Abstract
A set of 2-chloro-4-nitrophenyl glucosamino/xylosaminosides were synthesized and assessed as potential substrates in the context of glycosyltransferase-catalyzed formation of the corresponding UDP/TDP-α-D-glucosamino-/xylosaminosugars and single vessel transglycosylation reactions with a model acceptor. This study highlights a robust platform for aminosugar nucleotide synthesis and reveals OleD Loki as a proficient catalyst for U/TDP-aminosugar synthesis and utilization.
Keywords: glycobiology, enzyme, glycoside, glycosylation, glycorandomization, carbohydrate
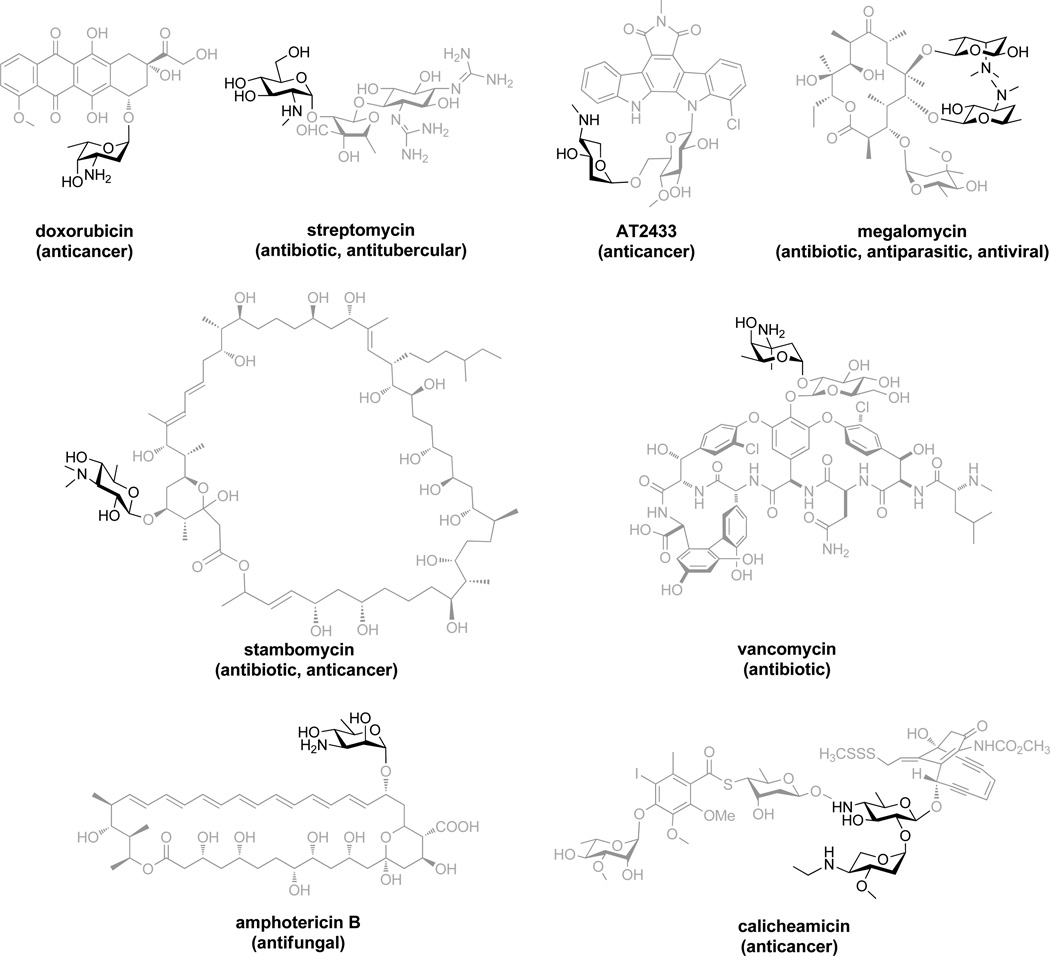
Aminosugars are ubiquitous in nature where they serve as functionally and/or structurally important building blocks for a range of biologically relevant glycoconjugates including peptidoglycans,[1] glycosaminoglycans,[2] aminoglycosides,[3] glycoproteins,[4] and glycosylated natural products (Scheme 1).[5] A unique feature of aminosugars is their enhanced solubility and potential for ionic interactions by virtue of the inherent positive charge under normal physiological conditions.[6] Within this context, aminosugar conjugation has been reported to improve the unconjugated parental compound’s basicity,[7] pharmacological properties,[8] and/or even alteration of mechanism.[9] Aminosugar conjugation can be accomplished via either chemical[10] or glycosyltransferase (GT)-catalyzed strategies,[11] the latter of which typically depends upon the availability of suitable aminosugar nucleotide donors. Yet, the reported syntheses of aminosugar nucleotides via chemical,[12] enzymatic,[13] or chemoenzymatic[14] strategies still typically are restricted to multi-step, low yielding processes and few, if any, are directly orthogonal to downstream GT-catalyzed reactions. A simple robust strategy directly compatible with downstream aminosugar nucleotide utilizing processes would therefore be considered advantageous.
scheme 1.
