Abstract
A customized metabolomics NMR database, termed 1H(13C)-TOCCATA, is introduced, which contains complete 1H and 13C chemical shift information on individual spin systems and isomeric states of common metabolites. Since this information directly corresponds to cross sections of 2D 1H–1H TOCSY and 2D 13C–1H HSQC-TOCSY spectra, it allows the straightforward and unambiguous identification of metabolites of complex metabolic mixtures at 13C natural abundance from these types of experiments. The 1H(13C)-TOCCATA database, which is complementary to the previously introduced TOCCATA database for the analysis of uniformly 13C-labeled compounds, currently contains 455 metabolites, and it can be used through a publicly accessible web portal. We demonstrate its performance by applying it to 2D 1H–1H TOCSY and 2D 13C–1H HSQC-TOCSY spectra of a cell lysate from E. coli, which yields a substantial improvement over other databases, as well as 1D NMR-based approaches, in the number of compounds that can be correctly identified with high confidence.
Over the past decade, NMR spectroscopy has become one of two main analytical techniques for metabolomics studies in the absence of extensive compound extraction and physical separation.1,2 The high-resolution information offered by NMR is the key for the identification and quantification of metabolites, which is the primary goal of most metabolomics studies.3 The retrieval of such information from one-dimensional (1D) NMR spectra of complex real-life mixtures can be very challenging because of the high frequency of overlapping resonances that belong to different compounds.4 Moreover, the lack of connectivity information on spins belonging to the same compound limits the combined use of multiple resonances as unique compound fingerprints.5 The availability of such connectivity information provides significant advantages for the identification and quantification of metabolites.6,7 Specifically, simultaneous searching of multiple peaks of a metabolite against a NMR database substantially improves the uniqueness and accuracy of the hits.8 For uncatalogued metabolites, connectivities provide information about chemical bonds and an opportunity for de novo elucidation of the backbone topology and the structure of metabolites.9 Once the connectivities are known, the accuracy of metabolite quantitation can be enhanced through coanalysis of multiple peaks of a given metabolite.10,11 Finally, connectivities can be used effectively for the deconvolution of complex mixtures using multidimensional NMR experiments.12
Although the use of multidimensional NMR experiments requires longer measurement times, it can overcome many of the limitations of 1D NMR.13 In a 2D NMR spectrum, cross-peaks belonging to spins whose resonances overlap in a 1D NMR spectrum are spread out along the indirect dimension thereby reducing the likelihood of peak overlap. A 2D 13C–1H HSQC spectrum,14 for example, provides excellent spectral dispersion along the indirect 13C dimension and allows the separation of many of the peaks that overlap in a 1D 1H NMR spectrum. Several NMR metabolomics databases and queries permit identification of peaks of 2D 13C–1H HSQC spectra.15−18 They all accept a list of the cross-peaks observed in the 2D 13C–1H HSQC spectrum of the mixture and perform a cross-peak by cross-peak match against the database entries. Although the introduction of the indirect 13C dimension increases the resolution in this approach, the lack of connectivity information between different 1H, 13C pairs belonging to the same molecule can cause ambiguities for peak annotation and metabolite identification analogous to 1D NMR.
Connectivity information between different resonances of a molecule is available in TOCSY spectra collected at long mixing times.19 Since TOCSY traces only correlate resonances with each other that belong to the same spin system, for molecules that have multiple spin systems or exist in multiple slowly interconverting isomeric forms, these traces represent only part of the entire 1D NMR spectrum. Therefore, their query against a NMR database consisting of entire 1D NMR spectra of metabolites leads to imperfect matches, carrying the risk of false interpretations.20 Because public NMR databases so far do not sort spins into individual spin systems or multiple slowly exchanging isomers for separate queries, we recently introduced a customized metabolite database, termed TOCCATA.20 This database is specifically geared toward the query of 13C TOCSY traces extracted from TOCSY experiments that directly employ magnetization transfer between 13C spins without the involvement of their attached protons. These experiments are 13C–13C CT-TOCSY,2113C–13C TOCSY,19 and even 13C–13C COSY22 after the user has established complete chemical shift lists of each spin system from a “COSY-walk” along directly coupled 13C spins. TOCCATA uses 13C chemical shift information for the reliable identification of metabolites, their isomeric states and spin systems. For a fully 13C labeled E. coli cell extract, querying with TOCCATA provided more than 30% improvement in matching accuracy over existing 1D 13C NMR web servers.20
TOCCATA can be generally used for metabolomics analysis of uniformly 13C labeled organisms such as bacteria, yeast, C. elegans, and plants. For more complex organisms, including humans, for which 13C labeling is not feasible, 1H-TOCSY-type experiments can be used instead, in particular 2D 1H–1H TOCSY and 2D 13C–1H HSQC-TOCSY.23 Here, we present a customized database for these types of experiments, which transfer magnetization by TOCSY via the 1H spins. In order to clearly distinguish between the new and the original TOCCATA database, we call the new database “1H(13C)-TOCCATA,” while we refer to the original database as “13C-TOCCATA.”
The new 1H(13C)-TOCCATA database stores the information content of TOCSY traces in the form of individual spin systems and/or multiple slowly exchanging isomers for separate queries. It therefore allows the querying of 1H TOCSY traces from 2D 1H–1H TOCSY spectra as well as the querying of 1H HSQC-TOCSY traces (rows) and 13C HSQC-TOCSY traces (columns) from 2D 13C–1H HSQC-TOCSY spectra. The performance of the new database is demonstrated for an E. coli cell lysate, which resulted in the accurate identification of over 50 metabolites from a single sample.
Results and Discussion
Generation of 1H(13C)-TOCCATA Database
The new 1H(13C)-TOCCATA database was derived primarily from the BMRB15 and HMDB17 metabolomics databases and it presently contains 455 compounds. From these 455 compounds, 219 contain a single spin system and adopt a single isomeric state, 199 compounds consist of more than one spin system in a single (isomeric) state, 24 compounds consist of a single spin system in multiple isomeric states, and 13 compounds consist of multiple states and multiple spin systems (Table S-1). This means that TOCSY traces of more than half of the metabolites in the new database, namely 236, cannot be matched with databases derived from 1D NMR data.
