The crystal structure of the C-terminal phosphotransferase domain of the bifunctional aminoglycoside acetyltransferase–phosphotransferase AAC(6′)-Ie-APH(2′′)-Ia has been determined to 2.3 Å resolution. The enzyme was crystallized as the Mg2GDP binary complex.
Keywords: AAC(6′)-Ie-APH(2′′)-Ia, aminoglycoside-resistance enzymes
Abstract
The bifunctional acetyltransferase(6′)-Ie-phosphotransferase(2′′)-Ia [AAC(6′)-Ie-APH(2′′)-Ia] is the most important aminoglycoside-resistance enzyme in Gram-positive bacteria, conferring resistance to almost all known aminoglycoside antibiotics in clinical use. Owing to its importance, this enzyme has been the focus of intensive research since its isolation in the mid-1980s but, despite much effort, structural details of AAC(6′)-Ie-APH(2′′)-Ia have remained elusive. The structure of the Mg2GDP complex of the APH(2′′)-Ia domain of the bifunctional enzyme has now been determined at 2.3 Å resolution. The structure of APH(2′′)-Ia is reminiscent of the structures of other aminoglycoside phosphotransferases, having a two-domain architecture with the nucleotide-binding site located at the junction of the two domains. Unlike the previously characterized APH(2′′)-IIa and APH(2′′)-IVa enzymes, which are capable of utilizing both ATP and GTP as the phosphate donors, APH(2′′)-Ia uses GTP exclusively in the phosphorylation of the aminoglycoside antibiotics, and in this regard closely resembles the GTP-dependent APH(2′′)-IIIa enzyme. In APH(2′′)-Ia this GTP selectivity is governed by the presence of a ‘gatekeeper’ residue, Tyr100, the side chain of which projects into the active site and effectively blocks access to the adenine-binding template. Mutation of this tyrosine residue to a less bulky phenylalanine provides better access for ATP to the NTP-binding template and converts APH(2′′)-Ia into a dual-specificity enzyme.
1. Introduction
Following the use of penicillin in the early 1940s, hundreds of antibiotics belonging to more than a dozen classes have been introduced into clinical practice. After seven decades of intensive use for the treatment of bacterial infections, antibiotic resistance has become a serious global problem (Bush et al., 2011 ▶). The continued spread of pathogens armed with multiple mechanisms capable of producing resistance to virtually all currently available antimicrobial agents poses a major threat to our ability to combat infectious diseases.
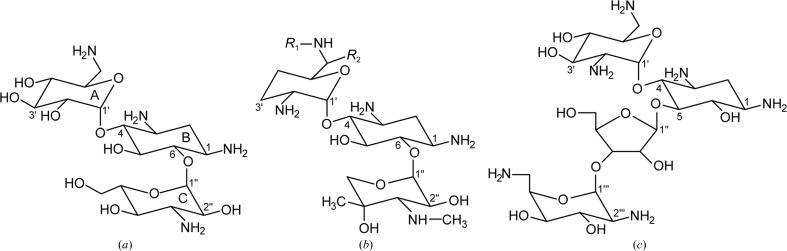
The aminoglycosides are antibiotics that are often used to treat serious life-threatening infections caused by bacteria resistant to other antimicrobial agents. The compounds consist of a central aminocyclitol ring (the B ring) with two or three substituted aminoglycan rings (A, C and, in some cases, D) attached at either the 4 and 5 positions or the 4 and 6 positions to give rise to 4,5-disubstituted and 4,6-disubstituted aminoglycosides, respectively (Fig. 1 ▶). As is the case with all other antibiotics, clinical use of aminoglycosides has been severely compromised by the spread of aminoglycoside-resistant clinical isolates. The major mechanism of aminoglycoside resistance is the production by bacteria of aminoglycoside-modifying enzymes (Shaw et al., 1993 ▶; Miller et al., 1997 ▶; Wright, 1999 ▶; Smith & Baker, 2002 ▶; Vakulenko & Mobashery, 2003 ▶; Ramirez & Tolmasky, 2010 ▶). The bacterial ribosome, which is the target for aminoglycosides, exhibits significantly diminished affinity for the modified antibiotics and they are unable to impair bacterial protein synthesis at clinically achievable concentrations (Moazed & Noller, 1987 ▶; Woodcock et al., 1991 ▶; Llano-Sotelo et al., 2002 ▶). Numerous representatives of three families of aminoglycoside-modifying enzymes, the aminoglycoside acetyltransferases (AACs), aminoglycoside nucleotidyltransferases (ANTs) and aminoglycoside phosphotransferases (APHs), have been identified in clinical isolates and aminoglycoside producers. There are several families of aminoglycoside phosphotransferases, with the two most important being the APH(3′) and APH(2′′) enzymes. They confer resistance to the aminoglycosides by phosphorylation of certain hydroxyl groups on the aminoglycan rings, and the enzymes are named according to the site at which they phosphorylate. The APH(3′) enzymes, for example, modify the 3′-hydroxyl on the A ring, whereas the APH(2′′) enzymes modify the aminoglycosides at the 2′′-hydroxyl on the C ring (Fig. 1 ▶). Amongst members of these families there are enzyme variants with differing substrate and nucleotide profiles, and these are designated by a roman numeral after the name of the enzyme. This naming convention is also used for the AAC and ANT enzymes.
Figure 1.
Aminoglycoside antibiotics. (a) Kanamycin A. (b) Gentamicin C, typically a mixture of types C1 (R 1 = R 2 = CH3), C1a (R 1 = R 2 = H) and C2 (R 1 = CH3, R 2 = H). (c) Neomycin.
The bifunctional aminoglycoside-modifying enzyme AAC(6′)-Ie-APH(2′′)-Ia is the single most important resistance enzyme for aminoglycoside antibiotics in Gram-positive bacteria such as staphylococci and enterococci, which are the causative agents of many life-threatening infections (Ferretti et al., 1986 ▶; Chow et al., 1997 ▶, 2001 ▶; Tsai et al., 1998 ▶; Chow, 2000 ▶; Kao et al., 2000 ▶; Tsuchizaki et al., 2006 ▶). It is broadly disseminated among clinical enterococcal and staphylococcal isolates and singlehandedly produces resistance to virtually all aminoglycoside antibiotics. The breadth of the aminoglycoside-resistance spectrum produced by the bifunctional enzyme is owing to the activity of both the N-terminal AAC(6′)-Ie acetyltransferase and the C-terminal APH(2′′)-Ia phosphotransferase enzymes, which are the products of a single fused gene with a single start and stop codon. Although separation of the AAC(6′)-Ie and APH(2′′)-Ia domains produces some impairment of the catalytic activity of the truncated acetyltransferase compared with the full-length enzyme, it leads to a fully active phosphotransferase enzyme (Boehr et al., 2004 ▶). Despite this enzyme being the most important with respect to acquired resistance to the aminoglycosides, its structure has remained elusive to date. Difficulties in obtaining diffraction-quality crystals of the entire AAC(6′)-Ie-APH(2′′)-Ia may result from significant differences in the optimal crystallization conditions for the acetyltransferase and phosphotransferase functionalities of this bifunctional enzyme, or to flexibility of the full-length enzyme, although a recent SAXS study on the full-length bifunctional enzyme suggest that it adopts a rigid conformation in solution (Caldwell & Berghuis, 2012 ▶).
There are four major APH(2′′) enzymes, and the structures of three of these have been determined: APH(2′′)-IIa (Young et al., 2009 ▶), APH(2′′)-IIIa (Smith et al., 2012 ▶) and APH(2′′)-IVa (Toth et al., 2010 ▶; Shi et al., 2011 ▶; Shakya et al., 2011 ▶). Here, we report the 2.3 Å resolution X-ray crystal structure of the Mg2GDP complex of the APH(2′′)-Ia phosphotransferase domain of the AAC(6′)-Ie-APH(2′′)-Ia bifunctional enzyme.
2. Experimental procedures
2.1. Cloning, protein expression and purification
Wild-type APH(2′′)-Ia was cloned, expressed and purified as described previously (Toth et al., 2009 ▶). The construct began at residue Met175 of the full-length bifunctional enzyme. This construct was renumbered such that this N-terminal residue was Met1. In order to study the effect of the substitution of Tyr100 by phenylalanine on the apparent affinity of ATP and GTP for their binding templates, we generated (by site-directed mutagenesis), expressed and purified the Y100F mutant enzyme [APH(2′′)-Ia Y100F] following the protocol described for wild-type APH(2′′)-Ia (Toth et al., 2009 ▶). The purity of the enzyme was estimated to be >95% as judged from SDS–PAGE analysis. The concentration was evaluated spectrophotometrically using the theoretical extinction coefficient ∊280 = 38 990 M −1 cm−1 and the Pierce BCA Protein Assay Kit (Thermo Scientific; Rockford, Illinois, USA). The enzyme was stored in liquid nitrogen.