Representative aminosugar-appended natural products where aminosugars are highlighted in darker shade.
We recently reported simple aromatic glycosides to serve as efficient donors in glycosyltransferase-catalyzed reactions for both sugar nucleotide formation (i.e., the ‘reverse’ of a conventional GT-catalyzed reaction) and transglycosylation wherein the use of 2-chloro-4-nitrophenyl glycoside donors also offered a convenient colorimetric screen to enable the directed evolution of enhanced GTs with broad substrate permissivity.[15] Herein we describe an interrogation of two of the most permissive glycosyltransferase prodigy from these prior studies (OleD TDP-16 – a P67T/S132F/A242L/Q268V variant;[15a,16] OleD Loki – a P67T/I112P/T113M/S132F/A242I variant[15b]) for their abilities to catalyze the production of variant aminopentose and/or aminohexose nucleotides in the presence of the corresponding 2-chloro-4-nitrophenyl aminosugar donors and TDP/UDP. This study reveals OleD Loki to catalyze the conversion of 6 out of 7 simple D-glucosamino- and D-xylosamino-glycoside donors into their corresponding UDP/TDP-aminosugar nucleotides and also to utilize 6-azido/acetylamino-D-glucoside donors. Using 4-methylumbelliferone as a model acceptor, this study also highlights the efficient nucleotide-mediated single vessel transglycosylation with 5 of the 6 representative OleD Loki aminosugar substrates. In addition to providing a convenient strategy for novel aminosugar nucleotides, this work also sets the stage to assess the potential for OleD-catalyzed aminosugar conjugation to a range of bioactive natural products and drugs.[17]
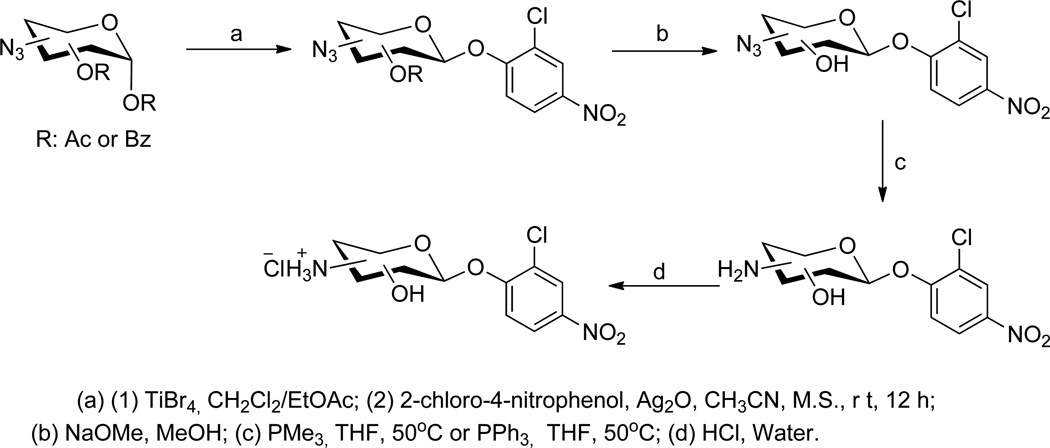
For this study, seventeen 2-chloro-4-nitrophenyl azidosugar and aminosugar glycoside donors were synthesized using a simple four-step synthesis (bromination, glycosylation,[18] deprotection, and reduction) from peracylated (acetyl or benzoyl) azidosugars with an overall average yield of 23%. The synthesized 2-chloro-4-nitrophenyl glucosamino/ xylosaminosides were subsequently converted to their corresponding hydrochloride salts in an average yield of 94% (Scheme 2, Table S1). In all cases, the corresponding 2-chloro-4-nitrophenyl glycosides were confirmed as the desired β-anomers. Additional analogs generated during synthetic methods development and included in this study include (2-chloro-4-nitrophenyl)-α-L/β-D-arabinoside (18, 18d), (2-chloro-4-nitrophenyl)-6-deoxy-6-N-acetylamino-β-D-glucoside (19), and (2-chloro-4-nitrophenyl)-2-deoxy-2-amino-α-D-glucoside (20) (Figure 1, Figure S1).
scheme 2.
Synthesis of 2-chloro-4-nitrophenyl aminosugar donors.