The new database is organized as follows. First, all 455 compounds were subdivided into their isomeric states, which were then further subdivided into individual spin systems. Each 1H chemical shift is stored together with the chemical shift of its directly attached 13C. This allows the extraction of complete 1D 1H TOCSY, 1D 1H HSQC-TOCSY, and 1D 13C HSQC-TOCSY traces for each spin system or isomeric state. 1D 1H TOCSY traces are used for the query of a 2D 1H–1H TOCSY spectrum, whereas 1D 1H and 13C HSQC-TOCSY traces are used to query cross sections along the direct and indirect dimensions of a 2D 13C–1H HSQC-TOCSY spectrum, respectively. It should be noted that in the absence of overlaps, the information content about a spin system in a 1D 1H TOCSY trace and a 1D 1H HSQC-TOCSY trace are the same.
1D 13C HSQC-TOCSY traces from 2D 13C–1H HSQC-TOCSY and 1D 13C TOCSY traces from 2D 13C–13C CT-TOCSY spectra are not necessarily the same, because in 13C–1H HSQC-TOCSY, the TOCSY-magnetization transfer is mediated by the 1H spins, whereas in 2D 13C–13C CT-TOCSY, the TOCSY magnetization is mediated by the 13C spins. This leads to distinct spectral differences for metabolites with nonprotonated carbons. Nonprotonated carbons are not displayed in 13C–1H HSQC-TOCSY spectra, but they appear in 2D 13C–13C CT-TOCSY spectra.9 Furthermore, a nonprotonated carbon may break up a molecule into two separate 13C traces in 13C–1H HSQC-TOCSY spectra, but not in 2D 13C–13C CT-TOCSY spectra. Hence, 1D 13C HSQC-TOCSY traces from 2D 13C–1H HSQC-TOCSY spectra cannot always be identified using our previous 13C-TOCCATA database20 with optimal accuracy, which explains the need to include 13C traces in the 1H(13C)-TOCCATA database. A comparison of the performance of 1H(13C)-TOCCATA and 13C-TOCCATA databases for the analysis of 2D 13C–1H HSQC-TOCSY spectra is provided in the section “Application of 1H(13C)-TOCCATA to E. coli Cell Lysate” (see below).
In our previous 13C-TOCCATA work, we used the fact that 1J(13C–13C) couplings are generally much larger than 2J(13C–13C) and 3J(13C–13C) couplings. Therefore, we divided a molecule into two (or more) spin systems when two carbons are separated by at least one noncarbon atom.20 For protons, this step requires modification, because neighboring protons that are still part of the same spin system are at least by two and three bonds apart. Hence, the spin system definition for protons is based on a contiguous spin network of 2J(1H–1H) and 3J(1H–1H) couplings. We observed that for most metabolites this rule agrees well with the cross-peak patterns of experimental 2D 1H–1H TOCSY spectra collected at a mixing time of ∼60–90 ms. However, there are some exceptions, such as in ring fragments of some of the metabolites, where four bond 4J(1H–1H) couplings can be quite strong,24 which creates additional cross-peaks in the 1H TOCSY spectra. Nicotinic acid is one of these exceptions, as is demonstrated in the Supporting Information Figure S-1: protons located in the structure of nicotinic acid at positions 4, 5, and 6 in Figure S-1A theoretically belong to the same spin system, while the proton located at position 3 (Figure S-1A) constitutes a separate spin system. However, the 1H–1H TOCSY spectrum of nicotinic acid taken from the BMRB (Figure S-1D) shows that protons located at positions 3, 4, 5, and 6 all belong to a single spin system. Therefore, the experimental verification of each 1H TOCSY spin system was required when assembling this customized database. For all 455 metabolites (Table S-1), spin system identification was based on the manual inspection of their 2D 1H–1H TOCSY spectra in the BMRB and HMDB. This definition of spin systems yielded a total of 846 different spin systems. A specifically designed web portal at http://spin.ccic.ohio-state.edu/index.php/toccata2/index allows querying of the 1H(13C)-TOCCATA database either using a 1H or 13C chemical shift list of a given spin system extracted from 1H–1H TOCSY and/or 13C–1H HSQC-TOCSY spectra.
The chemical shift assignments of all compounds in the new database were done manually by the extraction of spectral information from BMRB, HMDB, and the literature. Only NMR data of compounds dissolved in H2O/D2O at pH 7.0 or 7.4 were included in the new database. The new web server shares many of its querying features with the 13C-TOCCATA database. For instance, it allows users to specify the spectral range on which the database query should be performed by entering the most downfield and most upfield frequencies in parts per million (ppm). This feature can be used to eliminate potential mismatches arising from far off-resonance nuclei not detected in the TOCSY or HSQC-TOCSY experiment, but which are present in the database. Ideally, the number of query peaks is identical to the number of resonances of the best matching spin system. However, this is not always the case, because, e.g., a peak was missing in the query trace or because two multiplet components of the same resonance were assigned to two different chemical shifts. To facilitate the analysis of mismatches, the web server allows the user to specify a maximally tolerable mismatch Mmax, which is the absolute value of the difference between the number of query peaks and the number of resonances of the spin system in the database. If the user is confident that all query peaks were correctly identified, then a mismatch parameter Mmax = 0 should be entered (default value). The origin of a mismatch larger than zero should always be traced back in the original spectrum to prevent false identifications.
An important prerequisite for the querying of NMR chemical shifts is that they are properly referenced. Ideally, the chemical shifts are referenced against standard compounds, such as 4,4-dimethyl-4-silapentane-1-sulfonic acid (DSS) or tetramethylsilane (TMS). In the case that no standard was used, the web server permits the user to enter a chemical shift offset value (“Reference correction,” default 0.00 ppm) in order to reference a spectrum by uniformly increasing or decreasing the chemical shifts of all metabolite signals in the spectrum by the entered chemical shift offset. To find the minimum root-mean-square deviation (RMSD) for every metabolite, the matching algorithm performs an automated alignment with a tolerance of ±0.2 ppm for 1H and ±0.6 ppm for 13C before applying a weighted matching algorithm25 to find the best matching peak pairs from the query list and the database. Finally, the average chemical shift RMSD between input and database peak pairs is computed and used as a criterion for the identification of the best match, which will be then be returned to the user.
