Abstract
During progression of cervical cancer, human papillomavirus genomes and cellular tumor suppressor genes can become methylated. Toward a better understanding of these biomarkers, we studied 104 samples with HPV16, 18, 31, and 45 representing five pathological categories from asymptomatic infection to cancer. We grouped all samples by HPV type and pathology and measured the overall methylation of informative amplicons of HPV late genes and the cellular DAPK gene. Methylation of all four HPV types as well as of the DAPK gene is lowest in asymptomatic infection and increases successively in all four pathological categories during progression to cancer. 27 out of 28 cancer samples showed methylation both in the L2/L1 genes as well as in DAPK, but a much lower fraction in all other pathological categories. We discuss the problem to develop diagnostic tests based on complex methylation patterns that make it difficult to classify amplicons as “methylated” or “unmethylated”.
Keywords: Papillomaviruses, DAPK gene, Epigenetics, DNA methylation, Biomarker, Cancer progression
Introduction
Cervical cancer, premalignant cervical lesions and non-neoplastic HPV infections, i.e. atypical cells of undetermined significance (ASCUS) and cervical intraepithelial neoplasia (CIN), are diagnosed by cytology (Papanicolaou test, Pap test), colposcopic inspection, and histological examination of biopsies. These tests and procedures are successful at decreasing the incidence of cervical cancer, but their rate of false diagnoses is a matter of concern (Nanda et al., 2000; Stoler and Schiffman, 2001). Detection of the DNA of high-risk human papillomavirus (HPV) types (Munoz et al., 2003; Bernard et al., 2010), the primary cause of cervical cancer, has become a powerful criterion to amend these procedures, and has greatly increased the sensitivity of screening (Bulkmans et al., 2007; Mayrand et al., 2007; Naucler et al., 2007). However, since the fraction of women being infected by HPVs at some time of their lives (> 80%) vastly exceed the incidence rate of cervical cancer (about 1%), and since a positive HPV DNA test often indicates a transient infection rather than a developing cervical cancer, HPV DNA diagnosis alone is not sufficient to distinguish women with benign infections from those requiring intensive management. In order to prevent unnecessary procedures on patients with abnormal Pap smears who are not at risk for developing cervical cancer, gynecologic practice needs tests that are sensitive and specific to detect high-risk patients. Numerous attempts have been made to measure markers that change as the result of HPV-dependent carcinogenesis, but these tests are still of limited benefit (von Knebel Doeberitz, 2002).
The molecular mechanisms involved in the progression of asymptomatic or low-grade HPV infections to cervical cancer are yet poorly understood, but include the methylation of many of those cellular genes that are also epigenetically affected in cancers of other organ sites and without an HPV etiology. The search of clinically useful epigenetic biomarkers of cervical cancer that may allow risk stratification in patients began relatively recently, but this field of research expanded rapidly, and a review (Wentzensen et al., 2009) compared studies of more than 60 cellular genes. Unfortunately, this meta-analysis came to the conclusion that there is currently no single methylation marker that that has the appropriate performance to serve as cervical cancer biomarker. The reviewed studies point only to few genes, notably DAPK (death associated protein kinase 1) and RARB (retinoic acid receptor beta), which might be attractive targets of further evaluations. Notably, these two markers stood out in a large epidemiological study comparing a panel of twenty cellular methylation targets (Feng et al., 2005).
Independently of these studies of cellular genes, our group has investigated how methylation affects HPV genomes in different stages of cervical neoplastic disease (Kalantari et al., 2004, 2008a, 2010; Badal et al., 2004; Turan et al., 2006, 2007), and our findings have been confirmed and expanded by others (Brandsma et al., 2009; Fernandez et al., 2009; Sun et al., 2011; Clarke et al., 2012; Mirabello et al., 2012a). A recent review summarizes this field (Johannsen and Lambert, 2013). Methylation of HPV16 and 18 increases among viral infections progressing from asymptomatic infection through low-grade and high-grade disease and malignancy. This effect is particularly pronounced in the late genes L2 and L1, whose products are not required for neoplastic processes. Methylation may affect the whole viral genome, however, although methylation is a repression mechanism (Bird, 2002). This is possible since neoplastic cells normally contain numerous HPV genomes. As long as one single HPV genome is spared from methylation, it maintains the carcinogenic process, although the other HPV genomes in the same cell may be transcriptionally silenced by methylation (Van Tine et al., 2004). The exact trigger of HPV methylation is not well understood, but there is evidence that methylation correlates with recombination between the HPV genome and chromosomal DNA (Kalantari et al., 2008a, 2008b, 2010). Studies not related to methylation have shown that HPV genomes frequently integrate into the cellular DNA in cancer, but it is disputed whether this mechanism is only a frequent event or mechanistically necessary (Daniel et al., 1997; Ueda et al., 2003; Hudelist et al., 2004; Arias-Pulido et al., 2006; Kulmala et al., 2006; Briolat et al., 2007; Pett and Coleman, 2007; Häfner et al., 2008; Vinokurova et al., 2008; Campitelli et al., 2012; Xu et al., 2013). Foreign DNA that integrates into mammalian chromosomal DNA is known to be a preferred methylation target, and therefore a correlation between HPV recombination and HPV DNA methylation may have nothing to do with the properties of the HPV genome and the biology of the virus (Doerfler et al., 2001). There is evidence that integration of HPV genomes favors the carcinogenic process as it leads to increased E6 and E7 oncoproteins transcription by interference with negative feedback by E2 proteins (Tan et al., 1994); transcriptional induction by the nuclear matrix (Stünkel et al., 2000), and stabilized E6/E7 transcripts (Jeon et al., 1995; Häfner et al., 2008).
The study reported here had the primary goal to compare the methylation of HPV late genes with methylation of the DAPK promoter, and with histological or cytological diagnoses among high-risk patients that were referred to a colposcopy clinic based on abnormal cervical cytology. Based on the literature cited above, we considered DAPK the most promising among the cellular epigenetic markers and we intended to compare this diagnosis with that of the viral late gene methylation. Aside from HPV16 and HPV18, our study targeted HPV31 and HPV45, which had not yet been studied when this research was done, but has been reported since then (Wentzensen et al., 2012). Our research targeted the promoter region of the DAPK gene, and two or three amplicons of the L2 and L1 genes of the four high-risk HPV types.
Results
Sample identification, clinical diagnosis and evaluation of DNA methylation
The objective of this study was to establish the methylation of CpG dinucleotides in two or three segments of the L2 and L1 genes of HPV16, 18, 31, and 45, and compare it with the CpG methylation of the promoter of the cellular DAPK gene in order to analyze the viral and cellular epigenetic changes as potentially useful clinical progression markers of cervical cancer.
All samples of precursor lesions of cervical cancers and of asymptomatic HPV infection were selected based on the HPV typing of the DNA of consecutive patients of a colposcopy clinic of the University of California Irvine as described in the Materials and methods section. This cohort yielded 50 samples with HPV16, nine with HPV18, eleven with HPV31, and six with HPV45. As this cohort did not contain patients with invasive cancers, we complemented these samples with material from a Norwegian cervical cancer archive, namely 11 samples with HPV16, four samples with HPV18, four samples with HPV31, and six samples with HPV45.We also included the analysis of C33A and SiHa cells with HPV16, and HeLa cells with HPV18, and report these three cell lines as cancers.
