Abstract
Characterization of genetic admixture of populations in the Americas and the Caribbean is of interest for anthropological, epidemiological, and historical reasons. Asthma has a higher prevalence and is more severe in populations with a high African component. Association of African ancestry with asthma has been demonstrated. We estimated admixture proportions of samples from six trihybrid populations of African descent and determined the relationship between African ancestry and asthma and total serum IgE levels (tIgE). We genotyped 237 ancestry informative markers in asthmatics and nonasthmatic controls from Barbados (190/277), Jamaica (177/529), Brazil (40/220), Colombia (508/625), African Americans from New York (207/171), and African Americans from Baltimore/Washington, D.C. (625/757). We estimated individual ancestries and evaluated genetic stratification using Structure and principal component analysis. Association of African ancestry and asthma and tIgE was evaluated by regression analysis. Mean SD African ancestry ranged from 0.76 ± 0.10 among Barbadians to 0.33 ± 0.13 in Colombians. The European component varied from 0.14 ± 0.05 among Jamaicans and Barbadians to 0.26 ± 0.08 among Colombians. African ancestry was associated with risk for asthma in Colombians (odds ratio (OR) = 4.5, P = 0.001) Brazilians (OR = 136.5, P = 0.003), and African Americans of New York (OR: 4.7; P = 0.040). African ancestry was also associated with higher tIgE levels among Colombians (β = 1.3, P = 0.04), Barbadians (β = 3.8, P = 0.03), and Brazilians (β = 1.6, P = 0.03). Our findings indicate that African ancestry can account for, at least in part, the association between asthma and its associated trait, tIgE levels.
Keywords: African, asthma, ancestry
Introduction
Race and ethnicity are complex constructs incorporating social, cultural, and genetic factors. Populations that migrated from continental Africa as part of the African Diaspora were dispersed across the Americas, and their contemporary ancestors have varying proportions of African admixture depending upon local demographic history and patterns of admixture, geographical location, and other factors [Via et al., 2011; Wang et al., 2008]. Determination of the ancestral origin and extent of the hybridization is of great anthropological, epidemiological, and historical interest and affords opportunities to study the evolutionary biology of the ancestral as well as the admixed populations.
Differences in African or other ancestry proportions between cases and controls is a confounding factor in genetic association studies of admixed populations because differences in admixture proportions among subgroups can erroneously suggest association with disease instead of ancestry at loci where allele frequencies differ between the ancestral populations (e.g., population stratification) [Cardon and Palmer, 2003; Spielman et al., 1993; Ziv and Burchard, 2003]. This situation is typically addressed by genotyping ancestry informative markers (AIMs) to estimate the individual ancestral proportions for major putative parental populations in admixed groups, and even though multiple approaches can be used, the underlying concept it to incorporate those values represented either as proportions of admixture or eigenvalues derived from principal component analysis (PCA) in the model used to evaluate genetic associations [McKeigue, 2007; Patterson et al., 2006; Pritchard and Rosenberg, 1999].
Alternatively, admixed populations provide a unique opportunity for interrogating the relationship between ancestry components and those biological traits and diseases that have different prevalences or degrees of severity among specific ethnic groups [Winkler et al., 2010]. It is well established that the prevalence, morbidity, and mortality of allergic airway diseases, such as asthma, is disproportionately high among individuals of African descent in the United States [Akinbami et al., 2005, 2011; El-Ekiaby et al., 2006; Gold et al., 1993; Lang et al., 2009; Lester et al., 2001; Moorman et al., 2007; Nelson et al., 1997; Sarpong et al., 1996; Schwartz et al., 1990], in African countries and countries with African admixture (i.e., Latin America) [Pearce et al., 2007]. The Global Strategy for Asthma Management and Prevention report suggests that African ancestry itself is a risk factor [GINA, 2008]. The usual source of airway inflammation in asthma is an immunoglobulin E (IgE)-mediated reaction initiated by exposure to environmental allergens, and asthma is closely associated with serum total IgE (tIgE) levels [Burrows et al., 1989; Marsh et al., 1974]. Higher serum tIgE levels have also been observed among African Americans with persistent asthma compared to whites and other ethnic groups [Grant et al., 2000; Grundbacher and Massie, 1985]. The differences are noticeable even early in life as described in a children cohort selected from a multiethnic society, where black newborns had significantly higher concentrations of cord blood serum tIgE compared with whites newborns [Haus et al., 1988]. It is unknown to what extent this is due to the environment versus genetic factors; however, a number of genetic variants identified as risk variants for asthma and associated traits have been shown to have significantly higher frequencies in individuals of African descent compared to other racial and ethnic groups [Barnes et al., 2007; Caggana et al., 1999; Green et al., 1998; Maxwell et al., 2005; Ness et al., 2004]. Also, a significant ancestry association peak on chromosome 6q14 (rs1361549) was associated with asthma exclusively in African American subjects with local European admixture [Torgerson et al.].
Previously, we observed an association between a well-established AIM of African ancestry, the Duffy antigen receptor for chemokines (DARC), and risk of asthma and high tIgE levels in independent African-admixed populations [Vergara et al., 2008]. Analyzing 44 AIMs, Choudhry and colleagues also established that African ancestry in general increased the risk for asthma among Puerto Ricans with higher socioeconomic status compared to those with lower socioeconomic status [Choudhry et al., 2006]. Kumar et al. demonstrated that African ancestry was inversely related to forced expira-tory volume in 1 sec (FEV1) and forced vital capacity (FVC) in participants self-identified as African American in the Coro nary Artery Risk Development in Young Adults (CARDIA) study, the Health, Aging, and Body Composition (HABC) study, and the Cardiovascular Health Study (CHS) [Kumar et al., 2010], and in a recent publication, African ancestry was as predictor of early childhood wheezing in a prospective birth cohort of 1,034 children [Kumar et al., 2012]. Similarly, we estimated individual African ancestry using a panel of 52 AIMs in asthmatics and nonasthmatics from Cartagena (Colombia, South America), and determined that African ancestry was associated with a significant risk for asthma and high tIgE levels [Vergara et al., 2009]. Taking in account the relevance of replicating genetic associations [Chanock et al., 2007] and to expand our analyses and interrogate the strength of the association according to the degree of African ancestry in the population, we enlarged the AIMs panel to include 237 markers, and extended these studies to include additional, independent African Caribbean and African American populations from Colombia, Barbados, Jamaica, Brazil, and the United States. We estimated proportions of admixture of continental parental populations in those individuals and the relationship between African ancestry and asthma and serum tIgE levels.
Materials
Subjects
Barbadians
We included 467 Barbadian nonrelated individuals (190 cases and 277 nonasthmatic controls) from a study on the genetics of asthma as previously described [Barnes et al., 1996; Mathias et al., 2010] and in supplementary material and methods.
Jamaicans
A sample of 706 unrelated individuals (177 asthmatics, 529 nonasthmatic controls) was recruited from Kingston, St. Andrew, and St. Catherine, Jamaica as part of the Jamaican Adolescent Asthma Study as described in supplementary materials and methods and in references [Ashley et al., 1988; Tulloch-Reid et al., 2010]. All subjects provided informed written consent to participate as approved by the University of the West Indies, Mona campus.