In our experience, the database query is most accurate when Mmax = 0 and RMSD < 0.02 ppm for 1H and <0.2 ppm for 13C (default values). If none of the database entries satisfies the above criteria, the query returns “no match.” When multiple matches are returned, they are rank-ordered according to increasing RMSDs. Concise information about the number of isomeric states and spin systems of a compound is displayed for the top four returns.
Limitation of 1D 1H NMR Approaches for Metabolite Identification in E. coli
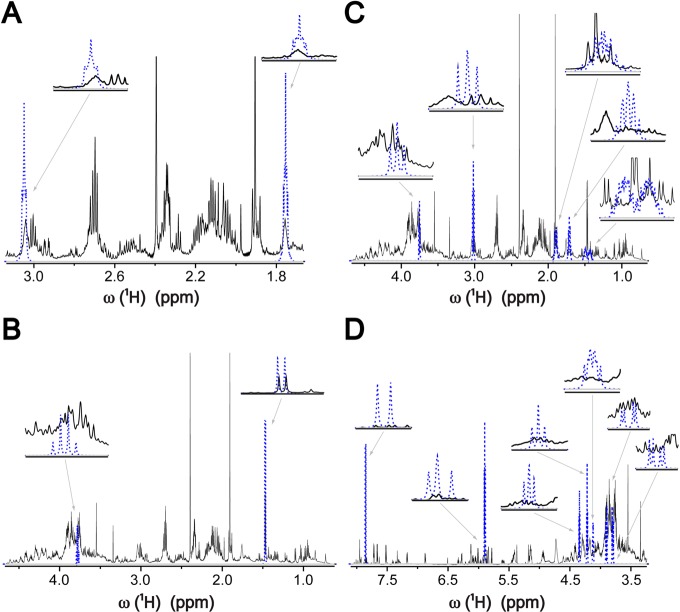
The standard 1D 1H NMR approach for metabolite identification relies on overlaying a 1D 1H NMR spectra of pure metabolites one by one with the experimental 1D 1H NMR mixture spectrum, which is implemented, for example, in the Chenomx NMR Suite software (Edmonton, AB, Canada), which is one of the most commonly used commercial software packages in the field. We acquired a 1D 1H NMR spectrum of E. coli cell lysate and tested the 1D 1H NMR approach by using the Chenomx software, which resulted in the observation of NMR peaks of 19 metabolites (Table S-2). However, for the majority of these metabolites, the identification was ambiguous, because of the strong peak overlaps in the 1D 1H NMR spectrum (Figure S-2), which resulted in the successful matching of only a subset of the peaks of a metabolite. For instance out of the 19 metabolites, 13 have multiple 1H signals, but only for putrescine and uracil all 1H signals can be unambiguously observed in the spectrum (Table S-2). The other 6 metabolites each possess a single 1H resonance, and single peak matching does not provide very high confidence unless the chemical shift position is unique such as is the case for fumarate. Overall, 19 metabolites represent only a small subset of the total number of metabolites in this same sample that could be unambiguously identified. Another problem we observed was the occurrence of numerous false positive identifications, which is consistent with observations reported by others.8 Figure 1 and Supporting Information Figure S-3 demonstrate the clear limitations of 1D 1H NMR for metabolite identification, at least for a spectrum of the complexity of the E. coli cell lysate. Putrescine, uracil, and fumarate can be identified with high confidence in the 1D 1H NMR spectrum. Alanine, valine, and nicotinic acid identifications are ambiguous, since not all of their peaks yield a good match. And finally, certain metabolites, such as lysine, uridine, malate, and ethanol, whose existence in the sample was verified by the use of multidimensional NMR spectra, could not be identified on the basis of 1D 1H NMR spectroscopy alone (Figure 1 and Figure S-3).
Figure 1.
Identification of metabolites by 1D 1H NMR spectral matching at the example of E. coli cell lysate using the Chenomx NMR software. Overlay of 1D 1H NMR spectra of metabolites from the Chenomx database (blue) on 1D 1H NMR spectrum of E. coli cell lysate (black). Putrescine (A) and alanine (B) possess at least one (partially) isolated peak in the lysate spectrum that matches a peak in the corresponding database spectrum. On the other hand, each of the peaks of lysine (C) and uridine (D) overlap with other peaks in the lysate spectrum, which makes their unambiguous identification impossible.
Application of 1H(13C)-TOCCATA to E. coli Cell Lysate
2D 1H–1H TOCSY
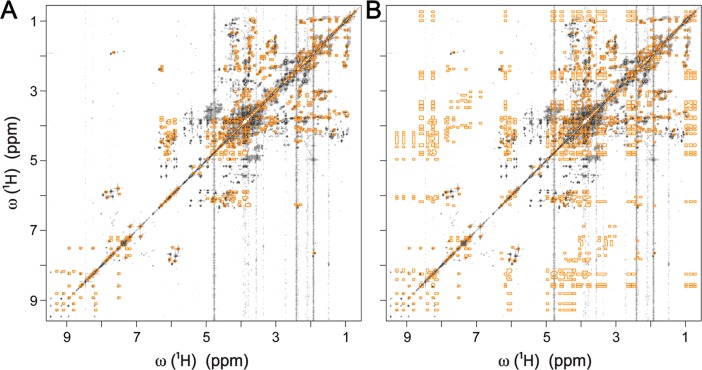
The 1H(13C)-TOCCATA database was first applied to the 1H–1H TOCSY spectrum of E. coli cell lysate (Figure S-4). A total of 45 1H TOCSY traces were extracted and identified in the 1H(13C)-TOCCATA database with the query results listed in Table 1. For 27 1H TOCSY traces, the query returned the correct compound as a single best hit. For the other 18 traces, on average 2.6 hits were returned with the top (i.e., best) hit always being the correct one. These 45 traces belong to 41 distinct metabolites. The TOCSY cross-peaks of these metabolites are shown by superimposing a 1H–1H TOCSY spectrum reconstructed from the 1H(13C)-TOCCATA database onto the experimental spectrum (Figure 2A). For comparison, Figure 2B shows the TOCSY spectrum reconstructed from entire 1D 1H NMR spectra. Since the 1D 1H NMR spectra do not discriminate between different spin systems or isomeric states, the reconstructed spectrum generates cross-peaks between all peaks of a metabolite, which leads to a large number of false positive cross-peaks (Figure 2B). For the same reason the querying of TOCSY traces against 1D NMR databases leads to a large number of mismatches. To compare the querying results using 1H(13C)-TOCCATA with 1D 1H NMR databases, we submitted the 45 1H TOCSY traces to the BMRB,15 MMCD,16 COLMAR,25 and HMDB17 databases for 1D 1H NMR querying. They identified 17, 20, 29, and 13 1H TOCSY traces correctly as first hit, respectively. The detailed performance of each of these databases for all 1H TOCSY traces can be found in Supporting Information Table S-3. In general, metabolites that exist as a single isomer and have a single spin system (e.g., valine, isoleucine, nicotinic acid etc.) were identified correctly by both 1D 1H NMR databases and the 1H(13C)-TOCCATA database. However, metabolites existing in multiple isomeric states and/or in multiple spin systems, including tyrosine, NADP+, coenzyme A, and adenosine, were almost always correctly identified only by the new database, 1H(13C)-TOCCATA. Overall, the new database provides ∼35% improvement over the best performing 1D 1H NMR query.
