Abstract

Small interfering RNA (siRNA)-based therapies have great promise in the treatment of a number of prevalent pulmonary disorders including lung cancer, asthma and cystic fibrosis. However, progress in this area has been hindered due to the lack of carriers that can efficiently deliver siRNA to lung epithelial cells, and also due to challenges in developing oral inhalation (OI) formulations for the regional administration of siRNA and their carriers to the lungs. In this work we report the ability of generation four, amine-terminated poly(amidoamine) (PAMAM) dendrimer (G4NH2)–siRNA complexes (dendriplexes) to silence the enhanced green fluorescent protein (eGFP) gene on A549 lung alveolar epithelial cells stably expressing eGFP. We also report the formulation of the dendriplexes and their aerosol characteristics in propellant-based portable OI devices. The size and gene silencing ability of the dendriplexes was seen not to be a strong function of the N/P ratio. Silencing efficiencies of up to 40% are reported. Stable dispersions of the dendriplexes encapsulated in mannitol and also in a biodegradable and water-soluble co-oligomer were prepared in hydrofluoroalkane (HFA)-based pressurized metered-dose inhalers (pMDIs). Their aerosol characteristics were very favorable, and conducive to deep lung deposition, with respirable fractions of up to 77%. Importantly, siRNA formulated as dendriplexes in pMDIs was shown to keep its integrity after the particle preparation processes, and also after long-term exposures to HFA. The relevance of this study stems from the fact that this is the first work to report the formulation of inhalable siRNA with aerosol properties suitable to deep lung deposition using pMDIs devices that are the least expensive and most widely used portable inhalers. This study is relevant because, also for the first time, it shows that siRNA–G4NH2 dendriplexes can efficiently target lung alveolar epithelial A549 cells and silence genes even after siRNA has been exposed to the propellant environment.
Keywords: pulmonary siRNA delivery, PAMAM dendrimers, siRNA dendriplexes, pressurized-metered dose inhalers (pMDIs), gene silencing, alveolar epithelium
Introduction
RNA interference (RNAi) promotes silencing of gene expression in a post-transcriptional manner.1,2 The delivery of exogenous double-stranded small interfering RNA (ds-siRNA, typically consisting of 20–27 base pairs),3 which targets the RNA-induced silencing complex (RISC) located in the cell cytoplasm, is considered a very promising therapeutic approach in the treatment of a broad range of diseases.4,5 RNAi-based therapies have also shown great potential in the treatment of pulmonary disorders including lung cancer,6 cystic fibrosis,7 asthma,8 chronic obstructive pulmonary disease (COPD),8 respiratory syncytial virus (RSV),9 and severe acute respiratory syndrome (SARS) viral infection.10 However, further progress on siRNA-based technologies that target lung ailments has been hindered by the lack of adequate carriers capable of overcoming the extra and intracellular barriers present in the lung tissue, and challenges in formulating siRNA and their carriers in portable oral inhalation (OI) devices for the local delivery of siRNA to the lungs.4,11,12
Poly(amidoamine) (PAMAM) dendrimers are highly monodispersed, hyperbranched polymers that contain easily functionalizable surface groups, and thus represent promising structures for the construction of effective drug carriers and gene transfer vehicles.13,14 Amine-terminated PAMAM dendrimers have their surface groups (primary amines) protonated at physiological conditions.15 They can thus efficiently bind to nucleic acids, forming nanoscale complexes termed dendriplexes16 that may offer opportunities to improve targeting of siRNA to the cell cytosol. Even though many publications have addressed the transfection of cells using dendrimer–DNA complexes,17 few reports have discussed the gene silencing (with siRNA3,18 or antisense oligonucleotide19) with PAMAM dendrimers in cells that are models of the lung alveolar epithelium.
Once a suitable carrier for the delivery of siRNA to the cytosol of lung cells is found, another important consideration is the ability to target the siRNA–carrier complex to the lungs. OI is a very promising route of administration for siRNA to the pulmonary epithelium since it provides a direct and noninvasive means of targeting the lungs.20 Pressurized metered-dose inhalers (pMDIs) are important devices within the context of siRNA delivery to the lungs as they are the least expensive and most widely used OI devices. pMDIs are portable, have long shelf life, are reliable, and can be used even in patients with compromised lung function as they are propellant based.21 However, due to formulations challenges, there has been no report in the literature to date of siRNA or siRNA–carrier systems formulated in pMDIs.
On the basis of the challenges and opportunities discussed above, we propose in this work the development of propellant-based OI formulations of siRNA–dendrimer complexes for the efficient silencing of genes of the lung epithelium. We report the preparation and characterization of siRNA–G4NH2 dendriplexes, and evaluate the in vitro gene knockdown efficiency in a model lung alveolar epithelial cell line. We propose different formulation strategies for the dendriplexes, and study their aerosol performance. The relevance of this study stems from the fact that this is the first work to report the formulation of inhalable siRNA with aerosol properties suitable to deep lung deposition in pMDIs, which are the least expensive and most widely used portable inhalers available in the market. This study is relevant because, also for the first time, it shows that siRNA–G4NH2 dendriplexes can efficiently target lung alveolar epithelial cells A549, and silence genes even after siRNA has been exposed to the propellant environment.
Experimental Section
Materials
Generation-four, amine-terminated poly(amidoamine) dendrimer (PAMAM G4NH2, 14,215 g × mol–1, 64 NH2 surface groups) was purchased from Dendritech Inc. (Midland, MI, U.S.A.) and provided in methanol (15.35 w/w). Methanol was removed with the help of a rotary evaporator (Buchi R-200). Double-stranded siRNA (sense: 5′-AAC UUC AGG GUC AGC UUG C dTdT-3′; antisense: 5′-GCA AGC UGA CCC UGA AGU U dTdT-3′) was purchased from Bioneer Inc. (Alameda, CA, U.S.A.). Double-stranded Dicer substrate siRNA (Ds-siRNA) targeting eGFP (sense: 5′-p ACC CUG AAG UUC AUC UGC ACC AC cg-3′; antisense: 5′-p CGG UGG UGC AGA UGA ACU UCA GGG UCA-3′) and a respective mismatch (both from Integrated DNA Technologies, IDT, Leuven, Belgium) were used for all gene knockdown experiments. Capital letters represent ribonucleotides, underlined bases depict 2′-O-methylribonucleotides, lower case letters represent 2′-deoxyribonucleotides, p and dTdT depict an additional phosphate at the 5′-end, and thymine overhangs, respectively. Deionized water (DI-water, resistivity of 18.2 MΩ × cm) was obtained from NANOpure DIamond UV ultrapure water system (Barnstead International, Dubuque, IA, U.S.A.) and treated with 0.1% v/v diethylpyrocarbonate (DEPC) overnight, following autoclaving. DEPC (high purity) and ethylenediaminetetraacetic acid (EDTA, 0.5 M sterile solution, pH 8, DEPC treated) were purchased from Amresco (Solon, OH, U.S.A.). A549 cells (CCL-185, passage 82)—human lung adenocarcinoma cell line, an in vitro model of Type II alveolar epithelium22—were sourced from ATCC (Manassas, VA, U.S.A.). Gibco Dulbecco’s Modified Eagle Medium (DMEM, high glucose, GlutaMAX, pyruvate), Gibco Penicillin-Streptomycin Liquid (AB), Quant-iT PicoGreen, and Lipofectamine 2000 Transfection Reagent were purchased from Invitrogen Life Technologies (Grand Island, NY, U.S.A.). Fetal Bovine Serum (FBS, Advantage, nonheat inactivated, S11050) was purchased from Atlanta Biologicals Inc. (Norcross, GA, U.S.A.). Trypsin EDTA 1X Corning Cellgro (0.25% trypsin, 2.21 mM EDTA in Hank’s Balanced Salt Solution, HBSS) was purchased from Mediatech Inc. (Manassas, VA, U.S.A.). CellTiter 96 AQueous Non-Radioactive Cell Proliferation Assay (MTS reagent powder) and TransFast Transfection Reagent were purchased from Promega Corporation (Madison, WI, U.S.A.). Phenazine methosulfate (PMS, 95%) was purchased from MP Biomedicals (Santa Ana, CA, U.S.A.). eGFP lentiviral particles (LVP-340, 1 × 107 IFU × mL–1, a puromycin gene under Rsv promoter allows the selection of transduced fluorescent positive cells) were purchased from GenTarget Inc. (San Diego, CA, U.S.A.). Puromycin dihydrochloride (10 mg × mL–1) was purchased from Toku-E (Bellingham, WA, U.S.A.). Cell culture flasks Cellstar (75 cm2), 24- and 96-well Costar cell culture microplates (flat bottom, tissue culture treated, polystyrene, sterile) were purchased from Corning Inc. Life Sciences (Tewksbury, MA, U.S.A.). Ninety-six-well microplate (black, flat bottom shape, polypropylene) was purchased from Greiner Bio One (Monroe, NC, U.S.A.). Ribonuclease A (RNase A, R5503, 43 U × mg–1 solid) from bovine pancreas, heparin sodium salt (H4784, 194 U × mg–1) from porcine intestinal mucosa, d-mannitol (minimum 98%), chitosan (CS, 100–300 kDa, 80% degree of deacetylation), 3,6-dimethyl-1,4-dioxane-2,5-dione (lactide, LA), tin(II) 2-ethylhexanoate (stannous octoate, SnOct2, 95%) were purchased from Sigma-Aldrich (St. Louis, MO, U.S.A.). RiboLock RNase Inhibitor (RI, EO0381, 40 U × μL–1) was purchased from Thermo Scientific (part of Thermo Fisher Scientific, Waltham, MA, U.S.A.). SeaKem LE Agarose was purchased from Lonza (Rockland, ME, U.S.A.). Ethidium bromide (98%, 10 mg × mL–1) was purchased from IBI Scientific (Peosta, IA, U.S.A.). Tris Base Ultrapure BioReagent (99.5%) was purchased from J. T. Baker (Avantor Performance Materials, Center Valley, PA, U.S.A.). Glacial acetic acid (100%), citric acid anhydrous (100%), phosphate buffered saline (PBS 10× solution), and microscope cover glasses (22 mm × 22 mm) were purchased from Fisher Scientific (part of Thermo Fisher Scientific, Waltham, MA, U.S.A.). Tris-HCl 1 M pH 8, and Tris-EDTA (TE) 20× pH 7.4 buffers were purchased from Boston BioProducts (Ashland, MA, U.S.A.). Dibasic sodium phosphate anhydrous (99%) was purchased from EMD Millipore (Billerica, MA, U.S.A.). Ethyl acetate (99.5%) was purchased from Mallinckrodt Chemicals (present Macron Chemicals, Avantor Performance Materials, Center Valley, PA, U.S.A.). Propellant HFA-277 (Dymel 227 ea/P) was a gift from DuPont (Wilmington, DE, U.S.A.). 2H,3H-perfluoropentane (HPFP, DuPont Vertrel XF) was purchased from TMC Industries Inc. (Waconia, MN, U.S.A.). Deuterated dimethyl sulfoxide (DMSO-d6, 99.9%) was purchased from Cambridge Isotope Laboratories Inc. (Tewksbury, MA, U.S.A.). Multi-75 silicon Atomic Force Microscopy (AFM) probes for force modulation and light tapping (75 kHz, 3 N × m–1) were purchased from Budget Sensors (distributed by NanoAndMore USA, Lady’s Island, SC, U.S.A.). Mica sheet was purchased from Ted Pella Inc. (Redding, CA, U.S.A.).
Preparation and Characterization of siRNA–G4NH2 Dendriplexes
Dendriplexes were formed by combining equal volumes (10–100 μL) of negatively charged siRNA (50–150 μg × mL–1, RNase free DI-water) and positively charged PAMAM G4NH2 (150–3000 μg × mL–1, 20 mM Tris-HCl pH 7.4 buffer RNase free).18,23 siRNA solution was added dropwise into the G4NH2 solution under vortex at 3000 rpm (VWR Digital Vortex Mixer) for 1 min. The siRNA–G4NH2 dispersions were kept at room temperature for at least 30 min before any experiments, in order to ensure complete formation of the dendriplexes.23 The G4NH2 concentration was varied in order to prepare dendriplexes at different N/P ratios—the molar ratio between the primary amine groups (N) from G4NH2 and phosphate groups (P) from siRNA.24
siRNA–G4NH2 dendriplex dispersions (minimum of five independent batches, n = 5) were quantitatively analyzed using PicoGreen assay25 in a Synergy 2 Microplate Reader (BioTek, VT) for the uncomplexed siRNA content, which was calculated using a linear calibration curve (siRNA concentration vs fluorescent units). PicoGreen is an ultrasensitive fluorescent nucleic acid stain used to quantify double-stranded DNA (ds-DNA)25 and ds-siRNA in solution.26 The siRNA complexation efficiency (CE) was calculated on the basis of the difference between the initial amount of siRNA added during the formation of the dendriplexes and the non-entrapped free siRNA remaining in the dendriplex dispersion, i.e., an indirect measurement of the siRNA content complexed with PAMAM G4NH2 within dendriplexes.