Most Californian patients were diagnosed prior to colposcopy by cytology and if medically indicated as part of the colposcopic examination by histology. Many of these diagnoses confirmed one another, e.g. patients with a low-grade squamous intraepithelial lesion (LSIL) by cytology were often found to have a cervical intraepithelial neoplasia grade I (CIN1) by histology. We sorted our samples according to these cytologic and histologic diagnoses, using in cases of discrepancy the higher grading of a lesion, i.e. a patient with LSIL and CIN3 is included in the category HSIL/CIN2-3. Our molecular data were based on analysis of cytological samples with the exception of cancer biopsies.
Methylation data were established for two or three, respectively, amplicons of the L2 and L1 genes of each HPV types, which had been found by us and others to be among the most highly methylated parts of the HPV genomes (Kalantari et al., 2004; Turan et al., 2006; Brandsma et al., 2009; Sun et al., 2011; Wentzensen et al., 2012), as well as for the promoter sequences of DAPK. Samples may contain cell and viral populations with diverse epigenetic states and histories. Many CpG residues in any particular genomic position can be completely methylated or unmethylated. Alternatively, a sample may contain molecules with mixtures of methylated and unmethylated CpGs in the same position (a sequencing output of overlapping C and T peaks). We report samples with mixtures of methylated and unmethylated CpGs as “methylated”, as they clearly contained HPV or DAPK populations with methylated CpGs.
Previous studies from our lab and others have shown that sporadic and low levels of CpG methylation occur in most HPV16 samples, including those derived from asymptomatic infection, low-grade lesions, and cell cultures with episomal HPV16 genomes. At this point no criterion exists to assign CpGs in any specific genomic position a diagnostically superior status, nor is it possible to define unequivocally a certain percentage of methylation as a diagnostically relevant threshold, making it difficult to classify individual samples as unambiguously “methylated” or “unmethylated”. As the principal output of our study, we therefore measured and reported the total number and percentage of methylated CpGs in all molecules that fall into any specific pathological category. All details of the methylation patterns of all amplicons are reported graphically, and we present statistical analyses as first steps to define quantitative criteria for the use of methylation data.
Methylation of the L2/L1 amplicons and the cellular DAPK promoter in samples containing HPV16
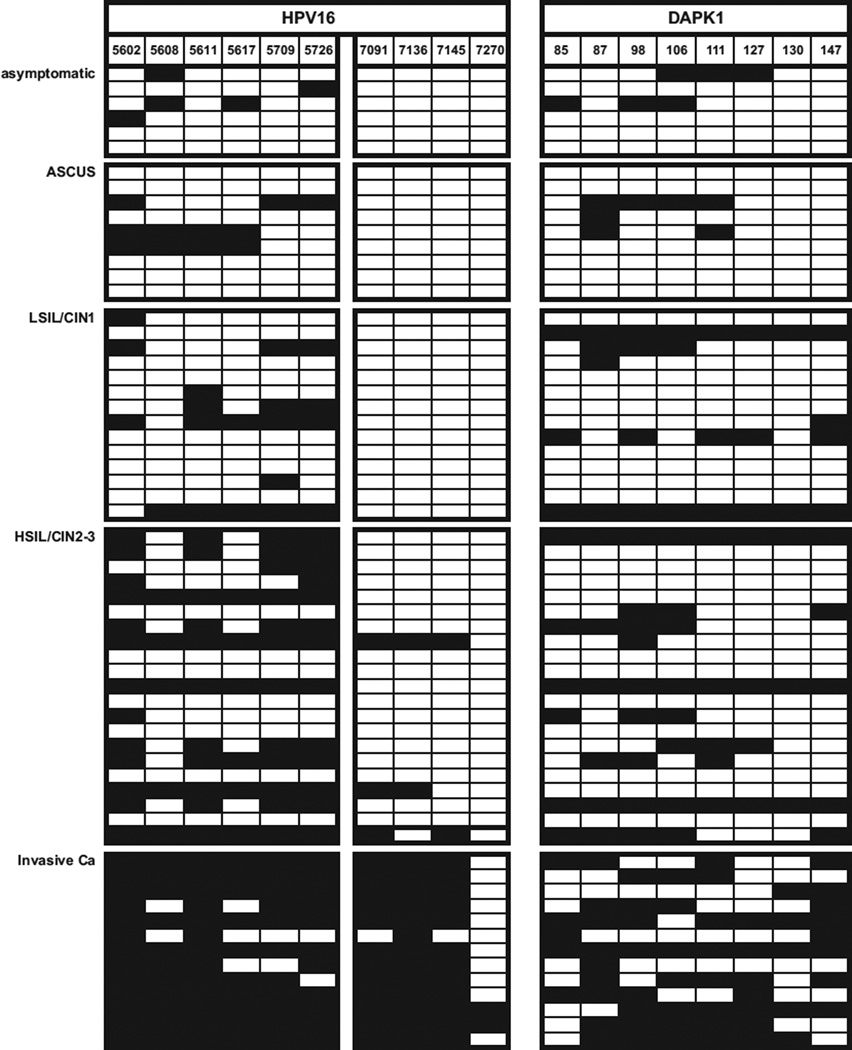
Sixty-three samples contained HPV16, and the methylation of their L2/L1 and DAPK methylation is shown in Fig. 1 and quantitatively summarized in Table 1. In HPV16, only 10–12.2% of all CpGs are methylated in asymptomatic infection and ASCUS (atypical squamous cells of undetermined significance). This fraction slightly increases in LSIL/CIN1 samples to 13.6%, in HSIL/CIN2-3 lesions to 31.9%, and in cancer to 83.1%. At the DAPK promoter, 9.7–12.5% of all CpGs are methylated in asymptomatic infection and ASCUS, and there is an increase in LSIL/CIN1 to 23.2%, in HSIL/CIN2-3 lesions to 27.4%, and in cancer to 54.8%. HPV16 methylation is in precancerous lesions higher in the L2/L1 segment (position 5602–5726) than in the 3′–L1 segment (position 7091–7270), which was the primary target of our previous published studies, and is clearly a superior target for future analyses. This is a novel observation, which suggests to address in the future studies preferentially the L2/L1 segment.
Fig. 1.
Methylation of the 5′ PCR amplicons of the HPV-16 L2 and L1 genes and the DAPK promoter in exfoliated cells of patients with abnormal Pap smears: direct sequencing of bisulfite treated and PCR amplified DNA. Each row identifies a patient in the corresponding pathological category, and each column a CpG dinucleotide in the respective genomic position of HPV16 and the DAPK gene. Black rectangles indicate methylation, white rectangles lack of methylation.
Table 1.
