the UCLA Neuroscience Genomics Core (http://www.semel.ucla.edu/ungc).
Footnotes
The authors have no potential conflicts of interest to disclose
References
- 1.Wang Q, Li W, Zhang Y, Yuan X, Xu K, Yu J, et al. Androgen Receptor Regulates a Distinct Transcription Program in Androgen-Independent Prostate Cancer. Cell. 2009;138:245–56. doi: 10.1016/j.cell.2009.04.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lin B, Wang J, Hong X, Yan X, Hwang D, Cho JH, et al. Integrated expression profiling and ChIP-seq analyses of the growth inhibition response program of the androgen receptor. PLoS One. 2009;4:e6589. doi: 10.1371/journal.pone.0006589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Massie CE, Lynch A, Ramos-Montoya A, Boren J, Stark R, Fazli L, et al. The androgen receptor fuels prostate cancer by regulating central metabolism and biosynthesis. EMBO J. 2011;30:2719–33. doi: 10.1038/emboj.2011.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tan PY, Chang CW, Chng KR, Wansa KDSA, Sung W-K, Cheung E. Integration of Regulatory Networks by NKX3-1 Promotes Androgen-Dependent Prostate Cancer Survival. Molecular and Cellular Biology. 2012;32:399–414. doi: 10.1128/MCB.05958-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sahu B, Laakso M, Ovaska K, Mirtti T, Lundin J, Rannikko A, et al. Dual role of FoxA1 in androgen receptor binding to chromatin, androgen signalling and prostate cancer. EMBO J. 2011;30:3962–76. doi: 10.1038/emboj.2011.328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang Q, Li W, Liu XS, Carroll JS, Janne OA, Keeton EK, et al. A hierarchical network of transcription factors governs androgen receptor-dependent prostate cancer growth. Mol Cell. 2007;27:380–92. doi: 10.1016/j.molcel.2007.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Massie CE, Adryan B, Barbosa-Morais NL, Lynch AG, Tran MG, Neal DE, et al. New androgen receptor genomic targets show an interaction with the ETS1 transcription factor. EMBO Rep. 2007;8:871–8. doi: 10.1038/sj.embor.7401046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baniwal SK, Khalid O, Sir D, Buchanan G, Coetzee GA, Frenkel B. Repression of Runx2 by androgen receptor (AR) in osteoblasts and prostate cancer cells: AR binds Runx2 and abrogates its recruitment to DNA. Mol Endocrinol. 2009;23:1203–14. doi: 10.1210/me.2008-0470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jia L, Berman BP, Jariwala U, Yan X, Cogan JP, Walters A, et al. Genomic androgen receptor-occupied regions with different functions, defined by histone acetylation, coregulators and transcriptional capacity. PLoS One. 2008;3:e3645. doi: 10.1371/journal.pone.0003645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blyth K, Cameron ER, Neil JC. The runx genes: gain or loss of function in cancer. Nat Rev Cancer. 2005;5:376–87. doi: 10.1038/nrc1607. [DOI] [PubMed] [Google Scholar]
- 11.Ito Y, George FVW, George K. RUNX Genes in Development and Cancer: Regulation of Viral Gene Expression and the Discovery of RUNX Family Genes. Advances in Cancer Research. 2008;99:33–76. doi: 10.1016/S0065-230X(07)99002-8. [DOI] [PubMed] [Google Scholar]
- 12.Chimge N-O, Frenkel B. The RUNX family in breast cancer: relationships with estrogen signaling. Oncogene. 2013;32:2121–30. doi: 10.1038/onc.2012.328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Komori T, Yagi H, Nomura S, Yamaguchi A, Sasaki K, Deguchi K, et al. Targeted Disruption of Cbfa1 Results in a Complete Lack of Bone Formation owing to Maturational Arrest of Osteoblasts. Cell. 1997;89:755–64. doi: 10.1016/s0092-8674(00)80258-5. [DOI] [PubMed] [Google Scholar]
- 14.Otto F, Thornell AP, Crompton T, Denzel A, Gilmour KC, Rosewell IR, et al. Cbfa1, a Candidate Gene for Cleidocranial Dysplasia Syndrome, Is Essential for Osteoblast Differentiation and Bone Development. Cell. 1997;89:765–71. doi: 10.1016/s0092-8674(00)80259-7. [DOI] [PubMed] [Google Scholar]