Brazilians
Asthmatic (N = 40) and nonasthmatic (N = 220) nonrelated subjects were recruited in five rural communities (Buri, Camarao, Genipapo, Sempre Viva, and Cobo) in the district of Conde, Bahia, located in the North East Coast of Brazil as described in detail elsewhere [Grant et al., 2011; Vergara et al., 2008] and in Supporting Information material and methods. Written consent was obtained from adult individuals or parent or guardian of children as approved by the Johns Hopkins Bayview Medical Center and Universidade de Federal de Bahia IRBs.
Colombians
A sample of 1,133 unrelated individuals (508 asthmatics and 625 nonasthmatic controls) were recruited from the Caribbean coastal city of Cartagena as described in detail elsewhere [Vergara et al., 2008, 2009] and in Supporting Information materials and methods. All subjects or their guardian/responsible adult gave written consent for their inclusion in the present study as approved by the Bioethics Committee of the School of Medicine of The University of Cartagena.
African Americans
Two samples of self-reported African Americans were included in this study. They were analyzed separately according to the recruiting site and place of residency of individuals. (1) A group of 1,382 individuals (625 asthma cases and 757 unrelated, unaffected controls) ascertained participating in Genomic Research on Asthma in the African Diaspora (GRAAD) recruited in the Baltimore-Washington D.C. metropolitan area and participants in the Baltimore Asthma Severity Study (BASS) residents of Baltimore City; (2) A second group comprising 378 individuals (207 asthmatic cases and 171 nonasthmatic controls) were recruited in New York and participated in The Reducing Emergency Asthma Care in Harlem (REACH) study. Details of these samples are described elsewhere [Ford et al., 2001; Mathias et al., 2007; Murray et al., 2010; Pesola et al., 2004] and in Supporting Information material and methods. The study protocols, recruitment procedures, and consent forms were approved by the Institutional Review Board of Columbia University, Johns Hopkins University, and Howard University.
tIgE Measurements
Sera were available in the Barbadian, Jamaican, Brazilian, Colombian, and African American samples from Baltimore-Washington, D.C. for measurements of tIgE as described previously [Vergara et al., 2008] and in supplementary Materials. For all datasets, tIgE values were log-transformed to reduce the skewness of the distribution. Measurements were not available for African Americans from New York and for participants of the BASS study.
Selection of AIMs and genotyping
We selected 237 AIMs distributed across 22 autosomes as presented in Supporting Information Table SI. The markers were a subset of 416 AIMs previously selected [Murray et al., 2010] from the 650Y panel based on a published list of AIMs [Cheng et al., 2009] showing large allelic frequency difference (δ) among the three putative parental populations (European, African, and Asians) using HapMap Phase 2 reference populations (CEU, YRI, CHB), respectively.
Due to limitations in the availability of Native Americans DNA samples, for this analysis, we used Asians as a proxy for Native Americans because they are closely related [Bamshad et al., 2004]. Genotypes on HapMap subjects were downloaded from HapMart (http://hapmart.hapmap.org/BioMart/martview), an extension of the HapMap data resource. A marker was considered informative if 0.30 difference of the allelic frequencies existed between any of two parental groups. Blood samples were collected from all subjects and genomic DNA was extracted using standard protocols. Samples were genotyped according to the manufacturer's protocol at the Center for Inherited Disease Research on an Illumina BeadStation 500G Golden Gate genotyping platform using a custom panel (GS0011473-OPA). Geno-typing plates were balanced by gender and asthma status and HapMap DNA samples and duplicates were included as genotyping controls.
Methods
Statistical Analysis
Comparisons of the demographic characteristics among asthmatics and nonasthmatics were performed by means of Student's t-test and χ2 test as needed using STATA software (StataCorp. 2009. Stata Statistical Software: Release 11. College Station, TX: StataCorp LP.). Arlequin version 3.11 software [Excoffier et al., 2005] was used to calculate allele and genotype frequency of the AIMs, departure of genotype frequencies from Hardy-Weinberg equilibrium (HWE), linkage disequilibrium between each pair of markers (as an indicator of stratification) independently for each sample set and genetic differentiation among the analyzed populations using FST genetic distances [Reynolds et al., 1983; Slatkin, 1995]. Average population admixture and individual admixture estimates for each sample set were determined using a model-based clustering method by grouping data for the total sample in three ancestral populations (K = 3) to reflect the admixture history in the Caribbean and North and South America with the software Structure (version 2.3.3, http://pritch.bsd.uchicago.edu/software) and assessment of stratification was done by using PCA using the smartpca program from the software package eigenstrat [Patterson et al., 2006] including the three putative ancestral populations as reference. We investigated the relationship between disease status and African ancestry by logistic regression analysis using disease status as the dependent variable and individual ancestry as independent variable, while adjusting for potential confounders (age, sex). To determine associations of African ancestry with tIgE levels, a linear regression model was used incorporating age, sex, and disease status as covariates. When needed, Glmperm and lmperm packages in R were used to perform permutation analysis to confirm the association results with asthma and tIgE, respectively. For the association of African ancestry and tIgE levels, meta-analysis P-values were computed using Stouffer's Z-score method [Stouffer et al., 1949].
Results
Subjects
Clinical and demographic characteristics of asthmatics and nonasthmatics in each sample are presented in Table I. Mean tIgE concentrations for each population were similar to those described previously [Grant et al., 2008; Vergara et al., 2009, 2008], wherein Brazilians showed higher tIgE levels than the other samples, most likely due to the high prevalence (83.5%) of extracellular parasitic disease in this population [de Jesus et al., 1993]. tIgE was significantly higher in asthmatics compared to nonasthmatics in all samples sets (P ≤ 0.001) except Brazilians (P = 0.64). Results about allelic frequencies, HWE, genetic differentiation, and genetic structure are presented as supplementary material.
Table I.
Descriptive characteristics of six African ancestry populations
| Male gender |
Age |
Total IgE† |
||||||
|---|---|---|---|---|---|---|---|---|
| Populations | N (%) | N (%) | P value | Mean (SE) | P value | Geom. mean (95%CI) | P value | |
| Barbadians | Asthmatics | 190 (40) | 67 (0.35) | 0.02 | 40.1 (0.79) | 0.0001 | 660.6 (389.0–1096.4) | 0.001 |
| Controls | 277 (59) | 128 (0.46) | 45.9 (0.71) | 239.8 (186.2–301.9) | ||||
| Jamaicans | Asthmatics | 177(25) | 80 (45) | 0.51 | 18.8 (0.04) | 0.7 | 213.8 (177.8–257.0) | 0.001 |
| Controls | 529 (75) | 256 (48) | 18.8 (0.03) | 120.2 (107.1–134.8) | ||||
| Brazilians | Asthmatics | 40 (15) | 12 (30.0) | 0.06 | 37.4 (3.30) | 0.07 | 2526.5 (1769.7–3606.9) | 0.6 |
| Controls | 220 (85) | 79 (35.9) | 43.6 (1.23) | 2294.1 (1957.6–2683.8) | ||||
| Colombians | Asthmatics | 508 (45) | 125 (24.6) | 0.001 | 31.6 (0.73) | 0.001 | 279.9 (247.1–318.0) | 0.001 |
| Controls | 625 (55) | 309 (49.4) | 36.7 (0.68) | 424.6 (380.2–468.8) | ||||
| African American (New York†) | Asthmatics | 207 (55) | 129 (62.3) | 0.001 | 40.3 (0.42) | 0.005 | NA | NA |
| Controls | 171 (45) | 73 (42.6) | 37.6 (0.87) | NA | ||||
| African Americans (Baltimore-Washington, D.C.)‡ | Asthmatics | 625 (45) | 343 (45) | 0.8 | 25.7 (0.62) | 0.001 | 400.21 (342.9–467.0) | 0.001 |
| Controls | 757 (54) | 323 (52) | 33.2 (0.51) | 187.4 (161.3–217.7) | ||||
NA: Not available
Plus-minus values are means ± SE. Log-transformed total IgE concentrations measured in ng/ml. REACH, Reducing Emergency Asthma Care in Harlem.