Figure 1.
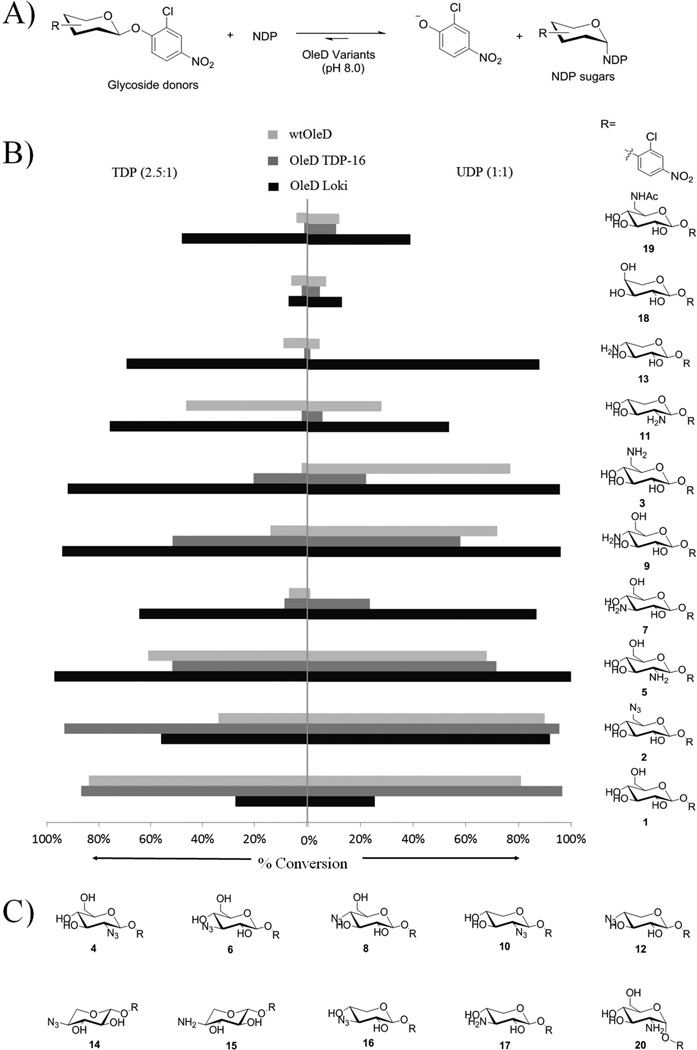
Comparison of catalysts in 2-chloro-4-nitrophenyl glycoside driven sugar nucleotide syntheses. A) General reaction scheme. B) Maximum observed percentage conversion of (U/T)DP to (U/T)DP-sugars by OleD Loki (dark), TDP-16 (light dark) and wtOleD (gray) (n ≥ 2, S. D. < 8%) (See Supplement methods). C) Additional 2-chloro-4-nitophenyl glycosides tested for which no products were observed within the sensitivity limits of the assay.
Three enzymes (wtOleD, OleD TDP-16, and OleD Loki) were selected to compare in the context of their potential to produce aminosugar nucleotides from the 2-chloro-4-nitrophenyl glycoside panel described above. Of these, TDP-16[16] and Loki[15b] are engineered variants of the Streptomyces antibioticus macrolide glucosyltransferase (wtOleD).[19] Standard conditions (10 µM OleD variant, 2 mM UDP or 5 mM TDP, 2 mM 2-chloro-4-nitophenyl glycoside, 50 mM Tris-HCl, pH 8.0, final volume of 100 µL, 25µC, 12 h followed by the RP HPLC analysis) were utilized to compare the turnover across the entire panel of enzyme/glycoside combinations. Figure 1 highlights the outcome of this cumulative study and reveals OleD Loki to have the broadest capacity for aminosugar conversion with all but one targeted free aminosugar donor (3-deoxy-3-amino-β-D-xyloside 17) leading to appreciable product (≥ 50%) in the presence of either UDP or TDP (Figure 1). An overall preference for glucosides (rank order of 6-NH2 ≈ 4-NH2 ≈ 2-NH2 > 3-NH2) over xylosides (rank order of 4-NH2 ≈ 2-NH2) was observed with no apparent difference between the donor free base and the corresponding hydrochloride salt (Table S2, Figure S2, S3, S4, S5). By comparison, both wtOleD and OleD TDP-16 were notably worse than OleD Loki with one exception (6-deoxy-6-azido-β-D-glucoside 2), a previously reported substrate of TDP-16,[15a] where TDP-16 was found to slightly outperform OleD Loki in this endpoint assay. In addition, OleD Loki displayed notable improvement with additional non-native donors beyond the scope of the targeted aminosugar series including 6-deoxy-6-N-acetylamino-β-D-glucoside 19 and slight improvement with α-L-arabinoside 18 - both analogs generated during the course of synthetic methods development. Intriguingly, both wtOleD and OleD TDP-16 outperformed OleD Loki with β-D-glucoside 1. As UDP-glucose is the native substrate of wtOleD,[19] this assessment suggests OleD Loki to offer a unique divergence in sugar specificity from wtOleD prodigy studied to date.