- 15.Pratap J, Wixted JJ, Gaur T, Zaidi SK, Dobson J, Gokul KD, et al. Runx2 Transcriptional Activation of Indian Hedgehog and a Downstream Bone Metastatic Pathway in Breast Cancer Cells. Cancer Research. 2008;68:7795–802. doi: 10.1158/0008-5472.CAN-08-1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Akech J, Wixted JJ, Bedard K, van der Deen M, Hussain S, Guise TA, et al. Runx2 association with progression of prostate cancer in patients: mechanisms mediating bone osteolysis and osteoblastic metastatic lesions. Oncogene. 2010;29:811–21. doi: 10.1038/onc.2009.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baniwal SK, Khalid O, Gabet Y, Shah RR, Purcell DJ, Mav D, et al. Runx2 transcriptome of prostate cancer cells: insights into invasiveness and bone metastasis. Mol Cancer. 2010;9:258. doi: 10.1186/1476-4598-9-258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Blyth K, Vaillant F, Jenkins A, McDonald L, Pringle MA, Huser C, et al. Runx2 in normal tissues and cancer cells: A developing story. Blood Cells, Molecules, and Diseases. 2010;45:117–23. doi: 10.1016/j.bcmd.2010.05.007. [DOI] [PubMed] [Google Scholar]
- 19.Onodera Y, Miki Y, Suzuki T, Takagi K, Akahira J-i, Sakyu T, et al. Runx2 in human breast carcinoma: its potential roles in cancer progression. Cancer Science. 2010;101:2670–5. doi: 10.1111/j.1349-7006.2010.01742.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chimge N-O, Baniwal S, Little G, Chen Y-b, Kahn M, Tripathy D, et al. Regulation of breast cancer metastasis by Runx2 and estrogen signaling: the role of SNAI2. Breast Cancer Research. 2011;13:R127. doi: 10.1186/bcr3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang H, Pan Y, Zheng L, Choe C, Lindgren B, Jensen ED, et al. FOXO1 Inhibits Runx2 Transcriptional Activity and Prostate Cancer Cell Migration and Invasion. Cancer Research. 2011;71:3257–67. doi: 10.1158/0008-5472.CAN-10-2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lim M, Zhong C, Yang S, Bell AM, Cohen MB, Roy-Burman P. Runx2 regulates survivin expression in prostate cancer cells. Lab Invest. 2009;90:222–33. doi: 10.1038/labinvest.2009.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chua C-W, Chiu Y-T, Yuen H-F, Chan K-W, Man K, Wang X, et al. Suppression of Androgen-Independent Prostate Cancer Cell Aggressiveness by FTY720: Validating Runx2 as a Potential Antimetastatic Drug Screening Platform. Clinical Cancer Research. 2009;15:4322–35. doi: 10.1158/1078-0432.CCR-08-3157. [DOI] [PubMed] [Google Scholar]
- 24.Yun SJ, Yoon HY, Bae SC, Lee OJ, Choi YH, Moon SK, et al. Transcriptional repression of RUNX2 is associated with aggressive clinicopathological outcomes, whereas nuclear location of the protein is related to metastasis in prostate cancer. Prostate Cancer Prostatic Dis. 2012;15:369–73. doi: 10.1038/pcan.2012.31. [DOI] [PubMed] [Google Scholar]
- 25.Khalid O, Baniwal SK, Purcell DJ, Leclerc N, Gabet Y, Stallcup MR, et al. Modulation of Runx2 Activity by Estrogen Receptor-α: Implications for Osteoporosis and Breast Cancer. Endocrinology. 2008;149:5984–95. doi: 10.1210/en.2008-0680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chimge N-O, Baniwal SK, Luo J, Coetzee S, Khalid O, Berman BPT, et al. Opposing Effects of Runx2 and Estradiol on Breast Cancer Cell Proliferation: In Vitro Identification of Reciprocally Regulated Gene Signature Related to Clinical Letrozole Responsiveness. Clinical Cancer Research 2012. 2012 Feb 1;18:901–11. doi: 10.1158/1078-0432.CCR-11-1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Frenkel B, Hong A, Baniwal SK, Coetzee GA, Ohlsson C, Khalid O, et al. Regulation of adult bone turnover by sex steroids. Journal of Cellular Physiology. 2010;224:305–10. doi: 10.1002/jcp.22159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Paredes R, Arriagada G, Cruzat F, Villagra A, Olate J, Zaidi K, et al. Bone-Specific Transcription Factor Runx2 Interacts with the 1α,25-Dihydroxyvitamin D3 Receptor To Up-Regulate Rat Osteocalcin Gene Expression in Osteoblastic Cells. Molecular and Cellular Biology. 2004;24:8847–61. doi: 10.1128/MCB.24.20.8847-8861.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kawate H, Wu Y, Ohnaka K, Takayanagi R. Mutual transactivational repression of Runx2 and the androgen receptor by an impairment of their normal compartmentalization. The Journal of Steroid Biochemistry and Molecular Biology. 2007;105:46–56. doi: 10.1016/j.jsbmb.2006.11.020. [DOI] [PubMed] [Google Scholar]
- 30.Baniwal SK, Little GH, Chimge N-O, Frenkel B. Runx2 controls a feed-forward loop between androgen and prolactin-induced protein (PIP) in stimulating T47D cell proliferation. Journal of Cellular Physiology. 2012;227:2276–82. doi: 10.1002/jcp.22966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McCarthy TL, Chang W-Z, Liu Y, Centrella M. Runx2 Integrates Estrogen Activity in Osteoblasts. Journal of Biological Chemistry. 2003;278:43121–9. doi: 10.1074/jbc.M306531200. [DOI] [PubMed] [Google Scholar]
