Summary
Genetically encoded, ratiometric, fluorescent biosensors can be used to quantitatively measure intracellular ion concentrations in living cells. We describe important factors to consider when selecting a Ca2+ or Zn2+ biosensor, such as the sensor’s dissociation constant (Kd’) and its dynamic range. We also discuss the limits of quantitative measurement using these sensors and reasons why a sensor may perform differently in different biological systems or subcellular compartments. We outline protocols for 1) quickly confirming sensor functionality in a new biological system, 2) calibrating a sensor to convert a sensor’s FRET ratio to ion concentration, and 3) titrating a sensor in living cells to obtain its Kd’ under different experimental conditions.
Keywords: Genetically encoded sensors, Fluorescence microscopy, Förster Resonance Energy Transfer, Live cell imaging, Zinc, Calcium, Quantitative, Ratiometric
1. Introduction
1.1 Sensor design
Fluorescent biosensors are valuable tools for observing dynamic changes in intracellular ion concentrations. We will briefly compare and contrast ratiometric to intensiometric sensors, describing how to select and use an appropriate genetically encoded, ratiometric, fluorescent, metal ion sensor in living cells. We will also discuss some important complexities and limitations of quantitative measurements using these sensors.
In practice, there are two major types of fluorescent biosensors for metal ions: “intensiometric” biosensors that change fluorescence intensity when bound to an ion and “ratiometric” biosensors that exhibit a shift in the absorption or emission spectra when bound to an ion. The fluorescence intensity of an intensiometric sensor is dependent on the sensor concentration in each cell and the path length (i.e. the thickness of a cell) in addition to the ion concentration; hence ratiometric biosensors are preferred for quantitative measurements. On the other hand, ratiometric biosensors have several experimental limitations, which include lower sensitivity (i.e. smaller dynamic range), a larger spectral bandwidth, and the need to acquire images with two combinations of fluorescence excitation and emission filters.
In this protocol, we focus on genetically encoded biosensors: proteins encoded by DNA that is introduced into living cells by transient transfection or viral transduction. In contrast to small molecule biosensors, which are chemically synthesized and are introduced to cells immediately before the experiment, genetically encoded biosensors are manufactured by the cell and become functional without further intervention by the investigator. Genetically encoded biosensors are readily targeted to subcellular locations by appending localization sequences to the DNA sequence, a major advantage when the investigator desires the ability to monitor ion concentrations in organelles, such as the ER, Golgi, nucleus, or mitochondria. Subcellular targeting to some locations like vesicles is hindered by the size of some sensors (typical ratiometric FRET sensors are 60-65 kDa).
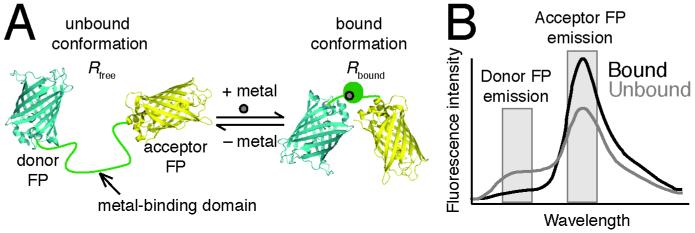
Almost all available genetically encoded, ratiometric, fluorescent metal ion sensors have the following design (see Fig. 1A): a donor fluorescent protein (FP) is attached to an acceptor FP by a linker containing the metal-binding domain. The chromophore of an FP forms auto-catalytically from three amino acid residues inside of a beta barrel. The biosensor undergoes a conformational change upon binding the metal ion, changing the distance and orientation between the two FPs and consequently the efficiency of Förster resonance energy transfer (FRET) (reviewed in (1, 2)). The change in FRET alters the emission spectra of the biosensor, decreasing the peak of donor FP emission and increasing the peak of acceptor FP emission (see Fig. 1B). The FRET ratio is defined as the ratio of the acceptor FP emission intensity upon donor excitation to the donor FP emission intensity. The FRET ratio can be converted to an ion concentration when three parameters are known: (1) the sensor affinity in terms of Kd’, (2) the FRET ratio in the absence of the ion (Rfree), and (3) the FRET ratio when the sensor is saturated with the ion (Rbound). The sensor affinity can be measured either in vitro or in situ (i.e. in cells) and is usually published in the literature, as discussed below. Rfree and Rbound are measured at the end of each experiment (see Section 3.2). Typically, Rfree is the minimum FRET ratio (Rmin) and Rbound is the maximum FRET ratio (Rmax), but sometimes the sensor response is inverted and the opposite is true (3).
Fig. 1.
Design of the Ca2+ and Zn2+ sensors used in this protocol. (A) A genetically encoded, fluorescent, ratiometric sensor contains a Ca2+ or Zn2+ binding domain fused to a donor FP (usually cyan FP) at is N-terminal end and an acceptor FP (usually yellow FP) at its C-terminal end. When Ca2+ or Zn2+ reversibly binds to its binding domain, the sensor changes conformation, which leads to a change in FRET efficiency. (B) shows how the change in FRET efficiency changes the FRET ratio, which is the ratio of acceptor FP to donor FP emission intensity upon donor FP excitation. Thus, the FRET ratio of the unbound sensor (Rfree) is distinct from that of the bound sensor (Rbound).