2.2. Enzyme kinetics
Kinetic parameters for the transfer of the γ-phosphate of ATP or GTP to tobramycin were evaluated spectrophotometrically at 340 nm by a coupled enzyme assay for both the wild-type and the Y100F mutant enzymes. The coupled assay reaction buffer was comprised of 100 mM HEPES pH 7.0, 10 mM MgCl2, 20 mM KCl, 2 mM phosphoenolpyruvate, 140 µM NADH, 15 U ml−1 pyruvate kinase and 20 U ml−1 lactate dehydrogenase in a total reaction volume of 250 µl. Tobramycin was used at a fixed saturating concentration of 200 µM, while the concentration of the NTPs (ATP and GTP) varied. Reaction was initiated by the addition of 120–240 nM enzyme. The steady-state kinetic parameters k cat and K m for ATP and GTP were determined by fitting the data to the Michaelis–Menten equation {v = V max[S]/(K m + [S]), where v is the initial velocity, [S] is the substrate concentration and V max is the maximum velocity} using GraphPad Prism 5 (GraphPad Software).
2.3. Crystallization
Initial coarse screens for apo APH(2′′)-Ia, binary complexes with Mg2GTP, Mg2GMPPNP, kanamycin and gentamicin, and ternary complexes with the same nucleotides and substrates were performed with commercially available sparse-matrix screens (Crystal Screen, Crystal Screen 2, PEG/Ion and PEG/Ion 2, Hampton Research) using the sitting-drop method. This resulted in a number of different conditions giving rise to crystals, with the best crystals being obtained for the binary APH(2′′)-Ia complex with Mg2GTP. Crystals were grown at 4 and 15°C in Intelli-Plates (Art Robbins Instruments) using a reservoir volume of 80 µl and drops comprising 1 µl protein complex and 1 µl reservoir solution. Selenomethionine-substituted (SeMet) APH(2′′)-Ia was also screened using the same set of crystallization screens and crystals were observed under identical conditions as for the native protein. The crystals of native and SeMet APH(2′′)-Ia were subsequently mounted in cryoloops, passed through a cryoprotectant solution containing crystallization buffer supplemented with 20% glycerol, flash-cooled in liquid nitrogen and stored in a sample cassette designed for use with the Stanford Automated Mounting (SAM) system (Cohen et al., 2002 ▶) for subsequent diffraction screening experiments.
2.4. Data collection and processing
Initial screening of the native crystals for diffraction quality was carried out on SSRL beamline BL7-1 using the Stanford Automated Mounting (SAM) robotic system (Cohen et al., 2002 ▶). Two conditions from Crystal Screen [Nos. 23 (0.2 M MgCl2, 0.1 M HEPES pH 7.5, 30% PEG 400) and 41 (0.1 M HEPES pH 7.5, 10% 2-propanol, 20% PEG 4000)] gave rise to diffraction-quality crystals of APH(2′′)-Ia. A complete data set was collected from a single crystal from condition No. 23 on SSRL beamline BL12-2 using a Pilatus 6M pixel-array detector. A total of 650 images were collected with an oscillation angle of 0.2° and an exposure time of 0.2 s per image. The data were processed with XDS (Kabsch, 2010a ▶) and were scaled using the program SCALA (Evans, 2006 ▶) from the CCP4 package (Winn et al., 2011 ▶). The crystal belonged to space group P21, with unit-cell parameters a = 90.68, b = 95.70, c = 91.15 Å, β = 104.9° (see Supporting Information1). Table 1 ▶ gives a summary of the data-collection statistics.
Table 1. APH(2′′)-Ia native and MAD data-collection and structure-solution statistics.
Values in parentheses are for the highest resolution shell.
| Native | Peak | Inflection | Remote | |
|---|---|---|---|---|
| Wavelength (Å) | 0.9795 | 0.97908 | 0.97920 | 0.91162 |
| Resolution limit (d min) (Å) | 36.2–2.30 (2.35–2.30) | 36.9–3.25 (3.33–3.25) | 37.0–3.35 (3.43–3.35) | 36.9–3.30 (3.38–3.30) |
| Observed reflections | 159151 | 126720 | 116808 | 122099 |
| Unique reflections to d min | 62555 | 25195 | 23122 | 24111 |
| R merge (%) | 3.8 (51.0) | 9.6 (77.4) | 8.6 (76.1) | 8.5 (76.8) |
| 〈I/σ(I)〉 | 13.6 (1.6) | 10.8 (2.0) | 12.3 (2.1) | 13.4 (2.1) |
| Average multiplicity | 2.5 | 5.0 | 5.0 | 5.0 |
| Completeness (%) | 93.3 (96.6) | 99.5 (96.1) | 99.5 (96.6) | 99.5 (95.4) |
| CC1/2 † | 99.9 (79.6) | 99.8 (73.7) | 99.8 (67.0) | 99.8 (71.8) |
| Wilson B ‡ (Å2) | 60.5 | |||
| MAD resolution limit (Å) | 3.5 | |||
| FOM from SOLVE | 0.61 | |||
| FOM from RESOLVE § | 0.71 | |||
| Residues autobuilt¶ | 742 (112) | |||
The SeMet APH(2′′)-Ia crystals were screened on SSRL beamline 9-2 and a single crystal was used to collect multiwavelength data for phasing. The crystal belonged to the same space group and had similar unit-cell parameters to the native crystals (P21; a = 91.1, b = 98.5, c = 93.2 Å, β = 105.3°). The data were collected at three wavelengths equivalent to the peak (0.97908 Å) and the inflection (0.97920 Å) of the selenium absorption edge and a remote energy (0.91162 Å). A total of 240 images were collected for each wavelength, with an oscillation range of 1°. The data were processed with XDS and scaled with XSCALE (Kabsch, 2010a ▶). Data-collection statistics are also given in Table 1 ▶.
2.5. Structure solution and refinement
The APH(2′′)-Ia structure was solved by multiwavelength anomalous diffraction (MAD) methods using the SeMet data to 3.3 Å resolution concurrently with molecular replacement (MR) using the native 2.3 Å resolution data and the APH(2′′)-IVa structure (PDB entry 3n4t; Toth et al., 2010 ▶) as the starting model. In the MAD solution, the program SOLVE (Terwilliger & Berendzen, 1999 ▶) gave the positions of all 20 Se atoms (five in each molecule), resulting in a mean figure of merit (FOM) of 0.61. Maximum-likelihood density modification and refinement of the phases and the fourfold noncrystallographic symmetry were carried out by RESOLVE (Terwilliger, 2000 ▶), giving a mean FOM of 0.71. Automated model building by RESOLVE resulted in 742 residues built out of 1220, of which 112 were placed according to the sequence into the electron-density map (Terwilliger, 2003 ▶). The initial crystallographic R factor prior to the first round of refinement was 0.41, with an R free of 0.50. For the MR calculation, the APH(2′′)-IVa model was converted to a pseudo-APH(2′′)-Ia model using the CCP4 program CHAINSAW (Stein, 2008 ▶) to truncate non-identical residues at the Cβ atom and leave identical residues intact. The program MOLREP (Vagin & Teplyakov, 2010 ▶) found a single solution comprising four molecules in the asymmetric unit with a weighted R factor of 0.54 and a score of 0.33. Initial cycles of refinement with REFMAC gave an R factor and an R free of 0.35 and 0.47, respectively, using data to 3.0 Å resolution. At this stage, comparison of the RESOLVE model with the model from MR showed that the same solution had been obtained by both methods. Use of the MR model allowed the remainder of the residues in the four independent molecules to be placed unequivocally into the density-modified electron density. Maximum-likelihood refinement with PHENIX (Adams et al., 2010 ▶), incorporating individual temperature-factor and TLS refinement at 2.3 Å resolution, and model building with Coot (Emsley & Cowtan, 2004 ▶) gave a final model with a crystallographic R factor of 0.1877 and an R free of 0.2467. Electron density observed in the experimentally phased electron-density maps (Supplementary Fig. S1) was modeled as GDP, and two residual peaks near the diphosphate moiety were modeled as magnesium ions in all four monomers in the asymmetric unit. The final model comprises 1183 amino-acid residues, four GDP molecules, eight magnesium ions and 211 water molecules. The overall average atomic displacement factor for all of the atoms included in the model (69.8 Å2) is approximately similar to the estimated B factor calculated from the Wilson plot (60.5 Å2; Table 1 ▶), and both are indicative of a significant degree of flexibility in the structure. The final refinement statistics are given in Table 2 ▶.