In the context of aminosugar nucleotide synthesis, this OleD catalyzed reversible reaction provides a noteworthy alternative to the synthesis of aminosugar nucleotides and compares favorably to prior precedent. For example, as comparison, prior chemical syntheses of the UDP-2-deoxy-2-amino-α-D-glucose and UDP-6-deoxy-6-amino-α-D-glucose from peracetylated azidosugar precursors required 6 steps with overall yields ranging from 4.5 – 20% and a lengthy (up to 5 days) key conjugation reaction between peracetylated azido-α-D-glucoside-1-phosphates and UMP-morpholidate.[20,21] The prior chemenzymatic syntheses of NDP-2-deoxy-2-amino-, 3-deoxy-3-amino-, 4-deoxy-4-amino-, and 6-deoxy-6-amino-α-D-glucose have also previously been accomplished via the use of an engineered α-D-glucose-1-phosphate thymidylyl-transferases (RmlA) with overall yields ranging from 5–24% (including up to 7 chemical transformations to provide the requisite aminosugar-α-1-phosphate substrates).[22] The current strategy affords the desired UDP/TDP-aminosugars in 7%–28% yield (including the simple four-step synthesis from peracylated azidosugars). Furthermore, given OleD Loki was evolved to also efficiently utilize ADP, CDP, and GDP,[15b] the current study suggests the potential to also employ OleD Loki for the corresponding syntheses of ADP-, CDP-, and/or GDP-aminosugars.
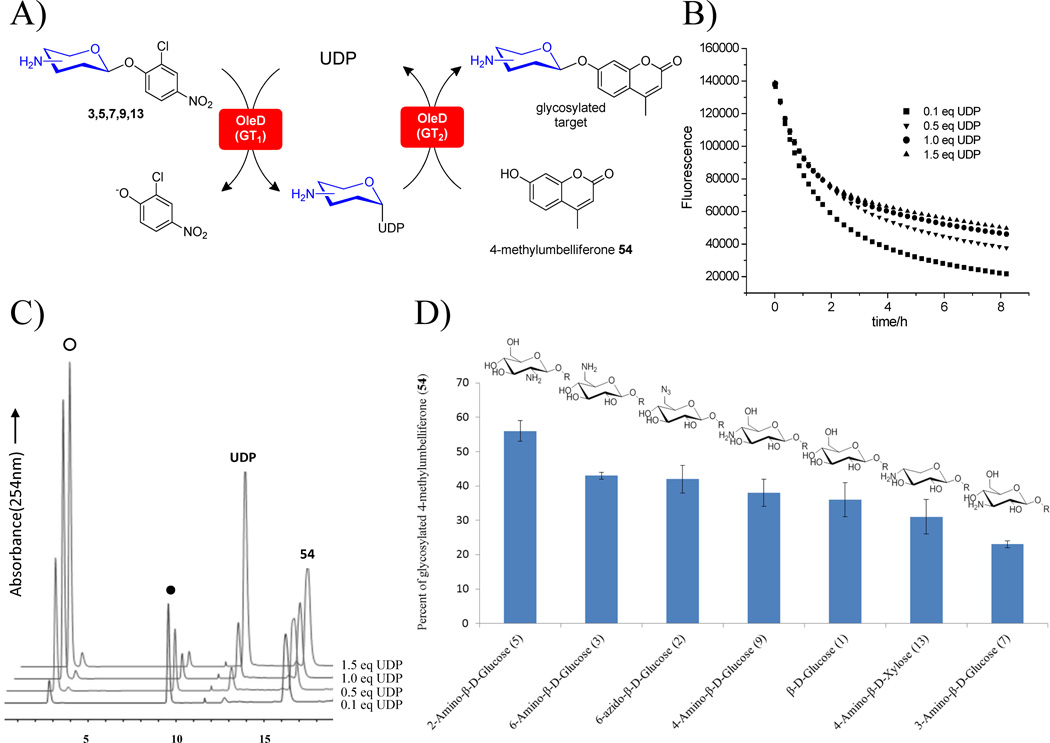
To assess the direct compatibility of this approach with a downstream coupled sugar nucleotide utilizing processes,[23] we examined the ability of the coupled OleD Loki-driven system to mediate the glycosylation of a model acceptor 4-methylumbelliferone 54 (Figure 2). The advantage of 4-methylumbelliferone as a surrogate acceptor is its inherent fluorescence. Specifically, glycosylation of the 4-methylumbelliferone C7-OH extinguishes fluorescence, thereby enabling a highly sensitive fluorescent-based continuous GT assay.[24] To set the stage for this assessment, the UDP concentration was first optimized in the context of the coupled reaction to afford the greatest transglycosylation output (i.e., the best 4-methyumbelliferone glycoside formation) in the presence of (2-chloro-4-nitrophenyl)-2-deoxy-2-amino-β-D-glucoside 5 as a representative aminosugar donor (Figure 2B, 2C). The optimization series [10 µM OleD Loki, 1 mM 4-methylumbelliferone 54, 1 mM 2-deoxy-2-amino-β-D-glucoside 5 and variant UDP (0.1 – 1.5 mM) in 50 mM Tris-HCl, pH 8.0 with a final volume of 100 µL] revealed 0.1 eq UDP as the optimal relative concentration to support the coupled transglycosylation process. Using this optimized protocol, the coupled system was subsequently examined in the context of seven 2-chloro-4-nitrophenyl glycoside donors including the five established aminosugar donors