Light Scattering (LS, Malvern ZetaSizer Nano ZS) was used to evaluate the hydrodynamic size (diameter) and zeta potential (ζ) of the siRNA–G4NH2 dendriplexes. Samples were diluted to 80 nM siRNA, and measurements were performed at 25 °C using refractive index, viscosity, and dielectric constant of the buffer (for size) or DI-water (for ζ). Dendriplexes were formed in RNase free DI-water for ζ, which was calculated using the Smoluchowski Model.24 A minimum of three independent batches (n = 3) were used to calculated an average size and ζ.
Scanning electron microscopy (SEM, Jeol/EO JSM-6510LV-LGS, 25 kV) and AFM (Pico SPM LE Molecular Imaging) were used to investigate the geometric size and morphology of the dendriplexes, which were formed in RNase-free DI-water. For SEM imaging, several drops of the siRNA–G4NH2 dispersion were deposited on a microscope cover glass, dried overnight, and sputter-coated with gold (Ernest Fullan) under vacuum for 40 s. The sizes of the dendriplexes were estimated from the SEM images using ImageJ 1.42q27 to confirm the size range determined by LS. The histogram of the measured diameters (>400 particles) was fit to a Gaussian distribution, from which average and standard deviation were calculated. For AFM imaging, 20 μL of siRNA–G4NH2 dendriplex dispersion was deposited on a freshly cleaved mica sheet and incubated at room temperature for 5 min28 to allow the binding of the dendriplexes onto the substrate. The substrate was then washed with few drops of DI-water, and dried with gentle air flow. AFM images were obtained using a Multi-75 silicon AFM probe in AC tapping mode.28
Gel Retardation Assay
The capacity of G4NH2 to form complexes with siRNA was also examined by gel electrophoresis. A known volume of the dendriplex dispersion (equivalent to 300 ng siRNA) was loaded in the slots of casted nondenaturing agarose gel (1.5% w/v in tris base, acetic acid, and EDTA (TAE) 1X pH 8.2 buffer) stained with ethidium bromide (0.5 μg × mL–1). The electrophoresis was performed at 60 V (E0160-VWR Mini Gel Electrophoresis) for 40 min, and the siRNA–dye migration was visualized under UV irradiation (FOTO/Analyst Investigator/Eclipse with UV Transilluminator Fotodyne Inc.). The images were recorded using the FOTO/Analyst PC Image software (v.5).
RNase Protection Assay
Three RNase protection assays were performed. (i) the determination of the minimal RNase A concentration needed to completely degrade free siRNA is described in the Supporting Information (SI). (ii) The effect of the N/P ratio on the protection efficiency against RNase A degradation was also evaluated. Briefly, siRNA–G4NH2 dendriplexes at different N/P ratios were formed in buffer as described earlier, and 6 μL dendriplex dispersions (equivalent to 250 ng siRNA) were incubated with 8 μL RNase A in TE 1X pH 8 buffer (0.162 μg RNase A per 1 μg siRNA) for 6 h at 37 °C; following the addition of RiboLock RNase inhibitor (RI, 1 μL = 40 U) and incubation for 30 min at 37 °C to stop degradation reaction.23 Dendriplexes were dissociated by adding 8 μL heparin (455 U per 1 μg siRNA) in TE 1X pH 8 buffer following incubation for 30 min at 37 °C. Samples were frozen at −20 °C overnight, and the integrity of siRNA was determined by gel electrophoresis at the same conditions as those applied for gel retardation assay. (iii) The third set of experiments consisted in determining the effect of RNase concentration on the siRNA integrity at a fixed N/P ratio. Briefly, siRNA–G4NH2 dendriplexes at N/P 5 were incubated with increased concentrations of RNase (0.35, 0.7, 1.0, 1.5, and 3.5 μg RNase A per 1 μg siRNA) for 6 h at 37 °C, following the treatments with RI and heparin, as described earlier. Appropriate controls (free siRNA and dendriplexes ± RNase A, and ± heparin treatments) were included in these experiments. Heparin decomplexation assay is described in SI.
In Vitro Release
siRNA–G4NH2 dendriplexes at different N/P ratios were formed in buffer as described earlier. A known volume of 0.1 M citrate/phosphate buffer (pH 5 or 7.4, to mimic intracellular endosomes/lysosomes and cytosol,29 respectively) was added to the dendriplexes dispersion (2.2 μg siRNA) to make 2 mL, and samples were incubated in a water bath at 37 °C. Aliquots of 50 μL were taken out at each time point, placed in a black 96-well microplate, and frozen at −20 °C. The collected samples were quantitatively analyzed using PicoGreen assay25 in Synergy 2 Microplate Reader (BioTek, VT) for the amount of siRNA released with time, which was calculated using a linear calibration curve (siRNA concentration vs fluorescent units) and taking into account the amount of free and complexed siRNA removed from the sample in each aliquot of 50 μL. Experiments were performed in triplicates (n = 3).
In Vitro Cytotoxicity
A549 cells (passages 5–8 from the original passage provided by ATCC) were seeded in 96-well microplate (5 × 103 cells per well) and cultured in 200 μL DMEM supplemented with 10% FBS and 1% AB (v/v) for 24 h at 37 °C and 5% CO2 (Thermo Scientific Incubator, NAPCO 8000WJ). For the cytotoxicity of free PAMAM G4NH2, cells were rinsed with PBS 1X buffer, the medium was replaced by 200 μL of regular culture medium containing increased concentrations of G4NH2 dendrimer (0–500 μM), and the cells were kept in the incubator for 48 h at 37 °C and 5% CO2. For the cytotoxicity of dendriplexes, cells were rinsed with PBS 1X buffer, and the medium was replaced by 100 μL DMEM containing increasing concentrations of siRNA–G4NH2 dendriplexes at N/P 30 (0–25 μM G4NH2, and 0–1.25 μM siRNA) formed in buffer as described earlier. Cells were kept in the incubator for 6 h at 37 °C and 5% CO2. They were then overlaid with 100 μL of regular culture medium (the dendriplexes were kept in contact with the cells) and incubated for 72 h at 37 °C and 5% CO2. The protocol for the in vitro cytotoxicity of dendriplexes was designed according to the in vitro gene knockdown experimental conditions, and therefore, slightly different from the protocol for dendrimer alone. The concentration ranges of G4NH2 and siRNA used in the cytotoxicity experiments reported here were designed in such way that the concentrations of G4NH2 and siRNA applied in all gene knockdown and aerosol characterization fall within that. Equally important, the incubation time in the cytotoxicity experiments falls within the usual time times (24–96 h) applied in gene knockdown experiments.30 The cell viability was assessed by MTS cell proliferation assay.31 Briefly, cells were rinsed with PBS 1X buffer, and 100 μL DMEM was added to the cells, followed by 20 μL MTS/PMS solution. Cells were incubated in this DMEM/MTS/PMS mixture for 4 h at 37 °C and 5% CO2. MTS is bioreduced into formazan (which is soluble in the culture medium) by dehydrogenase enzymes found in metabolically active cells.31 Absorbance of formazan at 490 nm was measured directly from the 96-well microplate (Molecular Devices, Spectra Max 250), and it is proportional to the number of living cells. Thus, cell viability (%) was calculated as the ratio between the absorbance of treated (incubated with free G4NH2 or dendriplexes) and untreated (G4NH2 and dendriplexes free) cells. Experiments were performed applying seven independent wells per each condition (n = 7).
In Vitro Gene Knockdown
In order to evaluate the knockdown efficiency achieved by siRNA-based dendriplexes, A549 cells stably expressing eGFP were developed as detailed in SI. After successfully establishing the eGFP A549 cell line, eGFP knockdown experiments were performed. We tested both a ds-siRNA targeting eGFP—the positive sequence, called siRNA(+), and an eGFP-mismatch ds-siRNA, the scramble or negative sequence called siRNA(−). Dendriplexes at different N/P ratios were formed between PAMAM G4NH2 and siRNA(+) and siRNA(−) as described earlier. Commercial transfection reagents (Lipofectamine 2000 and TransFast)30,32 were used as positive controls, and free siRNA (+ and −) was used as a negative control. All eGFP knockdown experiments were performed according to Lipofectamine 2000 protocol.30 First, eGFP stable A549 cells were seeded in a 75 cm2 cell culture flask and subcultured until approximately 90% confluence. The cell culture medium (DMEM supplemented with 10% FBS and 2.5 μg × mL–1 puromycin selective antibiotic) was changed every 2 days. Then, eGFP stable A549 cells (passages 10–20 from the original passage provided by ATCC) were seeded in a 24-well microplate (50,000 cells per well) and cultured in 500 μL DMEM supplemented with 10% FBS (v/v, no antibiotics) for 24 h at 37 °C and 5% CO2 (Thermo Scientific Incubator, NAPCO 8000WJ). Cells were rinsed with PBS 1X buffer, and 250 μL transfection medium (DMEM containing 80 nM of siRNA (+ or −) equivalent to 20 pmol per well) was added, and the transfection proceeded for 6 h at 37 °C and 5% CO2. The transfection medium was then replaced with 500 μL culture medium (DMEM + 10% FBS, no antibiotics), and the cells were returned to the incubator for 72 h at 37 °C and 5% CO2. Cells were resuspended in 1 mL PBS 1X buffer, and the median fluorescent intensity (MFI) was measured by flow cytometry (FACS, HWCRC 615 - BD LSR II Analyzer in Microscopy) with data from 10,000 to 20,000 cells. Untreated non-eGFP and eGFP stable A549 cells (no siRNA or dendriplexes) were used as further controls. The MFI level of eGFP expression in untreated eGFP stable A549 cells was taken as 100%. The % eGFP knockdown efficiency was calculated by correlating the eGFP expression of untreated stable eGFP A549 cells with that from transfected cells.
The effect of the propellant HFA-227 on the biological activity of the anti-eGFP ds-siRNA was also investigated. Briefly, a known amount of siRNA(+) dissolved in RNase free DI-water was placed in a pressure proof glass vial, frozen at −20 °C overnight, and lyophilized (Labconco Freeze Zone 1) at −47 °C and 0.055 mbar for 48 h. The vial was closed with a pressure valve (HiP, 15-11AF1) and propellant HFA-227 was filled into the vial with the help of a manual syringe pump (HiP, 50-6-15) and a home-built high-pressure filler. The siRNA(+) was stored in HFA-227 at 25 °C and saturation pressure of the propellant, simulating a pMDI formulation of free siRNA(+). After two months, the propellant HFA-227 was released by depressurization, and the siRNA(+) was allowed to dry at room temperature, following its dissolution in a known volume of RNase A free DI-water, making a siRNA(+) concentration of 10 nmol per 1 mL. As control, an aliquot of the lyophilized siRNA(+) was kept in the freezer at −20 °C instead of being stored in pressurized, liquid HFA. Gene knockdown experiments (as described earlier) were performed with the siRNA(+) to evaluate the effect of storage under a compressed HFA environment. All transfection experiments (siRNA as received, stored in HFA and freezer) were performed, at minimum, in triplicates (n = 3).
Preparation of Chitosan-g-Lactic Acid (CSLA) Microparticles Loaded with Dendriplexes
First, the biodegradable, water-soluble, and HFA-philic CSLA co-oligomer was synthesized according to our previous work.33 CSLA was composed by short segments of lactide [oligo(LA)] grafted onto low-Mw CS via ring-opening polymerization.33 Details about the synthesis and characterization can be found in SI. Emulsification diffusion33 was employed to prepare core–shell microparticles of CSLA loaded with siRNA–G4NH2 dendriplexes. Briefly, 15 mg CSLA was dissolved in 800 μL RNase free DI-water, and combined with 200 μL dispersion of siRNA–G4NH2 at N/P 10. The mixture (equivalent to 5–10 μg siRNA) was emulsified in 19 mL ethyl acetate using a sonication bath (VWR, P250D, set to 180 W) for 5 min at 15–20 °C making a water-in-oil (W/O) emulsion, which was quickly transferred to 180 mL ethyl acetate. Core–shell microparticles (CSLA as shell) loaded with siRNA–G4NH2 dendriplexes within their core were thus formed, collected via centrifugation (5000 rpm for 20 min), and dried in air-flow at room temperature.