Methylation of CpGs of HPV16, 18, 31 and 45 and DAPK amplicons and statistical evaluations.
| Methylated CpGs in L2/L1 | Percent of methylated CpG in L2/L1 per sample |
Methylated CpGs in DAPK | Percent of methylated CpGs in DAPK per sample |
|
|---|---|---|---|---|
| HPV16 (63 samples) | N/total | Mean (SE) | N/total | Mean (SE) |
| Asymptomatic | 5/50 (10.0%) | 8.3 (9.2) | 6/48 (12.5%) | 12.5 (12.7) |
| ASCUS | 11/90 (12.2%) | 12.2 (7.6) | 7/72 (9.7%) | 9.7 (10.4) |
| LSIL/CIN1 | 19/140(13.6%) | 13.6 (6.1) | 26/112 (23.2%) | 23.2 (8.3) |
| HSIL/CIN2-3 | 67/210 (31.9%) | 31.9 (4.9) | 46/168 (27.4%) | 27.4 (6.8) |
| Invasive cancer | 108/130 (83.1%) | 83.1 (6.3) | 57/104 (54.8%) | 58.7 (8.6) |
| p-value | p < 0.0005a | p < 0.0005b | p < 0.0005a | p=0.004b |
| HPV18 (14 samples) | ||||
| Asymptomatic | 0/28 (0.0%) | 0.0 (8.1) | 1/16 (6.3%) | 6.3 (15.1) |
| ASCUS | 5/42 (11.9%) | 11.9 (6.6) | 0/56 (0.0%) | 0.0 (12.3) |
| LSIL/CIN1 | 7/28 (25.0%) | 25.0 (8.1) | 3/16 (18.8%) | 18.8 (15.1) |
| HSIL/CIN2-3 | 21/28 (75.0%) | 75.0 (8.1) | 3/16 (18.8%) | 18.8 (15.1) |
| Invasive cancer | 56/70 (80.0%) | 81.4 (5.1) | 31/40 (77.5%) | 77.5 (9.6) |
| p-value | p < 0.0005a | p < 0.0005b | p < 0.0005a | p=0.004b |
| HPV31 (15 samples) | ||||
| Asymptomatic | 0/18 (0.0%) | 0.0 (16.1) | 0/8 (0.0%) | 0.0 (26.2) |
| ASCUS | 3/54 (5.6%) | 5.6 (9.3) | 1/24 (4.2%) | 4.2 (15.2) |
| LSIL/CIN1 | 11/54 (20.4%) | 20.4 (9.3) | 4/24 (16.7%) | 16.7 (15.2) |
| HSIL/CIN2-3 | 39/72 (54.2%) | 54.2 (8.1) | 3/32 (9.4%) | 9.4 (13.1) |
| Invasive cancer | 35/72 (48.6%) | 48.6 (8.1) | 20/32 (62.5%) | 62.5 (13.1) |
| p-value | p < 0.0005a | p=0.008b | p < 0.0005a | p=0.064b |
| HPV45 (12 samples) | ||||
| Asymptomatic | 0/18 (0.0%) | 0.0 (15.6) | 0/8 (0.0%) | 0.0 (28.0) |
| ASCUS | N/A | N/A | N/A | N/A |
| LSIL/CIN1 | 0/18 (0.0%) | 0.0 (15.6) | 1/8 (12.5%) | 12.5 (28.0) |
| HSIL/CIN2-3 | 59/72 (81.9%) | 81.9 (7.8) | 5/32 (15.6%) | 15.6 (14.0) |
| Invasive cancer | 102/108 (94.4%) | 94.4 (6.4) | 38/48 (79.2%) | 79.2 (11.4) |
| p-value | p < 0.0005a | p=0.001b | p < 0.0005a | p=0.021b |
SE: standard error.
Chi-square test for trend.
F-test.
In spite of the limits of classifying individual amplicons unambiguously as methylated or unmethylated, we note that all 13 cancers showed some methylation in L2/L1 as well as in DAPK, while only 14 and 11 out of 21 HSIL/CIN2-3 samples, respectively, showed methylation both in HPV16 and DAPK. The reverse can also be observed, as there were six HSIL/CIN2-3 samples that completely lacked methylation both in the HPV16 and DAPK, and one may speculate that these may be samples with a low propensity to progress. The mean percent methylation in L2/L1 samples increased significantly with disease grade from 8% for asymptomatic samples to 83% for invasive cancer (p < 0.001). For DAPK, percent methylation increased significantly from a mean of 13–59% (p=0.004).
Methylation of the L2/L1 amplicons and the cellular DAPK promoter in samples containing HPV18
Among 14 samples with HPV18 (Fig. 2), none of the two asymptomatic infections showed methylation in the HPV amplicons, while there were five out of 42 CpGs (11.9%) methylated in three samples with ASCUS, seven out of 28 CpGs (25%) in two samples with LSIL/CIN1, 21 out of 28 CpGs (75%) in two HSIL/CIN2-3 lesions, and to 56 out of 70 CpGs (80%) in cancer. For DAPK, the corresponding percentages were 6.3%, 0%, 18.8%, 18.8%, and 77.5% in the five pathological groups. All five HPV18 cancers showed some methylation both in L2/L1 and in DAPK, but only two out of nine samples in the other four pathological categories shared this property. Percent methylation in samples increased significantly with grade for L1/L2 (p < 0.001) and for DAPK (p=0.004).
Fig. 2.
Methylation of the 5′ PCR amplicons of the HPV-18 L2 and L1 genes and the DAPK promoter in exfoliated cells of patients with abnormal Pap smears: direct sequencing of bisulfite treated and PCR amplified DNA.
Methylation of the L2/L1 amplicons and the cellular DAPK promoter in samples containing HPV31
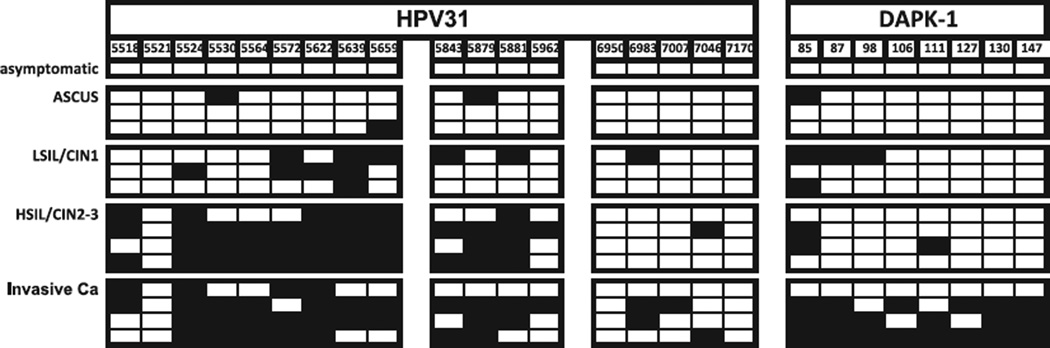
Among 15 samples with HPV31 (Fig. 3), there was no L2/L1 methylation in the only asymptomatic infection, three out of 54 CpGs (5.6%) were methylated in three samples with ASCUS, eleven out of 54 CpGs (20.4%) in three samples with LSIL/CIN1, 39 out of 72 CpGs (54.2%) in four HSIL/CIN2-3 lesions, and 35 out of 72 CpGs (48.6%) in four cancers. For DAPK, the corresponding percentages were 0%, 4.2%, 16.7%, 9.4%, and 62.5% in the five pathological groups. Three out of four cancer samples showed some methylation both in L2/L1 and DAPK, and five out of nine samples in the other four pathological categories. Percent methylation in samples increased significantly with grade for L1/L2 (p < 0.008). For DAPK, the increase in percent methylation with grade form 0% to 63% was not statistically significant (p=0.064).