BASS, Baltimore Asthma Severity Study; GRAAD, Genomic Research on Asthma in the African Diaspora.
Admixture Estimates
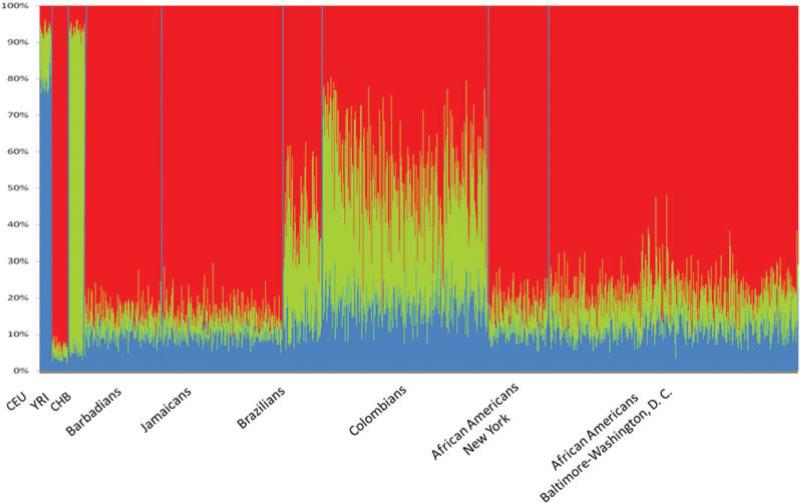
Average (±SD) population admixture estimate proportions are presented in Table II and individual admixture estimates are presented in Figure 1. The distribution of the African ancestry proportions are displayed as box plots in Supporting Information Figure S1. Mean (SD) population African ancestry was highest in West Indians (Jamaicans, 0.76 ± 0.10; Bar badians, 0.76 ± 0.10) and African Americans from New York (0.70 ± 0.15) and Baltimore-Washington D.C. (0.69 ±0.12) and lowest in the two South American samples (Brazilians, 0.45 ± 0.12; Colombians, 0.33 ± 0.13). The average Euro pean component ranged from a low of 0.14 ± 0.005 among Jamaicans and Barbadians to a high of 0.26 ± 0.08 among Colombians. The average Asian ancestry ranged from 0.08 ± 0.06 among Barbadians to 0.40 ± 0.13 among Colombians. There was a significant correlation between the first principal component and the estimates of individual African ancestry in each of the samples (Spearman's ρ > 0.98, P < 0.0001 for each pair of comparisons).
Table II.
African, European and Asian ancestry proportions for Barbadians, Jamaicans, Brazilians, Colombians and African Americans
| Ancestry proportions (Mean, SD) |
|||||||||
|---|---|---|---|---|---|---|---|---|---|
| African |
European |
Asian |
|||||||
| Population | Total | Asthmatics | Non asthmatic Controls |
Total | Asthmatics | Non asthmatic Controls |
Total | Asthmatics | Non asthmatic Controls |
| Barbadians | 0.76 (0.10) | 0.77 (0.11) | 0.77 (0.10) | 0.14(0.05) | 0.14(0.04) | 0.14 (0.05) | 0.08 (0.07) | 0.08 (0.08) | 0.09 (0.06) |
| Jamaicans | 0.76 (0.10) | 0.76 (0.09) | 0.77 (0.10) | 0.14(0.05) | 0.14(0.04) | 0.14 (0.05) | 0.09 (0.07) | 0.10(0.08) | 0.09 (0.08) |
| Brazilians | 0.45 (0.12) | 0.52 (0.12) | 0.45 (0.12) | 0.19(0.06) | 0.18 (0.07) | 0.19 (0.07) | 0.34 (0.12) | 0.29 (0.14) | 0.35 (0.13) |
| Colombians | 0.33 (0.13) | 0.35 (0.13) | 0.32 (0.14) | 0.26 (0.08) | 0.26 (0.08) | 0.26 (0.08) | 0.40 (0.13) | 0.39 (0.13) | 0.42 (0.13) |
| African Americans (New York) | 0.70 (0.15) | 0.71 (0.14) | 0.68 (0.16) | 0.15 (0.05) | 0.15 (0.05) | 0.15 (0.05) | 0.14 (0.12) | 0.14(0.13) | 0.16 (0.14) |
| African Americans (Baltimore-Washington, D.C.) | 0.69 (0.12) | 0.70 (0.14) | 0.70 (0.12) | 0.15 (0.05) | 0.15 (0.06) | 0.15 (0.05) | 0.14 (0.12) | 0.14 (0.11) | 0.15 (0.13) |
Figure 1.
Individual admixture estimates by Structure for Barbadians (N = 467), Jamaicans (N = 706), Brazilians (N = 260), Colombians (N = 1,133), African Americans from New York (N = 378) and African Americans from Baltimore-Washington, D.C. (N = 1,382). Included are the putative parental populations available in HapMap (CEU, CHB, and YRI).
Association of African Ancestry and Asthma and tIgE
Table III summarizes results of regression analyses of African ancestry and asthma and tIgE. African ancestry was associated with a significant risk for asthma among several populations. The odds ratio (OR) for asthma is associated with a unit increase in African admixture proportion (from 0 to 1). It was significant for Colombians (OR: 4.5; 95% CI: 1.79–11.27; P = 0.001), Brazilians (OR: 136.5; 95% CI: 5.4– 3405.6; P = 0.003), and African Americans of New York (OR: 4.7; 95% CI: 1.07–21.0; P = 0.04). In Brazilians, permutation analysis confirmed the association (P = 0.001). Principal component plots for each sample of each population are shown in Supporting Information Figure S2. The plotted individuals are discriminated by affection status. For the Jamaican and the two African American populations, cases and controls clustered together and there was no significant difference in the distribution of the first principal component among asthma cases and nonasthmatic controls for each sample. For Colombians and Brazilians and African Americans from NewYork, a significant difference was found when comparing the distribution of the first principal component among cases and nonasthmatic controls (P < 0.001).
Table III.
Results of regression analyses of African ancestry proportions and asthma and tIgE levels for Barbadians, Jamaicans, Brazilians, Colombians and two African American samples
| Population | Asthma (OR, 95% CI, P value)* | Total serum IgE levels (β, 95% CI, P value)** |
|---|---|---|
| Barbadians | 1.1 (0.16–6.9;0.9) | 3.8 (1.1–13.1;0.03) |
| Jamaicans | 0.8 (0.22–3.1;0.8) | 1.0 (0.7–1.4;0.9) |
| Brazilians | 136.5 (5.4–3406;0.003) | 1.6 (1.1–2.6;0.03) |
| Colombians | 4.5 (1.79–11.27;0.001) | 1.3 (1.01–1.7;0.04) |
| African Americans (New York) | 4.7(1.07–21.04;0.04) | NA# |
| African Americans (Baltimore-Washington, D.C.) | 1.1 (0.50–2.81;0.7) | 0.99 (0.6–1.7;0.9) |
Adjusted by age and sex
Adjusted by age, sex and asthma status.