Preparation of Mannitol Microparticles Loaded with Dendriplexes
siRNA–G4NH2 dendriplexes at N/P 10 were formed in buffer as described earlier, and 200 μL of a dendriplex dispersion (equivalent to 10 μg siRNA) was combined with 30 mg mannitol previously dissolved in 800 μL RNase free DI-water. The mixture was spray-dried (BUCHI Mini Spray Dryer B-290) using the following parameters: atomizing air flow = 473 L·h–1, aspiration = 70%, pump ratio = 5%, nozzle cleaner = 0, inlet temperature = 45 °C, outlet temperature = 30–33 °C. Nitrogen was the atomizing gas, and dry mannitol microparticles loaded with dendriplexes (white powder) were accumulated in the collection vessel at the end of the glass cyclone, from where they were collected to be used in further experiments.
Characterization of CSLA and Mannitol Microparticles Loaded with Dendriplexes
siRNA loading efficiency into microparticles was assessed by densitometry.7 Briefly, a known amount of particles was dissolved in 200 μL TE 1X pH 8 buffer and incubated at room temperature overnight, so that the water-soluble mannitol or CSLA shell could be broken down. Next, a known mass of heparin (equivalent to 455 U per 1 μg siRNA, based on the estimation that all siRNA was loaded into microparticles) was added to the mixture, which was vortexed until heparin dissolution. The system was incubated for 30 min at 37 °C, so that siRNA complexed with the G4NH2 could be released (see heparin decomplexation assay in SI). Samples were frozen at −20 °C overnight, and gel electrophoresis was performed at conditions applied for the gel retardation assay. The siRNA content encapsulated into the microparticles was quantified by densitometry using ImageJ 1.42q27 based on the electrophoresis images. Appropriate controls were employed—free siRNA mixed with heparin (positive control), and CSLA (or mannitol) mixed with heparin (no siRNA, negative control). Four independent batches (n = 4) of dendriplexes loaded into microparticles were used to calculate an average and standard deviation of the loading. In addition, the integrity of the siRNA after dendriplexes formation followed by encapsulation into CSLA or mannitol microparticles was also determined.
The size (diameter) of the microparticles was assessed by LS. Briefly, particles loaded with siRNA–G4NH2 at N/P 10 were dispersed in HPFP (2 mg per 1 mL) using a sonication bath, and measurements were performed at 25 °C using refractive index, viscosity, and dielectric constant of the HPFP, which is a model of propellant HFA that is liquid at ambient conditions.34 Next, HPFP was evaporated, and 1 mL RNase free DI-water was added to the microparticles to break down the CSLA shell (or mannitol) and release the siRNA from the dendriplexes. LS measurements were performed at 25 °C using refractive index, viscosity, and dielectric constant of the water, and thus, the hydrodynamic diameter of the dendriplexes was recorded, but at this time, in the presence of the CSLA (or mannitol) dissolved in the aqueous medium. SEM was used to investigate the geometrical size and morphology of the microparticles, which were dispersed in HPFP using a sonication bath. Several drops of the dispersion were deposited on a microscope cover glass, the HPFP was quickly evaporated by air flow, and the glass was sputter-coated with gold (Ernest Fullan) under vacuum for 40 s, following the SEM images.
Preparation of the pMDI Formulations and Evaluation of Their Physical Stability
A known mass of microparticles (CSLA or mannitol) encapsulating siRNA–G4NH2 dendriplexes (at N/P 10) was weighed into pressure proof glass vials (8412-B, West Pharmaceutical Services) and crimp-sealed (CroPharm, Inc.) with a 63 μL metering valves (EPDM, 3 M Drug Delivery Systems). A known volume of propellant HFA-227 was added with the help of a manual syringe pump (HiP, 50-6-15) and a home-built high pressure filler in order to make a 2 mg × mL–1 concentration.33 The particles were dispersed in the propellant using a sonication bath (VWR, P250D, set to 180 W, 15–20 °C).33 The physical stability was investigated via a sedimentation rate experiment, that is, by visually monitoring the quality of the dispersions as a function of time after stopping the mechanical energy from the sonication–digital images were taken according to the time.33 pMDI formulations of free siRNA–G4NH2 dendriplexes at N/P 10 were attempted, as described in SI. However, the free dendriplexes were found not to disperse in the propellant to any degree.
Aerosol Characterization of the pMDI Formulations
An eight-stage Andersen Cascade Impactor (ACI, Copley Scientific), fitted with a USP induction port and operated with a flow rate of 28.3 L × min–1 at 25 °C and 75% relative humidity,35 was used to evaluate the aerosol properties of the pMDI formulations, which were prepared as described earlier. Prior to each ACI test, the formulation was dispersed in the propellant with the help of a sonication bath for 10–15 min at 15–20 °C. After five actuations were fired to waste, the next 50–65 actuations (depending on the siRNA concentration in the formulation) were fired into the ACI, with an interval of 10 s between each actuation.33 After each test, the ACI was disassembled and had the actuator (AC), induction port (IP), and all stages rinsed with and kept in 20 mL RNase free DI-water for 6 h in order to break down the water-soluble CSLA shell (or mannitol) from the microparticles containing siRNA–G4NH2 dendriplexes. Samples were frozen at −20 °C overnight and lyophilized (Labconco Freeze Zone 1) at −47 °C and 0.055 mbar for 48 h. The collected powder was dissolved in 100 μL TE 1X buffer pH 8 and incubated with a known mass of heparin (equivalent to 455 U per 1 μg siRNA, as described in SI) for 30 min at 37 °C, in order to dissociate the siRNA from the PAMAM G4NH2. Samples were frozen (−20 °C) overnight, loaded into the slots of nondenaturing agarose gel following electrophoresis at conditions applied for gel retardation assay. The siRNA content in each ACI stage was quantified by densitometry using ImageJ 1.42q27 based on the electrophoresis images and appropriate controls (presence or absence of siRNA, as described earlier). The aerosol characteristics were thus calculated: (i) fine particle fraction [FPF: the siRNA content on the respirable stages of the ACI (from stage 3 to filter) over the total siRNA content released into the impactor (from IP to filter) excluding the siRNA remaining in the actuator];33 (ii) respirable fraction (RF: the siRNA content collected from stage 0 to filter over the total siRNA released into the impactor);36 (iii) % siRNA recovered, and siRNA content in a single puff dose; (iv) mass median aerodynamic diameter (MMAD); and (v) geometric standard deviation (GSD). MMAD and GSD were calculated as described in the literature.33 ACI experiments were performed in duplicates (n = 2).
Statistical Analysis
OriginPro 8 SR0 v8.0724 (B724) was used to perform statistical analysis. Data were compared using One-Way Analysis of Variance (ANOVA) followed by Tukey’s posthoc test. Means were considered statistically significant different with a p value <0.05.
Results and Discussion
Preparation and Characterization of siRNA–G4NH2 Dendriplexes
siRNA–G4NH2 dendriplexes at different N/P ratios were prepared and characterized with respect to their morphology, size and surface charge according to AFM, SEM and LS. The results are summarized in Table 1. Details of the characterization for one N/P ratio (N/P = 20), including AFM and SEM micrographs, are shown in Figure 1.
Table 1. Size of siRNA–G4NH2 Dendriplexes As a Function of the N/P Ratio As Determined by LS (hydrodynamic diameter) and SEM (geometric diameter)a.
| LS |
SEM | ||||
|---|---|---|---|---|---|
| N/P ratio | size (nm) | PDI | ζ (mV) | size (nm) | siRNA CE (%) |
| 5 | 267 ± 115 | 0.4 ± 0.2 | + 34 ± 9 | 285 ± 78 | 97.3 ± 0.6 |
| 10 | 246 ± 63 | 0.4 ± 0.2 | + 36 ± 7 | 285 ± 70 | 96.2 ± 1.5 |
| 20 | 262 ± 85 | 0.6 ± 0.2 | + 32 ± 4 | 207 ± 51 | 97.2 ± 0.6 |
| 30 | 254 ± 52 | 0.5 ± 0.1 | + 33 ± 3 | 257 ± 74 | 97.5 ± 0.8 |
Zeta potential (ζ) and siRNA complexation efficiency (CE, indirect measurement) are also shown. LS was performed with dendriplexes at 80 nM siRNA, and in 10 mM Tris-HCl pH 7.4 (for size) and pure water (for ζ). ImageJ was used to estimate the size of the dendriplexes from the SEM images: histograms of the measured diameters (> 400 particles) were fitted to Gaussian distributions, from which the average size and standard deviation was obtained.
Figure 1.
Size and morphology of siRNA–G4NH2 dendriplexes at N/P 20 as determined by LS (main distribution in the center), SEM (upper left inset), and AFM (lower left inset). Histogram and Gaussian fit to the diameter distribution obtained from SEM images (>400 particles) of the dendriplexes is also shown (upper right inset).
It was observed that the hydrodynamic diameter of the dendriplexes did not vary substantially with the N/P ratio. As measured by LS, the size average of all N/P ratios combined was 257 nm. This value was confirmed by SEM (size average of 258 nm), and since these techniques provide a good representation of the sizes of dendriplexes in solution, they were preferred to the sizes estimated from AFM images (∼100 nm). Discrepancies between the sizes of dendriplexes obtained from LS and AFM images have already been reported,28 and are likely due to sample preparation. As seen in the literature,28,37,38 the size of dendriplexes usually displays significant heterogeneities, as is the case for polyplexes prepared with other cationic polymers. This heterogeneity has been attributed to the electrostatic and entropic nature of the complexation process.37
The overall surface charge of the siRNA–G4NH2 dendriplexes were fairly similar, and did not show any specific trend as a function N/P either, with an average value of ∼34 mV, also in agreement with previous studies of dendriplexes with PAMAM dendrimers.38
Gel Retardation Assay
The ability of G4NH2 to form complexes with siRNA as a function of the N/P ratio was investigated by gel electrophoresis combined with PicoGreen assay. The results are summarized in Table 1 and Figure 2.
Figure 2.
siRNA complexation efficiency (indirect measurement) as a function of the N/P ratio, as quantified by PicoGreen Assay of residual free siRNA in the dispersion after preparation of the dendriplexes. Inset: Nondenaturing agarose gel electrophoresis of the corresponding dendriplexes: N/P 0.2 (lane 2), 0.5 (lane 3), 0.8 (lane 4), 1 (lane 5), 2 (lane 6), 3 (lane 7), 5 (lane 8), 10 (lane 9), 20 (lane 10), 30 (lane 11). Untreated siRNA control (300 ng) is shown in lane 1.
The complexation efficiency (CE) of G4NH2 to siRNA is seen to be low (<80%) at N/P < 1. However, almost all siRNA was complexed with the PAMAM G4NH2 dendrimer at N/P ratios >2 (siRNA CE > 95%). These quantitative results assessed by PicoGreen assay—measuring the uncomplexed siRNA remaining in solution after dendriplex formation, i.e., an indirect method of quantifying the siRNA CE—were confirmed by gel electrophoresis (inset in Figure 2). The siRNA band is seen to disappear in the gel as the N/P ratio increases, indicating that the siRNA was largely complexed with the PAMAM G4NH2 dendrimer and, thus, was unable to flow through the pores of the gel (compare lanes 2–11: siRNA–G4NH2 dendriplexes formed at N/P 0.2–30, with lane 1: untreated siRNA control). Therefore, these results reinforce the observation that highly positively charged PAMAM G4NH2 is able to condense siRNA into NPs very efficiently, since very little uncomplexed siRNA was found to be free in the supernatant after the complexation process (N/P > 2).
Protection of siRNA by PAMAM G4NH2 against RNase Degradation
siRNA is vulnerable to degradation by RNases,39 and one of the important potential advantages in using nanocarriers in siRNA therapeutic technologies is the ability to protect this frail cargo. Thus, in order to evaluate the ability of PAMAM G4NH2 to protect siRNA from RNase degradation, siRNA–G4NH2 dendriplexes were formed at several N/P ratios and incubated with the RNase A (the lowest concentration found to digest free siRNA completely −0.162 μg RNase A per 1 μg siRNA – as seen in the SI) for 6 h at 37 °C, following RNase inhibitor and heparin treatments, and gel electrophoresis so as to release and quantify the nondegraded siRNA. The results are presented in Figure 3. PAMAM G4NH2 is seen to efficiently protect siRNA from RNase A degradation upon complexation. As seen in Figure 3, for all tested N/P ratios, the siRNA released from the dendriplexes after being kept in contact with RNase A (lanes 3, 7, 11, and 15) was comparable to the siRNA extracted from the dendriplexes which were not exposed to RNase A (lanes 5, 9, 13, and 17 - positive controls, respectively).
Figure 3.