Fig. 3.
Methylation of the 5′ PCR amplicons of the HPV-31 L2 and L1 genes and the DAPK promoter in exfoliated cells of patients with abnormal Pap smears: direct sequencing of bisulfite treated and PCR amplified DNA.
Methylation of the L2/L1 amplicons and the cellular DAPK promoter in samples containing HPV45
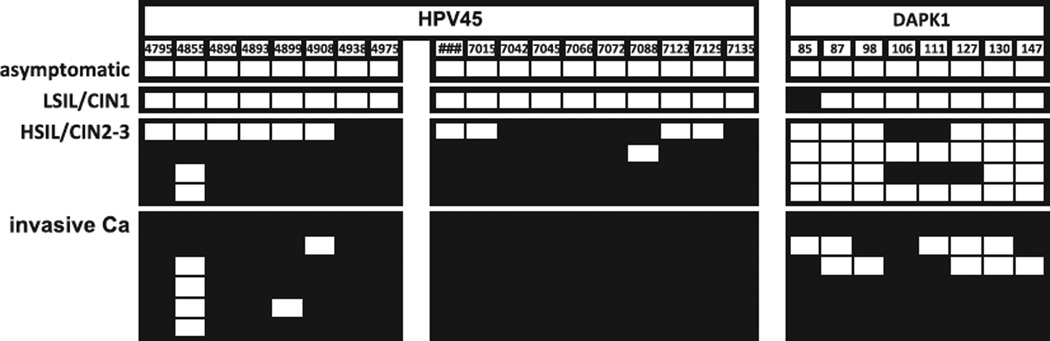
There were twelve samples with HPV45 (Fig. 4). The only asymptomatic infection was unmethylated in L2/L1. There was no sample with ASCUS. The only sample with LSIL/CIN1 had no methylation in the HPV45 amplicons. Four HSIL/CIN2-3 samples had 59 out of 72 CpGs (81.9%) methylated, and six cancer samples 102/108 CpGs (94.4%). For DAPK, the corresponding percentages were 0%, 12.5%, 15.6%, and 79.2% in the four pathological groups that contained at least one sample. (p < 0.001 for trend). All six cancer samples showed some methylation both in L2/L1 and DAPK, but only two of the six samples in the other four pathological categories.
Fig. 4.
Methylation of the 5′ PCR amplicons of the HPV-45 L2 and L1 gene2 and the DAPK promoter in exfoliated cells of patients with abnormal Pap smears: direct sequencing of bisulfite treated and PCR amplified DNA.
Separate consideration of DAPK methylation
In order to better evaluate the DAPK marker, we added the data for DAPK for infections of all four HPV types. For DAPK, methylation of all CpGs was 10% (asymptomatic infection), 5.3% (ASCUS), 17.5% (low-grade lesions), 23.2% (high-grade lesions), and 63.5% (cancer).
Sensitivity and specificity
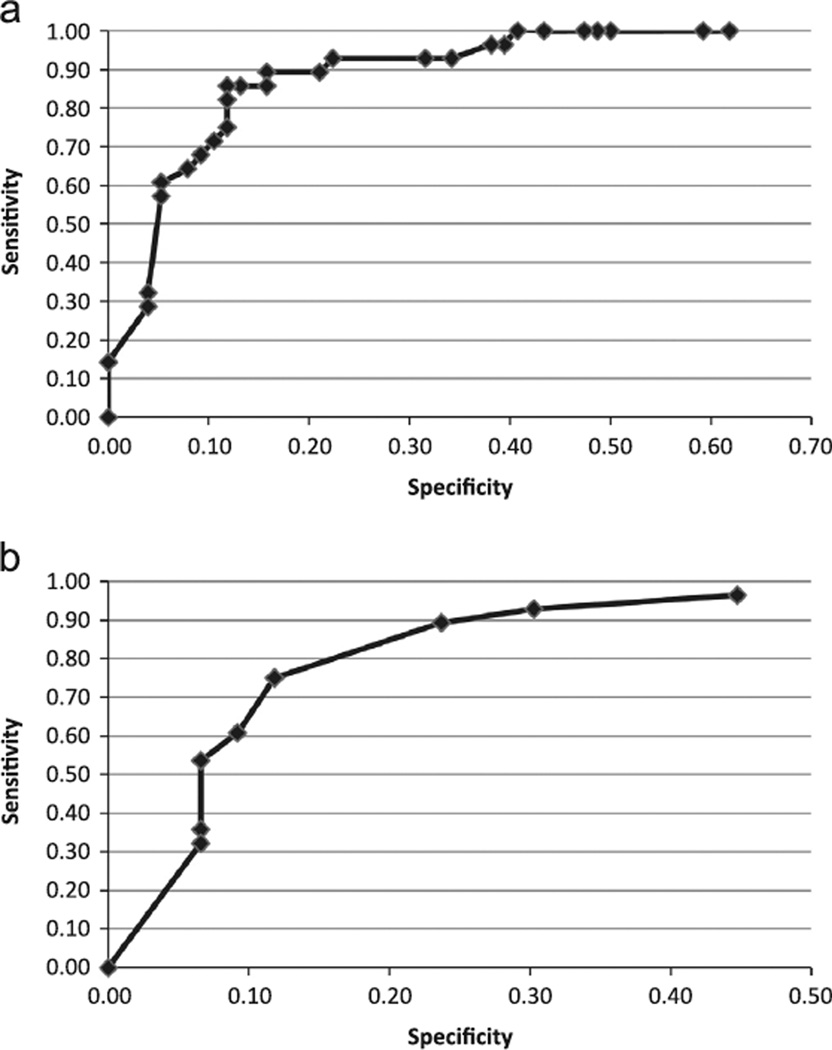
Our research is based on the hypothesis that strongly methylated samples identify molecular changes driving these lesions toward invasive cancer, irrespective of the cytological diagnosis. With the caveat of this hypothetical and molecular definition, 50% methylation for L1/L2 samples for all HPV types as a cutoff for detection of invasive cancer has good sensitivity at 89% and specificity at 84%. Sensitivity and specificity remain high (both 80–90%) for cutoffs between 50% and 69% methylation (Fig. 5A). When using DAPK methylation to discriminate invasive cancer from lesser grade disease, sensitivity is 89% when 25% methylation is used as a critical cutoff for cancer detection, specificity is lower at 76% (Fig. 5B). When the critical cutoff is increased to 38%, specificity increases to 88% with a loss of sensitivity (75%). Sensitivity and specificity for detection of HSIL/cancer vs. lesser grade disease in L1/L2 samples are 80% and 89% respectively for methylation ≥ 30%. Sensitivity for detection of HSIL/cancer in DAPK samples, however, is notably lower at 59% with specificity = 82% for methylation ≥ 24%.
Fig. 5.