Table 1. Metabolites Identified in 2D 1H–1H TOCSY Spectrum of E. coli Cell Lysate by Querying against the 1H(13C)-TOCCATA Databasea.
| RMSDb | Mc | shiftd | RMSD | M | shift | ||
|---|---|---|---|---|---|---|---|
| valine (4) | 0.002 | 0 | –0.016 | p-toluic acid (2) | 0.003 | 0 | –0.013 |
| lysine (5) | 0.004 | 0 | –0.018 | cytosine (2) | 0.000 | 0 | –0.015 |
| isoleucine (6) | 0.002 | 0 | –0.017 | propionic acid (2) | 0.000 | 0 | –0.015 |
| leucine (3) | 0.003 | 0 | –0.017 | ethanolamine (2) | 0.003 | 0 | –0.019 |
| proline (6) | 0.006 | 0 | –0.018 | n-acetyl-glutamate (4) | 0.008 | 0 | –0.015 |
| alanine (2) | 0.001 | 0 | –0.020 | citrulline (4) | 0.003 | 0 | –0.016 |
| ethanol (2) | 0.000 | 0 | –0.016 | cytidine (2) | 0.005 | 0 | –0.024 |
| arginine (5) | 0.003 | 0 | –0.013 | spermidine (2) | 0.001 | 0 | –0.015 |
| β-alanine (2) | 0.003 | 0 | –0.018 | 2-aminobutyrate (3) | 0.002 | 0 | –0.018 |
| γ-aminobutyrate (3) | 0.004 | 0 | –0.017 | threonine (3) | 0.002 | 0 | –0.020 |
| nicotinic acid (4) | 0.002 | 0 | –0.018 | uridine (6) | 0.008 | 0 | –0.016 |
| tyrosine (2) | 0.003 | 0 | –0.015 | N-α-acetyl-ornithine (4) | 0.005 | 0 | –0.004 |
| phenylalanine (3) | 0.002 | 0 | –0.009 | N-acetyl-glutamine (4) | 0.010 | 0 | –0.006 |
| uracil (2) | 0.001 | 0 | –0.009 | methionine-sulfoxide 1 (3) | 0.008 | 0 | –0.055 |
| lactate (2) | 0.002 | 0 | –0.019 | methionine-sulfoxide 2 (4) | 0.015 | 0 | –0.056 |
| phosphoenolpyruvate (2) | 0.005 | 0 | –0.029 | coenzyme A 1 (2) | 0.001 | 0 | –0.012 |
| putrescine (2) | 0.000 | 0 | –0.011 | coenzyme A 2 (2) | 0.001 | 0 | –0.007 |
| thymidine 1 (6) | 0.002 | 0 | –0.011 | pantothenate (2) | 0.001 | 0 | –0.016 |
| thymidine 2 (2) | 0.004 | 0 | –0.005 | glutamate (3) | 0.001 | 0 | –0.016 |
| 2-deoxycytidine 1 (2) | 0.001 | 0 | –0.013 | adenosine (6) | 0.008 | 0 | –0.010 |
| 2-deoxycytidine 2 (7) | 0.005 | 0 | –0.011 | adenosine-3-monophosphate (5) | 0.004 | 0 | –0.010 |
| NADP+ (4) | 0.003 | 0 | –0.018 | inosine (6) | 0.012 | 0 | –0.009 |
| tryptophan (4) | 0.003 | 0 | 0.008 |
The numbers behind certain compound names not in parentheses are used only when more than one spin systems of a metabolite is observed in the Table and they denote the different spin systems of the metabolite.
Chemical shift root-mean-square difference (in units of ppm) between the input and database chemical shifts.
Integer mismatch parameter, which is the absolute value of the difference between the number of input and database chemical shifts.
Amount by which the input chemical shifts were uniformly shifted (in ppm) so that the RMSD with respect to the database chemical shifts is minimized.
Figure 2.
Overlay of reconstructions of 1H–1H TOCSY spectra from databases (orange) with the experimental 1H–1H TOCSY spectrum of E. coli cell lysate (black). (A) The reconstruction of the TOCSY spectrum (orange) is based on spin-system information from the 1H(13C)-TOCCATA database. (B) The reconstruction of the TOCSY spectrum (orange) is based on entire 1D 1H NMR spectra from the BMRB database. A list with all 41 metabolites used for reconstruction in both panels is given in Table 1.