RNase protection assay (non-denaturing agarose gel electrophoresis) of the siRNA–G4NH2 dendriplexes as a function of the N/P ratio. Dendriplexes incubated in the absence (−) or presence (+) of the treatments: RNase A (0.162 μg per 1 μg siRNA) for 6 h at 37 °C, followed by 1 μL (40 U) RiboLock RNase inhibitor for 30 min at 37 °C to block RNase activity, and heparin (455 U per 1 μg siRNA) for 30 min at 37 °C to dissociate the siRNA from the dendrimer. Aqueous medium: TE buffer 1X pH 8. Untreated siRNA control (300 ng) before (lane 1) and after (lane 2) incubation with RNase A.
Next, siRNA–G4NH2 dendriplexes formed at N/P 5 were incubated with increasing concentrations of RNase A (0.35, 0.7, 1.0, 1.5, and 3.5 μg per 1 μg siRNA) for 6 h at 37 °C, following RNase inhibitor and heparin treatments, and gel electrophoresis, in order to probe the protection efficiency provided by the dendrimers at increasing RNase A concentrations. The results are shown in Figure 4 and again indicate that the siRNA was sufficiently protected from RNase degradation upon complexation with PAMAM G4NH2. Even at this relatively low N/P ratio (N/P of 5), the siRNA was well protected when kept in contact with very high concentrations of RNase A (3.5 μg per 1 μg siRNA); in Figure 4, compare the siRNA from dendriplexes incubated in presence (lane 20) and absence (lane 22) of RNase A.
Figure 4.
RNase protection assay (nondenaturing agarose gel electrophoresis) of the siRNA–G4NH2 dendriplexes (N/P 5) as a function of the RNase A concentration. Dendriplexes incubated in presence (+) or absence (−) of the treatments: RNase A (0.35, 0.7, 1.0, 1.5, and 3.5 μg per 1 μg siRNA, in lanes 4–7, 8–11, 12–15, 16–19, 20–23, respectively) for 6 h at 37 °C, followed by 1 μL (40 U) RiboLock RNase inhibitor for 30 min at 37 °C to block RNase activity, and heparin (455 U per 1 μg siRNA) for 30 min at 37 °C to dissociate the siRNA from the dendrimer. Aqueous medium: TE buffer 1X pH 8. Untreated siRNA control (250 ng) in lane 1, after incubation with heparin (lane 2) and 0.35 μg RNase A per 1 μg siRNA (lane 3).
In Vitro Release of siRNA from PAMAM G4NH2
The siRNA release from the dendriplexes was evaluated in vitro at 37 °C, in citrate/phosphate buffer at pHs 7.4 and 5, in order to mimic cytosol and endosomes/lysosomes, respectively.29 The results are shown in Figure 5, and indicate that the siRNA in vitro release profile from dendriplexes (N/P 10, 20, and 30) is highly dependent upon the pH. The release is much slower at acidic medium than at physiological pH. At low pHs (<5) all primary and tertiary amines from PAMAM G4NH2 are protonated,40 and consequently the electrostatic interactions between the positively charged dendrimer and negatively charged siRNA increase, which results in stronger binding and lower siRNA released at low pH. At physiological pH 7.4 the tertiary amines are deprotonated, and only the primary amines are protonated,40 which decreases the electrostatic interactions between siRNA and G4NH2, and as a result, the release of the siRNA increases at higher pH.
Figure 5.
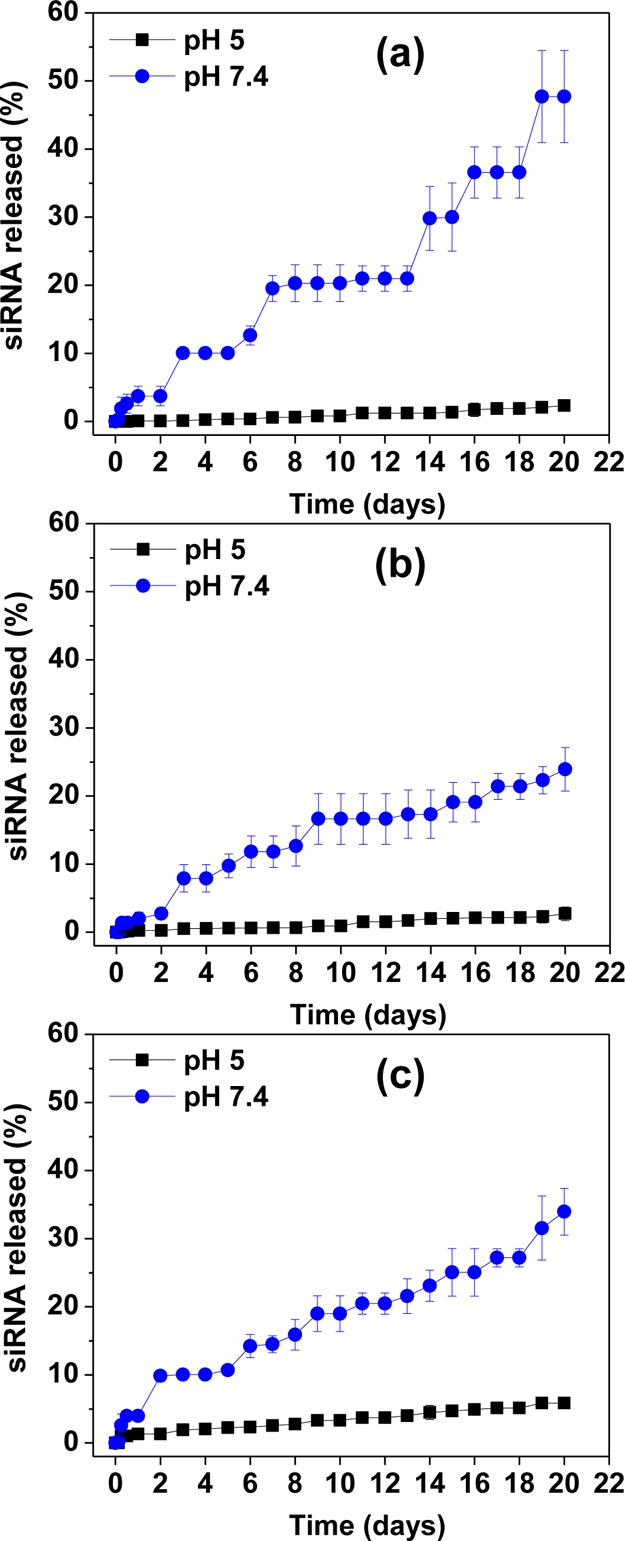
In vitro release of siRNA from dendriplexes in 0.1 M citrate/phosphate buffer at pH 5 and 7.4 (mimicking intracellular endosomes/lysosomes and cytosol, respectively), at 37 °C for dendriplexes at N/P ratio (a) 10; (b) 20; and (c) 30.
This degree of compactness observed at lower pHs is also manifested by the fact that no burst release is observed at pH 5.0. On the other hand, at physiological pH, a burst release was observed after 1–2 days, and it was ∼3.5% or 5.5 pmol—based on the initial loading of siRNA into dendriplexes. After 3–5 days, it was observed that 8–10% siRNA was released from the dendriplexes, which corresponds to 13–16 pmol. The results in Figure 5 show that 48, 24, and 34% of the total siRNA complexed with G4NH2 is released from the dendriplexes at N/P 10, 20, and 30, respectively, after 20 days in citrate/phosphate buffer pH 7.4 at 37 °C. While no literature results are available for the in vitro release of siRNA from PAMAM dendriplexes, we can contrast the results obtained here with the release of oligonucleotide (ON) from PEI polyplexes.41 At N/P 15 and 40 in PBS pH 7.4 and 37 °C, 55% ON was released from PEI after 20 days (i.e., ON was released faster from PEI at N/P 40 than N/P 15), a result somehow similar to that observed here–siRNA released slightly faster from G4NH2 at N/P 30 than N/P 20 in buffer pH 7.4. This is likely due to hydrophilization41 operated by G4NH2, i.e., dendriplexes formed at higher N/P have increased G4NH2 content, which can contribute for more rapid entry of water molecules into the dendriplexes. Water molecules can penetrate throughout the G4NH2 interior, cause the dendrimer structure to swell (due to the favorable interaction of water molecules with the primary and tertiary amines),42,43 and thus release the siRNA. However, the amount of siRNA released from dendriplexes at N/P 30 was statistically significant different from that at N/P 20 at 12 h and day 2 only. Since the in vitro release was conducted with 24 time points, we demonstrate that, in 92% of all time points, the siRNA released from dendriplexes at N/P 20 and 30 was not statistically significantly different (n = 3 for each time point, One-Way ANOVA followed by Tukey’s posthoc test, p value <0.05).
In Vitro Cytotoxicity of PAMAM G4NH2 and siRNA−G4NH2 Dendriplexes
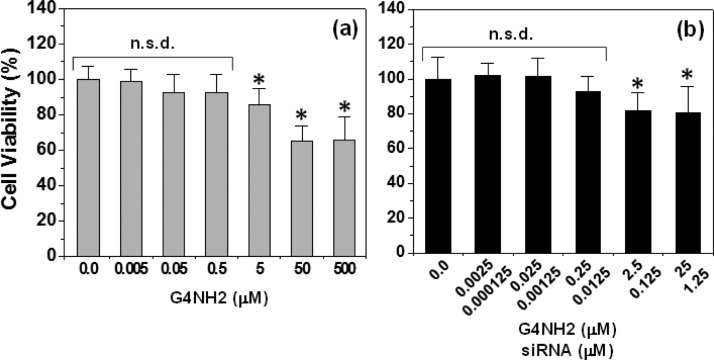
Cationic dendritic polymers, such as polypropyleneimine (PPI), poly-l-lysine (PLL), and PAMAM, may induce significant in vitro cytotoxicity due to the high density of cationic groups on their surface.44 There are many reports discussing the concentration- and generation-dependent toxicity of dendrimers,13,28,44−46 which is a critical factor to be considered when evaluating the potential of dendrimers as nanocarriers for siRNA delivery. Here, in vitro cytotoxicity studies were performed with A549 cells by incubating them with free PAMAM G4NH2, and siRNA–G4NH2 dendriplexes at N/P 30, according to the MTS cell proliferation assay.31 The cell viability results are shown in Figure 6.
Figure 6.
In vitro cytotoxicity of (a) PAMAM G4NH2 alone, and (b) siRNA–G4NH2 dendriplexes at N/P 30 in increasing concentration of G4NH2 (and thus siRNA – both concentrations shown). MTS assay on A549 lung alveolar cell line. * = statistically significantly different compared to untreated cells as control; and no statistical significant difference (n.s.d.) compared to untreated cells as control (n = 7, One-Way ANOVA followed by Tukey’s posthoc test, p value <0.05).
The results presented in Figure 6a indicate that the viability of A549 was >85% when the cells were in contact with PAMAM G4NH2 for 48 h at concentrations up to 5 μM. This result is in agreement with previous literature reporting that the viability of A549 cells was >80% for G4NH2 at low concentrations of 0.7 and 7 μM, as evaluated by MTT cell proliferation assay after 72 h of incubation.47 Several other works report different cytotoxicity results for PAMAM G4NH2,45,46,48 and thus, the cytotoxicity of G4NH2 is dependent upon concentration, cell type, and incubation time.49 PAMAM dendrimers are known to be cytotoxic by causing membrane rupture, substantially related to the formation of cavities in the cellular membrane.49 Toxicity and other dendritic properties (interactions, mechanisms of cellular uptake, and intracellular fate) are most likely governed by the surface groups.50
The toxic effects of siRNA–G4NH2 dendriplexes at N/P 30 (the highest N/P ratio applied in the gene knockdown experiments discussed next) to A549 were also investigated via MTS assay after 72 h of contact of dendriplexes with the cells. The results in Figure 6b indicate that the cytotoxicity of the dendriplexes at N/P 30 was low, even at the highest concentration studied–viability of A549 cells was ∼80% at 25 and 1.25 μM of G4NH2 and siRNA, respectively. In addition, the in vitro viability of A549 cells incubated with siRNA–G4NH2 dendriplexes at N/P 30 (72 h) is comparable to that with G4NH2 alone (48 h) at the same G4NH2 concentration, e.g., cell viability of ∼90% (Figure 6a and b) at 0.25 μM G4NH2; and ∼75% and 80% (Figures 6a and b, respectively) at 25 μM G4NH2. Moreover, the in vitro viability of A549 cells after incubation with 5 and 50 μM G4NH2 alone (48 h) was found not statistically significantly different from that with 25/1.25 μM G4NH2–siRNA at N/P 30 (72 h) (n = 7, One-Way ANOVA followed by Tukey’s posthoc test, p value <0.05). These results suggest that siRNA–G4NH2 dendriplexes did not become more toxic than G4NH2 alone.