Sensitivity and specificity of percent methylation for detection of cancer. (A) ROC curve for percentage of virus CpG methylation for detection of cells that according to our hypothesis have progressed to a cancerous state. (B) ROC curve for percentage of DAPK methylation for detection of cells that according to our hypothesis have progressed to a cancerous state.
Discussion
Our discovery of HPV16 and 18 methylation and their change during cervical carcinogenesis (Kalantari et al., 2004; Badal et al., 2004; Turan et al., 2006) have become generally confirmed and led to a major expansion of the available data base (Brandsma et al., 2009; Fernandez et al., 2009; Sun et al., 2011; Clarke et al., 2012; Mirabello et al., 2012a; Lorincz et al., 2013) and extension of this phenomenon to HPV31 and 45 (Wentzensen et al., 2012). It is now generally accepted that methylation is increased in high-grade lesions and cancer compared to asymptomatic infection or low-grade lesions, and that this mechanisms targets the late genes more than the early genes or the long control region. The question of the underlying mechanism has not become revisited after our reports of correlation between methylation and recombination between HPV genomes and cellular DNA (Kalantari et al., 2008a, 2008b, 2010), suggesting that transcriptionally inactive parts of the genome may become part of the heterochromatin like most exogenous DNA without relevance of the identity of the HPV genes (Doerfler et al., 2001). In contrast, endogenous tumor suppressor genes may be targeted at random by cellular de novo methylation, and cells with tumor suppressor genes inactivated by methylation may expand in number due the phenotypic consequences of the gene inactivation. While HPV methylation and cellular gene methylation are likely enzymatically related, the underlying mechanistic logic is clearly different.
Our research adds new aspects to the epigenetic profiling of HPV lesions by concomitantly analyzing the methylation of the late genes of four high-risk HPV-types HPV16, 18, 31, and 45 and that of a cellular gene, DAPK, one of the best cellular biomarkers for the progression of cervical cancer. It is also unique by including a substantial number of cervical cancers beyond the study of high-grade precursor lesions. Our study confirms that methylation of L2/L1 occurs in all four HPV types, and increases between all five pathological categories, asymptomatic infection, ASCUS, LSIL/CIN1, HSIL/CIN2-3 and cervical cancer, the biggest increases occurring in the two progression steps between LSIL/CIN1 and cancer. Interestingly, a similar gain of methylation occurs in the methylation of DAPK, in contrast to the data from others (Sun et al., 2011).
Our investigation is a pilot study of the potential of these biomarkers, as the numbers of samples, especially for HPV18, 31, and 45, is low. The value of biomarkers that are fully developed in invasive cancer and much less prevalent in patients with low-grade and high-grade lesion is at this point still debatable. It is our hypothesis that highly methylated LSIL/CIN1 and HSIL/CIN2-3 samples are those that have molecularly undergone changes that predestine them to develop into cancers. But the answer to the question of whether “high methylation” in general and which level of methylation in details identifies cells with an irrevocable propensity to grow into invasive cancer can only be resolved by future studies, in spite of support by other labs for the predictive value of HPV16 methylation (Mirabello et al., 2012b; Lorincz et al., 2013). The hypothesis needs further epidemiological and molecular evaluation. Epidemiological research could take the form of retrospective longitudinal studies, and ask whether cancer patients showed higher HPV L2/L1 and DAPK methylation in archival precancerous samples. Molecular research could reveal that the methylation is a mechanistic part of the etiological process. For example, as the normal life cycle of HPVs is irrevocably terminated by chromosomal recombination, methylation of the L2/L1 genes ensuing from recombination likely correlates with an interruption of E2 gene transcription and increased E6/E7 oncogene expression. On the other side, methylation of cellular tumor suppressor genes such as DAPK may suppress functions relevant for the maintenance of the non-cancerous state. We suggest that subsequent to such epidemiological and molecular confirmations the combination methylation analyses of HPV L2/L1 and cellular genes like DAPK and technical improvements by next-generation-sequencing will lead to clinically useful tests of early detection of cervical cancer progression.
Materials and methods
Clinical specimens and diagnoses
From October 2009 to May 2010 552 consecutive patients were enrolled who attended the University of California, Irvine Family Health Center Colposcopy Clinic in Santa Ana, California. Patients attending this clinic were considered to be a high-risk population as evidenced by abnormal referral cervical cytology. After securing patient's written informed consent, an additional Pap smear was obtained during the colposcopic examination and analyzed during our research. Cervical biopsies were obtained when indicated clinically but were not studied molecularly. Our study noted the cytological as well as histological (if applicable) diagnoses of all patients, but, if divergent, reports only the higher progression state (Figs. 1–4). The patients were triaged for their cytological and/or histological diagnoses as outlined in algorithms set forth by the American Society of Colposcopy and Cervical Pathology (ASCCP). All clinical evaluations were overseen by the Gynecologic Oncology Research Fellow (D.M.C.) and Attending Gynecologic Oncologist (K.S.T.). The molecular biologists (M.K., I.E.C.M., S.K., B. Y., and H.U.B.) were blinded to clinical information except to the enrollment criterion. The molecular data did not alter the clinical management of the patients. Exfoliated cells taken were taken from these patients by cotton brush, suspended in PreserveCyt solution, and DNA was prepared with the QIAamp DNA mini kit (Qiagen) according to the manufacturer's recommendations. This research had been approved by the Institutional Review Board of the University of California Irvine.
As this cohort did not contain any cancer patients, this sampling did not create a foundation for our objective to compare cervical cancer precursor lesions with cervical cancers. Therefore, we added to our study 26 cervical carcinomas from a collection of archival samples received from the Norwegian Cancer Registry, Oslo, by Dr. Bjoern Hagmar (Kalantari et al., 2004), as well as three widely used cervical cancer cell lines, SiHa, C33A and HeLa.
Typing of HPVs in DNA preparations from exfoliated cells
In order to identify samples with HPV16, 18, 31, and 45, all DNA preparation were amplified with degenerated primer pair MY09/11 targeting the L1 gene, followed by direct sequencing of the amplification products.
Bisulfite modification
DNA sequencing subsequent to bisulfite modification and PCR amplification (with or without additional cloning into Escherichia coli) is a sensitive technique to measure methylcytosines (Frommer et al., 1992). For bisulfite treatment, 50–1000 ng of sample DNA supplemented with 1 µg of salmon sperm DNA in 18 µl water were denatured with 2 µl 3 M NaOH and incubated at 37 °C. After denaturation, 278 µl 4.8 M sodium bisulfite and 2 µl 100 mM hydroquinone were added with the mixture being incubated in a thermal cycler for 20 cycles of 55 °C for 15 min and 95 °C for 30 s. The modified DNA was desalted with the QIAquick PCR purification protocol and desulfonated thereafter by adding 5.5 µl 3M NaOH and 5 µg glycogen prior to 15 min incubation at 37 °C. The DNA was precipitated with 5.6 µl of 3 M sodium acetate and 150 µl 100% ethanol and centrifuged. The pellet was washed with 70% ethanol and dissolved in 50 µl TE buffer (10 mM Tris–HCl; pH 8; and 1 mM EDTA).