Table 2. APH(2′′)-Ia refinement statistics.
| Resolution range (Å) | 36.2–2.30 |
| R work (%) | 18.52 |
| R free (%) | 24.49 |
| R all † (%) | 18.82 |
| No. of data used in refinement/for R free | 62509/3118‡ |
| No. of atoms | |
| Protein | 9775 |
| Solvent | 211 |
| Nucleotide | 120 |
| Average B (Å2) | |
| Overall | 69.8 |
| Monomers§ | 68.6/66.8/66.4/78.4 |
| Mg2GDP | 61.6 |
| Solvent | 64.8 |
| R.m.s.d., bonds (Å) | 0.009 |
| R.m.s.d., angles (°) | 1.20 |
| Ramachandran plot¶, residues in (%) | |
| Most favored regions | 89.2 |
| Additionally allowed regions | 10.3 |
| Generously allowed regions | 0.5 |
R all is the overall R factor using all unique reflections.
Representing 5% of the total unique data chosen randomly.
The B values given are for the four independent monomers in the asymmetric unit averaged over all protein atoms.
As defined in PROCHECK (Laskowski et al., 1993 ▶).
The atomic coordinates and structure factors for Mg2GDP-APH(2′′)-Ia have been deposited in the Protein Data Bank (Berman et al., 2000 ▶) with PDB code 4ork. Superpositions were performed using the SSM procedure (Krissinel & Henrick, 2004 ▶) as implemented in Coot (Emsley & Cowtan, 2004 ▶), and LSQKAB in the CCP4 suite (Winn et al., 2011 ▶). Figures were generated using PyMOL (DeLano, 2002 ▶).
3. Results and discussion
3.1. Crystal structure of APH(2′′)-Ia
The APH(2′′)-Ia structure was solved by MAD methods combined with MR. The structure was subsequently refined against native data at 2.3 Å resolution (Table 1 ▶) to a final R factor and R free of 0.1877 and 0.2469, respectively. The final refined APH(2′′)-Ia model consists of four independent molecules in the asymmetric unit (Supplementary Fig. S2 and Supporting Information): monomer A (299 residues: Asp6–Lys304), monomer B (296 residues: Asp6–Lys304 with three residues missing, Lys59, Lys60 and Ala61), monomer C (293 residues: Ala9–Lys304 with three residues missing, Asn58, Lys59 and Lys60) and monomer D (295 residues: Ala9–Asp305 with two residues missing, Thr57 and Asn58). The residues listed as missing had weak or no electron density and were therefore left out of the model.
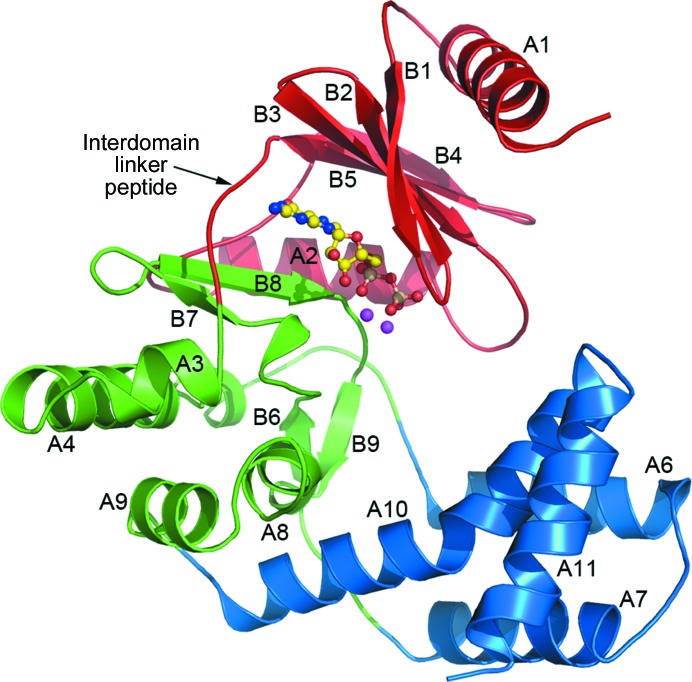
Each APH(2′′)-Ia molecule comprises two structural domains, the N-terminal domain (residues 1–100) and the C-terminal domain (residues 106–305), linked by a short interdomain hinge peptide (residues 101–106) (Fig. 2 ▶). The overall fold of APH(2′′)-Ia is similar to those of other APH enzymes for which structures are known, including APH(2′′)-IIa (Young et al., 2009 ▶), APH(2′′)-IIIa (Smith et al., 2012 ▶), APH(2′′)-IVa (Toth et al., 2010 ▶), APH(3′)-Ia (Stogios et al., 2013 ▶), APH(3′)-IIa (Nurizzo et al., 2003 ▶), APH(3′)-IIIa (Hon et al., 1997 ▶), APH(4)-Ia (Stogios et al., 2011 ▶) and APH(9)-Ia (Fong et al., 2010 ▶). The nucleotide-binding site is located in a cleft between the N-terminal and C-terminal domains. The C-terminal domain is further divided into two subdomains: the core subdomain (residues 106–147 and 194–257) and the helical subdomain (residues 148–193 and 258–305). Sequentially, the first part of the helical subdomain comprising residues 148–193 is essentially an insertion of two antiparallel α-helices (A6 and A7) into the core subdomain, with the second part (residues 258–305) also comprising two antiparallel α-helices (A10 and A11) which pack against the first antiparallel pair at an angle of almost 90° to create a four-helix bundle (Fig. 2 ▶). Structurally, this helical subdomain projects out from the side of the core subdomain such that the A10/A11 pair is on the inside of the bundle closer to the core subdomain, with the A6/A7 pair on the exterior of the molecule (Fig. 2 ▶). This creates a long deep cleft between the helical subdomain and the core subdomain; based upon the structures of other APH enzymes, this cleft houses the aminoglycoside-binding site.
Figure 2.
Ribbon representation of the APH(2′′)-Ia structure showing the three structural subdivisions: the N-terminal domain (red), the core subdomain (green) and the helical subdomain (blue). The location of the GDP molecule is shown as a yellow ball-and-stick representation and the two associated magnesium ions are represented as magenta spheres. The secondary-structure numbering is also given, and the interdomain linker peptide between the N-terminal domain and the core subdomain is indicated.
Superposition of Mg2GDP–APH(2′′)-Ia onto the three other APH(2′′) structures gives r.m.s.d.s of 2.1 Å for gentamicin–APH(2′′)-IIa (260–270 matching Cα positions), 3.0 Å for Mg2GDP–APH(2′′)-IIIa (264–270 matching Cα positions) and 1.9 Å for apo APH(2′′)-IVa (270–274 matching Cα positions). A summary of the results of all the superpositions described is given in Supplementary Table S1. Visual inspection of the superimposed molecules shows small movements of the N-terminal domain and the helical subdomain relative to the core subdomain (Supplementary Fig. S3). Superpositions based on the core subdomain only gave r.m.s.d.s of 1.0 Å for APH(2′′)-IIa, APH(2′′)-IIIa and APH(2′′)-IVa, and subsequent alignment of these superimposed structures based on the N-terminal domain and the helical subdomain gave a rough measure of the extent of movement of these domains relative to the core. Relative to APH(2′′)-IIa and APH(2′′)-IVa, which are both nucleotide-free structures, the APH(2′′)-Ia N-terminal domain has rotated as a rigid body closer to the core subdomain by between 4 and 6° (Supplementary Figs. S3a and S3c). This rotation is centered near residues Gly105 and Thr106 on the interdomain hinge peptide. In the case of the APH(2′′)-Ia/APH(2′′)-IIIa alignment, where there is a bound GDP in both cases, the nucleotide-binding cleft is closed to approximately the same degree in both molecules. There is, however, a small lateral movement of approximately 3–4 Å of the two N-terminal domains relative to each other (Supplementary Fig. S3b).