It has been reported in literature that J-774 cells (macrophage-like cell line) showed 90–95% viability after 24 h incubation with siRNA–G4NH2 dendriplexes at N/P 10 (0.05 μM siRNA) as measured by MTT assay.51 Here, at the same 0.05 μM siRNA concentration, A549 cells showed ∼85% viability. This result is interesting since the time of incubation and N/P ratio were both 3-fold higher than that reported in literature.51 Other work showed that the viability of A549 was reduced to 45% when the cells were incubated with G5NH2/ON (20 mer and 0.16 μM) dendriplexes at N/P 15 for 4 h, following measure by CellTiter-Blue.19 Here, at similar 0.125 μM RNA concentration, the viability of A549 cells was ∼80%. Thus, it can be seen that the in vitro cell viability resulting from the delivery of siRNA using PAMAM dendrimers is variable at some extent and depends on several factors—e.g., dendrimer surface groups and generation, physicochemical properties of the siRNA–dendrimer dendriplexes, assay, experimental conditions, cell type and passage, and certainly, some interlab variability can be observed as well.
The in vitro cytotoxicity results are necessary for the gene knockdown studies. When working in concentration regions of no toxicity, gene knockdown is not confounded with nonspecific toxicity. At the harshest conditions of the in vitro cytotoxicity studies, the cell viability of A549 was still ∼80% after 72 h of incubation with siRNA–G4NH2 dendriplexes at N/P 30 containing 25 and 1.25 μM G4NH2 and siRNA, respectively. In contrast, at the harshest condition of the in vitro gene knockdown experiments (discussed next), the A549 cells were incubated for 6 h only, with dendriplexes at N/P 30 containing 1.95 μM and 80 nM G4NH2 and siRNA, respectively. At these G4NH2 and siRNA concentrations, the viability of A549 cells was at least ∼90% (Figure 6b), and expected to be higher at smaller N/P ratios. It is worthwhile to mention that cellular debris (due to dead or damaged dying cells) was gated out during the FACS analyses to prevent any toxic effects of the G4NH2 alone and/or siRNA–G4NH2 dendriplexes to mask the true gene knockdown.
In Vitro Gene Knockdown of siRNA–G4NH2 Dendriplexes
FACS of A549 cells stably expressing eGFP was used to investigate the gene silencing efficiency of siRNA–G4NH2 dendriplexes. Lipofectamine 2000 (LF) and TransFast (TF) were the commercial transfection reagents selected as positive controls. TF is originally designed as a transfection reagent for DNA,32 but it has shown gene suppression of ∼60–90% when used to deliver siRNA in vitro.52 Free siRNA was used as the negative control. A549 cells that were not transduced (no eGFP) were used as reference. Cellular debris was gated out during FACS to prevent the small toxicity of the nanocarriers to mask true gene knockdown. Transfection of eGFP stable A549 cells was performed with ds-siRNA targeting eGFP, the positive sequence siRNA(+), and an eGFP-mismatch ds-siRNA, the nonrelevant negative sequence siRNA(−). The results are shown in Figure 7, and indicate that the gene knockdown efficiency of siRNA(+)–G4NH2 dendriplexes varied from 22 to 37%, and was intermediate between the positive controls (LF at 72% and TF at 70%), and free siRNA(+) (2.8%). The gene silencing efficiency of the eGFP cells treated with the commercial reagents and dendriplexes at N/P 5, 10, 20 and 30, was found to be statistically significantly different from that of eGFP cells treated with free siRNA(+) only. Additionally, no statistically significant difference was found when comparing the eGFP knockdown efficiency as a function of the N/P ratio (minimum n = 3, One-Way ANOVA followed by Tukey’s posthoc test, p value <0.05).
Figure 7.
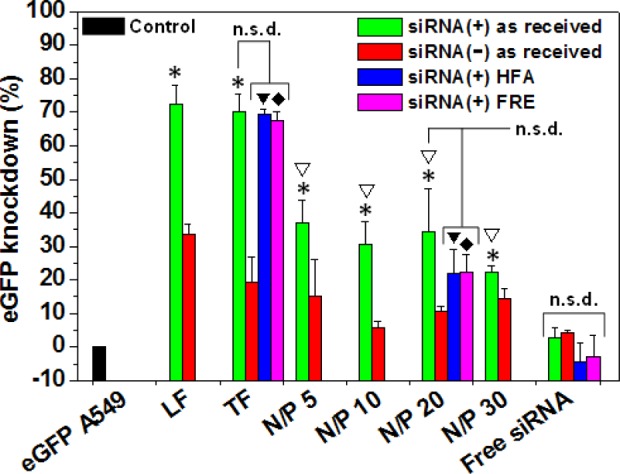
In vitro knockdown of eGFP expression in A549 cells stably expressing eGFP. siRNA–G4NH2 dendriplexes were prepared with siRNA as received from the supplier at N/P 5, 10, 20, and 30; with lyophilized siRNA stored in HFA-227 (HFA, at 25 °C and saturation pressure of the propellant) and freezer at −20 °C (FRE, at 253 K) for 2 months, at N/P 20. Specificity of the knockdown (siRNA(+) sequence, anti-eGFP) is maintained by comparison to effects with the siRNA(−) sequence (scramble). Lipofectamine 2000 (LF) and Transfast (TF) were the commercial transfection reagents used as positive controls, and free siRNA was the negative control. G4NH2 concentration at N/P 30 corresponds to 1.95 μM, and siRNA concentration in all systems was 80 nM. *,▼,⧫ = statistically significantly different compared to eGFP A549 cells treated with free siRNA(+) as received; and ∇ = no statistically significant difference (n.s.d.) among N/P 5, 10, 20, and 30 prepared with siRNA(+) as received (minimum n = 3, One-Way ANOVA followed by Tukey’s posthoc test, p value <0.05).
Few reports in literature have investigated the in vitro knockdown efficiency by siRNA or antisense oligonucleotide (AON) delivered via dendriplexes to A549 lung alveolar epithelial cells. Dendrosomes (dendriplexes formed between G5NH2 and AON incorporated into liposomes) have shown suppression of the PKC-α gene expression between ca. 27–74% at messenger RNA (mRNA) and protein levels, respectively.19 Dendriplexes formed between folate–PEG appended to cyclodextrin–G3NH2 conjugates and siRNA have provided luciferase knockdown ranging from ∼15–50%,3 and up to 80% with siRNA–G7NH2 dendriplexes.18 Such variability in the in vitro gene silencing efficiency is expected due to the differences in the target gene, assay to assess the knockdown, experimental conditions during transfection (e.g., N/P ratio, treatment times, siRNA and AON concentrations), generation of the PAMAM dendrimer, and presence of other molecules/structures in the carrier.
While no gene knockdown result has been reported for siRNA–G4NH2 dendriplexes on A549 cells, we contrast our results with the literature for other cell types. siRNA–G4NH2 dendriplexes at N/P 10 caused 12.5 and 22% inhibition of eGFP expression in J-774 (macrophage-like) and T98G (human glioblastoma) cell lines, respectively,51 and 10% eGFP knockdown in C-166 (mouse yolk sac embryo) cells.48 Thus, the eGFP silencing efficiency (∼22–37%) obtained with siRNA–G4NH2 dendriplexes on the A549 alveolar epithelial cells is comparable to other cell types and falls within the range reported for other PAMAM-based dendriplexes (with and without the help of other molecules/structures). Despite this low-to-moderate gene silencing in vitro, the proposed siRNA–G4NH2 dendriplexes have the potential to demonstrate gene silencing in vivo (following optimization, but such is not the focus of this work), since PAMAM-based nanocarreirs have shown promising results in the delivering siRNA in vivo, even if the gene silencing efficiency in vivo is lower compared to that observed in vitro.3,23,53 This is due to the siRNA potency, i.e., very small amounts of siRNA (once properly delivered to the target site) are shown sufficient for effective in vivo gene silencing.54−56
It is worth noticing a reduction in eGFP expression in A549 cells is also observed when the scramble siRNA(−) was delivered using LF (34%), TF (19.5%), and G4NH2 (6–15%): Figure 7. It has been shown that LF/siRNA(−) complexes cause measurable and undesirable gene silencing ∼25–50% in different cell types.57,58 This unwanted eGFP suppression is most likely due to off-target effects,59 which can be due to toxicity of nanocarrier,57 and also due to the similarity between the nucleotide sequence from the siRNA(−) and short motifs in mRNA and other unrelated genes not targeted during the transfection.59,60 Alterations in gene expression in vitro and in vivo have been also reported for several types of nanocarriers, including linear and branched PEI,61 PEG–PEI,61 polypropylenimine (PPI),62 diaminobutane (DAB).62 Off-target effects in RNAi are quite common and an issue of consideration when developing RNAi technologies,60 but may be hard to avoid,59 and are still not well understood.61
In Vitro Gene Knockdown of siRNA–G4NH2 Dendriplexes Exposed to Propellant HFA
In order to test whether the biological activity of the siRNA was preserved after the formulation in the pMDI (under HFA atmosphere), the gene knockdown activity of dendriplexes formed with siRNA(+) that was lyophilized and stored under HFA-227 at 25 °C and saturation pressure of the propellant for two months, was compared to a control (lyophilized but stored at −20 °C). Gene knockdown experiments were performed on eGFP A549 cells as discussed above, with dendriplexes having an N/P ratio of 20, and the results are shown in Figure 7. Even after storage conditions in propellant HFA, it can be observed that the siRNA(+) was still biologically active to silence the eGFP expression in A549 cells: ∼69% and 22% eGFP knockdown for TF and dendriplexes at N/P 20, respectively. Similar values were obtained with the control siRNA(+) that was not stored under HFA-227, but at −20 °C in freezer (FRE)—67% and 22% for TF and N/P 20, respectively. The gene silencing efficiency was found to be statistically significant between the eGFP cells treated with TF/siRNA(+) and dendriplexes at N/P 20–for both storages in HFA and FRE, when compared to cells treated with free siRNA(+) as received. Most importantly, the gene knockdown efficiency provided by dendriplexes at N/P 20 (and TF) prepared with siRNA(+) as received, stored in HFA and FRE, was found not to be statistically significantly different (minimum n = 3, One-Way ANOVA followed by Tukey’s posthoc test, p value <0.05). Collectively, these results indicate that the biological activity of the siRNA is kept after long-term exposure in propellant HFA.
Preparation and Characterization of Microparticles Loaded with Dendriplexes
There are major obstacles in the regional delivery of the dendriplexes to the lungs when using OI devices. While the dendriplexes are in the order of nanometers in size, the optimum aerosol size for deep lung deposition is in the micrometer range (0.5–5 μm).63 Particles with an aerodynamic diameter <0.5 μm may be exhaled, while those >5 μm tend to be deposited in the mouth and throat, ending up in the digestive tract. Moreover, alveolar macrophages are most selective to particles of 1–3 μm, and less efficient toward larger (≳6 μm) and smaller (≲260 nm) particles.12 In the case of pMDIs, another hurdle that needs to be overcome is the physical stabilization of the microparticle dispersions (suspension) in the low dielectric propellant HFA.64,65
It is necessary, therefore, to develop particle engineering strategies capable of forming stable suspension of the dendriplexes in the form of microparticles suitable for deep lung deposition and able to minimize the uptake by alveolar macrophages. We demonstrate in this work two such strategies. One consists in encapsulating the dendriplexes within mannitol microparticles, via spray drying; the other consists in using a CSLA co-oligomer shell that is formed during emulsification diffusion. Mannitol was chosen as it has high aqueous solubility, it is a generally recognized as a safe (GRAS) excipient widely used as bulking agent and nonactive carrier in dry powder inhalers.66 Mannitol particles alone also have shown to have less adhesion/cohesion in propellant HFA, slower sedimentation rates, and superior aerosol performance than other sugars, such as lactose.67 The CSLA co-oligomer was chosen because it is water-soluble and interfacially active at the W/O interface of the emulsion droplets (it migrates/resides at the interface formed between the small water droplets into the large ethyl acetate organic phase).65 During the emulsification diffusion, the dendriplexes are kept within the water emulsion droplets, and then encapsulated by the CSLA co-oligomer residing at the W/O interface of the emulsion droplets. Thus, the organic phase is not expected to be in contact with the siRNA of the dendriplexes and/or affect somehow the dendriplex aggregates within the CSLA microparticles. In addition, the CSLA co-oligomer is degradable and non-HFA soluble–i.e., the shell will not disintegrate in HFA but will break down when in contact with the fluid lining the lungs.65 The ester groups from the LA are HFA-philic,64,68 and microparticles formed with CSLA shell have shown enhanced physical stability in propellant HFAs.33,36,65 An important detail is that the nonactive ingredients used to form these microparticles (mannitol and CSLA) can readily dissolve in aqueous media65,66 as the fluid lining the lungs, thus releasing the nanosized dendriplexes, and consequently minimizing the alveolar macrophage uptake. In addition, it is also worthwhile to point out that we have previously shown that CSLA does not impact the gene transfer potential of CSLA-based microparticles as carriers for CS-pDNA complexes.33
The size and morphology of the CSLA and mannitol microparticles containing siRNA–G4NH2 dendriplexes at N/P 10 were evaluated by LS and SEM. The results are shown in Figure 8. Microparticles were loaded with dendriplexes at N/P 10 instead of N/P 5 and 20 (even though dendriplexes at N/P 5, 10, and 20 resulted in similar in vitro gene knockdown efficiency, as shown in Figure 7) because N/P 10 produced slightly smaller and more stable dendriplexes in solution (please refer to Table 1) than did N/P 5 and 20.