1.2 Understanding binding affinity and dynamic range
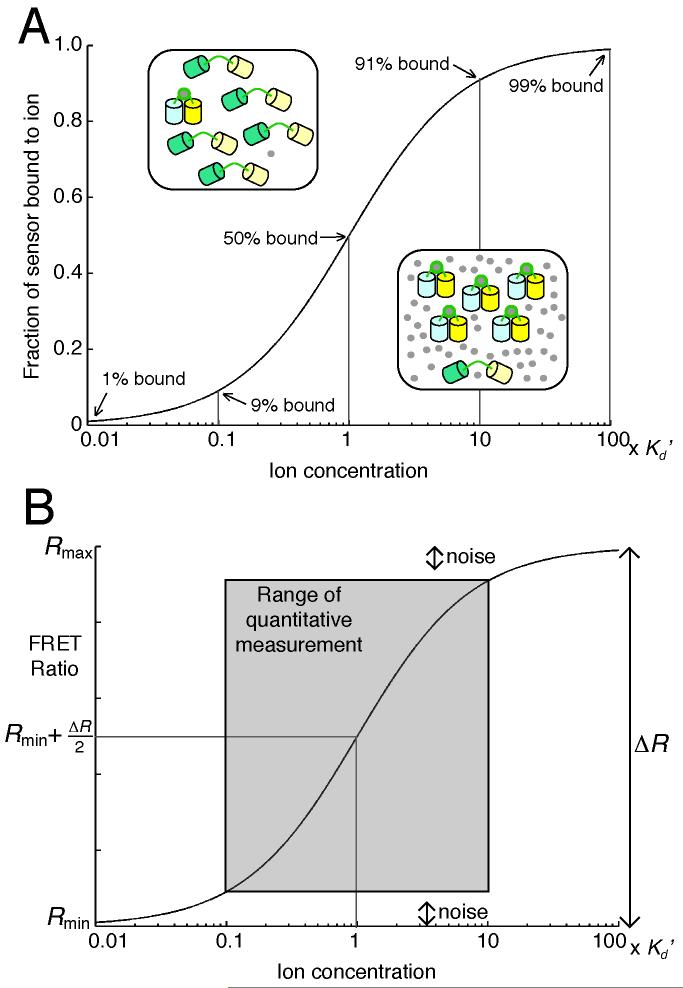
Every biosensor is sensitive to changes within a range of ion concentrations, which spans about two orders of magnitude. This range is largely determined by the sensor’s binding affinity for an ion (or ions) and its dynamic range (DR). The binding affinity is reported as the apparent dissociation constant (Kd’), which is equal to the ion concentration when 50% of the sensor is bound (see Fig. 2). Occasionally, when a sensor has multiple binding sites with different binding affinities, multiple Kd’ values are reported. A Kd’ is determined by fitting a binding curve to experimental data from a sensor titration experiment, in which the FRET ratio of the sensor is measured at different ion concentrations. The simplest binding equation is used to describe the titration data, even though it may not represent actual binding events (see Fig. 3). For example, the Ca2+ sensor D3cpV is fit to a single-site binding equation, even though each sensor binds to 4 Ca2+ ions (4). It is important to note that temperature, salt concentration, pH, and other factors can affect the Kd’, and that most titrations are performed in vitro with protein purified from a bacterial expression system. However, experiments performed in our group and others indicate that the Kd’ in vitro and in cells are often comparable (3, 5). This is something that each investigator can verify in his or her own experimental system using the protocol outlined in Section 3.3.
Fig. 2.
A sensor’s sensitivity to changes in Ca2+ or Zn2+ concentration is related to its dissociation constant (Kd’). (A) When the ion concentration ([ion]) is the same as the sensor’s Kd’, 50% of a population of sensors (typically 1-20 μM in cells) is bound to the ion. The fraction of sensor bound changes the most when the [ion] varies within 10-fold of the sensor’s Kd’. (B) The FRET ratio is a linear function of the fraction bound, and so the midpoint of Rmin and Rmax corresponds to an [ion] equal to the sensor’s Kd’. The SNR and the Kd’ limit the range of the [ion] that can be quantified by the sensor.
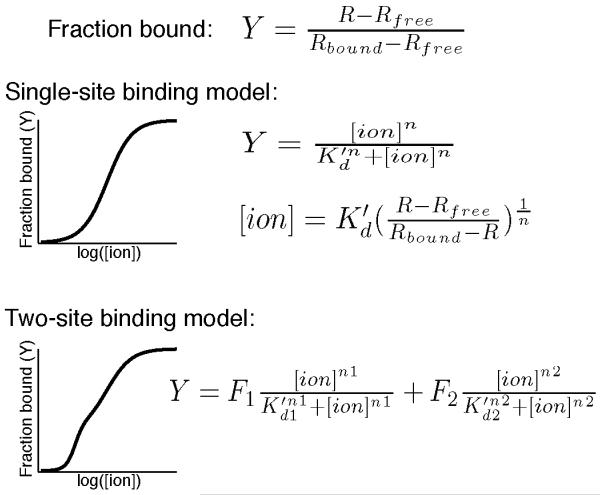
Fig. 3.
A sensor’s Kd’ is calculated by fitting experimental data to a single-site or two-site binding model. An example of each binding curve is shown next to the equation relating the [ion] to the fraction bound (Y), which is calculated from the FRET ratio (R), Rfree, and Rbound. When the single-site binding model is used, the [ion] is readily calculated using the displayed equation.
DR has many definitions in the literature, but it’s essentially an indicator of a sensor’s measurement sensitivity and its signal-to-noise ratio (SNR). Common definitions of DR include the fold-change in FRET ratio (DR=Rmax/Rmin); or, the maximum change in FRET ratio (DR= Rmax-Rmin). SNR is also described as the ratio of R to the standard deviation of R at baseline (6, 7). Several factors can significantly affect a sensor’s DR (or SNR), including the microscopy system used for measurement, the cell type expressing the sensor, and the subcellular location. Accordingly, reported DRs are most useful for relative comparisons and will not always be the same in across experimental systems. Moreover there are numerous examples of sensors exhibiting decreased DR in cells compared to that measured in vitro and hence researchers are encouraged to seek out and compare Rmax and Rmin values from in situ experiments, which are often reported graphically in publication figures.
A sensor is most sensitive to changes in ion concentration when it is close to 50% bound because a change in ion concentration near the Kd’ results in a greater change in fraction bound, which is proportional to the change in FRET ratio (see Fig. 2B). For example, if a sensor’s Kd’=1 μM, a change in ion concentration from 0.1 μM to 1 μM results in an increase from 9% to 50% fraction bound, whereas a change from 0.01 μM to 0.1 μM results an increase from 1% to 9% fraction bound. Consequently, changes in ion concentration close to the sensor’s Kd’ are more readily detected. In Fig. 2B, Rmin and Rmax correspond to 0% and 100% fraction bound, respectively. The variance of R, or noise, relative to the overall change in FRET ratio (ΔR), determines the range of fraction bound that a sensor can reliably report. Thus, a sensor with a larger SNR can be used to measure a greater range of ion concentrations.