Polymerase chain reactions, primers, and DNA sequencing of bisulfite treated DNA
Our previous studies have demonstrated that cervical smears contain mixtures of HPV genomes with diverse methylation patterns as well as unmethylated together with methylated DNA. This is to be expected since the collection of exfoliated cervical cells will sample HPV infected and uninfected epithelium as well as subsets of a lesion with different stages of disease. For the same reason, one should also expect a mixture of methylated and unmethylated CpGs at the promoter of DAPK. As the samples are processed by direct sequencing of the bisulfite treated and PCR amplified DNA (as opposed to cloning the reaction product and sequencing many independent clones), methylation signals (i.e. CpGs in bisulfite treated DNA) will often be overlaid by sequences indicating unmethylated CpGs (i.e. TpGs in bisulfite treated DNA). In our results, we scored all CpGs as well as jointly occurring CpG and TpG signals in bisulfite treated and PCR amplified DNA as representing methylated CpGs in the untreated DNA preparation.
Table 2 summarizes all primers that were used to amplify bisulfite treated sample DNA. As representative of the methylation of the late genes of HPV16, we targeted a segment spanning the 3′ end of L2 and the 5′ part of the L1 gene between the positions spanning six CpG dinucleotides at the genomic positions 5602, 5608, 5611, 5617, 5709, and 5726 of HPV16. We also analyzed the methylation status of CpGs at the positions 7091, 7136, 7145, and 7270, which had been the preferred target in all our previous studies. The two primer pairs for HPV18 spanned CpGs at the positions 6142, 6144, 6161, 6363, and 6366, as well as 7011, 7038, 7041, 7062, 7068, 7090, 7110, 7116, and 7122.
Table 2.
Polymerase chain reaction primers used to target bisulfite modified DNA in order to generate amplicons of the HPV16, 18, 31 and 45 L2 and L1 genes and the cellular DAPK genes, which bracket the CpG dinucleotides numbered in Figs. 1–4.
| HPV16-5526F | AGTTCCAGGGTCTCCACAAT |
| HPV16-5730R | GTGCGTGCAACATATTCATCCG |
| HPV16-7094F | AAAGCTACACCCACCACCTCAT |
| HPV16-7443R | AAATGGGCCTGGCGCTACAAA |
| 18mod7013-F | TAAATTTTTGGTTTAGGTTGGATTG |
| 18mod7188-R | AAAACATACAAACACAACAATAAATA |
| Bis18-6118F | GGGTTAAAGGTATTGTTTGTAAAT |
| Bis18-6393R | AAAATACCTAACAAAAAACTACTCAC |
| 31Bis-5475F | ATGGGGGTGATTTTTATTTGTATTT |
| 31Bis-5771R | ACCCTATATTATAATCCTAACACCTTTAAT |
| 31Bis-5801F | ATTTGGATTTTTTGATATATTTTTTTATAA |
| 31Bis-6993R | CATTCCCTATTATCAATACCAAAAC |
| 31Bis-6926F | AGATTTAGATTAGTTTTTATTGGGT |
| 31Bis-7281R | AAATATATATAAAAACAACATACACAACAC |
| 45Bis-4740F | GAGGTGTTTTAAATAGGGGAGGTAT |
| 45Bis-5017R | ATCCAAAAACTCATAAACTAAATTATCAAA |
| 45Bis-6965F | TTGATTTAAAGGAAAAATTTTTTTT |
| 45Bis-7290R | ATAACACCATACATACCACAAAACAC |
| DAPK1-1 | YGGAGGATAGTYGGATYTAGTTAA |
| DAPK1-2 | ACRAAAACACAACTAAAAAATAAATAAAAAAC |
Three primer pairs for the analysis of HPV31 spanned CpGs at the positions 5518, 5521, 5524, 5530, 5564, 5572, 5622, 5639 (first primer pair), 5843, 5879, 5881, and 5962 (second primer pair), and 6950, 6983, 7007, 7046 and 7170 (third primer pair). And two primer pairs for the analysis of HPV45 spanned CpGs at the positions 4795, 4855, 4890, 4893, 4899, 4908, 4938, and 4975, as well as 6990, 7015, 7042, 7045, 7066, 7072, 7088, 7123, 7129 and 7135.
The DAPK promoter was amplified with the primer pairs shown in Table 2 and included CpGs at position 85, 87, 98, 106, 111, 127, 130, and 147 (Narayan et al., 2003).
PCR was carried out in a 25 µl volume containing 0.2 mM of each of the four dNTPs, 10 pmol of each primer, 2 mM MgCl2 and 1 unit of AmpliTaqGold (Applied Biosystems, Foster City, California, USA). The PCR conditions were 94 °C for 1 min, followed by 40 cycles of 94 °C for 10 s, 54 °C for 30 s and 68 °C for 1 min with a final extension at 68 °C for 7 min. The presence of PCR products was verified by agarose gel electrophoresis. Confirmed amplicons were directly sequenced by Big Dye terminator v3.1 Cycle Sequencing with the same primers used for amplification (Applied Biosystems, Foster City, California, USA).
Statistical methods
The degree of methylation across grades of disease was compared using a chi-square test for trend for testing difference in the percent of all CpGs that were methylated and an F-test for comparing mean percent methylation per sample by grade. Combining all HPV types, sensitivity and specificity were calculated for detection of invasive cancer at different cutoffs for percent methylation, and ROC curves were constructed.
Acknowledgments
This research was supported by funding from University of California Irvine to H.U.B.