Figure 8.
Size and morphology of (a) mannitol and (b) CSLA engineered microparticles loaded with siRNA–G4NH2 dendriplexes at N/P 10 as determined by LS (main distribution on right) and SEM (lower left inset). Particles were dispersed in HPFP (2 mg × mL–1) to perform LS, and after that, the HPFP was evaporated, and 1 mL DI-water was added to dissolve the mannitol or CSLA shell, and LS was performed again, but at this time, the size of the dendriplexes released from the mannitol (or CSLA) was measured (upper left inset). Nondenaturing agarose gel electrophoresis (upper right inset) showing the integrity of siRNA after its release from mannitol (or CSLA) shell and upon incubation of the dendriplexes in aqueous heparin solution (455 U per 1 μg siRNA) for 30 min at 37 °C. Untreated siRNA (250 ng) as positive control in lane 1; mixture of G4NH2, mannitol (or CSLA) and heparin (but no siRNA) as negative control in lane 2; siRNA–G4NH2 dendriplexes at N/P 10 loaded into mannitol (or CSLA) microparticles after incubation with aqueous heparin in lane 3.
The hydrodynamic diameter of CSLA and mannitol microparticles measured by LS was found to be 2.0 ± 0.8 μm and 4.6 ± 0.9 μm, respectively, as shown in Figure 8; average and standard deviation were calculated based on eight (n = 8) independent batches. These results suggest that the spherical dendriplexes carrying microparticles are in the desired size range for appropriate deep lung deposition.67
It is worthwhile to mention that the size of the dendriplexes at N/P 10 was measured by LS after contacting the microparticles with aqueous solution, and it was found to be 236 ± 75 nm and 192 ± 38 nm for CSLA and mannitol, respectively. Size distribution by LS is shown as inset in Figure 8; average and standard deviation calculated based on five (n = 5) independent experiments. Thus, the hydrodynamic diameter of the dendriplexes at N/P 10 released from the shell are readily comparable to those (Table 1) before loading into CSLA or mannitol microparticles and are nonstatistically significantly different (n = 5, One-Way ANOVA followed by Tukey’s posthoc test, p value <0.05). This demonstrates that the process of forming the microparticles does not seem to induce any undesirable irreversible aggregation of the dendriplexes.
The siRNA loading efficiency (% siRNA encapsulated into microparticles from the initial content complexed with dendrimers) into CSLA and mannitol microparticles was measured via densitometry using ImageJ,27 based on the images obtained from gel electrophoresis (insets in Figure 8) using the protocol discussed in Experimental Section. The siRNA loading efficiency within the microparticles of 49.0 ± 8.4% was found for CSLA (230 ± 20 ng siRNA per 1 mg CSLA particles), and 25.4 ± 4.0% (229 ± 47 ng siRNA per 1 mg mannitol particles) for mannitol. The final siRNA yield was 65% and 50% (w/w) after emulsification diffusion and spray drying, considering the initial content of CSLA and mannitol, respectively.
The integrity of the siRNA after particle preparation was also observed using the same gel images (inset in Figure 8). The siRNA bands after particle preparation was found to be very similar (position in the gel) to the untreated siRNA control (compare lanes 1 and 3). The intensity of the band in lane 3, which corresponds to the siRNA released from microparticles, is weaker due to the fact that less siRNA is added to the gel (see loading efficiency above). Collectively, these results demonstrate the feasibility of encapsulating siRNA-based dendriplexes into CSLA and mannitol microparticles via emulsification diffusion and spray drying processes, respectively. The siRNA was successfully loaded into such microparticles (which showed appropriate size for formulation in pMDIs), it conserved its integrity, and the hydrodynamic diameter of the dendriplexes was kept after particle processing and shell dissolution.
Physical Stability of Microparticles Loaded with Dendriplexes in Propellant HFA
Sedimentation rate experiments of the microparticles containing dendriplexes at 25 °C and saturation pressure of the propellant were performed in order to evaluate their physical stability in HFA. Microparticles were weighed into pressure proof glass vials, crimp sealed with 63 μL metering valves, and a known volume of propellant HFA-227 was added to make 2 mg × mL–1. The particles were dispersed with the aid of a sonication bath for 30 min at 15–20 °C. Stability of the formulations was determined visually via sedimentation rate/flocculation as a function of the time after stopping the mechanical energy input from the sonication bath. Digital images were taken, and the results are presented as insets in Figure 9.
Figure 9.
Aerosol properties of pMDI formulations prepared with siRNA–G4NH2 dendriplexes at N/P 10 loaded into (a) mannitol and (b) CSLA microparticles. pMDI formulations at 2 mg particles (mannitol or CSLA) per 1 mL in HFA-227 at 25 °C, and saturation pressure of the propellant. siRNA concentration of 290–550 ng × mL–1 for pMDI formulations prepared with mannitol loaded with dendriplexes, and 420–505 ng × mL–1 for those prepared CSLA loaded with dendriplexes. AC, IP, and F refer to actuator, induction port and filter, respectively. Insets: Physical stability of freshly prepared pMDI formulations.
Dispersions of both CSLA and mannitol microparticles containing siRNA–G4NH2 dendriplexes at N/P 10 in propellant HFA-227 showed good physical stability. While some aggregation could be observed on the walls of the canister and in the bulk propellant, no strong flocculation/irreversible aggregation was observed (insets in Figure 9). Aggregates formed due to sedimentation could be easily redispersed by simple manual agitation. In the case of CSLA, the improved physical stability compared to dendriplexes alone, which are not dispersible in propellant HFA (see SI) arises due to the enhanced solvation of the ester groups from (LA chains) of CSLA by the propellant HFA.64,68 In the case of the mannitol, the reasons for the enhanced stability are less clear, but we expect to have an improved density matching to that of HFA-227 since spherical spray-dried mannitol microparticles produce densities ∼1.47–1.51 g × cm–3 (depending on the spray-drying conditions)69 and the density of HFA-227 is 1.39 at 25 °C.34 Moreover, the fact that the cohesive force for mannitol in HFA is relatively lower compared to other those of other sugars, such as lactose,67 indicates an enhanced solvation of that surface in the propellant, but this is a relative observation, and no quantitative comparison with the cohesive interaction between CSLA particles and those with mannitol is available.
Aerosol Performance of the pMDI Formulations with the Engineered Microparticles
An eight-stage ACI was used to quantify the aerosol properties of the HFA-based formulations containing the mannitol and CSLA engineered microparticles, with siRNA–G4NH2 dendriplexes (N/P 10) loaded within their core. As described earlier, free dendriplexes (negative control, with no mannitol or CSLA shell) did not disperse at all in propellant HFA-227 (the dendriplexes remained stuck onto the walls of the canister, and thus ACI tests could not even be performed). The siRNA content for the microparticle formulations was quantified by incubation of each ACI stage with RNase free DI-water (which broke down the mannitol or CSLA shell, thus releasing the dendriplexes to the aqueous solution) followed by freezing, lyophilization, heparin decomplexation assay (which dissociates the siRNA from the G4NH2), gel electrophoresis, and densitometry using the gel images, as described earlier. A summary of the results is shown in Table 2, and plotted as % in Figure 9.
Table 2. Aerosol Performance of pMDI Formulations Prepared with Mannitol and CSLA Engineered Microparticles Loaded with siRNA−G4NH2 Dendriplexes at N/P 10a,b.
| siRNA–G4NH2
dendriplexes loaded into microparticles of |
||
|---|---|---|
| stage | mannitol | CSLA |
| actuator (AC) | 74.3 ± 27.6 | 191.8 ± 13.7 |
| induction Port (IP) | 109.6 ± 87.4 | 403.1 ± 61.2 |
| stage 0 (9.0 - 10.0 μm) | 38.9 ± 0.4 | 72.2 ± 17.5 |
| stage 1 (5.8 - 9.0 μm) | 42.9 ± 14.1 | 44.0 ± 47.6 |
| stage 2 (4.7 - 5.8 μm) | 39.8 ± 22.2 | 87.5 ± 0.0 |
| stage 3 (3.3 - 4.7 μm) | 43.1 ± 2.2 | 43.7 ± 28.9 |
| stage 4 (2.1 - 3.3 μm) | 35.1 ± 8.6 | 112.1 ± 33.3 |
| stage 5 (1.1 - 2.1 μm) | 33.0 ± 6.9 | 108.5 ± 5.2 |
| stage 6 (0.7 - 1.1 μm) | 33.7 ± 5.6 | 75.6 ± 52.8 |
| stage 7 (0.7 - 0.4 μm) | 42.8 ± 12.9 | 99.7 ± 46.9 |
| filter (0.0–0.4 μm) | 23.1 ± 32.6 | 80.7 ± 17.6 |
| FPF (%) | 48.9 ± 5.7★ | 46.1 ± 2.5★ |
| RF (%) | 77.4 ± 9.9▲ | 64.3 ± 4.0▲ |
| recovery (%) | 28.0 ± 10.5° | 84.8 ± 0.1° |
| single puff dose (ng) | 9.5 ± 3.2∇ | 26.4 ± 0.7∇ |
| MMAD (μm) | 2.6 ± 0.5⧫ | 1.9 ± 0.7⧫ |
| GSD | 3.8 ± 0.4▼ | 3.7 ± 0.1▼ |
★, ▲, ⧫, ▼ = no statistically significant difference; ○, ∇ = statistically different (n = 2, One-Way ANOVA followed by Tukey’s posthoc test, p value <0.05).
pMDI formulations at 2 mg particles (mannitol or CSLA) per 1 mL in HFA-227 at 25°C, and saturation pressure of the propellant. siRNA concentration of 290–550 ng × mL–1 in pMDI formulations prepared with dendriplexes-loaded into mannitol, and 420–505 ng × mL–1 in those prepared CSLA. Results in ng siRNA ± deviation for n = 2 (two independent canisters) and 50–65 actuations each, from AC to Filter.
The FPF, an important aerosol characteristic that serves as a measure of the therapeutically beneficial portion of the inhaled mass of siRNA which would reach the lower respiratory tract,70 was determined from the ACI results. The FPF for mannitol and CSLA microparticles was determined to be 49% and 46%, respectively, which is excellent, falling within those of commercial HFA-based pMDIs for small-molecule therapeutics (30–55% on average)65,71 even though no optimization was attempted in our case. It is worth noticing that FPFs for both formulations were not significantly different (n = 2, One-Way ANOVA followed by Tukey’s posthoc test, p value <0.05). The respirable fraction (RF) (siRNA content collected from stage 0 to filter over the total siRNA released into the impactor36) was found to be slightly higher for pMDIs formulated with mannitol microparticles (77%) than those with CSLA (64%), but these results were not statistically significantly different from each other (n = 2, One-Way ANOVA followed by Tukey’s posthoc test, p value <0.05).
The MMAD and GSD are properties that characterize the particles in the aerosol spray. MMAD represents the aerodynamic diameter (AD) on a mass basis, and GSD is a measure of the spread of particle size around this median.72 As seen in Table 2, the MMAD and GSD of the CSLA and mannitol microparticles loaded with siRNA–G4NH2 dendriplexes were not statistically significantly different from each other: MMAD of 1.9 and 2.6 μm, and GDS of 3.7 and 3.8 on average, respectively (n = 2, One-Way ANOVA followed by Tukey’s posthoc test, p value <0.05). These results indicate that microparticles from both strategies, mannitol and CSLA, would be able to deliver siRNA to the deep lungs.
The MMAD (by ACI) and hydrodynamic diameter (by LS) of CSLA microparticles were similar to each other (1.9 and 2.0 μm on average, respectively), but different for mannitol microparticles (2.6 and 4.6 μm on average, respectively). Differences between aerodynamic and hydrodynamic diameters of particles are somehow expected because the aerodynamic diameter is a property that depends on shape, density, and size (geometric diameter) of the particles.73,74 For spherical particles with density of 1, the geometric and aerodynamic diameters should be the same, provided that there is no aggregation.74 Hydrodynamic diameter is expected to be slightly larger than geometric diameter75 due to the presence of a solvation layer around the particle.