The MMCD,16 HMDB,17 and Metabominer8 databases are, at least in part, derived from 1H–1H TOCSY experiments. The major difference between these databases and 1H(13C)-TOCCATA is the database organization for the TOCSY peaks of each metabolite. In the MMCD, HMDB, and Metabominer databases, the 2D 1H–1H TOCSY spectrum of each isolated metabolite is stored as a single entry. Therefore, each database entry consists of all TOCSY peaks of a metabolite. By contrast, in 1H(13C)-TOCCATA each such entry is subdivided into a molecule’s isomeric states and spin systems. This is because the TOCSY spectrum only displays correlations (connectivities) between spins in the same spin system, but not between spins belonging to different spin systems or resonances belonging to different isomeric states. Therefore, for such molecules the query of individual TOCSY traces against these databases will cause mismatches.
We queried the 45 1H TOCSY traces of the E. coli cell lysate against the 2D 1H–1H TOCSY databases of MMCD, HMDB, and Metabominer. They correctly identified only 20, 20, and 24 1H TOCSY traces as first hits, respectively, which illustrates the benefits of 1H(13C)-TOCCATA. The detailed performance of each database for all 1H TOCSY traces is given in Supporting Information Table S-4.
2D 13C–1H HSQC-TOCSY
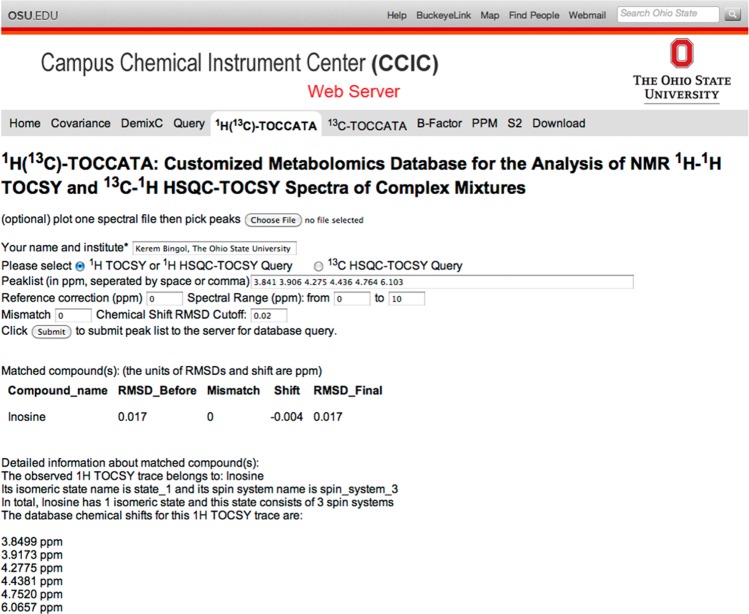
The second application of the 1H(13C)-TOCCATA database was performed using a 13C–1H HSQC-TOCSY spectrum of E. coli cell lysate (Figure S-5). A total of 38 13C and 1H HSQC-TOCSY trace pairs were extracted from the spectrum. For each pair, the 1H chemical shift list was queried against the 1H(13C)-TOCCATA database (using a 0.02 ppm RMSD cutoff and Mmax = 0) independently of the querying of the 13C chemical shift list (using a 0.2 ppm RMSD cutoff and Mmax = 0). Figures 3 and S-6 each represent a screenshot of the query result of the web server of one of these trace pairs using the new database. In Figure 3, querying of the peak list of a 1H HSQC-TOCSY trace (row) extracted from a 2D HSQC-TOCSY spectrum results in a single hit, corresponding to the ribose ring of inosine. When on the same web page, the box for “13C HSQC-TOCSY Query” is selected, the default values for the “Spectral Range (ppm)” and “Chemical Shift RMSD Cutoff” are automatically updated for 13C nuclei, and the 13C chemical shifts extracted from the 13C HSQC-TOCSY (column) trace of the pair is entered. In Figure S-6, the query for the corresponding 13C peak list yields a single hit, which is also the ribose ring of inosine. Therefore, both traces independently identify inosine as the compound belonging to this pair of HSQC-TOCSY traces. The query results of all such pairs are compiled in Table 2. Overall, 23 13C HSQC-TOCSY traces are identified as a single, correct hit. For the remaining traces, the querying of 11 13C HSQC-TOCSY traces yield the correct metabolite as the top hit (from an average of 2.9 hits). For the remaining 4 13C HSQC-TOCSY traces, ambiguities among the top hits could be resolved after querying the corresponding 1H HSQC-TOCSY chemical shifts whereby the correct hit turned out to always be the top one (Table S-5). The total set of 38 1H and 13C HSQC-TOCSY traces belong to 33 different metabolites.
Figure 3.
Screenshot of the 1H(13C)-TOCCATA web server. The peak list of a 1H HSQC-TOCSY trace from the 2D 13C–1H HSQC-TOCSY spectrum is queried against the database. Query returns the best matching compound (in this case ribose ring of inosine) with the chemical shift root-mean-square difference (rmsd) before and after a uniform shift of −0.004 ppm was applied. A mismatch number M = 0 indicates that the number of query peaks and database peaks for inosine were the same.
Table 2. Metabolites Identified in 2D 13C–1H HSQC-TOCSY Spectrum of E. coli Cell Lysate by Querying against the 1H(13C)-TOCCATA Databasea.