for aminosugar nucleotide synthesis (amino-β-D-glucosides 3, 5, 7, 9 and 4-deoxy-4-amino-β-D-xyloside 13), 6-deoxy-6-azido-β-D-glucoside 2 and β-D-glucoside 1 (Figure 2D). Overall, the trends for transglycosylation generally paralleled that for sugar nucleotide formation highlighted in Figure 1 with a general preference for hexose over pentose congeners and 2-/6-amination favored over the corresponding 3-/4-substitution. Importantly, this assessment confirms that the OleD-catalyzed NDP-aminosugar production strategy can be directly coupled to the downstream sugar nucleotide-utilizing applications.
Figure 2.
Single enzyme coupled UDP-mediated transglycosylation reaction of 4-methylumbelliferone 54 with 2-chloro-4-nitrophenyl glucosamino/xylosaminosides. A) General reaction scheme. B) Time course of 5 fluorescence as a measure of reaction progress over 8 h with different UDP concentrations. C) HPLC chromatogram of the final reaction mixtures from panel c after 8 h where the solid circle (●) denotes the 54 glycoside product and (○) represents UDP-2-deoxy-2-amino-α-D-glucose. D) Percentage conversion to 54 glycoside products via OleD Loki-catalyzed NDP-mediated transglycosylation containing 0.1 mM UDP, 1 mM 54, and 1 mM 2-chloro-4-nitrophenyl glycoside donors (1, 2, 3, 5, 7, 9, 13) (n ≥ 2. S. D. < 5%).
Inspired by the ability of diverse simple ‘activated’ donors to modulate the thermodynamics of GT-catalyzed reactions, this work highlights the first systematic interrogation of the most proficient/permissive OleD variants in the context of aminosugar nucleotide formation and utilization. This study revealed OleD Loki to slightly outperform OleD TDP-16 in nearly all standard endpoint assays conducted and to serve as an efficient catalyst for the production of 12 out of 14 targeted UDP/TDP-α-D-glucosamino-/xylosaminosugars from a series of simple 2-chloro-4-nitrophenyl glucosamino/xylosaminoside donors. In addition, OleD Loki also enabled the subsequent production of the corresponding set of model 4-methylumbelliferone glucosamino/xylosaminosides in a series of model UDP-mediated transglycosylation reactions. As such, this work notably highlights an efficient platform for UDP-/TDP-(and potentially ADP-/GDP-/CDP-) aminosugar production that is directly orthogonal to subsequent sugar nucleotide-dependent reactions relevant to a range of glycoconjugation and glycobiology applications.
Supplementary Material
Acknowledgements
We thank the School of Pharmacy Analytical Instrumentation Center (University of Wisconsin-Madison) for analytical support and Dr. Pauline Peltier-Pain and Dr. Richard W. Gantt for valuable consultation.This work was supported by NIH R37 AI52218 (JST) and the National Center for Advancing Translational Sciences (UL1TR000117).