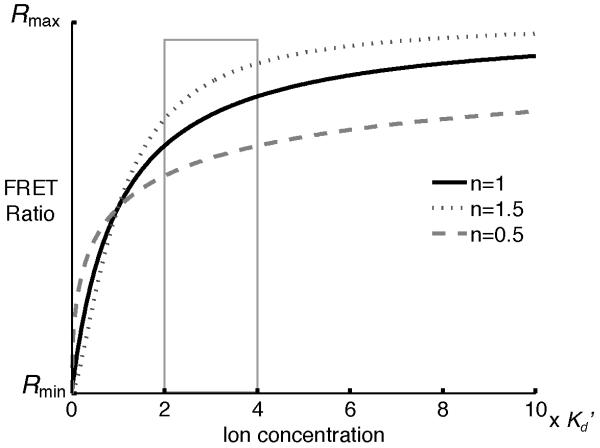
Binding cooperativity of multiple ions to one sensor is reported as the Hill coefficient (n or nH). The n value affects the steepness of the binding curve: positive cooperativity results in a Hill coefficient greater than 1 and a steeper curve (see Fig. 4). Positive cooperativity also increases the sensitivity of measurement near the Kd’ but decreases the range of ion concentration that the sensor can report. In Fig. 4, a change in ion concentration from 1 × Kd’ to 2 × Kd’ results in a larger change in R when n=1.5 than when n=0.5.
Fig. 4.
The value of the Hill coefficient (n) does not affect the sensor’s Kd’ if the data are fit using the equations in Fig. 3. n affects the change in the FRET ratio resulting from a change in the [ion]. For example, if the [ion] changes from 2 × Kd’ to 4 × Kd’, the change in FRET ratio is greater when n is larger.
1.3 Choosing a sensor for a specific application
Currently, many biosensors are available for Ca2+ or Zn2+ imaging, so the investigator must attempt to choose the one best suited to the experimental system. The Kd’ and the DR are the most important factors to consider, along with the other factors discussed below, but often the best sensor is revealed by empirical testing of multiple sensors (8).
First, fluorescence properties of the donor and acceptor FPs affect each sensor. The cyan FP and yellow FP donor-acceptor pair is by far the most common. Single or multiple amino acid mutations in an FP can change its excitation and/or emission wavelengths, brightness, maturation time, or photostability, and so there are several different cyan FPs and yellow FPs with slightly different fluorescence properties (9, 10). A word of caution regarding yellow FPs is that their fluorescence is quenched by acid and hence it may be challenging to use a sensor containing a yellow FP in an acidic compartment. Circularly permuted (cp) variants of FPs are constructed by changing the N- and C-termini to different loops within the FP. Although circular permutation does not significantly change the fluorescence properties, the incorporation of a cp FP into a sensor can change the DR by altering the relative orientation of the two FPs in the bound and/or unbound conformation of the sensor (4, 11). In addition, circular permutation can affect the pH sensitivity by changing the pKa of the chromophore, and rearrangement of cysteine residues can affect its sensitivity to oxidizing organelles. We have found that sensors containing a cpVenus FP have decreased DR and SNR in the ER due to the low signal from the cpVenus FP. Red-green FRET pairs are preferred in acidic compartments, such as the secretory pathway, since sensors containing yellow FPs can be much dimmer at low pH (< 6.5). The substitution of different FPs into a sensor can alter its Kd’ and DR in unpredictable ways, so the investigator should evaluate each sensor’s performance in situ before performing important experiments.
Replacement or mutation of the binding domain also alters the sensor’s properties. To monitor increases in ion concentration, it’s useful to pick a sensor that is ~20% saturated at baseline, whereas a sensor that is ~50% saturated is better for comparing differences in resting ion concentrations in different cells or different environmental conditions. Cameleon-Nano sensors have lower Kd’s and are better for quantitative measurement of cytosolic Ca2+ in some cell types (6), whereas D1ER is preferred for ER measurement because Ca2+ levels are high in the ER and the Kd’ of D1ER is much higher than other cameleons (12). Table 1 and 2 summarize Kd’s and DRs of some current Ca2+ and Zn2+ sensors, respectively. Table 3 summarizes sensors used in specific subcellular compartments. In addition to a sensor’s Kd’ and DR, the investigator may consider properties such as sensitivity to other biologically relevant metal ions, pH, redox balance, salt concentration, or sensor concentration. It is also important to consider the on and off rates of ion binding when attempting to observe dynamic changes in ion concentrations (6-8, 13).
Table 1.
Selected ratiometric Ca2+ biosensors
| Sensor | Ca2+- responsive elements |
Kd’ for Ca2+ |
Hill coefficient |
Comments | References | |
|---|---|---|---|---|---|---|
| Yellow Cameleon series |
YC2.60 | CaM, M13p | 93.5 nM | 2.7 | (6, 11) | |
| Yellow Cameleon series |
YC3.60 | CaM E104Q, M13p |
215 nM, 779 nM |
3.6, 1.2 | High dynamic range |
(6, 11) |
| Yellow Cameleon Nano series |
YC-Nano50 | CaM, M13p | 52.5 nM | 2.5 | Optimized for detecting subtle cytosolic Ca2+ transients in living organisms |
(6) |
| D-family Cameleons |
D1 | mCaM, mM13p |
0.8 μM, 60 μM |
1.18, 1.67 | Does not bind endogenous CaM; optimized for ER |
(12) |
| D-family Cameleons |
D3cpV | mCaM, mM13p |
0.6 μM | 0.74 | Does not bind endogenous CaM; optimized for cytosol & mitochondria |
(4) |
| TroponinC family |
TN-XXL | mTpC | 800 nM | 1.5 | Optimized for imaging of neurons; fast response |
(13) |
Table 2.