At a first glance, it seems unusual that the APH(2′′)-Ia/APH(2′′)-IIIa alignment gives rise to the largest r.m.s.d. for the entire structure (3.0 Å), given that these are the two enzymes which both use GTP exclusively. Inspection of the superposition shows that the main source of this higher r.m.s.d. lies in the relative orientations of the helical subdomains. In APH(2′′)-IIIa the helical subdomain appears to have moved downwards by approximately 5 Å relative to the core subdomain when the molecule is viewed as in Supplementary Fig. S3(b). When the two structures are superimposed based upon the helical subdomain only, it was found that the A6/A7 helix pair superimpose better than the A10/A11 pair. Upon inspection of the resulting alignment the reason becomes clear: in APH(2′′)-Ia the long A10 helix is bent significantly at the center near residue Tyr273, with the two halves of the helix making an angle of approximately 45°. In this regard the APH(2′′)-Ia enzyme closely resembles the structures of APH(2′′)-IIa and APH(2′′)-IVa, which both have a bent A10 helix. In contrast, the corresponding helix in APH(2′′)-IIIa is essentially straight (Supplementary Fig. S3b). Moreover, the reorientation of the helical subdomain in APH(2′′)-Ia relative to APH(2′′)-IIIa is not necessarily a rigid-body movement, in that the two sets of antiparallel helices appear to move differently with respect to each other. This is also evident to a certain extent in the superpositions with APH(2′′)-IIa and APH(2′′)-IVa. When the four helices are superimposed as a single rigid group, the r.m.s.d.s are greater than when the two sets of helices are aligned separately (Supplementary Table S1). This differential flexibility of the helical subdomain was also observed in the comparison of three different apo forms of APH(2′′)-IVa (Toth et al., 2010 ▶), and may contribute to the variations in substrate specificity observed in the four APH(2′′) enzymes.
3.2. The nucleotide-binding site
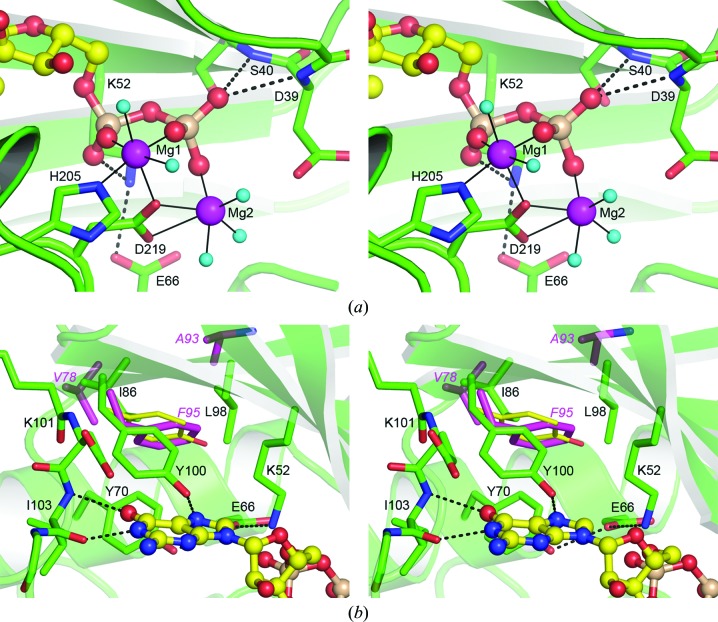
The Mg2GDP is bound in the cleft between the N-terminal domain and the core subdomain, interacting primarily with residues from the N-terminal domain and the interdomain linker peptide. Although the nucleotide was added as the Mg2GTP complex, the observed electron density showed the presence of a diphosphate, which suggests that hydrolysis of the γ-phosphate had occurred. The diphosphate group is anchored by several hydrogen-bonding interactions. One of the β-phosphate O atoms accepts two hydrogen-bonding interactions from the amide N atoms of Asp39 and Ser40 from the loop between β-strands B1 and B2 (Fig. 3 ▶ a). This loop is structurally homologous with the glycine-rich G-loop found in the protein kinases, which folds down over the active site to interact with the nucleotide. Although conformational changes in this loop have been observed in the kinases in response to nucleotide binding (Aimes et al., 2000 ▶; Madhusudan et al., 1994 ▶; Narayana et al., 1997 ▶), and also in the APH(3′) enzymes (Burk et al., 2001 ▶; Nurizzo et al., 2003 ▶), this loop adopts an identical conformation in the four APH(2′′) enzymes irrespective of the presence or absence of nucleotide in the binding cleft (Young et al., 2009 ▶; Toth et al., 2010 ▶; Shi et al., 2011 ▶; Smith et al., 2012 ▶). The α-phosphate makes a hydrogen bond to the side chain of Lys52 from strand B3 (Fig. 3 ▶ a). This lysine residue, which is conserved in almost all kinases and phosphotransferases known to date, forms an electrostatic interaction with a conserved glutamate residue (Glu66) on helix A2. This salt bridge serves to hold the lysine residue in place so that it can interact with the triphosphate of either ATP or GTP and orient it during catalysis (Shi et al., 2013 ▶). Two magnesium ions are coordinated to the diphosphate moiety and bridge between the diphosphate and residues from two loops on the core subdomain (Fig. 3 ▶ a). The first magnesium (Mg1) is octahedrally coordinated to the α- and β-phosphate groups, His205, Asp219 and two water molecules, and the second (Mg2) binds to one β-phosphate O atom, both O atoms of the Asp219 side chain and three water molecules. The aspartate residue (Asp219) is highly conserved in all kinases and phosphotransferases and lies on the loop between strands B8 and B9 in a segment with a consensus sequence motif GxIDxG. In the protein kinases this loop is generally referred to as the activation segment (Johnson, 2009 ▶; Taylor & Kornev, 2011 ▶). The neighboring loop between strands B6 and B7 with consensus sequence HxDxxxxN (the Brenner sequence; Brenner, 1987 ▶) is known as the catalytic loop in the protein kinases, in which the aspartate at the third position is the catalytic residue. The aspartate serves to correctly orient the substrate hydroxyl group (Kim & Mobashery, 2005 ▶) and accepts a proton once cleavage of the phosphorus–oxygen bond has begun (Valiev et al., 2003 ▶). The highly conserved asparagine at the C-terminal end of the Brenner sequence is involved in magnesium coordination. The presence of histidine (His205) at this position in APH(2′′)-Ia is highly unusual amongst the kinases and is a point of distinction between the APH(2′′) enzymes and other phosphotransferases and kinases. APH(2′′)-Ia, APH(2′′)-IIIa and APH(2′′)-IVa all have a histidine in this location coordinating to Mg1, and only APH(2′′)-IIa has the canonical asparagine.
Figure 3.
The APH(2′′)-Ia nucleotide-binding site. (a) Stereoview of the binding site showing the interactions which anchor the GDP (yellow ball-and-stick representation) in APH(2′′)-Ia (green). The two magnesium ions (Mg1 and Mg2) are shown as magenta spheres, with their coordinating water molecules shown as small cyan spheres. Hydrogen bonds are shown as dashed lines and the magnesium coordinate bonds as thin solid lines. The amino-acid residues involved in nucleotide binding and magnesium coordination are indicated. (b) Stereoview of the secondary hydrophobic binding pocket in APH(2′′)-Ia. The gatekeeper residue Tyr100 is shown in the center of the figure. The residues which make up the secondary hydrophobic pocket are indicated for APH(2′′)-Ia (green sticks) and APH(2′′)-IVa (magenta sticks). The location of the gatekeeper Phe95 in APH(2′′)-IVa is shown bound in the pocket formed by the less bulky residues Val78 and Ala93 (residue labels in magenta). The Tyr100 side chain, were it to swing into the pocket, is shown as thin yellow sticks and the severe steric clashes that this side chain would make with the pocket residues can be seen.