NA = Not available. The odds ratio (OR) for asthma is associated with a unit increase in African admixture proportion (from 0 to 1).
Of the five samples with tIgE data available, African ancestry was associated with tIgE levels in Colombians (β: 1.3; 95% CI: 1.01–1.7; P = 0.04), Barbadians (β: 3.8; 95% CI: 1.1–13.1; P = 0.030), and Brazilians (β: 1.6; 95% CI: 1.1–2.6; P = 0.03; Permutation P = 0.05). No relationship between African ancestry and asthma or tIgE was detected among the Jamaicans or African American of Baltimore-Washington, D.C. No relationship between ancestry and asthma or tIgE was detected among the Jamaicans or African American of Baltimore-Washington, D.C. No associations were observed between European ancestry and asthma and/or tIgE in any of the datasets. Supporting Information Figure S4 graphically shows the effect (β) and 95% CI for the association of African ancestry and tIgE levels in each one of the analyzed population and the P-value calculated for all populations. The P-value of the meta-analysis of the five analyzed populations was statistically significant (P = 0.019).
Discussion
In this study, we determined the admixture proportion of six samples from populations of African descent representing geographical regions across Continental America and two islands in the Caribbean, for which the African Diaspora figured prominently in the demographic history of these populations. We also investigated the association of African ancestry and asthma and tIgE levels including a sample of considerable size for each population and a comprehensive panel of AIMs distributed across the genome.
We included only self-reported unrelated individuals in each of the analyzed populations to minimize the potential effect of relatedness in calculations of allelic frequencies of the AIMs and consequently in the ancestry estimates and association analysis. Also, African Americans from Baltimore-Washington DC, African Americans from New York and Barbadians included in this study were previously analyzed using a Genome Wide Association Study (GWAS) panel of and all individuals identified as related by IBD analysis were removed from the dataset prior to this analysis [Mathias et al., 2010]. Additional cryptic relatedness could be elucidated by a larger panel of markers in the other populations but we do not think it would have a significant effect on our association analysis. We observed a wide range of average proportions of parental populations across the samples and a significant association of African ancestry and asthma and serum tIgE levels. To date, measured by number of populations included, sample size per populations, and number of AIMs, this is one of the largest studies to analyze ancestry proportions and the relationship with asthma and tIgE in populations of the Americas and the Caribbean.
Although each one of the analyzed populations has a unique history, they share certain characteristics regarding to the dynamics of the admixture process. Historical records indicate that there were generally three parental populations, West Africans, Europeans, and Native Americans. The process of admixture extended across Latin America and the Caribbean during the 15th through 19th centuries, including the arrival of Europeans (predominantly Spanish, Portuguese, and British) and the extinction of Native Americans converging with the arrival and expansion of African slaves, largely from West Africa during the 16th and 19th centuries. Nevertheless, the analyzed populations underwent diverse patterns of admixture given the location of settlement of the parental populations approximately 500 years ago [Curtin, 1969; Salzano, 2002; Sanchez-Albornoz, 1974; Thomas 1997].
In several previous genetic studies evaluating samples from the same contemporary countries of continental America included in the current study, ancestry estimates vary by region, as displayed in Supporting Information Table SIII. Several factors contributed to make up the current genetic pool including admixture, genetic drift, migration, and recent genetic flow [Winkler et al., 2010]. Asian ancestry estimates, which are a surrogate for Native American ancestry are the lowest of the three parental components in Barbadians, Jamaicans, and African Americans. Asian ancestry estimates obtained with this set of AIMs showed a large variation, and, we conclude these estimates may well be erroneous rather than indicative of admixture in contrast to estimates for the other two parental populations. It is well known that the Na tive American population was decimated and exterminated along the whole continent either by disease or labor [De las Casas, 1985; Simon, 1953; Williams, 1984]. For instance, there was a rapid annihilation of the Arawak population following colonization of the Spaniards from 1494 to 1655 in Jamaica; a similar history was described for the Caribs and Arawaks in Barbados [Rouse, 1992] and for the Caribs and Taironas in Cartagena [Meisel and Aguilera, 1997].
We found a predominant proportion of African ancestry and a low proportion of European ancestry in the populations of the Caribbean Islands. According to the records, the number of African slaves brought to work in the sugar plantation in Barbados was around 900,000 [Pepin, 2005] and outnumbered European settlers by a proportion of 20:1 [Simms et al., 2010]. Europeans were also decimated by disease and the predominant population that finally contributed to the contemporary genetic pool was of African origin [Benn-Torres et al., 2008; Knight, 1997]. A history that is very similar to that described in the island of Jamaica [Benn-Torres et al., 2008]. In continental America, a highly diverse genetic makeup has been noted previously including territories of Colombia and Brazil (Supporting Information Table SIII). Local history, diversity of the geography, as well as social and economic factors may have played a crucial role in the dynamic of admixture after the first admixture event. African admixture is especially pronounced in the coastal states, which underwent a social history similar to the rest of the Caribbean, whereby African slaves were imported as agricultural labor. In Colombia, a higher African ancestry has been detected in those places that were port of entry for slaves (Cartagena, Choco) or located in the route of transportation of slaves trough the inner country and those regions close to the original sugar cane plantations [Rojas et al., 2010]. A high variation in admixture proportions has also been observed in different regions of Brazil among and within studies, however there has been a pattern observed indicating higher European ancestry in southern Brazil compared to the north as well as isolated populations of predominant African descent that do not follow this pattern [Pena et al., 2009; Santos et al., 2010]. If we compare our results to previous ones in the included populations (Supporting Information Table SIII), the estimates obtained are slightly different to those described before which can be attributable to the markers chosen, the samples analyzed, and the statistical methods used in the studies. Nevertheless, we consider that the admixture estimates obtained using this panel of markers is accurate. The current estimates are very consistent with those obtained with a significant higher number of markers in the same individuals of Barbados and African Americans of Baltimore-Washington DC [Murray et al., 2010] and the estimates obtained for the Colombian population were different compared with those obtained in the same individuals using a panel of 52 AIMs [Vergara et al., 2009]. These patterns of variation in ancestry across countries seem to be a common feature across the Americas even in small islands as Puerto Rico [Via et al., 2011]. This has large repercussion for genetic association studies in Latin American populations, given the high probability of population stratification, even in samples from a single country or a geographic region, and highlights the relevance of correcting by using ancestry estimates in any genetic association study in these populations.
We observed a pattern correlating the geographic proximity of the samples and the genetic differentiation values observed with two independent statistical approaches (FST values and PCAs, Supporting Information Table SII and Figure S2). Brazilians and Colombians tended to cluster together (FST: 0.02, P = 0.001), close to the CEU samples and further apart from the West African cluster. In contrast, African Americans, Barbadians, and Jamaicans clustered very closely to each other and to the YRI samples, which is consistent with the low level of genetic differentiation detected by the FST statistic in those groups.