Selected ratiometric Zn2+ biosensors
| Sensor | Zn2+- responsive elements |
Kd’ for Zn2+ |
Hill coefficient |
Comments | References | |
|---|---|---|---|---|---|---|
| eCALWY family |
eCALWY4 | Atox1 and the WD4 domain of ATP7B |
630 pM | 1 | Optimized for cytosol |
(3) |
| ZinCh family |
eZinCh | Zn2+- coordinating residues on CFP and YFP connected by a flexible linker |
8.2 μM | 1 | Targeted to vesicles by fusion to VAMP2 |
(3, 21) |
| Zap family | ZapCY1 | Zap | 2.53 pM |
1 | Optimized for ER, Golgi, and mitochondria; high dynamic range |
(5) |
| Zap family | ZapCY2 | mZap | 811 pM | 0.44 | Optimized for cytosol |
(5) |
Table 3.
Sensors targeted to subcellular locations
1.4 Outline of procedures described in this protocol
The first step in using a biosensor is to confirm it functions in the biological system used in the experiment, even though most published sensors function to some degree in different cell types and subcellular compartments (see Section 3.1). Next, calibration of the sensor at the end of an experiment, in single cells, enables the investigator to convert the sensor’s FRET ratio to the ion concentration (see Section 3.2). It is essential to obtain accurate Kd’ values, which are used to convert experimental data into ion concentrations. Experimental conditions that significantly differ from those used in a published sensor titration (typically, at pH 7.0-7.4 and 20-25°C), can change the Kd’, and so a protocol for titrating a sensor in living cells (in situ) can be found in Section 3.3. This protocol also describes how to use metal-chelate buffer systems to accurately maintain the free Ca2+ or Zn2+ concentrations at sub-μM concentrations.
2. Materials
2.1 Materials for preparation of cells
Glass-bottom imaging dishes: available commercially or constructed using 35 mm polystyrene cell culture dishes, SYLGARD 184 silicone elastomer kit (Dow Corning), 18 mm × 18 mm No. 1 glass coverslips, and an industrial-strength 0.375 in. hole punch (Roper Whitney) (see Note 1).
Mammalian expression plasmid encoding the biosensor of choice
Transfection reagent, such as Lipofectamine (Life Technologies) or TransIT-LT1 (Mirus Bio)
Cells and cell culture media
2.2 Reagents for cellular imaging
Prepare all solutions using Chelex-treated water and use metal-free, plastic containers to store solutions. Metal-free containers and consumables can be purchased, or metal can be removed from plastic containers and consumables by washing them with dilute acid.
Chelex 100 sodium form, 50-100 mesh (Sigma Aldrich)
Chelex-treated double deionized H2O: mix Chelex with autoclaved water in a large plastic container (~3-4 L) on a stir plate for at least 18 hours. Let the Chelex settle to the bottom of the container for several hours. Use a bottle-top filter to remove all Chelex from the water, and store the filtered water in a plastic container.
For Ca2+ measurement, HEPES-buffered Hanks balanced salt solution (HHBSS) with and without Ca2+: dilute 10X HBSS (Life technologies) with water and supplement with 20 mM HEPES and 16.8 mM D-glucose. To prepare Ca2+-free HHBSS, dilute 10X Ca2+-free, Mg2+-free HBSS (Life Technologies) and supplement with 16.8 mM D-glucose and 20 mM HEPES. For each solution, adjust pH to 7.4 with 1 M sodium hydroxide.
For Zn2+ measurement, phosphate-free HHBSS with and without Ca2+ and Mg2+ (see Note 6): 1.26 mM calcium chloride, 5.4 mM potassium chloride, 1.1 mM magnesium chloride, 137 mM sodium chloride, 16.8 mM D-glucose, 20 mM HEPES, pH 7.4. Mix all components in 900 mL of water. Add 0.3 g of sodium hydroxide to around pH 7, and then adjust with 1 M sodium hydroxide solution to pH 7.4. Add water to 1L and use a 0.22 μm pore filter to sterilize. Omit calcium chloride and magnesium chloride to make phosphate-, Ca2+-, and Mg2+-free HHBSS.
2.3 Chemicals for Ca2+ and Zn2+ perturbations (see Table 4 and Note 2)
Table 4.
Chemicals used for Ca2+ and Zn2+ calibrations
| Chemical | Working concentration |
Effect | Comments and References |
|---|---|---|---|
| Ionomycin Bromo-A23187 |
5-10 μM 0.1 – 20 μM |
Ca2+ ionophore | |
| TPEN | 50-150 μM | Zn2+ depletion | Membrane-permeable Highest affinity for Zn2+, but also chelates other transition metal ions |
| EDTA EGTA |
2-5 mM 2-5 mM |
Zn2+ or Ca2+ depletion |
|
|
S. aureus α toxin Digitonin Alamethicin Saponin |
variable 10-20 μM 50 μg/mL 0.01-0.1% |
Membrane permeabilization |
A variety of permeabilization conditions have been used for Zn2+ calibrations. (3, 5, 23) |
| Pyrithione | 1-500 μM | Zn2+ ionophore | Lower concentrations may result in a more stable Rbound |
0.5 M EGTA stock solution, pH 7.4: dissolve EGTA in 1M NaOH and adjust pH to 8.0 to solubilize.
1 M calcium chloride stock solution in water
1 mM ionomycin stock solution in DMSO: Add 141 μL of 100% DMSO to 1 mg of ionomycin free acid (EMD Millipore, CAS number 56092-81-0) to yield a 10 mM stock solution. Mix well. Make 5 μL aliquots and store at −20°C. Dilute 10 mM stock solution to 1 mM before use.
3 mM digitonin stock solution in ethanol: dissolve digitonin in 100% DMSO. Aliquot & store at −20°C (see Note 3).
25 mM TPEN (N, N, N’, N’-tetrakis-(2-pyridylmethyl)-ethylenediamine) stock solution in 100% DMSO: Make 50-100 μL aliquots and store at −20°C.
500 μM pyrithione (2-Mercaptopyridine N-oxide sodium salt) stock solution in 100% DMSO: Make 50-100 μL aliquots and store at −20°C.