At the other end of the nucleotide-binding site, the guanine base is anchored to the interdomain hinge peptide by two hydrogen bonds to the main chain of Ile103 (Fig. 3 ▶ b). The guanine O6 atom accepts a hydrogen bond from the Ile103 N atom, and the guanine N1 atom donates a hydrogen bond to the Ile103 carbonyl O atom. There is an additional hydrogen-bonding interaction between the guanine N7 atom and the Oη atom of Tyr100 (Fig. 3 ▶ b). The main chain of Ile103 forms one part of a dual-specificity nucleotide-binding motif that was initially recognized in casein kinase II (Niefind et al., 1999 ▶) and is also present in the APH(2′′) phosphotransferases (Toth et al., 2010 ▶; Smith et al., 2012 ▶). Briefly, a nucleotide-binding scaffold composed of three residues on the interdomain hinge peptide forms two overlapping specificity templates for adenine and guanine. The adenine-specific binding site comprises a carbonyl O atom from the residue at the N-terminus of the interdomain hinge peptide (arbitrarily designated as residue N) and an amide N atom two residues further along (residue N + 2). This binding mode is observed in the APH(3′)-IIIa and APH(2′′)-IIa structures in complex with ADP and ATP analogs (Burk et al., 2001 ▶; Young et al., 2009 ▶). Superimposed on this binding site is a guanine-specific binding template comprising the amide N atom and the carbonyl O atom of the N + 2 residue, as observed in the GDP complex of APH(2′′)-IIIa (Smith et al., 2012 ▶) and here in the GDP complex of APH(2′′)-Ia. The guanine-binding modes in APH(2′′)-Ia and APH(2′′)-IIIa are essentially identical, although in the APH(2′′)-Ia structure there is no third hydrogen bond to a water molecule as described for APH(2′′)-IIIa (Smith et al., 2012 ▶). The recent structures of adenine and guanine APH(2′′)-IVa complexes confirm that the two overlapping binding modes are equally accessible in the same protein molecule (Shi & Berghuis, 2012 ▶).
Based upon the APH(2′′)-IIIa structure (Smith et al., 2012 ▶) and the two nucleoside–APH(2′′)-IVa structures (Shi & Berghuis, 2012 ▶), it was established that the identity of the amino acid preceding residue N of the nucleotide-binding scaffold was critical in determining whether the adenine-specific template was accessible, thus determining the co-substrate specificity of the APH(2′′) enzymes. This residue (N − 1) is referred to as the ‘gatekeeper’ residue (Smith et al., 2012 ▶). In the case of the GTP-exclusive enzymes APH(2′′)-Ia and APH(2′′)-IIIa this residue is a tyrosine, whereas in APH(2′′)-IIa and APH(2′′)-IVa, which have an almost equal preference for ATP and GTP, this gatekeeper residue is a methionine and a phenylalanine, respectively. It was suggested that the tyrosine gatekeeper residue was likely to be the reason why the APH(2′′)-Ia enzyme was an exclusively GTP-utilizing phosphotransferase (Bhattacharya et al., 2013 ▶). In APH(2′′)-IIa, although the gatekeeper Met85 [APH(2′′)-IIa residue numbering] side chain projects into the binding cleft, it does not interfere with the positioning of an adenine. In APH(2′′)-IVa the gatekeeper Phe95 [APH(2′′)-IVa residue numbering] is in a different conformation which has the phenyl ring in an adjacent secondary hydrophobic pocket (Bhattacharya et al., 2013 ▶; Toth et al., 2010 ▶).
In APH(2′′)-Ia, the Tyr100 side chain projects into the nucleotide-binding cleft in roughly the same orientation as the Met85 side chain in APH(2′′)-IIa, yet takes up significantly more room than the less bulky methionine and thus effectively blocks access to the adenine-binding template. The situation is almost identical to that described for APH(2′′)-IIIa (Smith et al., 2012 ▶). The importance of Tyr100 to the nucleotide selectivity of APH(2′′)-Ia was tested by the generation of the Y100F mutant. In this mutant there is an eightfold increase in K m for GTP and a decrease of almost one order of magnitude in the catalytic activity (Table 3 ▶), reflecting the loss of the hydrogen-bonding interaction between the N7 atom of the guanine ring and the phenolate O atom of the tyrosine. Furthermore, there is a modest fourfold decrease in K m for ATP and a fivefold increase in catalytic activity (Table 3 ▶), which may indicate that the loss of the phenolate O atom either creates some additional room in the binding site such that an ATP can interact with the adenine-specificity template or confers a degree of freedom of movement on the phenylalanine side chain which the tyrosine side chain did not possess, enabling the phenyl ring to move away from the adenine-binding site and thus open access for an incoming ATP.
Table 3. Kinetic parameters for wild-type APH(2′′)-Ia and the Y100F mutant.
Tobramycin was used as the substrate.
| Nucleotide | K m (µM) | k cat (s−1) | k cat/K m (M −1 s−1) | |
|---|---|---|---|---|
| APH(2′′)-Ia wild type | ATP | 1100 ± 100 | 0.25 ± 0.01 | (2.3 ± 0.2) × 102 |
| GTP | 1.9 ± 0.1 | 0.24 ± 0.01 | (1.3 ± 0.1) × 105 | |
| APH(2′′)-Ia Y100F | ATP | 337 ± 34 | 0.33 ± 0.01 | (1.0 ± 0.1) × 103 |
| GTP | 15 ± 1.4 | 0.57 ± 0.01 | (3.8 ± 0.3) × 104 |
In the wild-type APH(2′′)-Ia enzyme, in order for ATP to bind in the nucleotide-binding site the Tyr100 side chain would clearly have to move. However, the Tyr100 side chain appears to be rigidly constrained in its current orientation by the surrounding protein structure, flanked on one side by the side chain of Lys52 and on the other side by the main chain of Lys101 (residue N in the nucleotide-binding scaffold; Fig. 3 ▶ b). These flanking residues serve to lock the phenolate ring in place, such that it has very limited degrees of movement. An almost 10° rotation of the side chain in a direction which would move the tyrosine towards the secondary hydrophobic pocket would bring the Oη atom within a van der Waals nonbonded contact distance (approximately 3.2 Å) of the Cδ atom of Lys52. Moreover, in order for the tyrosine side chain to fully swing past the lysine, were the latter to remain in its current location, the Oη atom would have to approach to within 2.0 Å of the Lys52 side chain, which does not seem plausible. A rotation of less than 4° in the other direction would bring the side of the phenolate ring within nonbonded contact distance of the carbonyl O atom of Lys101. Although it is possible that the side chain of Lys52 could move aside to allow the Tyr100 side chain to pass, this also seems to be highly unlikely since the movement of the former would be restricted owing to the electrostatic interaction with Glu66. Moreover, upon nucleotide binding the additional hydrogen-bonding interactions with the α- and β-phosphate groups would further anchor the lysine in its currently observed position.
Is it possible that the gatekeeper residue does not need to swing at all to provide access to the adenine-specific template, but can occupy the secondary hydrophobic pocket during protein folding? This may well be what has happened in APH(2′′)-IVa, although there is currently no way to distinguish between these two possibilities. However, analysis of the secondary hydrophobic pocket adjacent to the gatekeeper residue in the four APH(2′′) enzymes does give us an indication as to whether this pocket is accessible at all. In APH(2′′)-Ia the secondary hydrophobic pocket is composed of the side chains of Tyr70, Ile86 and Leu98 (Fig. 3 ▶ b). The spatial orientation of a tyrosine side chain modeled into this site in approximately the same orientation as the Phe95 side chain in APH(2′′)-IVa suggests that there may not be enough room for the incorporation of its bulky side chain. Severe clashes between the phenolate ring and the Ile86 and Leu98 side chains would ensue (Fig. 3 ▶ b) such that it is highly unlikely that the tyrosine side chain could occupy any other position than where it is currently observed. By contrast, in APH(2′′)-IVa the corresponding secondary hydrophobic pocket residues [Val78 and Ala93; APH(2′′)-IVa residue numbering] are less bulky, and this opens the secondary pocket significantly in this enzyme such that the phenylalanine gatekeeper can be readily accommodated. Structural analysis of the F95Y mutant of APH(2′′)-IVa showed that the bulkier tyrosine could also be accommodated in the larger secondary pocket (Shi & Berghuis, 2012 ▶), and two independent kinetic analyses of F95Y mutants of APH(2′′)-IVa demonstrate the ability of this mutant to utilize both GTP and ATP as co-substrates (Shi & Berghuis, 2012 ▶; Bhattacharya et al., 2013 ▶).
3.3. Substrate binding
The substrate-binding site of the APH enzymes is located in the deep cleft formed between the core subdomain and the helical subdomain, typically comprising residues from both sides of the binding site and from helix A10, which forms the lower end of the cleft. It was found that in the gentamicin–APH(2′′)-IIa complex, the central aminocyclitol ring (also called the B ring) packs against the core subdomain and is anchored by a network of hydrogen-bonding contacts (Young et al., 2009 ▶). The A ring sits on helix A10 and interacts with residues from the helical subdomain. A similar mode of substrate binding is seen in the two independent structures of the kanamycin–APH(2′′)-IVa complex (Shi et al., 2011 ▶; Shakya et al., 2011 ▶). The majority of the protein–substrate interactions are with the A and B rings, with the C ring essentially free. Conversely, in the APH(3′) structures the kanamycin is bound ‘back-to-front’, such that the A and C rings are near the core subdomain with the B ring projecting across the binding cleft.