The most significant difference in both strategies came in the form of the total siRNA recovered and the siRNA content in a single puff dose (n = 2, One-Way ANOVA followed by Tukey’s posthoc test, p value <0.05). While the fraction recovered in the mannitol formulation was only 28%, 85% was recovered in the CSLA formulation. The single puff dose in the mannitol formulation was only 9.5 ng, while that for CSLA engineered microparticles was 26 ng. These results indicate that the strategy of encapsulating siRNA–G4NH2 into CSLA microparticles using the emulsification diffusion technique seems to have greater efficiency for delivering siRNA to the lungs. Collectively, the results shown here indicate that pMDI formulations from both microparticle engineering strategies proposed produce aerosols conducive to deep deposition of siRNA to the lungs. However, in terms of total siRNA recovered and the amount of siRNA actuated from the pMDI, the CSLA strategy displayed a much greater efficiency.
Conclusions
In this work we demonstrate, for the first time, the ability to formulate siRNA in pMDIs. This carries great weight as pMDIs are the least expensive and most widely used portable oral inhalation devices available in the market today and also because of the tremendous promise of therapeutic siRNA in treating lung diseases, as indicated by the many ongoing clinical trials where the lungs are the target tissue.11,76 We show that complexes between siRNA and positively charged PAMAM dendrimers (G4NH2) can successfully silence the production of genes in a model alveolar epithelial cell (A549), and aerosols of pMDIs formulated with those nanosized complexes encapsulated either in a GRAS component (mannitol) or a biodegradable co-oligomer (CSLA) have high respirable fractions (up to 77%), and fine particle fractions (∼50%), which are comparable to those of highly optimized commercial pMDIs for small-molecule therapeutics. We also show that, even after exposure to the propellant HFA used in pMDIs, the biological activity of the siRNA (gene silencing) is still maintained. Moreover, the dendriplexes are shown to preserve their characteristics after particle engineering, a necessary feature to reproduce its gene silencing ability after delivery to deep lungs. This work is highly complementary to other studies that have demonstrated the possibility of delivering siRNA to the lungs using dry powder inhalers77,78 and nebulizers,79 which are other important tools available to the clinicians to target medically important lung disorders along with pMDIs.
Acknowledgments
This work was supported in part by NSF (CBET Grant # 0933144), and by Nano@WSU (seed support). Denise S. Conti was partially supported through a scholarship from the Graduate School at Wayne State University (WSU). The authors thank DuPont for the propellant HFA-227, 3M Drug Delivery Systems for the metering valves, and West Pharmaceutical Services for the canisters. At WSU, we would like to thank Dr. Oupicky (for the access to the Synergy 2 Microplate Reader) and Dr. Merkel (for kindly providing the anti-eGFP and mismatch Ds-siRNA) at College of Pharmacy & Health Sciences; Dr. Chow (Chemistry), Dr. Mao and Dr. Matthew (Chemical Engineering and Materials Science) for access to LS and Spectra Max 250 UV plate reader; Dr. Pile (Biological Sciences) for access to the FOTO/Analyst Investigator/Eclipse with UV Transilluminator Fotodyne; Dr. Jessica Back and Mr. Eric Van Buren (Microscopy, Imaging and Cytometry Resources Core) for help with the flow cytometry analysis. The MICR Core is supported, in part, by NIH CenterGrant P30CA022453 to The Karmanos Cancer Institute, and the Perinatology Research Branch of the National Institutes of Child Health and Development, both at WSU.
Supporting Information Available
siRNA degradation by RNase A, heparin decomplexation assay, development of eGFP stable A549 cell line, characterization of CSLA co-oligomer, and pMDI formulation of free siRNA–G4NH2 dendriplexes. This material is available free of charge via the Internet at http://pubs.acs.org.
Author Present Address
† Science Group, Office of Generic Drugs (OGD), Center for Drug Evaluation and Research (CDER), US Food and Drug Administration (FDA), 10903 New Hampshire Avenue, WO75, Room 1711B, Silver Spring, MD 20993, United States.
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Pecot C. V.; Calin G. A.; Coleman R. L.; Lopez-Berestein G.; Sood A. K. RNA interference in the clinic: Challenges and future directions. Nat. Rev. Cancer 2011, 11159–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siomi H.; Siomi M. C. On the road to reading the RNA-interference code. Nature 2009, 4577228396–404. [DOI] [PubMed] [Google Scholar]
- Arima H.; Yoshimatsu A.; Ikeda H.; Ohyama A.; Motoyama K.; Higashi T.; Tsuchiya A.; Niidome T.; Katayama Y.; Hattori K.; Takeuchi T. Folate-PEG-appended dendrimer conjugate with α-cyclodextrin as a novel cancer cell-selective siRNA delivery carrier. Mol. Pharmaceutics 2012, 992591–2604. [DOI] [PubMed] [Google Scholar]
- Lam J. K.-W.; Liang W.; Chan H.-K. Pulmonary delivery of therapeutic siRNA. Adv. Drug Delivery Rev. 2012, 6411–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao K.; Huang L. Nonviral methods for siRNA delivery. Mol. Pharmaceutics 2008, 63651–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taratula O.; Garbuzenko O. B.; Chen A. M.; Minko T. Innovative strategy for treatment of lung cancer: Targeted nanotechnology-based inhalation co-delivery of anticancer drugs and siRNA. J. Drug Targeting 2011, 1910900–914. [DOI] [PubMed] [Google Scholar]
- Caci E.; Melani R.; Pedemonte N.; Yueksekdag G.; Ravazzolo R.; Rosenecker J.; Galietta L. J.; Zegarra-Moran O. Epithelial sodium channel inhibition in primary human bronchial epithelia by transfected siRNA. Am. J. Respir. Cell Mol. Biol. 2009, 402211–216. [DOI] [PubMed] [Google Scholar]
- Séguin R. M.; Ferrari N. Emerging oligonucleotide therapies for asthma and chronic obstructive pulmonary disease. Expert Opin. Invest. Drugs 2009, 18101505–1517. [DOI] [PubMed] [Google Scholar]
- Zamora M. R.; Budev M.; Rolfe M.; Gottlieb J.; Humar A.; DeVincenzo J.; Vaishnaw A.; Cehelsky J.; Albert G.; Nochur S.; Gollob J. A.; Glanville A. R. RNA interference therapy in lung transplant patients infected with respiratory syncytial virus. Am. J. Respir. Crit. Care Med. 2011, 1834531–538. [DOI] [PubMed] [Google Scholar]
- Li B.-j.; Tang Q.; Cheng D.; Qin C.; Xie F. Y.; Wei Q.; Xu J.; Liu Y.; Zheng B.-j.; Woodle M. C.; Zhong N.; Lu P. Y. Using siRNA in prophylactic and therapeutic regimens against SARS coronavirus in Rhesus macaque. Nat. Med. 2005, 119944–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita Y.; Takeshita F.; Kuwano K.; Ochiya T. RNAi therapeutic platforms for lung diseases. Pharmaceuticals 2013, 62223–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkel O. M.; Zheng M.; Debus H.; Kissel T. Pulmonary gene delivery using polymeric nonviral vectors. Bioconjugate Chem. 2011, 2313–20. [DOI] [PubMed] [Google Scholar]
- Heiden T. C. K.; Dengler E.; Kao W. J.; Heideman W.; Peterson R. E. Developmental toxicity of low generation PAMAM dendrimers in zebrafish. Toxicol. Appl. Pharmacol. 2007, 225170–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paleos C. M.; Tsiourvas D.; Sideratou Z. Molecular engineering of dendritic polymers and their application as drug and gene delivery systems. Mol. Pharmaceutics 2007, 42169–188. [DOI] [PubMed] [Google Scholar]
- Albertazzi L.; Storti B.; Marchetti L.; Beltram F. Delivery and subcellular targeting of dendrimer-based fluorescent pH sensors in living cells. J. Am. Chem. Soc. 2010, 1325118158–18167. [DOI] [PubMed] [Google Scholar]
- Kesharwani P.; Gajbhiye V.; Jain N. K. A review of nanocarriers for the delivery of small interfering RNA. Biomaterials 2012, 33297138–7150. [DOI] [PubMed] [Google Scholar]
- Shakhbazau A.; Isayenka I.; Kartel N.; Goncharova N.; Seviaryn I.; Kosmacheva S.; Potapnev M.; Shcharbin D.; Bryszewska M. Transfection efficiencies of PAMAM dendrimers correlate inversely with their hydrophobicity. Int. J. Pharm. 2010, 3831–2228–235. [DOI] [PubMed] [Google Scholar]
- Zhou J.; Wu J.; Hafdi N.; Behr J.-P.; Erbacher P.; Peng L. PAMAM dendrimers for efficient siRNA delivery and potent gene silencing. Chem. Commun. 2006, 2006222362. [DOI] [PubMed] [Google Scholar]
- Movassaghian S.; Moghimi H. R.; Shirazi F. H.; Koshkaryev A.; Trivedi M. S.; Torchilin V. P. Efficient down-regulation of PKC-α gene expression in A549 lung cancer cells mediated by antisense oligodeoxynucleotides in dendrosomes. Int. J. Pharm. 2013, 4411–282–91. [DOI] [PubMed] [Google Scholar]
- Durcan N.; Murphy C.; Cryan S.-A. Inhalable siRNA: Potential as a therapeutic agent in the lungs. Mol. Pharmaceutics 2008, 54559–566. [DOI] [PubMed] [Google Scholar]
- Zhang J.; Wu L.; Chan H.-K.; Watanabe W. Formation, characterization, and fate of inhaled drug nanoparticles. Adv. Drug Delivery Rev. 2011, 636441–455. [DOI] [PubMed] [Google Scholar]
- Foster K. A.; Oster C. G.; Mayer M. M.; Avery M. L.; Audus K. L. Characterization of the A549 cell line as a type II pulmonary epithelial cell model for drug metabolism. Exp. Cell Res. 1998, 2432359–366. [DOI] [PubMed] [Google Scholar]
- Tang Y.; Li Y.-B.; Wang B.; Lin R.-Y.; van Dongen M.; Zurcher D. M.; Gu X.-Y.; Banaszak Holl M. M.; Liu G.; Qi R. Efficient in vitro siRNA delivery and intramuscular gene silencing using PEG-modified PAMAM dendrimers. Mol. Pharmaceutics 2012, 961812–1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra M. K.; Gérard H. C.; Whittum-Hudson J. A.; Hudson A. P.; Kannan R. M. Dendrimer-enabled modulation of gene expression in Chlamydia trachomatis. Mol. Pharmaceutics 2012, 9, 413–421. [DOI] [PubMed] [Google Scholar]
- Molecular Probes - Invitrogen Detection Technologies. Protocol for Quant-iT PicoGreen ® dsDNA Reagent and Kits; 2008; MP 07581, 1–7; http://www.lifetechnologies.com/us/en/home/brands/molecular-probes.html.
- Bedi D.; Musacchio T.; Fagbohun O. A.; Gillespie J. W.; Deinnocentes P.; Bird R. C.; Bookbinder L.; Torchilin V. P.; Petrenko V. A. Delivery of siRNA into breast cancer cells via phage fusion protein-targeted liposomes. Nanomed. Nanotechnol. Biol. Med. 2011, 73315–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasband W. S.ImageJ; U. S. National Institutes of Health: Bethesda, Maryland, USA. http://imagej.nih.gov/ij/1997–2012. [Google Scholar]
- Nomani A.; Haririan I.; Rahimnia R.; Fouladdel S.; Gazori T.; Dinarvand R.; Omidi Y.; Azizi E. Physicochemical and biological properties of self-assembled antisense/poly(amidoamine) dendrimer nanoparticles: The effect of dendrimer generation and charge ratio. Int. J. Nanomed. 2010, 5, 359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavignac N.; Nicholls J. L.; Ferruti P.; Duncan R. Poly(amidoamine) conjugates containing doxorubicin bound via an acid-sensitive linker. Macromol. Biosci. 2009, 95480–487. [DOI] [PubMed] [Google Scholar]
- Invitrogen by Life Technologies. Protocol for Lipofectamine 2000 2006; pp 1–4.
- Promega Corporation. Protocol for CellTiter 96® aqueous non-radioactive cell proliferation assay, Part # TB169 2009; pp1–16.
- Promega Corporation. Protocol for TransFast Transfection Reagent, Part # TB260 2009; pp 1–17.