References
- Arias-Pulido H, Peyton CL, Joste NE, Vargas H, Wheeler CM. Human papillomavirus type 16 integration in cervical carcinoma in situ and in invasive cervical cancer. J. Clin. Microbiol. 2006;44:1755–1762. doi: 10.1128/JCM.44.5.1755-1762.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badal S, Badal V, Calleja-Macias IE, Kalantari M, Chuang LSH, Li BFL, Bernard HU. The human papillomavirus-18 genome is efficiently targeted by cellular DNA methylation. Virology. 2004;324:483–492. doi: 10.1016/j.virol.2004.04.002. [DOI] [PubMed] [Google Scholar]
- Bernard HU, Burk RD, Chen Z, van Doorslaer K, zur Hausen H, de Villiers EM. Classification of papillomaviruses based on 189 PV types and proposal of taxonomic amendments. Virology. 2010;401:70–79. doi: 10.1016/j.virol.2010.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- Brandsma JL, Sun Y, Lizardi PM, Tuck DP, Zelterman D, Haines GK, Martel M, Harigopal M, Schofield K, Neapolitano M. Distinct human papillomavirus type 16 methylomes in cervical cells at different stages of premalignancy. Virology. 2009;389:100–107. doi: 10.1016/j.virol.2009.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briolat J, Dalstein V, Saunier M, Joseph K, Caudroy S, Pretet JL, Birembau P, Clavel C. HPV prevalence, viral load and physical state of HPV-16 in cervical smears of patients with different grades of CIN. Int. J. Cancer. 2007;121:2198–2204. doi: 10.1002/ijc.22959. [DOI] [PubMed] [Google Scholar]
- Bulkmans NW, Berkhof J, Rozendaal L, van Kemenade FJ, Boeke AJ, Bulk S, Voorhorst FJ, Verheijen RH, van Groningen K, Boon ME, Ruitinga W, van Ballegooijen M, Snijders PJ, Meijer CJ. Human papillomavirus DNA testing for the detection of cervical intraepithelial neoplasia grade 3 and cancer: 5-year follow-up of a randomised controlled implementation Trial. Lancet. 2007;370:1764–1772. doi: 10.1016/S0140-6736(07)61450-0. [DOI] [PubMed] [Google Scholar]
- Campitelli M, Jeannot E, Peter M, Lappartient E, Saada S, de la Rochefordiere A, Fourchote V, Alran S, Petrow P, Cottu P, Pierga JY, Lantz O, Couturier J, Sastre-Garau X. Human papillomavirus mutational insertion: specific marker of circulating tumor DNA in cervical cancer patients. PLOS ONE. 2012;7:e43393. doi: 10.1371/journal.pone.0043393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke MA, Wentzensen N, Mirabello L, Ghosh A, Wacholder S, Harari A, Lorincz A, Schiffman M, Burk RD. Human papillomavirus DNA methylation as a potential biomarker for cervical cancer. Cancer Epidemiol. Biomark. Prev. 2012;21:2125–2137. doi: 10.1158/1055-9965.EPI-12-0905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel B, Rangarajan A, Mukherjee G, Vallikad E, Krishna S. The link between integration and expression of human papillomavirus type 16 genomes and cellular changes in the evolution of cervical intraepithelial neoplastic lesions. J. Gen. Virol. 1997;78:1095–1101. doi: 10.1099/0022-1317-78-5-1095. [DOI] [PubMed] [Google Scholar]
- Doerfler W, Remus R, Muller K, Heller H, Hohlweg U, Schubbert R. The fate of foreign DNA in mammalian cells and organisms. Dev. Biol. 2001;106:89–97. [PubMed] [Google Scholar]
- Feng Q, Balasubramanian A, Hawes SE, Toure P, Sow PS, Dem A, Dembele B, Critchlow CW, Xi L, Lu H, McIntosh MW, Young AM, Kiviat NB. Detection of hypermethylated genes in women with and without cervical neoplasia. J. Natl. Cancer Inst. 2005;97:273–282. doi: 10.1093/jnci/dji041. [DOI] [PubMed] [Google Scholar]
- Fernandez AF, Rosales C, Lopez-Nieva P, Graña O, Ballestar E, Ropero S, Espada J, Melo SA, Lujambio A, Fraga MF, Pino I, Javierre B, Carmona FJ, Acquadro F, Steenbergen RD, Snijders PJ, Meijer CJ, Pineau P, Dejean A, Lloveras B, Capella G, Quer J, Buti M, Esteban JI, Allende H, Rodriguez-Frias F, Castellsague X, Minarovits J, Ponce J, Capello D, Gaidano G, Cigudosa JC, Gomez-Lopez G, Pisano DG, Valencia A, Piris MA, Bosch FX, Cahir-McFarland E, Kieff E, Esteller M. The dynamic DNA methylomes of double-stranded DNA viruses associated with human cancer. Genome Res. 2009;19:438–451. doi: 10.1101/gr.083550.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frommer M, McDonald LE, Millar DS, Collis CM, Watt F, Grigg GW, Molloy PL, Paul CL. A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc. Natl. Acad. Sci. USA. 1992;89:1827–1831. doi: 10.1073/pnas.89.5.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Häfner N, Driesch C, Gajda M, Jansen L, Kirchmayr R, Runnebaum IB, Dürst M. Integration of the HPV16 genome does not invariably result in high levels of viral oncogene transcripts. Oncogene. 2008;27:1610–1617. doi: 10.1038/sj.onc.1210791. [DOI] [PubMed] [Google Scholar]
- Hudelist G, Manavi M, Pischinger KI, Watkins-Riedel T, Singer CF, Kubista E, Czerwenka KF. Physical state and expression of HPV DNA in benign and dysplastic cervical tissue: different levels of viral integration are correlated with lesion grade. Gynecol. Oncol. 2004;92:873–880. doi: 10.1016/j.ygyno.2003.11.035. [DOI] [PubMed] [Google Scholar]
- Jeon S, Allen-Hoffmann BL, Lambert PF. Integration of human papillomavirus type 16 into the human genome correlates with a selective growth advantage of cells. J. Virol. 1995;69:2989–2997. doi: 10.1128/jvi.69.5.2989-2997.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johannsen E, Lambert PF. Epigenetics of human papillomaviruses. Virology. 2013;445:205–212. doi: 10.1016/j.virol.2013.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalantari M, Calleja-Macias IE, Tewari D, Hagmar B, Barrera-Saldana HA, Wiley DJ, Bernard HU. Conserved methylation patterns of human papillomavirus-16 DNA in asymptomatic infection and cervical neoplasia. J. Virol. 2004;78:12762–12772. doi: 10.1128/JVI.78.23.12762-12772.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalantari M, Lee D, Calleja-Macias IE, Lambert P, Bernard HU. Effects of cellular differentiation, chromosomal integration, and 5′-aza-2′-deoxycyticine treatment on human papillomavirus-16 DNA methylation in cultured cell lines. Virology. 2008a;374:292–303. doi: 10.1016/j.virol.2007.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalantari M, Villa LL, Calleja-Macias IE, Bernard HU. Human papillomavirus-16 and 18 in penile carcinomas: DNA methylation, chromosomal recombination, and genomic variation. Int. J. Cancer. 2008b;123:1832–1840. doi: 10.1002/ijc.23707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalantari M, Chase DM, Tewari KS, Bernard HU. Recombination of Human Papillomavirus-16 and Host DNA in exfoliated cervical cells: a pilot study of L1 gene methylation and chromosomal integration as biomarkers of carcinogenic progression. J. Med. Virol. 2010;82:311–320. doi: 10.1002/jmv.21676. [DOI] [PubMed] [Google Scholar]