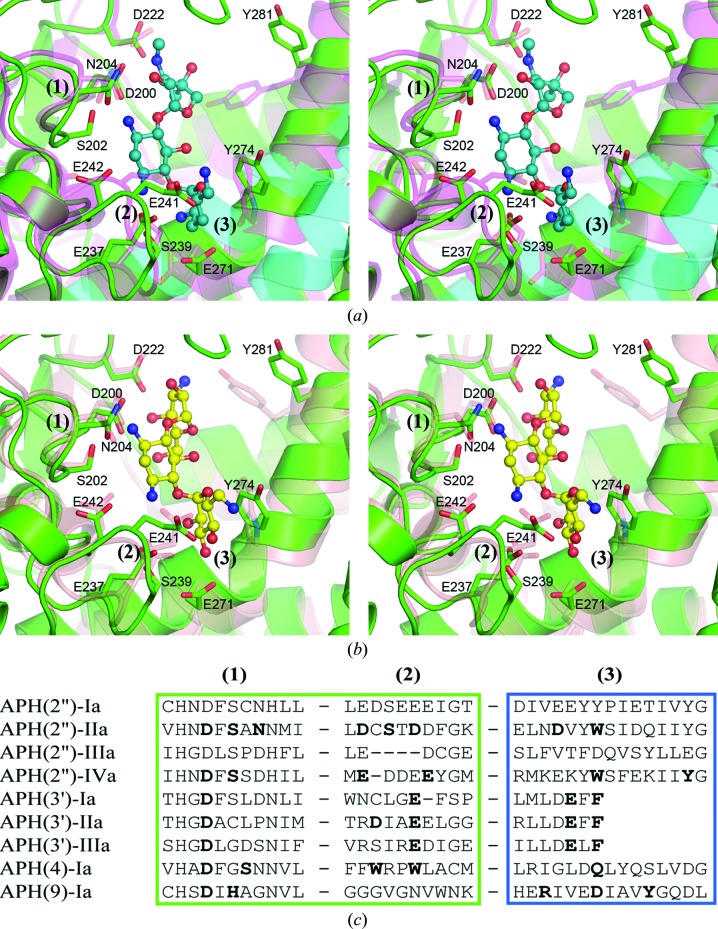
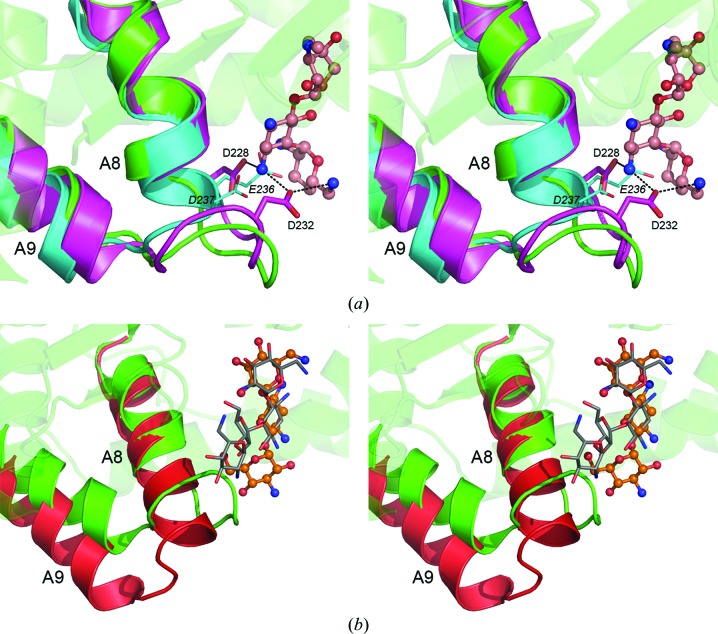
Analysis of the putative aminoglycoside-binding site of APH(2′′)-Ia shows that all of the residues responsible for gentamicin and kanamycin binding in APH(2′′)-IIa and APH(2′′)-IVa, respectively, are conserved, including the catalytic aspartate (Asp200), five other acidic residues (Asp222, Glu237, Glu241, Glu242 and Glu271) and Ser202, Asn204, Ser239, Tyr274 and Tyr281 (Figs. 4 ▶ a and 4 ▶ b). The majority of the substrate-binding residues cluster in three structural motifs: (1) the N-terminal end of the catalytic loop between strands B6 and B7 (Asp200–Asn204); (2) a loop between helices A8 and A9 (Glu237–Glu242), which could almost be viewed as an extension to helix A8, although it comprises eight residues and bends the polypeptide into a wide circle (Fig. 2 ▶); and (3) residues from the inner face of helix A10 (Glu271–Tyr274). Fig. 4 ▶(c) shows a sequence alignment of the three binding motifs for the aminoglycoside phosphotransferases. The conformation of the catalytic loop motif (1) is conserved in all of the known APH structures (Figs. 4 ▶ a and 4 ▶ b), even though the sequence between the catalytic aspartate and the magnesium-binding asparagine (or histidine) is variable (Fig. 4 ▶ c). A predominance of acidic residues, particularly in motif (2), is evident for the APH(2′′) enzymes, and this is understandable since the active site of these broad-spectrum aminoglycoside kinases has evolved to accommodate a large variety of substrates containing a significant number of positively charged amino side groups. Although there is currently no structural information on the mode of aminoglycoside binding to APH(2′′)-Ia, analysis of the binding cleft would suggest that this enzyme would bind substrates in a manner similar to that observed in APH(2′′)-IIa and APH(2′′)-IVa.
Figure 4.
The substrate-binding site in the APH enzymes. (a) Stereoview of the superposition of APH(2′′)-Ia (green) and APH(2′′)-IIa (magenta, partially transparent). The gentamicin bound to APH(2′′)-IIa is shown in a blue/dull green ball-and-stick representation. The residues which interact with gentamicin are shown as partially transparent magenta sticks. The corresponding residues in APH(2′′)-Ia are shown as green sticks and their residue numbers are given. Helices A10 and A11 from APH(2′′)-IIIa are shown as partially transparent cyan coils. (b) Stereoview of the superposition of APH(2′′)-Ia (green) and APH(2′′)-IVa (salmon, partially transparent). The kanamycin bound to APH(2′′)-IVa is shown in a yellow ball-and-stick representation. The residues which interact with kanamycin are shown as partially transparent salmon sticks. The corresponding residues in APH(2′′)-Ia are shown as green sticks. The three substrate-binding motifs (1), (2) and (3) are indicated in both (a) and (b). (c) Partial sequence alignment of the three substrate-binding motifs identified in the APH(2′′) enzymes. The corresponding sequences from three APH(3′) enzymes, APH(4)-Ia and APH(9)-Ia are also aligned. Residues highlighted in bold are known to interact with substrate molecules based upon the known structures of enzyme–substrate complexes.