- Conti D. S.; Bharatwaj B.; Brewer D.; da Rocha S. R. P. Propellant-based inhalers for the non-invasive delivery of genes via oral inhalation. J. Controlled Release 2012, 1573406–417. [DOI] [PubMed] [Google Scholar]
- Rogueda P. G. A. HPFP, a model propellant for pMDIs. Drug Dev. Ind. Pharm. 2003, 29139–49. [DOI] [PubMed] [Google Scholar]
- U.S. Department of Health and Human Services. Guidance for Industry - Metered dose inhaler (MDI) and dry powder inhaler (DPI) drug products - chemistry, manufacturing, and controls documentation; Food and Drug Administration (FDA), Center for Drug Evaluation and Research (CDER): Rockville, MD: 199862
- Bharatwaj B.; Wu L.; Whittum-Hudson J. A.; da Rocha S. R. P. The potential for the noninvasive delivery of polymeric nanocarriers using propellant-based inhalers in the treatment of Chlamydial respiratory infections. Biomaterials 2010, 31287376–7385. [DOI] [PubMed] [Google Scholar]
- Antipina M. N.; Gainutdinov R. V.; Rachnyanskaya A. A.; Tolstikhina A. L.; Yurova T. V.; Khomutov G. B. Studies of nanoscale structural ordering in planar DNA complexes with amphiphilic mono- and polycations. Surf. Sci. 2003, 532–535, 1025–1033. [Google Scholar]
- Jensen L. B.; Pavan G. M.; Kasimova M. R.; Rutherford S.; Danani A.; Nielsen H. M.; Foged C. Elucidating the molecular mechanism of PAMAM-siRNA dendriplex self-assembly: Effect of dendrimer charge density. Int. J. Pharm. 2011, 4162410–418. [DOI] [PubMed] [Google Scholar]
- Juliano R.; Bauman J.; Kang H.; Ming X. Biological barriers to therapy with antisense and siRNA oligonucleotides. Mol. Pharmaceutics 2009, 63686–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y.; Bryantsev V. S.; Diallo M. S.; Goddard Iii W. A. PAMAM dendrimers undergo pH responsive conformational changes without swelling. J. Am. Chem. Soc. 2009, 13182798–2799. [DOI] [PubMed] [Google Scholar]
- De Rosa G.; Quaglia F.; Bochot A.; Ungaro F.; Fattal E. Long-term release and improved intracellular penetration of oligonucleotide-polyethylenimine complexes entrapped in biodegradable microspheres. Biomacromolecules 2003, 43529–536. [DOI] [PubMed] [Google Scholar]
- Kolhe P.; Misra E.; Kannan R. M.; Kannan S.; Lieh-Lai M. Drug complexation, in vitro release and cellular entry of dendrimers and hyperbranched polymers. Int. J. Pharm. 2003, 2591–2143–160. [DOI] [PubMed] [Google Scholar]
- Maiti P. K.; Çağın T.; Lin S.-T.; Goddard W. A. Effect of solvent and pH on the structure of PAMAM dendrimers. Macromolecules 2005, 383979–991. [Google Scholar]
- Jain K.; Kesharwani P.; Gupta U.; Jain N. K. Dendrimer toxicity: Let’s meet the challenge. Int. J. Pharm. 2010, 3941–2122–142. [DOI] [PubMed] [Google Scholar]
- Jevprasesphant R.; Penny J.; Jalal R.; Attwood D.; McKeown N. B.; D’Emanuele A. The influence of surface modification on the cytotoxicity of PAMAM dendrimers. Int. J. Pharm. 2003, 2521–2263–266. [DOI] [PubMed] [Google Scholar]
- Li C.; Liu H.; Sun Y.; Wang H.; Guo F.; Rao S.; Deng J.; Zhang Y.; Miao Y.; Guo C.; Meng J.; Chen X.; Li L.; Li D.; Xu H.; Wang H.; Li B.; Jiang C. PAMAM nanoparticles promote acute lung injury by inducing autophagic cell death through the Akt-TSC2-mTOR signaling pathway. J. Mol. Cell Biol. 2009, 1137–45. [DOI] [PubMed] [Google Scholar]
- Kannan S.; Kolhe P.; Raykova V.; Glibatec M.; Kannan R. M.; Lieh-Lai M.; Bassett D. Dynamics of cellular entry and drug delivery by dendritic polymers into human lung epithelial carcinoma cells. J. Biomater. Sci., Polym. Ed. 2004, 15, 311–330. [DOI] [PubMed] [Google Scholar]
- Biswas S.; Deshpande P. P.; Navarro G.; Dodwadkar N. S.; Torchilin V. P. Lipid modified triblock PAMAM-based nanocarriers for siRNA drug co-delivery. Biomaterials 2013, 3441289–1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavan G. M.; Posocco P.; Tagliabue A.; Maly M.; Malek A.; Danani A.; Ragg E.; Catapano C. V.; Pricl S. PAMAM dendrimers for siRNA delivery: Computational and experimental insights. Chem.—Eur. J. 2010, 16267781–7795. [DOI] [PubMed] [Google Scholar]
- Kim Y.; Klutz A. M.; Jacobson K. A. Systematic investigation of polyamidoamine dendrimers surface-modified with poly(ethylene glycol) for drug delivery applications: Synthesis, characterization, and evaluation of cytotoxicity. Bioconjugate Chem. 2008, 1981660–1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez A. P.; Romero E. L.; Morilla M. J. Ethylendiamine core PAMAM dendrimers/siRNA complexes as in vitro silencing agents. Int. J. Pharm. 2009, 3801–2189–200. [DOI] [PubMed] [Google Scholar]
- Wang X.-L.; Xu R.; Lu Z.-R. A peptide-targeted delivery system with pH-sensitive amphiphilic cell membrane disruption for efficient receptor-mediated siRNA delivery. J. Controlled Release 2009, 1343207–213. [DOI] [PubMed] [Google Scholar]
- Liu X.; Liu C.; Laurini E.; Posocco P.; Pricl S.; Qu F.; Rocchi P.; Peng L. Efficient delivery of sticky siRNA and potent gene silencing in a prostate cancer model using a generation 5 triethanolamine-core PAMAM dendrimer. Mol. Pharmaceutics 2012, 93470–481. [DOI] [PubMed] [Google Scholar]
- Li S.-D.; Chen Y.-C.; Hackett M. J.; Huang L. Tumor-targeted delivery of siRNA by self-assembled nanoparticles. Mol. Ther. 2007, 161163–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zintchenko A.; Philipp A.; Dehshahri A.; Wagner E. Simple modifications of branched PEI lead to highly efficient siRNA carriers with low toxicity. Bioconjugate Chem. 2008, 1971448–1455. [DOI] [PubMed] [Google Scholar]
- Gu S.; Rossi J. J. Uncoupling of RNAi from active translation in mammalian cells. RNA 2005, 11138–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkel O. M.; Mintzer M. A.; Librizzi D.; Samsonova O.; Dicke T.; Sproat B.; Garn H.; Barth P. J.; Simanek E. E.; Kissel T. Triazine dendrimers as nonviral vectors for in vitro and in vivo RNAi: The effects of peripheral groups and core structure on biological activity. Mol. Pharmaceutics 2010, 74969–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng M.; Librizzi D.; Kılıç A.; Liu Y.; Renz H.; Merkel O. M.; Kissel T. Enhancing in vivo circulation and siRNA delivery with biodegradable polyethylenimine-graft-polycaprolactone-block-poly(ethylene glycol) copolymers. Biomaterials 2012, 33276551–6558. [DOI] [PubMed] [Google Scholar]
- Tschuch C.; Schulz A.; Pscherer A.; Werft W.; Benner A.; Hotz-Wagenblatt A.; Barrionuevo L.; Lichter P.; Mertens D. Off-target effects of siRNA specific for GFP. BMC Mol. Biol. 2008, 9160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naito Y.; Yamada T.; Matsumiya T.; Ui-Tei K.; Saigo K.; Morishita S. dsCheck: Highly sensitive off-target search software for double-stranded RNA-mediated RNA interference. Nucleic Acids Res. 2005, 33web serverW589–W591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkel O. M.; Beyerle A.; Beckmann B. M.; Zheng M.; Hartmann R. K.; Stöger T.; Kissel T. H. Polymer-related off-target effects in non-viral siRNA delivery. Biomaterials 2011, 3292388–2398. [DOI] [PubMed] [Google Scholar]
- Akhtar S.; Benter I. Toxicogenomics of non-viral drug delivery systems for RNAi: Potential impact on siRNA-mediated gene silencing activity and specificity. Adv. Drug Delivery Rev. 2007, 592–3164–182. [DOI] [PubMed] [Google Scholar]
- Patton J. S.; Byron P. R. Inhaling medicines: Delivering drugs to the body through the lungs. Nat. Rev. Drug Discovery 2007, 6167–74. [DOI] [PubMed] [Google Scholar]
- Peguin R. P. S.; Wu L.; da Rocha S. R. P. The ester group: How hydrofluoroalkane-philic is it?. Langmuir 2007, 23168291–8294. [DOI] [PubMed] [Google Scholar]
- Wu L.; Bharatwaj B.; Panyam J.; da Rocha S. Core-shell particles for the dispersion of small polar drugs and biomolecules in hydrofluoroalkane propellants. Pharm. Res. 2008, 252289–301. [DOI] [PubMed] [Google Scholar]
- Kho K.; Hadinoto K. Effects of excipient formulation on the morphology and aqueous re-dispersibility of dry-powder silica nano-aggregates. Colloids Surf. A 2010, 3591–371–81. [Google Scholar]
- Young P. M.; Adi H.; Patel T.; Law K.; Rogueda P.; Traini D. The influence of micronised particulates on the aerosolisation properties of pressurised metered dose inhalers. J. Aerosol Sci. 2009, 404324–337. [Google Scholar]
- Wu L.; Peguin R. P. S.; da Rocha S. R. P. Understanding solvation in hydrofluoroalkanes: Ab Initio calculations and chemical force microscopy. J. Phys. Chem. B 2007, 111288096–8104. [DOI] [PubMed] [Google Scholar]
- Elversson J.; Millqvist-Fureby A. Particle size and density in spray drying - Effects of carbohydrate properties. J. Pharm. Sci. 2005, 9492049–2060. [DOI] [PubMed] [Google Scholar]
- Mitchell J. P.; Nagel M. W.; Wiersema K. J.; Doyle C. C. Aerodynamic particle size analysis of aerosols from pressurized metered-dose inhalers: Comparison of Andersen 8-stage cascade impactor, next generation pharmaceutical impactor, and model 3321 aerodynamic particle sizer aerosol spectrometer. AAPS PharmSciTech 2003, 441–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCabe J. C.; Koppenhagen F.; Blair J.; Zeng X.-M. ProAir® HFA delivers warmer, lower-impact, longer-duration plumes containing higher fine particle dose than Ventolin® HFA. J. Aerosol Med. Pulm. Drug Delivery 2012, 252104–109. [DOI] [PubMed] [Google Scholar]
- LeBelle M.; Pike R. K.; Graham S. J.; Ormsby E. D.; Bogard H. A. Metered-dose inhalers I: Drug content and particle size distribution of beclomethasone dipropionate. J. Pharm. Biomed. Anal. 1996, 147793–800. [DOI] [PubMed] [Google Scholar]
- Yang W.; Peters J. I.; Williams R. O. III. Inhaled nanoparticles - A current review. Int. J. Pharm. 2008, 3561–2239–247. [DOI] [PubMed] [Google Scholar]
- Crowder T. M.; Rosati J. A.; Schroeter J. D.; Hickey A. J.; Martonen T. B. Fundamental effects of particle morphology on lung delivery: Predictions of Stokes’ law and the particular relevance to dry powder inhaler formulation and development. Pharm. Res. 2002, 193239–245. [DOI] [PubMed] [Google Scholar]
- Gu J.; Su S.; Li Y.; He Q.; Shi J. Hydrophilic mesoporous carbon nanoparticles as carriers for sustained release of hydrophobic anti-cancer drugs. Chem. Commun. 2011, 4772101–2103. [DOI] [PubMed] [Google Scholar]
- Burnett J. C.; Rossi J. J.; Tiemann K. Current progress of siRNA/shRNA therapeutics in clinical trials. Biotechnol. J. 2011, 691130–1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen D. M. K.; Cun D.; Maltesen M. J.; Frokjaer S.; Nielsen H. M.; Foged C. Spray drying of siRNA-containing PLGA nanoparticles intended for inhalation. J. Controlled Release 2010, 1421138–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen D. K.; Jensen L. B.; Koocheki S.; Bengtson L.; Cun D.; Nielsen H. M.; Foged C. Design of an inhalable dry powder formulation of DOTAP-modified PLGA nanoparticles loaded with siRNA. J. Controlled Release 2012, 1571141–148. [DOI] [PubMed] [Google Scholar]
- Huth S.; Schmidt C.; Rudolph C.; Rosenecker J. Analysis of the stability and functionality of siRNA after nebulization of siRNA polyplexes. Mol. Ther. 2006, 13S1S272–S272. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.