| RMSDb | Mc | shiftd | RMSD | M | shift | ||
|---|---|---|---|---|---|---|---|
| valine 1H (4) | 0.002 | 0 | –0.018 | phosphoenolpyruvate 1H (2) | 0.007 | 0 | –0.033 |
| valine 13C (4) | 0.015 | 0 | –0.090 | phosphoenolpyruvate 13C (1) | 0.000 | 0 | –0.562 |
| lysine 1H (5) | 0.002 | 0 | –0.016 | serine 1H (2) | 0.001 | 0 | –0.019 |
| lysine 13C (5) | 0.110 | 0 | –0.162 | serine 13C (2) | 0.018 | 0 | –0.102 |
| malate 1H (3) | 0.002 | 0 | –0.020 | methanol 1H (1) | 0.000 | 0 | –0.014 |
| malate 13C (2) | 0.012 | 0 | –0.127 | methanol 13C (1) | 0.000 | 0 | –0.182 |
| alanine 1H (2) | 0.002 | 0 | –0.016 | glycine 1H (1) | 0.000 | 0 | –0.018 |
| alanine 13C (2) | 0.021 | 0 | –0.129 | glycine 13C (1) | 0.000 | 0 | –0.162 |
| leucine 1H (5) | 0.003 | 0 | –0.014 | succinate 1H (1) | 0.000 | 0 | –0.020 |
| leucine 13C (5) | 0.156 | 0 | –0.200 | succinate 13C (1) | 0.000 | 0 | –0.053 |
| threonine 1H (3) | 0.004 | 0 | –0.020 | N-acetyl-alanine 1H (2) | 0.005 | 0 | –0.019 |
| threonine 13C (3) | 0.034 | 0 | –0.062 | N-acetyl-alanine 13C (2) | 0.040 | 0 | –0.198 |
| β-alanine 1H (2) | 0.004 | 0 | –0.013 | acetic acid 1H (1) | 0.000 | 0 | –0.014 |
| β-alanine 13C (2) | 0.044 | 0 | –0.078 | acetic acid 13C (1) | 0.000 | 0 | –0.124 |
| uracil 1H (2) | 0.000 | 0 | –0.008 | putrescine 1H (2) | 0.001 | 0 | –0.012 |
| uracil 13C (2) | 0.046 | 0 | –0.033 | putrescine 13C (2) | 0.005 | 0 | –0.099 |
| tyrosine 1 1H (3) | 0.003 | 0 | –0.028 | thymidine 1 1H (2) | 0.004 | 0 | –0.006 |
| tyrosine 1 13C (2) | 0.014 | 0 | 0.084 | thymidine 1 13C (2) | 0.040 | 0 | –0.101 |
| tyrosine 2 1H (2) | 0.003 | 0 | –0.017 | thymidine 2 1H (6) | 0.004 | 0 | –0.010 |
| tyrosine 2 13C (2) | 0.049 | 0 | –0.097 | thymidine 2 13C (5) | 0.020 | 0 | –0.143 |
| phenylalanine 1 1H (3) | 0.003 | 0 | –0.021 | cytidine 1H (2) | 0.007 | 0 | –0.019 |
| phenylalanine 1 13C (2) | 0.030 | 0 | –0.090 | cytidine 13C (2) | 0.015 | 0 | –0.212 |
| phenylalanine 2 1H (3) | 0.004 | 0 | –0.005 | dTMP 1 1H (2) | 0.002 | 0 | –0.015 |
| phenylalanine 2 13C (3) | 0.015 | 0 | –0.059 | dTMP 1 13C (2) | 0.035 | 0 | –0.085 |
| arginine 1H (4) | 0.003 | 0 | –0.008 | dTMP 2 1H (5) | 0.020 | 0 | –0.036 |
| arginine 13C (4) | 0.088 | 0 | –0.069 | dTMP 2 13C (5) | 0.127 | 0 | –0.017 |
| γ-aminobutyrate 1H (3) | 0.003 | 0 | –0.015 | uridine 1 1H (6) | 0.005 | 0 | –0.008 |
| γ-aminobutyrate 13C (3) | 0.034 | 0 | –0.089 | uridine 1 13C (5) | 0.054 | 0 | –0.097 |
| aspartate 1H (3) | 0.003 | 0 | –0.012 | uridine 2 1H (2) | 0.010 | 0 | –0.004 |
| aspartate 13C (2) | 0.015 | 0 | –0.094 | uridine 2 13C (2) | 0.010 | 0 | –0.127 |
| glutamate 1H (3) | 0.001 | 0 | –0.011 | adenosine 1H (6) | 0.006 | 0 | –0.010 |
| glutamate 13C (3) | 0.048 | 0 | –0.042 | adenosine 13C (5) | 0.008 | 0 | –0.056 |
| lactate 1H (2) | 0.000 | 0 | –0.014 | inosine 1H (6) | 0.017 | 0 | –0.004 |
| lactate 13C (2) | 0.019 | 0 | –0.081 | inosine 13C (5) | 0.049 | 0 | –0.113 |
| nicotinic acid 1H (4) | 0.003 | 0 | –0.013 | glutathione reduced 1H (3) | 0.008 | 0 | –0.010 |
| nicotinic acid 13C (4) | 0.043 | 0 | –0.094 | glutathione reduced 13C (3) | 0.036 | 0 | –0.166 |
| fumarate 1H (1) | 0.000 | 0 | –0.012 | cystathionine 1H (3) | 0.006 | 0 | –0.023 |
| fumarate 13C (1) | 0.000 | 0 | –0.097 | cystathionine 13C (3) | 0.147 | 0 | –0.398 |
The numbers behind certain compound names that are not in parentheses are used only when more than one spin systems of a metabolite is observed in the Table and they denote the different spin systems of the metabolite. “1H” and “13C” labels behind compound names indicates whether the queried trace is a 1H HSQC-TOCSY trace or 13C HSQC-TOCSY trace.
Chemical shift root-mean-square difference (in units of ppm) between the input and database chemical shifts.
Integer mismatch parameter, which is the absolute value of the difference between the number of input and database chemical shifts.
Amount by which the input chemical shifts were uniformly shifted (in ppm) so that the RMSD with respect to the database chemical shifts is minimized.