In comparison to the other three APH(2′′) enzymes, APH(2′′)-IIIa presents something of an anomaly with regard to its substrate-binding site. Two major differences between APH(2′′)-IIIa and the three other members of the APH(2′′) family are the straight helix A10, as noted above, and a conformational difference in the substrate-binding motif (2) between helices A8 and A9. The superposition of this motif in APH(2′′)-Ia, APH(2′′)-IIa and APH(2′′)-IIIa is shown in Fig. 5 ▶(a). In the latter, rather than forming an elongated loop, the polypeptide has a deletion of four residues in such a way that helix A8 leads directly into helix A9. However, two acidic residues [Glu236 and Asp237 in APH(2′′)-IIIa numbering] are located in essentially the same positions as Asp228 and Asp232 in APH(2′′)-IIa, residues which interact with the gentamicin B and C rings (Young et al., 2009 ▶), and it would be expected that they might interact with amino groups on the aminoglycoside substrates in a similar manner as seen in the complexes of APH(2′′)-IIa and APH(2′′)-IVa (Shakya et al., 2011 ▶; Shi et al., 2011 ▶; Young et al., 2009 ▶). Structural motif (2) is also present in other aminoglycoside phosphotransferases. In the three known APH(3′) structures, motif (2) comprises an additional turn of α-helix at the C-terminus of helix A8 (Fig. 5 ▶ b), which appears to open the side of the substrate-binding pocket in comparison to APH(2′′)-Ia. Despite the elongated loop being absent in the APH(3′) enzymes, there are still between one and three acidic residues projecting into the binding site to interact with the substrate (depending upon the enzyme; Fong & Berghuis, 2002 ▶, 2009 ▶; Nurizzo et al., 2003 ▶; Stogios et al., 2013 ▶), and some of these acidic residues are involved in substrate binding (Fig. 4 ▶ c). The differences in this motif (2) region of the APH(3′) binding site, coupled with the observation that the substrates are bound ‘back-to-front’ (which places the C ring against the core subdomain), could facilitate the binding of the larger four-ringed aminoglycosides such as neomycin (Fig. 1 ▶; Fong & Berghuis, 2002 ▶). The narrow-spectrum APH(4)-Ia and APH(9)-Ia enzymes also have a variation of the extended loop seen in APH(2′′)-Ia, but in both cases the loop does not contain any acidic residues and is comprised mostly of hydrophobic residues (Fig. 4 ▶ c). In the hygromycin–APH(4)-Ia complexes, two tryptophan residues [Trp235 and Trp238; APH(4)-Ia numbering] from the motif (2) loop contribute to interactions with the substrate by aromatic stacking and hydrogen-bonding interactions (Stogios et al., 2011 ▶; Iino et al., 2013 ▶). In the spectinomycin–APH(9)-Ia complex this motif (2) loop plays no role in binding, with all the hydrogen-bonding contributions coming from motifs (1) and (3) (Fong et al., 2010 ▶).
Figure 5.
The structure of the A8–A9 loop in the APHs. (a) Superposition of APH(2′′)-Ia (green, partially transparent), APH(2′′)-IIa (magenta) and APH(2′′)-IIIa (cyan). Only helices A8 and A9 are shown for the latter two enzymes for clarity. Gentamicin from APH(2′′)-IIa (Young et al., 2009 ▶) is shown in a pink ball-and-stick representation. APH(2′′)-IIIa lacks the extended A8–A9 loop that the other three APH(2′′) enzymes possess. (b) Superposition of APH(2′′)-Ia (green, partially transparent) and APH(3′)-IIIa (red). The latter enzyme also lacks the extended A8–A9 loop. Kanamycin (orange ball-and-stick representation) and neomycin (gray sticks) are shown in the location observed in complexes with APH(3′)-IIIa (Fong & Berghuis, 2002 ▶).
4. Conclusions
The bifunctional aminoglycoside-modifying enzyme AAC(6′)-Ie-APH(2′′)-Ia is the single most important resistance enzyme for aminoglycoside antibiotics in Gram-positive bacteria. The structure of the GTP-dependent aminoglycoside phosphotransferase APH(2′′)-Ia domain has been solved to 2.3 Å resolution as the Mg2GDP complex. The GTP selectivity of APH(2′′)-Ia is driven primarily by the presence of the tyrosine ‘gatekeeper’ residue Tyr100. Structural comparisons with other APH(2′′) enzymes indicate that although an adenine-specificity template does exist with the same overall conformation in APH(2′′)-Ia, superimposed upon the guanine-recognition site, the side chain of Tyr100 prevents the binding of ATP by blocking the adenine-binding site in a similar fashion to that observed recently for the only other GTP-specific APH(2′′) enzyme, APH(2′′)-IIIa (Smith et al., 2012 ▶). Moreover, the tyrosine side chain is unable to be repositioned to allow ATP access as it is locked in place by the surrounding amino-acid residues. The identity of the gatekeeper residue is vitally important in nucleotide selectivity, as demonstrated by mutational studies on the three other APH(2′′) enzymes. The generation of the Y92A mutant of APH(2′′)-IIIa switched the enzyme from being GTP-exclusive to being equally capable of binding GTP and ATP with affinities matching APH(2′′)-IVa (Smith et al., 2012 ▶). This indicates that removal of the phenolate side chain gives access to the adenine-specificity template and the mutant is able to bind ATP efficiently. The recent kinetic analysis of the M85Y mutant of APH(2′′)-IIa had the opposite effect, switching the enzyme to a GTP-exclusive variant with a more than 300-fold increase in the K m for ATP and a dramatic decrease in catalytic efficiency by three orders of magnitude (Bhattacharya et al., 2013 ▶). Here, we observe an interesting phenomenon with the Y100F mutant of APH(2′′)-Ia, the effect of which is not as clear as the aforementioned mutations, yet which overall produces a mutant enzyme capable of conferring resistance to the aminoglycosides. The resultant decrease in the K m for ATP produces an enzyme which is now capable of using this nucleotide almost as efficiently as the APH(2′′)-IVa enzyme (Toth et al., 2009 ▶). Given that the K m values for both nucleotides are tenfold or more lower than the estimated cellular concentrations of both GTP and ATP, this mutant enzyme would be expected to utilize either nucleotide as a co-substrate.
Supplementary Material
Supporting Information.. DOI: 10.1107/S1399004714005331/be5260sup1.pdf
PDB reference: APH(2′′)-Ia, 4ork
Acknowledgments
This work was supported by a grant from the National Institutes of Health (AI057393 to SBV). Portions of this research were carried out at the Stanford Synchrotron Radiation Lightsource, a national user facility operated by Stanford University on behalf of the US Department of Energy, Office of Basic Energy Sciences. The SSRL Structural Molecular Biology Program is supported by the Department of Energy (BES, BER) and by the National Institutes of Health (NCRR, BTP, NIGMS). The project described was also supported by Grant No. 5 P41 RR001209 from the NCRR, a component of the National Institutes of Health.
Footnotes
Supporting information has been deposited in the IUCr electronic archive (Reference: BE5260).
References
- Adams, P. D. et al. (2010). Acta Cryst. D66, 213–221.
- Aimes, R. T., Hemmer, W. & Taylor, S. S. (2000). Biochemistry, 39, 8325–8332. [DOI] [PubMed]
- Berman, H. M., Westbrook, J., Feng, Z., Gilliland, G., Bhat, T. N., Weissig, H., Shindyalov, I. N. & Bourne, P. E. (2000). Nucleic Acids Res. 28, 235–242. [DOI] [PMC free article] [PubMed]
- Bhattacharya, M., Toth, M., Smith, C. A. & Vakulenko, S. B. (2013). Antimicrob. Agents Chemother. 57, 3763–3766. [DOI] [PMC free article] [PubMed]
- Boehr, D. D., Daigle, D. M. & Wright, G. D. (2004). Biochemistry, 43, 9846–9855. [DOI] [PubMed]
- Brenner, S. (1987). Nature (London), 329, 21. [DOI] [PubMed]
- Burk, D. L., Hon, W. C., Leung, A. K.-W. & Berghuis, A. M. (2001). Biochemistry, 40, 8756–8764. [DOI] [PubMed]
- Bush, K. et al. (2011). Nature Rev. Microbiol. 9, 894–896. [DOI] [PMC free article] [PubMed]
- Caldwell, S. J. & Berghuis, A. M. (2012). Antimicrob. Agents Chemother. 56, 1899–1906. [DOI] [PMC free article] [PubMed]
- Chow, J. W. (2000). Clin. Infect. Dis. 31, 586–589.
- Chow, J. W., Kak, V., You, I., Kao, S. J., Petrin, J., Clewell, D. B., Lerner, S. A., Miller, G. H. & Shaw, K. J. (2001). Antimicrob. Agents Chemother. 45, 2691–2694. [DOI] [PMC free article] [PubMed]
- Chow, J. W., Zervos, M. J., Lerner, S. A., Thal, L. A., Donabedian, S. M., Jaworski, D. D., Tsai, S., Shaw, K. J. & Clewell, D. B. (1997). Antimicrob. Agents Chemother. 41, 511–514. [DOI] [PMC free article] [PubMed]
- Cohen, A. E., Ellis, P. J., Miller, M. D., Deacon, A. M. & Phizackerley, R. P. (2002). J. Appl. Cryst. 35, 720–726. [DOI] [PMC free article] [PubMed]
- DeLano, W. L. (2002). PyMOL http://www.pymol.org.