In this study, we found that African ancestry conferred a significant risk for asthma and higher levels of serum tIgE in comparison with non-African ancestries. The associations were not due to potential confounders as gender and age. We previously observed the association of African ancestry with asthma and tIge levels in family-based association analyses investigating the relationship between a unique polymorphism in the gene encoding the DARC, a well-known marker of African ancestry as conferring risk for asthma and tIgE in a similar group of populations [Vergara et al., 2008]. In subsequent analyses, we investigated a Colombian case/control database and genotyped a set of 52 AIMs with similar results (OR for asthma associated with African admixture was 2.97 (95% CI: 1.08–8.08) and for tIgE 1.9 (95% CI: 1.17– 3.12) [Vergara et al., 2009]. In this case, we replicated our findings with the same direction of the effect for asthma and tIgE in Brazilians and in Colombians (this time using a larger dataset and a higher number of AIMs), for asthma in African Americans from New York, and for tIgE in Barbadians. When comparing our results to those described in other admixed populations, we found that outcomes varied according to the populations and phenotype analyzed, design of the studies, and covariates included. Our results are highly consistent with those recently published for African Americans of the REACH cohort, where it was determined an OR of 1.16 (95% CI: 1.06–1.28; P = 0.002) for the risk of asthma per 10% increase of African ancestry after correcting by age, gender, and study site [Flores et al., 2012]. Similarly, in a Puerto Rican population, asthma risk increased with African ancestry among individuals with higher socioeconomic status [Choudhry et al., 2006] and European ancestry conferred risk for asthma in those from lower socioeconomic status [Choudhry et al., 2006]. In Mexican Americans, higher European ancestry was significantly associated with more severe asthma as measured by both FEV1 and by a clinical assessment of severity among Mexican Americans [Salari et al., 2005]. There are some differences between Salari et al.’s study and the current study, however, that may explain the discrepancy in the results, including the analyzed phenotypes (FEV1 vs. asthma/IgE) and the ancestries analyzed (European compared to Native American vs. African compared to European/Asian). Moreover, among Mexican Americans, the African component is low and not evaluable. In terms of IgE, even though the phenotype is not identical, our result are similar to those described by Kumar et al., who determined that African ancestry was associated with increased odds of peanut and milk specific IgE levels of ≥5 kUA/L in children from an urban multiethnic birth cohort [Kumar et al., 2011].
Individual genetic ancestries as determined by AIMs have been also described as an important determinant of variation of multiple traits and/or susceptibility to other diseases. Among African American, Hispanic American, and European American children, a greater African admixture proportion has been associated with lower fat mass, lower total abdominal adipose tissue, lower intraabdominal adipose tissue, lower subcutaneous abdominal adipose tissue, and higher bone mineral content, after adjusting for socioeconomic status, sex, age, height, race/ethnicity, and pubertal status [Cardel et al., 2010]. Percentage of African ancestry was also inversely associated with function measurements (FEV1, FVC) in three independent cohorts of African Americans across a wide range of ages [Kumar et al., 2010]. In our study, we observed no associations between African ancestry and asthma in the African American group from Baltimore-Washington, D.C, Barbadians, and Jamaicans. The lack of association with asthma observed these populations could be attributed to the low variance of the African ancestry proportions in these populations (Supporting Information Figure S1) and low proportions of European and Asian ancestries, which in turn underpowered the comparisons. Among African Americans from the Baltimore-Washington D.C. metropolitan area, the sample size had power of only 0.25 to detect an OR of 4, given the European ancestry proportions and the case/control relation in those populations [Gauderman, 2006]. Power was even lower for Jamaicans and Barbadians, where a considerably larger sample size (N = 2,757 for Barbados and N = 2,182 for Jamaica) would be required to adequately evaluate the effect. Environmental factors, effect of socioeconomic status, different strategies for recruiting that potentially determine light differences in the asthma phenotype can also account for the lack of association in those groups.
The replication of our results in several populations with the same direction of the effect provide evidence that genetic factors contained in the African genetic component of hybrid populations contribute to ethnic disparities for asthma, and that studying populations of West African descent should help to identify additional genetic risk factors associated with asthma susceptibility. For example, Choudhry et al. performed admixture mapping for asthma including Puerto Rican individuals with moderate-to-severe asthma and nonasthmatic controls and identified chromosomal regions 5q23.3 and 13q13.3 as potential regions harboring genes for asthma in this population [Choudhry et al., 2008]. Differences in association in candidate genes for asthma according to ancestry have also been identified, indicating that specific and maybe different genetic risk factors account for asthma susceptibility in African descent populations compared with European descent populations [Barnes et al., 2007; Baye et al.]. We are aware of several caveats in our study design, one of which is the use of Asian populations as representative of Native American populations. Another limitation is the lack of adjustment by socioeconomic status, a potentially critical confounder. Even though the Colombian samples used in this study were selected using the same approach as a previous report, in which the association of African ancestry and asthma remained after correcting for socioeconomic status [Vergara et al., 2009], we did not have access to the same level of socioeconomic status in the other populations in this study, and were therefore unable to evaluate the effect of this potential confounder. In summary, among populations of African descent distributed in different areas of the Americas and the Caribbean, African ancestry confers a significant risk for asthma and high tIgE levels. Population structure is an important confounding factor that can be efficiently corrected with this comprehensive set of markers in any future association studies. These populations are an excellent tool to examine loci associated with asthma and tIgE by admixture mapping.
Acknowledgments
We thank all the individuals participating in this study. We thank Jane Romm, Roxann Ashworth, Alan Scott, Kim Doheny, Jie Zhang and Corinne Boehm, members of the Center for Inherited Disease Research for the valuable help with genotyping as well as Joseph Potee and Branch Coleman III for preparation of the samples. We also thank Patricia Oldewurtel for technical assistance.
Contract grant sponsor: National Institutes of Health; Contract grant number: HL087699; Contract grant sponsor:The National Heart, Lung, and Blood Institute (NHLBI). Contract grant sponsor: COLCIENCIAS; Contract grant number:680-2009. Contract grant sponsor: Health Research Council. Contract grant sponsor: Caribbean Cardiac Society. Contract grant sponsor: University Hospital of West Indies. Contract grant sponsor: The Cultural Arts Sports and Education (CHASE) Fund. Contract grant sponsor: National Health Fund. Contract grant sponsor:National Health Fund Phase 2, 2007–2008. Contract grant sponsor: Mary Beryl Patch Turnbull Scholar Program. Contract grant sponsor: Brazilian National Research Council (CNPq).