- 32.van der Deen M, Akech J, Wang T, FitzGerald TJ, Altieri DC, Languino LR, et al. The cancer-related Runx2 protein enhances cell growth and responses to androgen and TGFbeta in prostate cancer cells. J Cell Biochem. 2010 Mar 1;109:828–37. doi: 10.1002/jcb.22463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Little GH, Noushmehr H, Baniwal SK, Berman BP, Coetzee GA, Frenkel B. Genome-wide Runx2 occupancy in prostate cancer cells suggests a role in regulating secretion. Nucleic Acids Research. 2012;40:3538–47. doi: 10.1093/nar/gkr1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang Y, Liu T, Meyer C, Eeckhoute J, Johnson D, Bernstein B, et al. Model-based Analysis of ChIP-Seq (MACS) Genome Biology. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Barski A, Cuddapah S, Cui K, Roh T-Y, Schones DE, Wang Z, et al. High-Resolution Profiling of Histone Methylations in the Human Genome. Cell. 2007;129:823–37. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 36.Tepper CG, Boucher DL, Ryan PE, Ma A-H, Xia L, Lee L-F, et al. Characterization of a Novel Androgen Receptor Mutation in a Relapsed CWR22 Prostate Cancer Xenograft and Cell Line. Cancer Research. 2002;62:6606–14. [PubMed] [Google Scholar]
- 37.Thomas MA, Preece DM, Bentel JM. Androgen regulation of the prostatic tumour suppressor NKX3. 1 is mediated by its 3′ untranslated region. Biochem J. 2010;425:575–83. doi: 10.1042/BJ20091109. [DOI] [PubMed] [Google Scholar]
- 38.Balbín M, López-Otín C. Hormonal Regulation of the Human Pepsinogen C Gene in Breast Cancer Cells. Journal of Biological Chemistry. 1996;27:15175–81. doi: 10.1074/jbc.271.25.15175. [DOI] [PubMed] [Google Scholar]
- 39.Wu K, Gore C, Yang L, Fazli L, Gleave M, Pong R-C, et al. Slug, a Unique Androgen-Regulated Transcription Factor, Coordinates Androgen Receptor to Facilitate Castration Resistance in Prostate Cancer. Molecular Endocrinology. 2012;26:1496–507. doi: 10.1210/me.2011-1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Peinado H, Olmeda D, Cano A. Snail, Zeb and bHLH factors in tumour progression: an alliance against the epithelial phenotype? Nat Rev Cancer. 2007;7:415–28. doi: 10.1038/nrc2131. [DOI] [PubMed] [Google Scholar]
- 41.Yilmaz M, Christofori G. EMT, the cytoskeleton, and cancer cell invasion. Cancer and Metastasis Reviews. 2009;28:15–33. doi: 10.1007/s10555-008-9169-0. [DOI] [PubMed] [Google Scholar]
- 42.Brubaker KD, Vessella RL, Brown LG, Corey E. Prostate cancer expression of runt-domain transcription factor Runx2, a key regulator of osteoblast differentiation and function. The Prostate. 2003;56:13–22. doi: 10.1002/pros.10233. [DOI] [PubMed] [Google Scholar]
- 43.Bowen C, Bubendorf L, Voeller HJ, Slack R, Willi N, Sauter G, et al. Loss of NKX3. 1 Expression in Human Prostate Cancers Correlates with Tumor Progression. Cancer Research. 2000;60:6111–5. [PubMed] [Google Scholar]
- 44.Steffan JJ, Koul HK. Prostate derived ETS factor (PDEF): A putative tumor metastasis suppressor. Cancer Letters. 2011;310:109–17. doi: 10.1016/j.canlet.2011.06.011. [DOI] [PubMed] [Google Scholar]
- 45.Emadi Baygi M, Soheili Z-S, Essmann F, Deezagi A, Engers R, Goering W, et al. Slug/SNAI2 regulates cell proliferation and invasiveness of metastatic prostate cancer cell lines. Tumor Biology. 2010;31:297–307. doi: 10.1007/s13277-010-0037-5. [DOI] [PubMed] [Google Scholar]
- 46.Liu YN, Yin JJ, Abou-Kheir W, Hynes PG, Casey OM, Fang L, et al. MiR-1 and miR-200 inhibit EMT via Slug-dependent and tumorigenesis via Slug-independent mechanisms. Oncogene. 2013;32:296–306. doi: 10.1038/onc.2012.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tomlins SA, Mehra R, Rhodes DR, Cao X, Wang L, Dhanasekaran SM, et al. Integrative molecular concept modeling of prostate cancer progression. Nat Genet. 2007;39:41–51. doi: 10.1038/ng1935. [DOI] [PubMed] [Google Scholar]
- 48.Liu Y-N, Abou-Kheir W, Yin JJ, Fang L, Hynes P, Casey O, et al. Critical and Reciprocal Regulation of KLF4 and SLUG in Transforming Growth Factor β-Initiated Prostate Cancer Epithelial-Mesenchymal Transition. Molecular and Cellular Biology. 2012;32:941–53. doi: 10.1128/MCB.06306-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.