Footnotes
Supporting information for this article is available on the WWW under http://dx.doi.org/10.1002/cbic.20xxxxxxx
References
- 1.a) Van. Heijenoort J. Glycobiology. 2001;11:25R–36R. doi: 10.1093/glycob/11.3.25r. [DOI] [PubMed] [Google Scholar]; b) Gust B, Eitel K, Tang X. Biol. Chem. 2013;394:251–259. doi: 10.1515/hsz-2012-0274. [DOI] [PubMed] [Google Scholar]
- 2.a) Kadokawa J. Chem. Rev. 2011;111:4308–4345. doi: 10.1021/cr100285v. [DOI] [PubMed] [Google Scholar]; b) DeAngelis PL, Liu J, Linhardt RJ. Glycobiology. 2013;23:764–777. doi: 10.1093/glycob/cwt016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.a) Kudo F, Kawabe K, Kuriki H, Eguchi T, Kakinuma K. J. Am. Chem. Soc. 2005;127:1711–1718. doi: 10.1021/ja044921b. [DOI] [PubMed] [Google Scholar]; b) Park SR, Park JW, Ban YH, Sohng JK, Yoon YJ. Nat. Prod. Rep. 2013;30:11–20. doi: 10.1039/c2np20092a. [DOI] [PubMed] [Google Scholar]
- 4.a) Schmaltz RM, Hanson SR, Wong C–H. Chem. Rev. 2011;111:4259–4307. doi: 10.1021/cr200113w. [DOI] [PubMed] [Google Scholar]; b) Iwashkiw JA, Vozza NF, Kinsella RL, Feldman MF. Mol. Microbiol. 2013;89:14–28. doi: 10.1111/mmi.12265. [DOI] [PubMed] [Google Scholar]
- 5.a) Thorson JS, Hosted TJ, Jiang J, Biggins JB, Ahlert J. Curr. Org. Chem. 2001;5:139–167. [Google Scholar]; b) Flatt PM, Mahmud T. Nat. Prod. Rep. 2007;24:358–392. doi: 10.1039/b603816f. [DOI] [PubMed] [Google Scholar]; c) Timmons SC, Thorson JS. Curr. Opin. Chem. Biol. 2008;12:297–305. doi: 10.1016/j.cbpa.2008.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Mahmud T. Curr. Opin. Chem. Biol. 2009;13:161–170. doi: 10.1016/j.cbpa.2009.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]; e) Lin CI, McCarty RM, Liu H–w. Chem. Soc. Rev. 2013;42:4377–4407. doi: 10.1039/c2cs35438a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.a) La Ferla BL, Airoldi C, Zona C, Orsato A, Cardona F, Merlo S, Sironi E, D’Orazio G, Nicotra F. Nat. Prod. Rep. 2011;28:630–648. doi: 10.1039/c0np00055h. [DOI] [PubMed] [Google Scholar]; b) Das I, Desire J, Manvar D, Baussanne I, Pandey VN, Decout J–L. J. Med. Chem. 2012;55:6021–6032. doi: 10.1021/jm300253q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pedersen CM, Olsen J, Brka AB, Biols M. Chem. Eur. J. 2011;17:7080–7086. doi: 10.1002/chem.201100020. [DOI] [PubMed] [Google Scholar]
- 8.a) Weymouth-Wilson AC. Nat. Prod. Rep. 1997;14:99–110. doi: 10.1039/np9971400099. [DOI] [PubMed] [Google Scholar]; b) Matsushima Y, Nakayama T, Fujita M, Bhandari R, Eguchi T, Shindo K, Kakinuma K. J. Antibiot. 2001;54:211–219. doi: 10.7164/antibiotics.54.211. [DOI] [PubMed] [Google Scholar]; c) Fu X, Albermann C, Jiang J, Liao J, Zhang C, Thorson JS. Nat. Biotechnol. 2003;21:1467–1469. doi: 10.1038/nbt909. [DOI] [PubMed] [Google Scholar]; d) Lin Y, Jones GB, Hwang G, Kappen L, Goldberg IH. Org. Lett. 2005;7:71–74. doi: 10.1021/ol0400591. [DOI] [PubMed] [Google Scholar]; e) Siitonen V, Claesson M, Patrikainen P, Aromaa M, Mantsala P, Schneider G, Metsa-Ketela M. Chembiochem. 2012;13:120–128. doi: 10.1002/cbic.201100637. [DOI] [PubMed] [Google Scholar]
- 9.a) Croatt MP, Carreira EM. Org. Lett. 2011;13:1390–1393. doi: 10.1021/ol2000765. [DOI] [PubMed] [Google Scholar]; b) Bai L, Ho H, Ma D, Fu HYW, Jiang Z. PloS. ONE. 2013;8:e53962. doi: 10.1371/journal.pone.0053962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.a) Galonic DP, Gin DY. Nature. 2007;446:1000–1007. doi: 10.1038/nature05813. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Boltje TJ, Buskas T, Boons GJ. Nat. Chem. 2009;1:611–622. doi: 10.1038/nchem.399. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Wagner GK, Pesnot T, Field RA. Nat. Prod. Rep. 2009;26:1172–1194. doi: 10.1039/b909621n. [DOI] [PubMed] [Google Scholar]; d) Fraser-Reid B, Lopez JC. Top. Curr. Chem. 2011;301:1–29. doi: 10.1007/128_2010_105. [DOI] [PubMed] [Google Scholar]; e) Zhang J, Ponomareva LV, Marchillo K, Zhou M, Andes DR, Thorson JS. J. Nat. Prod. 2013;76:1627–1636. doi: 10.1021/np4003096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.a) Davis BG, Boyer V. Nat. Prod. Rep. 2001;18:618–640. doi: 10.1039/b003667f. [DOI] [PubMed] [Google Scholar]; b) Gantt RW, Peltier-Pain P, Thorson JS. Nat. Prod. Rep. 2011;28:1811–1853. doi: 10.1039/c1np00045d. [DOI] [PubMed] [Google Scholar]; c) Sanchez-Moreno I, Oroz-Guinea I, Iturrate L, Garcia-Junceda E. Comprehensive Chirality. 2012;7:430–453. [Google Scholar]; d) Davids T, Schmidt M, Bottcher D, Bornscheuer UT. Curr. Opin. Chem. Biol. 2013;17:215–220. doi: 10.1016/j.cbpa.2013.02.022. [DOI] [PubMed] [Google Scholar]
- 12.a) Sala RF, MacKinnon SL, Palcic MM, Tanner ME. Carbohydrate Res. 1998;306:127–136. doi: 10.1016/s0008-6215(97)10033-7. [DOI] [PubMed] [Google Scholar]; b) Weiwer M, Sherwood T, Green DE, Chen M, DeAngelis PL, Liu J, Linhardt RJ. J. Org. Chem. 2008;73:7631–7637. doi: 10.1021/jo801409c. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Danac R, Ball L, Gurr SJ, Rairbanks AJ. Carbohydrate Res. 2008;343:1012–1022. doi: 10.1016/j.carres.2008.01.031. [DOI] [PubMed] [Google Scholar]; d) Wagner GK, Pesnot T, Field RA. Nat. Prod. Rep. 2009;26:1172–1194. doi: 10.1039/b909621n. [DOI] [PubMed] [Google Scholar]
- 13.a) Hong L, Zhao Z, Melancon CE, III, Zhang H, Liu H–w. J. Am. Chem. Soc. 2008;130:4954–4967. doi: 10.1021/ja0771383. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Gu X, Glushka J, Lee SG, Bar-Peled M. J. Biol. Chem. 2010;285:24825–24833. doi: 10.1074/jbc.M110.125872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.a) Jiang J, Albermann C, Thorson JS. ChemBioChem. 2003;4:443–446. doi: 10.1002/cbic.200200566. [DOI] [PubMed] [Google Scholar]; b) Moretti R, Thorson JS. J. Biol. Chem. 2007;282:16942–16947. doi: 10.1074/jbc.M701951200. [DOI] [PubMed] [Google Scholar]; c) Jakeman DL, Young JL, Huestis MP, Peltier P, Daniellou R, Nugier-Chauvin C, Ferrieres V. Biochemistry. 2008;47:8719–8725. doi: 10.1021/bi800978u. [DOI] [PubMed] [Google Scholar]; d) Williams GJ, Gantt RW, Thorson JS. Curr. Opin. Chem. Biol. 2008;12:556–564. doi: 10.1016/j.cbpa.2008.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]; e) Guan W, Cai L, Fang J, Wu B, Wang PG. Chem. Commun. 2009;45:6976–6978. doi: 10.1039/b917573c. [DOI] [PubMed] [Google Scholar]; f) Mizanur RM, Pohl NL. Org. Biol. Chem. 2009;7:2135–2139. doi: 10.1039/b822794b. [DOI] [PubMed] [Google Scholar]; g) Guan W, Cai L, Wang PG. Chem. Eur. J. 2010;16:13343–13345. doi: 10.1002/chem.201002315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.a) Gantt RW, Peltier-Pain P, Cournoyer WJ, Thorson JS. Nat. Chem. Biol. 2011;7:685–691. doi: 10.1038/nchembio.638. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Gantt RW, Peltier-Pain P, Singh S, Zhou M, Thorson JS. Proc. Nat. Acad. Soc. USA. 2013;110:7648–7653. doi: 10.1073/pnas.1220220110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Williams GJ, Yang J, Zhang C, Thorson JS. ACS Chem. Biol. 2011;6:95–100. doi: 10.1021/cb100267k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.a) Gantt RW, Goff RD, Williams GJ, Thorson JS. Angew. Chem. Int. Ed. 2008;47:8889–8892. doi: 10.1002/anie.200803508. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Zhou M, Thorson JS. Org. Lett. 2011;13:2786–2788. doi: 10.1021/ol200977u. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Zhou M, Hamza A, Zhan C–G, Thorson JS. J. Nat. Prod. 2013;76:279–286. doi: 10.1021/np300890h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.For Koenigs-Knorr reaction that using Ag2O, see: Takeo K. Carbohydrate Res. 1977;59:258–260. Takeo K. Carbohydrate Res. 1979;77:131–140. Gouliaras C, Lee D, Chan L, Taylor MS. J. Am. Chem. Soc. 2011;133:13926–13929. doi: 10.1021/ja2062715. Lichtenthaler FW. Chem. Rev. 2011;111:5569–5609. doi: 10.1021/cr100444b. Monch B, Gebert A, Emmerling F, Becker R, Nehls I. Carbohydrate Res. 2012;352:186–190. doi: 10.1016/j.carres.2012.01.002.