References
- Akinbami LJ, Rhodes JC, Lara M. Racial and ethnic differences in asthma diagnosis among children who wheeze. Pediatrics. 2005;115(5):1254–1260. doi: 10.1542/peds.2004-0897. [DOI] [PubMed] [Google Scholar]
- Akinbami LJ, Moorman JE, Liu X. Asthma prevalence, health care use, and mortality: United States, 2005–2009. 2011:1–14. [PubMed] [Google Scholar]
- Ashley D, McCaw-Binns A, Foster-Williams K. The perinatal morbidity and mortality survey of Jamaica 1986–1987. Paediatr Perinat Epidemiol. 1988;2(2):138–1347. doi: 10.1111/j.1365-3016.1988.tb00194.x. [DOI] [PubMed] [Google Scholar]
- Bamshad M, Wooding S, Salisbury BA, Stephens JC. Deconstructing the relationship between genetics and race. Nat Rev Genet. 2004;5(8):598–609. doi: 10.1038/nrg1401. [DOI] [PubMed] [Google Scholar]
- Barnes K, Neely J, Duffy D, Freidhoff L, Breazeale D, Schou C, Naidu R, Levett P, Renault B, Kucherlapati R. Linkage of asthma and total serum IgE concentration to markers on chromosome 12q: evidence from Afro-Caribbean and Caucasian populations. Genomics. 1996;37(1):41–50. doi: 10.1006/geno.1996.0518. others. [DOI] [PubMed] [Google Scholar]
- Barnes K, Grant A, Hansel N, Gao P, Dunston G. African Americans with asthma: genetic insights. Proc Am Thorac Soc. 2007;4(1):58–68. doi: 10.1513/pats.200607-146JG. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baye TM, Butsch Kovacic M, Biagini Myers JM, Martin LJ, Lindsey M, Patterson TL, He H, Ericksen MB, Gupta J, Tsoras AM. Differences in candidate gene association between European ancestry and African American asthmatic children. PLoS One. 6(2):e16522. doi: 10.1371/journal.pone.0016522. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benn-Torres J, Bonilla C, Robbins CM, Waterman L, Moses TY, Hernandez W, Santos ER, Bennett F, Aiken W, Tullock T. Admixture and population stratification in African Caribbean populations. Ann Hum Genet. 2008;72(1):90–98. doi: 10.1111/j.1469-1809.2007.00398.x. others. [DOI] [PubMed] [Google Scholar]
- Burrows B, Martinez FD, Halonen M, Barbee RA, Cline MG. Association of asthma with serum IgE levels and skin-test reactivity to allergens. N Engl J Med. 1989;320(5):271–277. doi: 10.1056/NEJM198902023200502. [DOI] [PubMed] [Google Scholar]
- Caggana M, Walker K, Reilly AA, Conroy JM, Duva S, Walsh AC. Population-based studies reveal differences in the allelic frequencies of two functionally significant human interleukin-4 receptor polymorphisms in several ethnic groups. Genet Med. 1999;1(6):267–271. doi: 10.1097/00125817-199909000-00005. [DOI] [PubMed] [Google Scholar]
- Cardel M, Higgins PB, Willig AL, Keita AD, Casazza K, Gower BA, Fernandez JR. African genetic admixture is associated with body composition and fat distribution in a cross-sectional study of children. Int J Obes (Lond) 2010;35(1):60–65. doi: 10.1038/ijo.2010.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardon LR, Palmer LJ. Population stratification and spurious allelic association. Lancet. 2003;361(9357):598–604. doi: 10.1016/S0140-6736(03)12520-2. [DOI] [PubMed] [Google Scholar]
- Chanock SJ, Manolio T, Boehnke M, Boerwinkle E, Hunter DJ, Thomas G, Hirschhorn JN, Abecasis G, Altshuler D, Bailey-Wilson JE. Replicating genotype-phenotype associations. Nature. 2007;447(7145):655–660. doi: 10.1038/447655a. others. [DOI] [PubMed] [Google Scholar]
- Cheng CY, Kao WH, Patterson N, Tandon A, Haiman CA, Harris TB, Xing C, John EM, Ambrosone CB, Brancati FL. Admixture mapping of 15,280 African Americans identifies obesity susceptibility loci on chromosomes 5 and X. PLoS Genet. 2009;5(5):e1000490. doi: 10.1371/journal.pgen.1000490. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhry S, Burchard EG, Borrell LN, Tang H, Gomez I, Naqvi M, Nazario S, Torres A, Casal J, Martinez-Cruzado JC. Ancestry-environment interactions and asthma risk among Puerto Ricans. Am J Respir Crit Care Med. 2006;174(10):1088–1093. doi: 10.1164/rccm.200605-596OC. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhry S, Taub M, Mei R, Rodriguez-Santana J, Rodriguez-Cintron W, Shriver MD, Ziv E, Risch NJ, Burchard EG. Genome-wide screen for asthma in Puerto Ricans: evidence for association with 5q23 region. Hum Genet. 2008;123(5):455–468. doi: 10.1007/s00439-008-0495-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtin P. The Atlantic Slave Trade: A Census. Madison, WI. University of Wisconsin Press; The publisher's location is Madison, WI. USA: 1969. [Google Scholar]
- de Jesus AM, Almeida RP, Bacellar O, Araujo MI, Demeure C, Bina JC, Dessein AJ, Carvalho EM. Correlation between cell-mediated immunity and degree of infection in subjects living in an endemic area of schistosomiasis. Eur J Immunol. 1993;23(1):152–158. doi: 10.1002/eji.1830230125. [DOI] [PubMed] [Google Scholar]
- De las Casas B. Brevísima Relación de la Destrucción de las Indias. Sarpe; Madrid: 1985. pp. 99–100. [Google Scholar]
- El-Ekiaby A, Brianas L, Skowronski ME, Coreno AJ, Galan G, Kaeberlein FJ, Seitz RE, Villaba KD, Dickey-White H, McFadden ER., Jr. Impact of race on the severity of acute episodes of asthma and adrenergic responsiveness. Am J Respir Crit Care Med. 2006;174(5):508–513. doi: 10.1164/rccm.200603-431OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Excoffier L, Laval G, Schneider S. Arlequin ver 3.0: an integrated software package for population genetics data analysis. Evol Bioinform Online. 2005;1:47–50. [PMC free article] [PubMed] [Google Scholar]
- Flores C, Ma SF, Pino-Yanes M, Wade MS, Perez-Mendez L, Kittles RA, Wang D, Papaiahgari S, Ford JG, Kumar R. African ancestry is associated with asthma risk in African Americans. PLoS One. 2012;7(1):e26807. doi: 10.1371/journal.pone.0026807. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford JG, Meyer IH, Sternfels P, Findley SE, McLean DE, Fagan Jk, Richardson L. Patterns and predictors of asthma related emergency department use. Chest. 2001;120:1129–1135. doi: 10.1378/chest.120.4.1129. [DOI] [PubMed] [Google Scholar]
- Gauderman WJ MJ. QUANTO 1.1: A computer program for power and sample size calculations for genetic-epidemiology studies. 2006 Available at http://hydra.usc.edu/gxe.
- GINA Global Strategy for Asthma Management and Prevention. 2008 Available from: www.ginasthma.com.