- Emsley, P. & Cowtan, K. (2004). Acta Cryst. D60, 2126–2132. [DOI] [PubMed]
- Evans, P. (2006). Acta Cryst. D62, 72–82. [DOI] [PubMed]
- Ferretti, J. J., Gilmore, K. S. & Courvalin, P. (1986). J. Bacteriol. 167, 631–638. [DOI] [PMC free article] [PubMed]
- Fong, D. H. & Berghuis, A. M. (2002). EMBO J. 21, 2323–2331. [DOI] [PMC free article] [PubMed]
- Fong, D. H. & Berghuis, A. M. (2009). Antimicrob. Agents Chemother. 53, 3049–3055. [DOI] [PMC free article] [PubMed]
- Fong, D. H., Lemke, C. T., Hwang, J., Xiong, B. & Berghuis, A. M. (2010). J. Biol. Chem. 285, 9545–9555. [DOI] [PMC free article] [PubMed]
- French, S. & Wilson, K. (1978). Acta Cryst. A34, 517–525.
- Hon, W.-C., McKay, G. A., Thompson, P. R., Sweet, R. M., Yang, D. S. C., Wright, G. D. & Berghuis, A. M. (1997). Cell, 89, 887–895. [DOI] [PubMed]
- Iino, D., Takakura, Y., Fukano, K., Sasaki, Y., Hoshino, T., Ohsawa, K., Nakamura, A. & Yajima, S. (2013). J. Struct. Biol. 183, 76–85. [DOI] [PubMed]
- Johnson, L. N. (2009). Q. Rev. Biophys. 42, 1–40. [DOI] [PubMed]
- Kabsch, W. (2010a). Acta Cryst. D66, 125–132. [DOI] [PMC free article] [PubMed]
- Kabsch, W. (2010b). Acta Cryst. D66, 133–144. [DOI] [PMC free article] [PubMed]
- Kao, S. J., You, I., Clewell, D. B., Donabedian, S. M., Zervos, M. J., Petrin, J., Shaw, K. J. & Chow, J. W. (2000). Antimicrob. Agents Chemother. 44, 2876–2879. [DOI] [PMC free article] [PubMed]
- Kim, C. & Mobashery, S. (2005). Bioorg. Chem. 33, 149–158. [DOI] [PubMed]
- Krissinel, E. & Henrick, K. (2004). Acta Cryst. D60, 2256–2268. [DOI] [PubMed]
- Laskowski, R. A., MacArthur, M. W., Moss, D. S. & Thornton, J. M. (1993). J. Appl. Cryst. 26, 283–291.
- Llano-Sotelo, B., Azucena, E. F., Kotra, L. P., Mobashery, S. & Chow, C. S. (2002). Chem. Biol. 9, 455–463. [DOI] [PubMed]
- Madhusudan, Trafny, E. A., Xuong, N.-H., Adams, J. A., Ten Eyck, L. F., Taylor, S. S. & Sowadski, J. M. (1994). Protein Sci. 3, 176–187. [DOI] [PMC free article] [PubMed]
- Miller, G. H., Sabatelli, F. J., Hare, R. S., Glupczynski, Y., Mackey, P., Shlaes, D., Shimizu, K. & Shaw, K. J. (1997). Clin. Infect. Dis. 24, S46–S62. [DOI] [PubMed]
- Moazed, D. & Noller, H. F. (1987). Nature (London), 327, 389–394. [DOI] [PubMed]
- Narayana, N., Cox, S., Xuong, N.-H., Ten Eyck, L. F. & Taylor, S. S. (1997). Structure, 5, 921–935. [DOI] [PubMed]
- Niefind, K., Pütter, M., Guerra, B., Issinger, O. G. & Schomburg, D. (1999). Nature Struct. Biol. 6, 1100–1103. [DOI] [PubMed]
- Nurizzo, D., Shewry, S. C., Perlin, M. H., Brown, S. A., Dholakia, J. N., Fuchs, R. L., Deva, T., Baker, E. N. & Smith, C. A. (2003). J. Mol. Biol. 327, 491–506. [DOI] [PubMed]
- Ramirez, M. S. & Tolmasky, M. E. (2010). Drug Resist. Updat. 13, 151–171. [DOI] [PMC free article] [PubMed]
- Shakya, T., Stogios, P. J., Waglechner, N., Evdokimova, E., Ejim, L., Blanchard, J. E., McArthur, A. G., Savchenko, A. & Wright, G. D. (2011). Chem. Biol. 18, 1591–1601. [DOI] [PubMed]
- Shaw, K. J., Rather, P. N., Hare, R. S. & Miller, G. H. (1993). Microbiol. Rev. 57, 138–163. [DOI] [PMC free article] [PubMed]
- Shi, K. & Berghuis, A. M. (2012). J. Biol. Chem. 287, 13094–13102. [DOI] [PMC free article] [PubMed]
- Shi, K., Caldwell, S. J., Fong, D. H. & Berghuis, A. M. (2013). Front. Cell. Infect. Microbiol. 3, 1–17. [DOI] [PMC free article] [PubMed]
- Shi, K., Houston, D. R. & Berghuis, A. M. (2011). Biochemistry, 50, 6237–6244. [DOI] [PubMed]
- Smith, C. A. & Baker, E. N. (2002). Curr. Drug Targets Infect. Dis. 2, 143–160. [DOI] [PubMed]
- Smith, C. A., Toth, M., Frase, H., Byrnes, L. J. & Vakulenko, S. B. (2012). J. Biol. Chem. 287, 12893–12903. [DOI] [PMC free article] [PubMed]
- Stein, N. (2008). J. Appl. Cryst. 41, 641–643.
- Stogios, P. J., Shakya, T., Evdokimova, E., Savchenko, A. & Wright, G. D. (2011). J. Biol. Chem. 286, 1966–1975. [DOI] [PMC free article] [PubMed]
- Stogios, P. J., Spanogiannopoulos, P., Evdokimova, E., Egorova, O., Shakya, T., Todorovic, N., Capretta, A., Wright, G. D. & Savchenko, A. (2013). Biochem. J. 454, 191–200. [DOI] [PMC free article] [PubMed]
- Taylor, S. S. & Kornev, A. P. (2011). Trends Biochem. Sci. 36, 65–77. [DOI] [PMC free article] [PubMed]
- Terwilliger, T. C. (2000). Acta Cryst. D56, 965–972. [DOI] [PMC free article] [PubMed]
- Terwilliger, T. C. (2003). Acta Cryst. D59, 38–44. [DOI] [PMC free article] [PubMed]
- Terwilliger, T. C. & Berendzen, J. (1999). Acta Cryst. D55, 849–861. [DOI] [PMC free article] [PubMed]
- Toth, M., Chow, J. W., Mobashery, S. & Vakulenko, S. B. (2009). J. Biol. Chem. 284, 6690–6696. [DOI] [PMC free article] [PubMed]
- Toth, M., Frase, H., Antunes, N. T., Smith, C. A. & Vakulenko, S. B. (2010). Protein Sci. 19, 1565–1576. [DOI] [PMC free article] [PubMed]
- Tsai, S. F., Zervos, M. J., Clewell, D. B., Donabedian, S. M., Sahm, D. F. & Chow, J. W. (1998). Antimicrob. Agents Chemother. 42, 1229–1232. [DOI] [PMC free article] [PubMed]
- Tsuchizaki, N., Ishino, K., Saito, F., Ishikawa, J., Nakajima, M. & Hotta, K. (2006). J. Antibiot. 59, 229–233. [DOI] [PubMed]
- Vagin, A. & Teplyakov, A. (2010). Acta Cryst. D66, 22–25. [DOI] [PubMed]
- Vakulenko, S. B. & Mobashery, S. (2003). Clin. Microbiol. Rev. 16, 430–450. [DOI] [PMC free article] [PubMed]
- Valiev, M., Kawai, R., Adams, J. A. & Weare, J. H. (2003). J. Am. Chem. Soc. 125, 9926–9927. [DOI] [PubMed]
- Winn, M. D. et al. (2011). Acta Cryst. D67, 235–242.
- Woodcock, J., Moazed, D., Cannon, M., Davies, J. & Noller, H. F. (1991). EMBO J. 10, 3099–3103. [DOI] [PMC free article] [PubMed]
- Wright, G. D. (1999). Curr. Opin. Microbiol. 2, 499–503. [DOI] [PubMed]
- Young, P. G., Walanj, R., Lakshmi, V., Byrnes, L. J., Metcalf, P., Baker, E. N., Vakulenko, S. B. & Smith, C. A. (2009). J. Bacteriol. 191, 4133–4143. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information.. DOI: 10.1107/S1399004714005331/be5260sup1.pdf
PDB reference: APH(2′′)-Ia, 4ork