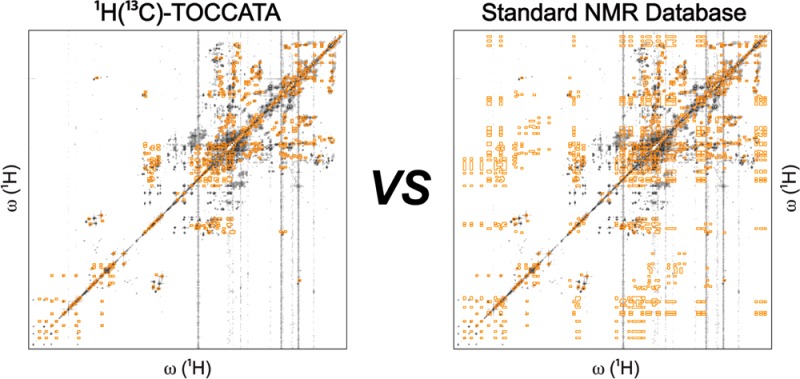
The cross-peaks of these metabolites are shown by superimposing a 13C–1H HSQC-TOCSY spectrum reconstructed from the 1H(13C)-TOCCATA database onto the experimental spectrum (Figure S-7A). For comparison, Figure S-7B shows the 13C–1H HSQC-TOCSY spectrum reconstructed from entire 1D NMR spectra revealing a large number of false positive cross-peaks, similar to Figure 2B. To compare the performance of 1H(13C)-TOCCATA with 1D NMR databases, we submitted the 38 1H and 13C HSQC-TOCSY traces for 1D NMR querying. BMRB, MMCD, COLMAR, and HMDB identified 16, 4, 25, and 27 13C HSQC-TOCSY traces correctly as best hits, respectively. The detailed query performance for each database for all 1H and 13C HSQC-TOCSY traces can be found in Supporting Information Table S-5. Similar to 1H–1H TOCSY results, metabolites existing in multiple isomeric states and/or multiple spin systems can be identified by the new database with very high accuracy and efficiency. 1H(13C)-TOCCATA provides ∼21% improvement over the best-performing 1D 13C NMR query.
The MMCD16 database also allows the querying of chemical shifts extracted from 13C–1H HSQC-TOCSY. Again, this database does not group the HSQC-TOCSY peaks into different spin systems and/or different isomeric states. To compare the 1H(13C)-TOCCATA with the MMCD 2D 13C–1H HSQC-TOCSY NMR database, we queried 38 13C–1H HSQC-TOCSY sets of peaks against the MMCD database. It allowed the identification of 20 HSQC-TOCSY peak lists correctly as first hits (with the “H_tol” and “C_tol” parameters set to 0.05 ppm and 0.2 ppm, respectively).
Finally, 38 13C HSQC-TOCSY traces were queried against our original 13C-TOCCATA database20 developed for uniformly 13C-labeled metabolites. Not surprisingly, those cell lysate metabolites that possess nonprotonated carbons, namely tyrosine, phenylalanine, nicotinic acid, phosphoenolpyruvate, and nucleic acid portions of thymidine, cytidine, dTMP, and uridine could not be identified in the 13C-TOCCATA database when using a mismatch parameter Mmax = 0. Therefore, the querying of 13C HSQC-TOCSY traces from 2D 13C–1H HSQC-TOCSY is best performed with the 1H(13C)-TOCCATA database, while querying of 13C TOCSY traces from 13C–13C CT-TOCSY is optimal when using the 13C-TOCCATA database.
The 1H(13C)-TOCCATA database can also be applied to 13C-labeled samples dependent on the type of TOCSY experiment chosen for metabolite identification. The analysis of 1H–1H TOCSY and 13C–1H HSQC-TOCSY spectra of 13C-labeled samples is best performed with the new database 1H(13C)-TOCCATA, whereas for the analysis of the 13C–13C CT-TOCSY spectrum of 13C labeled samples the original 13C-TOCCATA database is best suited.
In this study, 21 metabolites were identified in both 13C–1H HSQC-TOCSY and 1H–1H TOCSY spectra. An additional 12 metabolites were identified only in 13C–1H HSQC-TOCSY, but not in 1H–1H TOCSY, because their signals strongly overlapped in the 1H–1H TOCSY spectrum, as for example in the case of serine and N-acetyl-alanine. Additionally, 20 metabolites were only identified in 1H–1H TOCSY, but not in 13C–1H HSQC-TOCSY, because their signals were below the detection limit of the 13C–1H HSQC-TOCSY spectrum such as in the case of ethanolamine and proline. Therefore, in total, 53 different metabolites could be positively identified in E. coli by using the 1H(13C)-TOCCATA database.
Conclusions
Accurate and unambiguous identification of the metabolites in biological samples is a key step for downstream metabolomics analysis. In the past, 1D 1H NMR spectra have often been the first choice for this task despite the fact that they frequently suffer from severe spectral overlaps. For a quite complex, real-world metabolomics sample, such as an E. coli cell lysate, we find that this approach produces correct identifications for only a small subset the compounds that can be achieved by 2D NMR methods with many additional false positive identifications. Therefore, in metabolomics studies, acquisition of at least one 2D NMR experiment for unambiguous compound identification, such as a 1H–1H TOCSY or 13C–1H HSQC-TOCSY experiment, is highly beneficial when combined querying against the customized 1H(13C)-TOCCATA database. As metabolomics databases continue to grow, the chances that two compounds have very similar NMR properties increases. This requires customized databases that take full advantage of the specific appearance of NMR information in the raw spectra, as does 1H(13C)-TOCCATA, for the unambiguous identification of a large number of mixture components. It should be noted that 1H(13C)-TOCCATA can be applied to NMR data collected at variable magnetic field strengths as only the chemical shift information on each peak is utilized provided that strong coupling effects are not dominant.
Although the acquisition of 2D NMR experiments takes more time, recent advances in 2D NMR methodology, including covariance NMR26 for TOCSY-type spectra, nonuniform sampling for HSQC,27,28 single-scan and ultrafast HSQC,29 and approaches with shortened recovery delays between scans30,31 are expected to help decrease the measurement time making the use of multidimensional NMR methods increasingly practicable also for applications involving multiple metabolic samples.
Materials and Methods
An extract from E. coli DH5α strain was prepared, and 1D 1H, 2D 1H–1H TOCSY, and 2D 13C−1H HSQC-TOCSY data sets were collected as described in the Supporting Information.
Acknowledgments
This work was supported by the National Institutes of Health (grant R01 GM 066041 and SECIM (Southeast Center for Integrated Metabolomics) grant U24 DK097209-01A1).