- Gold DR, Rotnitzky A, Damokosh AI, Ware JH, Speizer FE, Ferris BG, Jr, Dockery DW. Race and gender differences in respiratory illness prevalence and their relationship to environmental exposures in children 7 to 14 years of age. Am Rev Respir Dis. 1993;148(1):10–18. doi: 10.1164/ajrccm/148.1.10. [DOI] [PubMed] [Google Scholar]
- Grant EN, Lyttle CS, Weiss KB. The relation of socioeconomic factors and racial/ethnic differences in US asthma mortality. Am J Public Health. 2000;90(12):1923–1925. doi: 10.2105/ajph.90.12.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant AV, Araujo MI, Ponte EV, Oliveira RR, Cruz AA, Barnes KC, Beaty TH. High heritability but uncertain mode of inheritance for total serum IgE level and Schistosoma mansoni infection intensity in a schistosomiasis-endemic Brazilian population. J Infect Dis. 2008;198(8):1227–1236. doi: 10.1086/591946. [DOI] [PubMed] [Google Scholar]
- Grant AV, Araujo MI, Ponte EV, Oliveira RR, Cruz AA, Barnes KC, Beaty TH. Polymorphisms in IL10 are associated with total Immunoglobulin E levels and Schistosoma mansoni infection intensity in a Brazilian population. Genes Immun. 2011;12(1):46–50. doi: 10.1038/gene.2010.50. [DOI] [PubMed] [Google Scholar]
- Green SL, Gaillard MC, Song E, Dewar JB, Halkas A. Polymorphisms of the beta chain of the high-affinity immunoglobulin E receptor (Fcepsilon RI-beta) in South African black and white asthmatic and nonasthmatic individuals. Am J Respir Crit Care Med. 1998;158(5 Pt 1):1487–1492. doi: 10.1164/ajrccm.158.5.9707099. [DOI] [PubMed] [Google Scholar]
- Grundbacher FJ, Massie FS. Levels of immunoglobulin G, M, A, and E at various ages in allergic and nonallergic black and white individuals. J Allergy Clin Immunol. 1985;75(6):651–658. doi: 10.1016/0091-6749(85)90089-2. [DOI] [PubMed] [Google Scholar]
- Haus M, Heese HD, Weinberg EG, Potter PC, Hall JM, Malherbe D. The influence of ethnicity, an atopic family history, and maternal ascariasis on cord blood serum IgE concentrations. J Allergy Clin Immunol. 1988;82(2):179–189. doi: 10.1016/0091-6749(88)90997-9. [DOI] [PubMed] [Google Scholar]
- Knight F. The Slave Societies of the Caribbean. UNESCO Publishing/Macmillan Education Ltd; London: 1997. [Google Scholar]
- Kumar R, Seibold MA, Aldrich MC, Williams LK, Reiner AP, Colangelo L, Galanter J, Gignoux C, Hu D, Sen S. Genetic ancestry in lung-function predictions. N Engl J Med. 2010;363(4):321–330. doi: 10.1056/NEJMoa0907897. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar R, Tsai HJ, Hong X, Liu X, Wang G, Pearson C, Ortiz K, Fu M, Pongracic JA, Bauchner H. Race, ancestry, and development of food-allergen sensitization in early childhood. Pediatrics. 2011;128(4):e821–e829. doi: 10.1542/peds.2011-0691. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar R, Tsai HJ, Hong X, Gignoux C, Pearson C, Ortiz K, Fu M, Pongracic JA, Burchard EG, Bauchner H. African ancestry, early life exposures, and respiratory morbidity in early childhood. Clin Exp Allergy. 2012;42(2):265–274. doi: 10.1111/j.1365-2222.2011.03873.x. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang DM, Polansky M, Sherman MS. Hospitalizations for asthma in an urban population: 1995–1999. Ann Allergy Asthma Immunol. 2009;103(2):128–133. doi: 10.1016/S1081-1206(10)60165-2. [DOI] [PubMed] [Google Scholar]
- Lester LA, Rich SS, Blumenthal MN, Togias A, Murphy S, Malveaux F, Miller ME, Dunston GM, Solway J, Wolf RL. Ethnic differences in asthma and associated phenotypes: collaborative study on the genetics of asthma. J Allergy Clin Immunol. 2001;108(3):357–362. doi: 10.1067/mai.2001.117796. others. [DOI] [PubMed] [Google Scholar]
- Marsh DG, Bias WB, Ishizaka K. Genetic control of basal serum immunoglobulin E level and its effect on specific reaginic sensitivity. Proc Nat Acad Sci. 1974;71:3588–3592. doi: 10.1073/pnas.71.9.3588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathias RA, Grant A, Wilson AF, Beaty T, Barnes K. Genome-wide association study of asthma-related quantitative traits in populations of African descent. Genet Epidemiol. 2007;31(6):638–638. [Google Scholar]
- Mathias RA, Grant AV, Rafaels N, Hand T, Gao L, Vergara C, Tsai YJ, Yang M, Campbell M, Foster C. A genome-wide association study on African-ancestry populations for asthma. J Allergy Clin Immunol. 2010;125(2):336–346. e4. doi: 10.1016/j.jaci.2009.08.031. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxwell TJ, Ameyaw MM, Pritchard S, Thornton N, Folayan G, Githang'a J, Indalo A, Tariq M, Mobarek A, Evans DA. Beta-2 adrenergic receptor genotypes and haplotypes in different ethnic groups. Int J Mol Med. 2005;16(4):573–580. others. [PubMed] [Google Scholar]
- McKeigue P. Population admixture and stratification in genetic epidemiology. In: Balding DJ, Bishop M, Cannings C, editors. Handbook of Statistical Genetics. Third ed. Wiley; West Sussex, England: 2007. pp. 1190–1213. [Google Scholar]
- Meisel A, Aguilera M. Cartagena de Indias en 1777: Un Análisis Demográfico. Biblioteca Virtual Banco de la Republica. Colombia; Bogotá, Colombia: 1997. [Google Scholar]
- Moorman JE, Rudd RA, Johnson CA, King M, Minor P, Bailey C, Scalia MR, Akinbami LJ. National surveillance for asthma–United States, 1980–2004. MMWR Surveill Summ. 2007;56(8):1–54. [PubMed] [Google Scholar]
- Murray T, Beaty TH, Mathias RA, Rafaels N, Grant AV, Faruque MU, Watson HR, Ruczinski I, Dunston GM, Barnes KC. African and non-African admixture components in African Americans and an African Caribbean population. Genet Epidemiol. 2010;34(6):561–568. doi: 10.1002/gepi.20512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson DA, Johnson CC, Divine GW, Strauchman C, Joseph CL, Ownby DR. Ethnic differences in the prevalence of asthma in middle class children. Ann Allergy Asthma Immunol. 1997;78(1):21–26. doi: 10.1016/S1081-1206(10)63365-0. [DOI] [PubMed] [Google Scholar]
- Ness RB, Haggerty CL, Harger G, Ferrell R. Differential distribution of allelic variants in cytokine genes among African Americans and White Americans. Am J Epidemiol. 2004;160(11):1033–1038. doi: 10.1093/aje/kwh325. [DOI] [PubMed] [Google Scholar]
- Patterson N, Price AL, Reich D. Population structure and eigenanalysis. PLoS Genet. 2006;2(12):e190. doi: 10.1371/journal.pgen.0020190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce N, Ait-Khaled N, Beasley R, Mallol J, Keil U, Mitchell E, Robertson C. Worldwide trends in the prevalence of asthma symptoms: phase III of the International Study of Asthma and Allergies in Childhood (ISAAC). Thorax. 2007;62(9):758–766. doi: 10.1136/thx.2006.070169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pena SD, Bastos-Rodrigues L, Pimenta JR, Bydlowski SP. DNA tests probe the genomic ancestry of Brazilians. Braz J Med Biol Res. 2009;42(10):870–876. doi: 10.1590/s0100-879x2009005000026. [DOI] [PubMed] [Google Scholar]