- Kulmala SM, Syrjänen SM, Gyllensten UB, Shabalova IP, Petrovichev N, Tosi P, Syrjänen KJ, Johansson BC. Early integration of high copy HPV16 detectable in women with normal and low grade cervical cytology and histology. J. Clin. Pathol. 2006;59:513–517. doi: 10.1136/jcp.2004.024570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorincz AT, Brentnall AR, Vasiljevic N, Scibior-Bentkowska D, Castanon A, Fiander A, Powell N, Tristram A, Cuzick J, Sasieni P. HPV16 L1 and L2 DNA methylation predicts high-grade cervical intraepithelial neoplasia in women with mildly abnormal cervical cytology. Int. J. Cancer. 2013;133:637–644. doi: 10.1002/ijc.28050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayrand MH, Duarte-Franco E, Rodrigues I, Walter SD, Hanley J, Ferenczy A, Ratnam S, Coutlée F, Franco EL. Human papillomavirus DNA versus Papanicolaou screening tests for cervical cancer. N. Engl. J. Med. 2007;357:1589–1597. doi: 10.1056/NEJMoa071430. [DOI] [PubMed] [Google Scholar]
- Mirabello L, Schiffman M, Ghosh A, Rodriguez AC, Vasiljevic N, Wentzensen N, Herrero R, Hildesheim A, Wacholder S, Scibior-Bentkowska D, Burk RD, Lorincz AT. Elevated methylation of HPV16 DNA is associated with the development of high grade cervical intraepithelial neoplasia. Int. J. Cancer. 2012a:1412–1422. doi: 10.1002/ijc.27750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirabello L, Sun C, Ghosh A, Rodriguez AC, Schiffman M, Wentzensen N, Hildesheim A, Herrero R, Wacholder S, Lorincz A, Burk RD. Methylation of human papillomavirus type 16 genome and risk of cervical precancer in a Costa Rican population. J. Natl. Cancer Inst. 2012b;104:556–565. doi: 10.1093/jnci/djs135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz N, Bosch FX, de Sanjosé S, Herrero R, Castellsagué X, Shah KV, Snijders PJF, Meijer CJLM. Epidemiological classification of human papillomavirus types associated with cervical cancer. N. Engl. J. Med. 2003;348:518–527. doi: 10.1056/NEJMoa021641. [DOI] [PubMed] [Google Scholar]
- Nanda K, McCrory DC, Myers ER, Bastian LA, Hasselblad V, Hickey JD, Matchar DB. Accuracy of the Papaniolaou test in screening for and follow-up of cervical cytologic abnormalities: a sytematic review. Ann. Int. Med. 2000;132:810–819. doi: 10.7326/0003-4819-132-10-200005160-00009. [DOI] [PubMed] [Google Scholar]
- Narayan G, Arias-Pulido H, Koul S, Vargas H, Zhang FF, Villella J, Schneider A, Terry MB, Mansukhani M, Murty VV. Frequent promoter methylation of CDH1, DAPK, RARB, and HIC1 genes in carcinoma of the cervix uteri: Its relationship to clinical outcome. Mol. Cancer. 2003;2:24. doi: 10.1186/1476-4598-2-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naucler P, Ryd W, Törnberg S, Strand A, Wadell G, Elfgren K, Rådberg T, Strander B, Johansson B, Forslund O, Hansson BG, Rylander E, Dillner J. Human papillomavirus and Papanicolaou tests to screen for cervical cancer. N. Engl. J. Med. 2007;357:1589–1597. doi: 10.1056/NEJMoa073204. [DOI] [PubMed] [Google Scholar]
- Pett M, Coleman N. Integration of high-risk human papillomavirus: a key event in cervical carcinogenesis? J. Pathol. 2007;212:356–367. doi: 10.1002/path.2192. [DOI] [PubMed] [Google Scholar]
- Stünkel W, Huang Z, Tan SH, O'Connor M, Bernard HU. Nuclear matrix attachment regions of human papillomavirus-16 repress or activate the E6 promoter depending on the physical state of the viral DNA. J. Virol. 2000;74:2489–2501. doi: 10.1128/jvi.74.6.2489-2501.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoler MH, Schiffman M. Interobserver reproducibility of cervical cytologic and histologic interpretations: realistic estimates from the ASCUS-LSIL Triage Study. J. Am. Med. Assoc. 2001;285:1500–1505. doi: 10.1001/jama.285.11.1500. [DOI] [PubMed] [Google Scholar]
- Sun C, Reimers LL, Burk RD. Methylation of HPV16 genome CpG sites is associated with cervical precancer and cancer. Gynecol. Oncol. 2011;121:59–63. doi: 10.1016/j.ygyno.2011.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan SH, Leong LEC, Walker PA, Bernard HU. The human papillomavirus type 16 transcription factor E2 binds with low cooperativity to two flanking binding sites and represses the E6 promoter through displacement of Sp1 and TFIID. J. Virol. 1994;68:6411–6420. doi: 10.1128/jvi.68.10.6411-6420.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turan T, Kalantari M, Calleja-Macias IE, Villa LL, Cubie HA, Cuschieri K, Skomedal H, Barrera-Saldana HA, Bernard HU. Methylation of the human papillomavirus-18 L1 gene: a biomarker of neoplastic progression? Virology. 349:175–183. doi: 10.1016/j.virol.2005.12.033. [DOI] [PubMed] [Google Scholar]
- Turan T, Kalantari M, Cuschieri K, Cubie HA, Skomedal H, Bernard HU. High-throughput detection of human papillomavirus-18 L1 gene methylation, a candidate biomarker for the progression of cervical neoplasia. Virology. 2007;361:185–191. doi: 10.1016/j.virol.2006.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda Y, Enomoto T, Miyatake T, Ozaki K, Yoshizaki T, Kanao H, Ueno Y, Nakashima R, Shroyer KR, Murata Y. Monoclonal expansion with integration of high-risk type human papillomaviruses is an initial step for cervical carcinogenesis: association of clonal status and human papillomavirus infection with clinical outcome in cervical intraepithelial neoplasia. Lab. Invest. 2003;83:1517–1527. doi: 10.1097/01.lab.0000092234.68751.83. [DOI] [PubMed] [Google Scholar]
- Van Tine BA, Kappes JC, Banerjee NS, Knops J, Lai L, Steenbergen RD, Meijer CL, Snijders PJ, Chatis P, Broker TR, Moen PT, Jr, Chow LT. Clonal selection for transcriptionally active viral oncogenes during progression to cancer. J. Virol. 2004;78:11172–11186. doi: 10.1128/JVI.78.20.11172-11186.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinokurova S, Wentzensen N, Kraus I, Klaes R, Driesch C, Melsheimer P, Kisseljov F, Dürst M, Schneider A, von Knebel Doeberitz M. Type-dependent integration frequency of human papillomavirus genomes in cervical lesions. Cancer Res. 2008;68:307–313. doi: 10.1158/0008-5472.CAN-07-2754. [DOI] [PubMed] [Google Scholar]
- von Knebel Doeberitz M. New markers for cervical dysplasia to visualise the genomic chaos created by aberrant oncogenic papillomavirus infections. Eur. J. Cancer. 2002;38:2229–2242. doi: 10.1016/s0959-8049(02)00462-8. [DOI] [PubMed] [Google Scholar]
- Wentzensen N, Sherman ME, Schiffman M, Wang SS. Utility of methylation markers in cervical cancer early detection: appraisal of the state-of-the-science. Gynecol. Oncol. 2009;112:293–299. doi: 10.1016/j.ygyno.2008.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wentzensen N, Sun C, Ghosh A, Kinney W, Mirabello L, Wacholder S, Shaber R, Lamere B, Clarke M, Lorincz AT, Castle PE, Schiffman M, Burk RD. Methylation of HPV18, HPV31, and HPV45 Genomes and Cervical Intraepithelial Neoplasia Grade 3. J. Natl. Cancer Inst. 2012;104:1738–1749. doi: 10.1093/jnci/djs425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu B, Chotewutmontri S, Wolf S, Clos U, Schmitz M, Dürst M, Schwarz E. Multiplex identification of human papillomavirus 16 DNA integration sites in cervical carcinomas. PLOS ONE. 2013;8:e66693. doi: 10.1371/journal.pone.0066693. [DOI] [PMC free article] [PubMed] [Google Scholar]