Supporting Information Available
Additional tables and figures can be found as Supporting Information, including tables with all compounds of the 1H(13C)-TOCCATA database and a comparison of the performance of different databases. This material is available free of charge via the Internet at http://pubs.acs.org
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Nicholson J. K.; Holmes E.; Kinross J. M.; Darzi A. W.; Takats Z.; Lindon J. C. Nature 2012, 491, 384–392. [DOI] [PubMed] [Google Scholar]
- Lenz E. M.; Wilson I. D. J. Proteome Res. 2007, 6, 443–458. [DOI] [PubMed] [Google Scholar]
- Bingol K.; Brüschweiler R. Anal. Chem. 2014, 86, 47–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis I. A.; Schommer S. C.; Hodis B.; Robb K. A.; Tonelli M.; Westler W. M.; Sussman M. R.; Markley J. L. Anal. Chem. 2007, 79, 9385–9390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinette S. L.; Ajredini R.; Rasheed H.; Zeinomar A.; Schroeder F. C.; Dossey A. T.; Edison A. S. Anal. Chem. 2011, 83, 1649–1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandusky P.; Raftery D. Anal. Chem. 2005, 77, 2455–2463. [DOI] [PubMed] [Google Scholar]
- Zhang F.; Brüschweiler R. Angew. Chem., Int. Ed. 2007, 46, 2639–2642. [DOI] [PubMed] [Google Scholar]
- Xia J.; Bjorndahl T. C.; Tang P.; Wishart D. S. BMC Bioinf. 2008, 9, 507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bingol K.; Zhang F.; Bruschweiler-Li L.; Brüschweiler R. J. Am. Chem. Soc. 2012, 134, 9006–9011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu K.; Westler W. M.; Markley J. L. J. Am. Chem. Soc. 2011, 133, 1662–1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bingol K.; Zhang F.; Bruschweiler-Li L.; Brüschweiler R. Anal. Chem. 2013, 85, 6414–6420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bingol K.; Brüschweiler R. Anal. Chem. 2011, 83, 7412–7417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Q. N.; Issaq H. J.; Jiang Q.; Li Q.; Muschik G. M.; Waybright T. J.; Lou H.; Dean M.; Uitto J.; Veenstra T. D. J. Proteome Res. 2008, 7, 630–639. [DOI] [PubMed] [Google Scholar]
- Bodenhausen G.; Ruben D. J. Chem. Phys. Lett. 1980, 69, 185–189. [Google Scholar]
- Ulrich E. L.; Akutsu H.; Doreleijers J. F.; Harano Y.; Ioannidis Y. E.; Lin J.; Livny M.; Mading S.; Maziuk D.; Miller Z.; Nakatani E.; Schulte C. F.; Tolmie D. E.; Wenger R. K.; Yao H. Y.; Markley J. L. Nucleic Acids Res. 2008, 36, D402–D408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Q.; Lewis I. A.; Hegeman A. D.; Anderson M. E.; Li J.; Schulte C. F.; Westler W. M.; Eghbalnia H. R.; Sussman M. R.; Markley J. L. Nat. Biotechnol. 2008, 26, 162–164. [DOI] [PubMed] [Google Scholar]
- Wishart D. S.; Knox C.; Guo A. C.; Eisner R.; Young N.; Gautam B.; Hau D. D.; Psychogios N.; Dong E.; Bouatra S.; Mandal R.; Sinelnikov I.; Xia J. G.; Jia L.; Cruz J. A.; Lim E.; Sobsey C. A.; Shrivastava S.; Huang P.; Liu P.; Fang L.; Peng J.; Fradette R.; Cheng D.; Tzur D.; Clements M.; Lewis A.; De Souza A.; Zuniga A.; Dawe M.; Xiong Y. P.; Clive D.; Greiner R.; Nazyrova A.; Shaykhutdinov R.; Li L.; Vogel H. J.; Forsythe I. Nucleic Acids Res. 2009, 37, D603–D610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chikayama E.; Sekiyama Y.; Okamoto M.; Nakanishi Y.; Tsuboi Y.; Akiyama K.; Saito K.; Shinozaki K.; Kikuchi J. Anal. Chem. 2010, 82, 1653–1658. [DOI] [PubMed] [Google Scholar]
- Braunschweiler L.; Ernst R. R. J. Magn. Reson. 1983, 53, 521–528. [Google Scholar]
- Bingol K.; Zhang F.; Bruschweiler-Li L.; Brüschweiler R. Anal. Chem. 2012, 84, 9395–9401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eletsky A.; Moreira O.; Kovacs H.; Pervushin K. J. Biomol. NMR 2003, 26, 167–179. [DOI] [PubMed] [Google Scholar]
- Rance M.; Sorensen O. W.; Bodenhausen G.; Wagner G.; Ernst R. R.; Wüthrich K. Biochem. Biophys. Res. Commun. 1983, 117, 479–485. [DOI] [PubMed] [Google Scholar]
- Lerner L.; Bax A. J. Magn. Reson. 1986, 69, 375–380. [Google Scholar]
- Friebolin H.Basic One- and Two-Dimensional NMR Spectroscopy; Wiley-VCH: Weinheim, Germany, 2005. [Google Scholar]
- Robinette S. L.; Zhang F.; Bruschweiler-Li L.; Brüschweiler R. Anal. Chem. 2008, 80, 3606–3611. [DOI] [PubMed] [Google Scholar]
- Brüschweiler R.; Zhang F. J. Chem. Phys. 2004, 120, 5253–5260. [DOI] [PubMed] [Google Scholar]
- Hyberts S. G.; Heffron G. J.; Tarragona N. G.; Solanky K.; Edmonds K. A.; Luithardt H.; Fejzo J.; Chorev M.; Aktas H.; Colson K.; Falchuk K. H.; Halperin J. A.; Wagner G. J. Am. Chem. Soc. 2007, 129, 5108–5116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rai R. K.; Sinha N. Anal. Chem. 2012, 84, 10005–10011. [DOI] [PubMed] [Google Scholar]
- Giraudeau P.; Shrot Y.; Frydman L. J. Am. Chem. Soc. 2009, 131, 13902–13903. [DOI] [PubMed] [Google Scholar]
- Schanda P.; Brutscher B. J. Am. Chem. Soc. 2005, 127, 8014–8015. [DOI] [PubMed] [Google Scholar]
- Schulze-Sünninghausen D.; Becker J.; Luy B. J. Am. Chem. Soc. 2014, 136, 1242–1245. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.