475 μM zinc chloride or zinc sulfate in phosphate-free HHBSS, pH 7.4. Mix well before use (see Note 4).
Metal-chelate buffer stock solutions (see Section 3.3).
2.4 Equipment for cellular imaging
Epifluorescence microscope equipped with a mercury or xenon arc lamp and power supply; excitation and emission filter wheels; Lambda SC Smart Shutter controller; cooled CCD camera; 20X, 40X, 60X and/or 100X plan apochromatic objective (depending on application). Other microscopy systems, such as two-photon confocal, FLIM, and spectral imaging, can also be used to monitor FRET, but they will not be discussed in this protocol.
Neutral density filters
Excitation (x) and emission (m) filter sets and dichroic mirrors: CFPx and FRETx 430/24, YFPx 495/10, CFPm 470/24, YFPm and FRETm 535/25, CFP and FRET dichroic 450, YFP dichroic 515. Sputter-coated/ET or brightline filters that provide high transmission will give rise to the brightest images.
Microscopy software: MetaFluor (Molecular Devices), NIS Elements (Nikon), Micro-Manager (open-source), or equivalent.
Software for image processing: MetaFluor (Molecular Devices), NIS Elements (Nikon), ImageJ (NIH), Fiji (open-source), MATLAB (Mathworks), Excel (Microsoft), or equivalent.
Pipets or perfusion system
3. Methods
3.1 Testing sensor functionality
Plate cells on several imaging dishes and transfect them with a plasmid encoding the biosensor of choice (see Note 5).
Image cells 24-72 hours after transfection, or when sensor expression and cell density are appropriate for the experiment.
Gently remove cell culture media from the imaging dish and replace with 2 mL of HHBSS (for Ca2+ sensors) or phosphate-free HHBSS (for Zn2+ sensors) (see Note 6). Wash the dish 3-5 times to remove all cell culture media.
Place the imaging dish on the microscope stage. Identify and focus on cells.
Verify that the sensor is correctly localized to the subcellular compartment. This can be performed using dyes such as MitoTracker, a different fluorescent protein targeted by a different localization tag to the same organelle, or visually if the morphology of the compartment is unique. Alternatively, cells can be fixed, and immunofluorescence can be used to confirm correct localization. It is unnecessary to have 100% transfection efficiency or 100% correct localization because analysis will be performed on single cells.
Set up acquisition parameters. Limit fluorescent light exposure during acquisition to prevent photobleaching of the sensor. Acquire one set of images that includes a FRET, a donor, an acceptor, and a brightfield/DIC image. Confirm that the fluorescence intensity of the biosensor is about 2.5-8 times the background intensity and does not saturate the camera. Many software programs controlling image acquisition will allow you to select regions of interest and calculate the ratio of two images or regions of interest during acquisition. This function is helpful because it allows you to observe the sensor response during the experiment.
Begin the experiment by acquiring images for at least 5 minutes (if you have added a dye for colocalization purposes, start this step with a different imaging dish and repeat steps 3 and 4). Confirm that the sensor responds to increases and decreases of the metal ion of interest by using typical chemical perturbations (see Table 4). Note 7 describes the procedure we use to change imaging solutions manually (i.e. without a perfusion system). You may need to try several different conditions in several imaging dishes. In this protocol, it is sufficient to observe relative changes, whereas quantitative measurement in the next two protocols requires careful and systematic chemical perturbations of metal ion concentration. Remember to wash the cells 3-5 times with HHBSS (Ca2+ sensors) or phosphate-free HHBSS (Zn2+ sensors) between chemical perturbations and re-focus if necessary.
After image acquisition, calculate the average FRET ratio of each cell using the full spectrum unprocessed images (usually stored as .tiff files), not images that have been autoscaled and saved as new images (usually .jpg or .png files). This image analysis workflow is illustrated in Fig. 5. Define regions of interest (ROI) and measure the mean intensity of the same ROI in each set of images (CFP, FRET, and YFP) using image analysis software. Subtract the mean background intensity from each ROI’s mean intensity in every image. Calculate the FRET ratio by dividing the ROI’s background-subtracted mean intensity in the FRET image by that in the CFP image at each time point. Plot the FRET ratio over time and confirm that the sensor responds to perturbations of metal ion concentration. In addition, plot each ROI’s mean intensity in the YFP image over time to observe potential photobleaching.
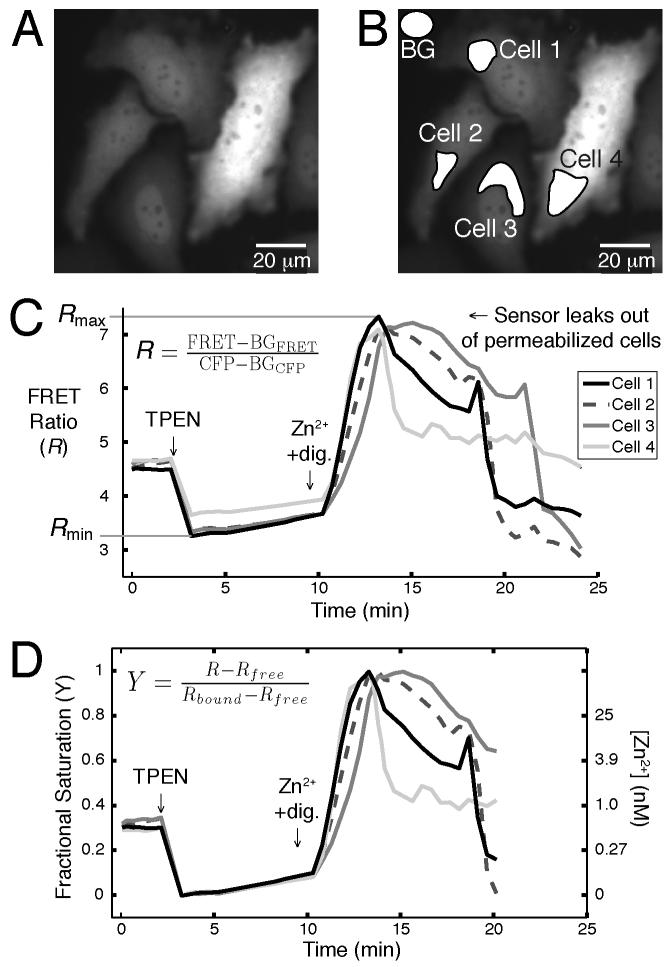
Fig. 5.