- Pepin J. From the Old World to the New World: an ecologic study of population susceptibility to HIV infection. Trop Med Int Health. 2005;10(7):627–639. doi: 10.1111/j.1365-3156.2005.01441.x. [DOI] [PubMed] [Google Scholar]
- Pesola GR, Xu F, Ahsan H, Sternfels P, Meyer IH, Ford JG. Predicting asthma morbidity in Harlem emergency department patients. Acad Emerg Med. 2004;11(9):944–950. doi: 10.1197/j.aem.2004.03.020. [DOI] [PubMed] [Google Scholar]
- Pritchard JK, Rosenberg NA. Use of unlinked genetic markers to detect population stratification in association studies. Am J Hum Genet. 1999;65(1):220–228. doi: 10.1086/302449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds J, Weir BS, Cockerham CC. Estimation of the coancestry coefficient: basis for a short-term genetic distance. Genetics. 1983;105(3):767–779. doi: 10.1093/genetics/105.3.767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojas W, Parra MV, Campo O, Caro MA, Lopera JG, Arias W, Duque C, Naranjo A, Garcia J, Vergara C. Genetic make up and structure of Colombian populations by means of uniparental and biparental DNA markers. Am J Phys Anthropol. 2010;143(1):13–20. doi: 10.1002/ajpa.21270. others. [DOI] [PubMed] [Google Scholar]
- Rouse I. The Tainos: Rise & Decline of the People Who Greeted Columbus. Yale University Press; New Haven: 1992. [Google Scholar]
- Salari K, Choudhry S, Tang H, Naqvi M, Lind D, Avila PC, Coyle NE, Ung N, Nazario S, Casal J. Genetic admixture and asthma-related phenotypes in Mexican American and Puerto Rican asthmatics. Genet Epidemiol. 2005;29(1):76–86. doi: 10.1002/gepi.20079. others. [DOI] [PubMed] [Google Scholar]
- Salzano FB, Bortolini MC. The Evolution and Genetics of Latin American Populations. Cambridge Univ. Press; Cambridge UK: 2002. [Google Scholar]
- Sanchez-Albornoz N. The Population of Latin America: A History Berkeley. Univ. California Press; 1974. [Google Scholar]
- Santos NP, Ribeiro-Rodrigues EM, Ribeiro-Dos-Santos AK, Pereira R, Gusmao L, Amorim A, Guerreiro JF, Zago MA, Matte C, Hutz MH. Assessing individual interethnic admixture and population substructure using a 48-insertion-deletion (INSEL) ancestry-informative marker (AIM) panel. Hum Mutat. 2010;31(2):184–190. doi: 10.1002/humu.21159. others. [DOI] [PubMed] [Google Scholar]
- Sarpong SB, Hamilton RG, Eggleston PA, Adkinson NF., Jr Socioeconomic status and race as risk factors for cockroach allergen exposure and sensitization in children with asthma. J Allergy Clin Immunol. 1996;97(6):1393–1401. doi: 10.1016/s0091-6749(96)70209-9. [DOI] [PubMed] [Google Scholar]
- Schwartz J, Gold D, Dockery DW, Weiss ST, Speizer FE. Predictors of asthma and persistent wheeze in a national sample of children in the United States. Association with social class, perinatal events, and race. Am Rev Respir Dis. 1990;142(3):555–562. doi: 10.1164/ajrccm/142.3.555. [DOI] [PubMed] [Google Scholar]
- Simms TM, Rodriguez CE, Rodriguez R, Herrera RJ. The genetic structure of populations from Haiti and Jamaica reflect divergent demographic histories. Am J Phys Anthropol. 2010;142(1):49–66. doi: 10.1002/ajpa.21194. [DOI] [PubMed] [Google Scholar]
- Simon F. Noticias Historiales de las Conquistas de Tierra Firme en las Indias Occidentales (1625) Kelly; Bogotá: 1953. pp. 207–210. [Google Scholar]
- Slatkin M. A measure of population subdivision based on microsatellite allele frequencies. Genetics. 1995;139(1):457–462. doi: 10.1093/genetics/139.1.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spielman RS, McGinnis RE, Ewens WJ. Transmission test for linkage disequilibrium: the insulin gene region and insulin-dependent diabetes mellitus (IDDM). Am J Hum Genet. 1993;52(3):506–516. [PMC free article] [PubMed] [Google Scholar]
- Stouffer SA, Suchman EA, Devinney LC, Star SA, Williams RM. Adjustment during Army Life. Princeton University Press; Princeton: 1949. The American Soldier. [Google Scholar]
- Thomas H. The Slave Trade. The Story of the Atlantic Slave Trade. Estados Unidos: Simon and Schuster; New York: 1997. pp. 1440–1870. [Google Scholar]
- Torgerson DG, Capurso D, Ampleford EJ, Li X, Moore WC, Gignoux CR, Hu D, Eng C, Mathias RA, Busse WW. Genome-wide ancestry association testing identifies a common European variant on 6q14.1 as a risk factor for asthma in African American subjects. J Allergy Clin Immunol. 130(3):622–629. doi: 10.1016/j.jaci.2012.03.045. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tulloch-Reid MK, Ferguson TS, Younger NO, Van den Broeck J, Boyne MS, Knight- Madden JM, Samms-Vaughan ME, Ashley DE, Wilks RJ. Appropriate waist circumference cut points for identifying insulin resistance in black youth: a cross sectional analysis of the 1986 Jamaica birth cohort. Diabetol Metab Syndr. 2010;2:68–73. doi: 10.1186/1758-5996-2-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vergara C, Tsai YJ, Grant AV, Rafaels N, Gao L, Hand T, Stockton M, Campbell M, Mercado D, Faruque M. Gene encoding Duffy antigen/receptor for chemokines is associated with asthma and IgE in three populations. Am J Respir Crit Care Med. 2008;178(10):1017–1022. doi: 10.1164/rccm.200801-182OC. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vergara C, Caraballo L, Mercado D, Jimenez S, Rojas W, Rafaels N, Hand T, Campbell M, Tsai YJ, Gao L. African ancestry is associated with risk of asthma and high total serum IgE in a population from the Caribbean Coast of Colombia. Hum Genet. 2009;125(5-6):565–579. doi: 10.1007/s00439-009-0649-2. others. [DOI] [PubMed] [Google Scholar]
- Via M, Gignoux CR, Roth LA, Fejerman L, Galanter J, Choudhry S, Toro-Labrador G, Viera-Vera J, Oleksyk TK, Beckman K. History shaped the geographic distribution of genomic admixture on the Island of Puerto Rico. PLoS One. 2011;6(1):e16513. doi: 10.1371/journal.pone.0016513. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Ray N, Rojas W, Parra M, Bedoya G, Gallo C, Poletti G, Mazzotti G, Hill K, Hurtado AM. Geographic patterns of genome admixture in Latin American Mestizos. PLoS Genetics. 2008;4(3):e1000037–e1000037. doi: 10.1371/journal.pgen.1000037. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams E. The History of the Caribbean. Vintage Books; New York: 1984. From Columbus to Castro. [Google Scholar]
- Winkler CA, Nelson GW, Smith MW. Admixture mapping comes of age*. Annu Rev Genomics Hum Genet. 2010;11(1):65–89. doi: 10.1146/annurev-genom-082509-141523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziv E, Burchard EG. Human population structure and genetic association studies. Pharmacogenomics. 2003;4(4):431–441. doi: 10.1517/phgs.4.4.431.22758. [DOI] [PubMed] [Google Scholar]