- 19.a) Hernandez C, Olano C, Mendez C, Salas JA. Gene. 1993;134:139–140. doi: 10.1016/0378-1119(93)90189-a. [DOI] [PubMed] [Google Scholar]; b) Quiros LM, Carbajo RJ, Brana AF, Salas JA. J. Biol. Chem. 2000;275:11713–11720. doi: 10.1074/jbc.275.16.11713. [DOI] [PubMed] [Google Scholar]
- 20.Masuko S, Bera S, Green DE, Weiwer M, Liu J, DeAngelis PL, Linhardt RJ. J. Org. Chem. 2012;77:1449–1456. doi: 10.1021/jo202322k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Losey HC, Jiang J, Biggins JB, Oberthur M, Ye X–Y, Thorson JS, Walsh CT. Chem. Biol. 2002;9:1305–1314. doi: 10.1016/s1074-5521(02)00270-3. [DOI] [PubMed] [Google Scholar]
- 22.Jiang J, Biggins JB, Thorson JS. Angew. Chem. Int. Ed. 2001;40:1502–1505. [PubMed] [Google Scholar]
- 23.a) Lougheed B, Ly HD, Wakarchuk WW, Withers SG. J. Biol. Chem. 1999;274:37717–37722. doi: 10.1074/jbc.274.53.37717. [DOI] [PubMed] [Google Scholar]; b) Minami A, Kakinuma K, Eguchi T. Tetrahedron Lett. 2005;46:6187–6190. [Google Scholar]; c) Zhang C, Griffith BR, Fu Q, Albermann C, Fu X, Lee I, Li L, Thorson JS. Science. 2006;313:1291–1294. doi: 10.1126/science.1130028. [DOI] [PubMed] [Google Scholar]; d) Melancon CE, Thibodeaux CJ, Liu H–w. ACS Chem. Biol. 2006:1499–1504. doi: 10.1021/cb600365q. [DOI] [PubMed] [Google Scholar]; e) Borisova SA, Zhang C, Takahashi H, Zhang H, Wong AW, Thorson JS, Liu H-w. Angew. Chem. Int. Ed. 2006;45:2748–2753. doi: 10.1002/anie.200503195. [DOI] [PubMed] [Google Scholar]; f) Zhang C, Albermann C, Fu X, Thorson JS. J. Am. Chem. Soc. 2006;128:16420–16421. doi: 10.1021/ja065950k. [DOI] [PubMed] [Google Scholar]; g) Zhang, Fu Q, Albermann C, Li L, Thorson JS. ChemBioChem. 2007;8:385–390. doi: 10.1002/cbic.200600509. [DOI] [PubMed] [Google Scholar]; h) Bode HB, Muller R. Angew. Chem. Int. Ed. 2007;46:2147–2150. doi: 10.1002/anie.200604671. [DOI] [PubMed] [Google Scholar]; i) Lairson LL, wakarchuk WW, Withers SG. Chem Commun. 2007:365–367. doi: 10.1039/b614636h. [DOI] [PubMed] [Google Scholar]; j) Zhang C, Moretti R, Jiang J, Thorson JS. ChemBioChem. 2008;9:2506–2541. doi: 10.1002/cbic.200800349. [DOI] [PMC free article] [PubMed] [Google Scholar]; k) Peltier-Pain P, Marchillo K, Zhou M, Andes DR, Thorson JS. Org. Lett. 2012;14:5086–5089. doi: 10.1021/ol3023374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.a) Williams GJ, Zhang C, Thorson JS. Nat. Chem. Biol. 2007;3:657–662. doi: 10.1038/nchembio.2007.28. [DOI] [PubMed] [Google Scholar]; b) Gantt RW, Thorson JS. Methods Enzymol. 2012;516:345–360. doi: 10.1016/B978-0-12-394291-3.00009-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.