A typical analysis of a Zn2+ sensor calibration is shown. Regions of interest (ROIs), including a background (BG) region, are selected in the acquired FRET and CFP images in (A) and (B). The mean intensity of each region is exported into a spreadsheet (or equivalent). The mean intensity of the BG region (BGFRET or BGCFP) is subtracted from the mean intensity of each cell’s ROI (FRET or CFP), and the equation in (C) is used to calculate the FRET ratio (R). (D) shows a plot of the sensor’s fractional saturation (Y) over time and indicates the Zn2+ concentrations corresponding to Y.
3.2 Calibration of a Ca2+ or Zn2+ biosensor in living cells for quantitative measurement
Plate and transfect cells as in the previous protocol.
Image cells 24-72 hours after transfection or when appropriate for the experiment. We find that if HeLa cells are at 80-90% density, they are less likely to detach from the imaging dish during chemical perturbations.
Gently remove cell culture media from the imaging dish and replace with 2 mL of HHBSS (use phosphate-free HHBSS in all steps if measuring Zn2+). Wash the dish 3-5 times to remove all cell culture media.
Secure the imaging dish on the microscope stage and focus the objective using brightfield illumination. Fine-tune the focus using fluorescence excitation and emission. Limiting fluorescent light exposure before calibration prevents photobleaching and phototoxicity.
Set up the image acquisition parameters. Again, limit fluorescent light exposure during acquisition by using neutral density filters. Acquire one set of images that includes a FRET, a donor, an acceptor, and a brightfield/DIC image. Confirm that the fluorescence intensity of the biosensor is about 2.5-8 times the background intensity and does not saturate the camera. If these requirements are not met, change the acquisition parameters and/or select different cells. This step can be tedious, but it is essential for accurate and repeatable quantitative measurements.
At this point, make sure that the acquired images are being saved as raw image files and that you are recording the time of each acquisition. Acquire one set of images every 20-60 seconds for about 5 minutes. Be sure that the FRET ratio, defined as the intensity of the biosensor in the FRET image divided by its intensity in the donor image, is stable during this time. Most software can plot the FRET ratio of a ROI as images are acquired. If the FRET ratio decreases (or increases), change the acquisition parameters to decrease light exposure (for example, decrease the frequency of image acquisition, decrease the exposure time of each acquisition, etc.). If the FRET ratio does not stabilize after the adjustment of acquisition parameters, change the imaging buffer, select new cells, or go back to Step 3 and start over with a new dish.
Begin the experiment by acquiring images for at least 5 minutes. If the purpose of this experiment is to measure the baseline metal ion concentration, move on to the next step. Otherwise, acquire images while performing any desired environmental or chemical perturbations of the cells. Note 7 describes the procedure we use to change imaging solutions manually (i.e. without a perfusion system).
Measure the FRET ratios of the fully unbound (Rfree) and saturated (Rbound) Ca2+ sensor (skip to the next step for Zn2+ sensors). Wash cells with Ca2+-free HHBSS before adding EGTA and ionomycin to remove Ca2+ from the sensor (final concentrations: 5 μM ionomycin and 3 mM EGTA), and acquire images every 20-30 seconds until the FRET ratio stabilizes (see Note 9). Wash cells 3-5 times with HHBSS. The FRET ratio should increase as Ca2+ from the HHBSS enters cells. Saturate the sensor by adding 5 μM ionomycin and 10 mM CaCl2 to HHBSS. These conditions should be optimized because adding too much Ca2+, too quickly, can lead to cell death before Rbound is reached.
Measure the FRET ratios of the fully unbound (Rfree) and saturated (Rbound) Zn2+ sensor. Add TPEN to a final concentration of 150 μM in phosphate-free HHBSS (see Notes 7 and 9). Wash cells 3-5 times with Ca2+-, Mg2+-, and phosphate-free HHBSS (see Note 8). Acquire 2-3 sets of images to ensure that the cells are in focus and change the acquisition interval to ≤ 20 seconds before proceeding because obtaining Rbound usually results in rapid cell death. Saturate the sensor with Zn2+ appropriately (see Table 5). We use 10 μM digitonin/100 μM Zn2+ to determine Rbound of a cytosolic Zn2+ sensor.
Calculate the FRET ratio of individual cells over time (see Step 8 in Section 3.1 and Fig. 5) and determine Rmin, Rmax, and ΔR for each cell (see Note 10). Convert single-cell FRET ratios to fractional saturation, which is equal to (R–Rmin)/ΔR. Solve for the ion concentration using the appropriate equation (see Fig. 3).
Table 5.
Metal-chelate buffers for sub-μM Ca2+ and Zn2+ solutions
| Chelator | Ca2+ buffering range at pH 7.4 |
Zn2+ buffering range at pH 7.4 |
Zn2+ buffering range at pH 7.4 with competing metal |
|---|---|---|---|
| EDTA | 0.002 to 0.18 μM | 0.003 to 0.3 pM | |
| EGTA | 0.006 to 0.5 μM | 0.15 to 15 nM | 9.7 to 1340 nM (with 2 mM Sr2+) 2 to 134 μM (with 2 mM Ca2+) |
| HEDTA | 0.3 to 25 μM | 0.15 to 15 pM | 0.05 to 7.5 nM (with 2 mM Ca2+) |
| NTA | 20 to 1670 μM |
3.3 Measurement of sensor affinity in situ with metal-chelate buffers
Select the appropriate metal-chelate buffer(s) for the desired free Ca2+ or Zn2+ concentration(s) (see Table 5 and Note 11). Some general references for creating different metal-chelate buffers are (14), (15) and (16). Detailed descriptions of Zn2+-chelate buffers used in publications are found in the supplementary methods of (5) and (3).
Prepare two different 0.1 M stock solutions for each buffer. For a single metal-chelate buffer, these solutions are: (1) metal and chelator, and (2) chelator only. For a mixed metal-chelate buffer, these solutions are: (1) Metal #2 plus a fixed amount of Metal #1 plus the chelator, and (2) a fixed amount of Metal #1 plus the chelator. Note that Metal #1 is the competing metal while Metal #2 is the metal to be buffered (e.g. Ca2+ or Zn2+). The “pH titration method” (14) should be used to prepare stock solution (1) because it is extremely important to make a 1:1 solution of metal:chelator (see Note 12).
Mix the two stock solutions in appropriate proportions to buffer at specific ion concentrations. The stock solution concentration of 0.1 M can be diluted 100X to the 1 mM working concentration in Ca2+- and Mg2+-free HHBSS (for buffered Zn2+ solutions, use Ca2+-, Mg2+-, and phosphate-free HHBSS).
Measure the FRET ratio of the sensor in living cells when they are permeabilized in the presence of a defined Ca2+ or Zn2+ concentration. Follow the protocol in Section 3.2 to measure Rfree in single cells. Wash the cells 3-5 times with Ca2+- and Mg2+-free HHBSS, and then replace it with a metal-chelate buffered HHBSS before permeabilizing the cells.
Calculate the FRET ratio of individual cells over time (see Step 8 in Section 3.1 and Fig. 5) and determine Rmin and Rfinal for each cell. Plot Rfinal –Rfree vs. the ion concentration and fit data to a binding expression (see Fig. 3) using the least squares method.
Acknowledgements
Financial support was provided by the Signaling and Cell Cycle Regulation Training Grant (NIH T32 GM08759) to J.G.P. and NIH GM084027 and Alfred P. Sloan Fellowship to A.E.P.
Footnotes
To construct a glass bottomed imaging dish, punch a hole in the middle of the cell culture dish and place it bottom-up. Use an 18-gauge needle and syringe to apply SYLGARD 184 around the rim of the punched hole, and place a coverslip on top. Let the glue cure, and then sterilize the dish with ethanol and UV light.
The appropriate chemical perturbations to reach Rmin or Rmax will depend on the cell type, the biosensor, and the organelle to which it is targeted. For example, in HeLa cells transfected with a mitochondrial Zn2+ sensor, saturating them with 10 μM ZnCl2 instead of 100 μM ZnCl2 results in a higher Rmax measurement. Try a few different perturbations and use the one that gives consistent results.
For most applications, digitonin is purified by crystallization in ethanol and made freshly before use. We find it sufficient to use commercially purified digitonin for sensor calibrations.
While zinc chloride and zinc sulfate are very soluble in water, zinc hydroxide readily precipitates out of solution, decreasing the concentration of free Zn2+. At pH 7.4 and room temperature, up to 475 μM Zn2+ is soluble in water. Use the Ksp of zinc hydroxide, which is 3.0 × 10−17 at room temperature, to calculate the maximum solubility of Zn2+ at the desired pH.
Optimal cell plating density and transfection conditions will differ for each cell type and biosensor. For example, we transfect each imaging dish of HeLa cells with 5 μl of Mirus TransIT-LT1 transfection reagent, and 0.5 μg of plasmid DNA encoding a mitochondria-targeted Zn2+ sensor or 1.25 μg of plasmid DNA encoding the same Zn2+ sensor targeted to the cytosol. In our experience, it is worthwhile to identify conditions that minimize cell toxicity, promote expression levels detectable by the microscope, and maximize correct localization of biosensors targeted to organelles.
Optically clear salt solutions are preferred for cellular imaging because the autofluorescence of Phenol Red and serum in complete media increases background fluorescence. In addition, components of complete media, such as amino acids and proteins, will bind metal ions and affect sensor calibrations. Phosphate-free imaging media is used for Zn2+ sensor calibrations because Zn3(PO4)2 precipitates and changes the free Zn2+ concentration.
When working manually (i.e. without a perfusion chamber or a “perfect-focus” system), we find that the following procedure for adding chemicals to the imaging dish minimizes changes in focus. Remove 1 mL of HHBSS from the dish and pipet it into a 1.5 mL tube. Make a 2X solution, containing the chemical perturbation, using this aliquot of HHBSS. Gently pipet the 2X solution into the imaging dish and mix gently.
Failure to use Ca2+-free imaging buffer during cell permeabilization will cause rapid cell death.
The FRET ratio will initially increase after the addition of ionomycin/EGTA due to the permeabilization of the ER and release of Ca2+ stores. It may take up to 10 minutes to reach Rmin. High affinity Zn2+ sensors, such as ZapCY1, have a very slow off rate. It can take up to 40 minutes to reach Rmin. An alternative is to monitor the FRET ratio for 15-20 minutes, fit the data to an exponential decay, and calculate Rmin.
Sometimes a cell will have an unusually low dynamic range. This could be due to proteolysis, misfolding, or oxidation of the sensor. These data are difficult to interpret and may have to be discarded.
For a sensor titration, these concentrations should vary ~10-fold above and below the expected Kd’, and about 10 evenly-spaced concentrations within this range should be used. Metal-chelate buffers consist of a chelator (e.g. EDTA), the buffered ion (Ca2+ or Zn2+), and sometimes a competing ion (e.g. Sr2+). The addition of a competing ion effectively increases the buffering range of the chelator, and other competing metals like Mg2+ can affect the metal-chelate buffer. The final concentration of the chelator must be much greater than the free ion concentration, just as in a pH buffer, where the concentration of the weak acid/conjugate base is much greater than the H3O+ concentration.
The pH affects the protonation state of chelators like EDTA, EGTA, and HEEDTA, and as a result affects the buffering range of the metal-chelate solution. Refer to (14) and (17) to re-calculate free ion concentrations at different pH.
The pH titration method (14) can be used to make solutions of polyprotic acid chelators (EDTA, EGTA, HEEDTA) and metal ions, where the concentrations of each are verified to be within 0.5% of each other. For example, when Ca2+ binds EGTA, two protons are released, causing a drop in the pH of the solution. Therefore, the change in pH upon Ca2+ addition (ΔpH/ΔCa2+) decreases when there are equal concentrations of Ca2+ and EGTA.
References
- 1.Lakowicz JR. Principles of Fluorescence Spectroscopy. Springer; New York: 2006. [Google Scholar]
- 2.Zhang J, Campbell RE, Ting AY, et al. Creating new fluorescent probes for cell biology. Nat Rev Mol Cell Biol. 2002;3:906–918. doi: 10.1038/nrm976. [DOI] [PubMed] [Google Scholar]
- 3.Vinkenborg JL, Nicolson TJ, Bellomo EA, et al. Genetically encoded FRET sensors to monitor intracellular Zn2+ homeostasis. Nat Meth. 2009;6:737–740. doi: 10.1038/nmeth.1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Palmer A, Giacomello M, Kortemme T, et al. Ca2+ indicators based on computationally redesigned calmodulin-peptide pairs. Chemistry & Biology. 2006;13:521–530. doi: 10.1016/j.chembiol.2006.03.007. [DOI] [PubMed] [Google Scholar]
- 5.Qin Y, Dittmer PJ, Park JG, et al. Measuring steady-state and dynamic endoplasmic reticulum and Golgi Zn2+ with genetically encoded sensors. Proc Natl Acad Sci USA. 2011;108:7351–7356. doi: 10.1073/pnas.1015686108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Horikawa K, Yamada Y, Matsuda T, et al. Spontaneous network activity visualized by ultrasensitive Ca2+ indicators, yellow Cameleon-Nano. Nat Meth. 2010;7:729–732. doi: 10.1038/nmeth.1488. [DOI] [PubMed] [Google Scholar]
- 7.Tian L, Hires SA, Mao T, et al. Imaging neural activity in worms, flies and mice with improved GCaMP calcium indicators. Nat Meth. 2009;6:875–881. doi: 10.1038/nmeth.1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yamada Y, Michikawa T, Hashimoto M, et al. Quantitative comparison of genetically encoded Ca indicators in cortical pyramidal cells and cerebellar Purkinje cells. Front. Cell. Neurosci. 2011;5:18. doi: 10.3389/fncel.2011.00018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Newman RH, Fosbrink MD, Zhang J. Genetically encodable fluorescent biosensors for tracking signaling dynamics in living cells. Chem Rev. 2011;111:3614–3666. doi: 10.1021/cr100002u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davidson MW, Campbell RE. Engineered fluorescent proteins: innovations and applications. Nat Meth. 2009;6:713–717. doi: 10.1038/nmeth1009-713. [DOI] [PubMed] [Google Scholar]
- 11.Nagai T, Yamada S, Tominaga T, et al. Expanded dynamic range of fluorescent indicators for Ca(2+) by circularly permuted yellow fluorescent proteins. Proc Natl Acad Sci USA. 2004;101:10554–10559. doi: 10.1073/pnas.0400417101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Palmer AE, Jin C, Reed JC, et al. Bcl-2-mediated alterations in endoplasmic reticulum Ca2+ analyzed with an improved genetically encoded fluorescent sensor. Proc Natl Acad Sci USA. 2004;101:17404–17409. doi: 10.1073/pnas.0408030101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mank M, Santos AF, Direnberger S, et al. A genetically encoded calcium indicator for chronic in vivo two-photon imaging. Nat Meth. 2008;5:805–811. doi: 10.1038/nmeth.1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tsien R, Pozzan T. Measurement of cytosolic free Ca2+ with quin2. Meth Enzymol. 1989;172:230–262. doi: 10.1016/s0076-6879(89)72017-6. [DOI] [PubMed] [Google Scholar]
- 15.Cheng KL, Ueno K, Imamura T. CRC handbook of organic analytical reagents. CRC Press; Boca Raton, Fla: 1982. [Google Scholar]
- 16.Smith RM, Martell AE, Motekaitis RJ. NIST critically selected stability constants of metal complexes database. NIST; Gaithersburg, Md: 2004. [Google Scholar]
- 17.Kao JPY, Li G, Auston DA. Chapter 5 - Practical Aspects of Measuring Intracellular Calcium Signals with Fluorescent Indicators. Methods in Cell Biology. 2010;99:113–152. doi: 10.1016/B978-0-12-374841-6.00005-0. [DOI] [PubMed] [Google Scholar]
- 18.Dean KM, Qin Y, Palmer A. Visualizing metal ions in cells: An overview of analytical techniques, approaches, and probes. BBA - Molecular Cell Research. 2012:1–10. doi: 10.1016/j.bbamcr.2012.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Palmer A, Tsien R. Measuring calcium signaling using genetically targetable fluorescent indicators. Nature Protocols. 2006;1:1057–1065. doi: 10.1038/nprot.2006.172. [DOI] [PubMed] [Google Scholar]
- 20.Tian L, Hires SA, Looger L. Imaging neuronal activity with genetically encoded calcium indicators. Cold Spring Harbor Protocols. 2012;2012 doi: 10.1101/pdb.top069609. [DOI] [PubMed] [Google Scholar]
- 21.Evers TH, Appelhof MA, de Graaf-Heuvelmans PT, et al. Ratiometric detection of Zn(II) using chelating fluorescent protein chimeras. J Mol Biol. 2007;374:411–425. doi: 10.1016/j.jmb.2007.09.021. [DOI] [PubMed] [Google Scholar]
- 22.Emmanouilidou E, Teschemacher AG, Pouli AE, et al. Imaging Ca2+ concentration changes at the secretory vesicle surface with a recombinant targeted cameleon. Curr Biol. 1999;9:915–918. doi: 10.1016/s0960-9822(99)80398-4. [DOI] [PubMed] [Google Scholar]
- 23.Dittmer P, Miranda J, Gorski J, et al. Genetically Encoded Sensors to Elucidate Spatial Distribution of Cellular Zinc. Journal of Biological Chemistry. 2009;284:16289–16297. doi: 10.1074/jbc.M900501200. [DOI] [PMC free article] [PubMed] [Google Scholar]