NAF-1 has been shown to be related with human health and disease, is upregulated in epithelial breast cancer and suppression of its expression significantly suppresses tumor growth. It is shown that replacement of the single His ligand with Cys resulted in dramatic changes to the properties of its 2Fe-2S clusters without any global crystal structural changes.
Keywords: autophagy, redox potential, 2Fe–2S cluster transfer and cluster stability, breast cancer, WFS2, longevity
Abstract
NAF-1 is an important [2Fe–2S] NEET protein associated with human health and disease. A mis-splicing mutation in NAF-1 results in Wolfram Syndrome type 2, a lethal childhood disease. Upregulation of NAF-1 is found in epithelial breast cancer cells, and suppression of NAF-1 expression by knockdown significantly suppresses tumor growth. Key to NAF-1 function is the NEET fold with its [2Fe–2S] cluster. In this work, the high-resolution structure of native NAF-1 was determined to 1.65 Å resolution (R factor = 13.5%) together with that of a mutant in which the single His ligand of its [2Fe–2S] cluster, His114, was replaced by Cys. The NAF-1 H114C mutant structure was determined to 1.58 Å resolution (R factor = 16.0%). All structural differences were localized to the cluster binding site. Compared with native NAF-1, the [2Fe–2S] clusters of the H114C mutant were found to (i) be 25-fold more stable, (ii) have a redox potential that is 300 mV more negative and (iii) have their cluster donation/transfer function abolished. Because no global structural differences were found between the mutant and the native (wild-type) NAF-1 proteins, yet significant functional differences exist between them, the NAF-1 H114C mutant is an excellent tool to decipher the underlying biological importance of the [2Fe–2S] cluster of NAF-1 in vivo.
1. Introduction
NAF-1 (nutrient-deprivation autophagy factor 1; synonyms ERIS and Miner1) is a member of a newly discovered family of iron–sulfur (Fe–S) proteins encoded by the cisd gene family that is defined by a unique CDGSH amino-acid sequence in the Fe–S cluster binding domain of its members (Conlan et al., 2009 ▶; Hou et al., 2007 ▶; Lin et al., 2007 ▶, 2011 ▶; Paddock et al., 2007 ▶). Since its initial discovery (Wiley et al., 2007 ▶; Amr et al., 2007 ▶), interest in NAF-1 has increased because the gene encoding this protein is located in a region of chromosome 4 associated with neuronal development (Boucquey et al., 2006 ▶) and because it has recently been shown to be critical for the maintenance of skeletal muscle (Chang et al., 2012 ▶) and for promoting longevity (Chen et al., 2009 ▶, 2010 ▶). Moreover, a transcriptional splicing error which deletes the NEET domain leads to a rare but severe disease called Wolfram Syndrome type 2 (WFS2; Amr et al., 2007 ▶), which is associated with hearing deficiencies, severe blindness, diabetes and a decreased life expectancy. NAF-1 is localized to both the ER (Amr et al., 2007 ▶) and the mitochondrial outer membrane (OMM; Chen et al., 2009 ▶), as well as being involved in the interaction between these two organelles (Wiley et al., 2013 ▶). NAF-1 has been functionally implicated in cell autophagy, possibly as a mediator of Bcl-2 antagonism of Beclin 1-dependent autophagy at the surface of the ER (Chen et al., 2010 ▶).
Interestingly, levels of NAF-1 mRNA are increased significantly in many different human cancer cells (Cortes et al., 2011 ▶; Stelzer et al., 2011 ▶). In addition, the NAF-1 protein accumulates in human epithelial breast cancer cells (Sohn et al., 2013 ▶). Suppressing NAF-1 expression levels by shRNA in breast cancer cells results in significantly decreased cell proliferation and tumor growth, decreased mitochondrial performance, uncontrolled accumulation of iron and ROS in mitochondria and activation of autophagy (Sohn et al., 2013 ▶). In a previous study, we solved the crystal structure of native NAF-1 (previously designated as Miner1) to 2.1 Å resolution (Conlan et al., 2009 ▶) and found that it is a homodimeric protein in which each protomer harbors one [2Fe–2S] cluster bound by an unusual 3-Cys-1-His coordination geometry. The structure bears a similar backbone fold to its paralog mitoNEET, yet NAF-1 has different surface properties (Conlan et al., 2009 ▶; Paddock et al., 2007 ▶; Tamir et al., 2013 ▶). These differences in the surface properties of NAF-1 lead to distinct protein partners (Tamir et al., 2014 ▶) and thereby different biological functions.
Cluster transfer from NAF-1 to the acceptor apo-ferredoxin (apo-Fd) protein occurs when the [2Fe–2S] clusters are oxidized to a [2Fe–2S]2+ state and the free cysteines of the acceptor protein are reduced to the free thiol form (Zuris et al., 2011 ▶; Tamir et al., 2013 ▶). The addition of glutathione (GSH), a physiological reducing agent, is effective in preparing the apo acceptor protein for cluster acceptance. NAF-1 is a cluster/Fe transfer donor and addition of excess NAF-1 to cells leads to iron accumulation in mitochondria. The anti-type II diabetes drug pioglitazone and the anti-diabetic and longevity-promoting natural product resveratrol stabilize the [2Fe–2S] clusters of NAF-1, preventing their release and preventing iron overload in mitochondria (Tamir et al., 2013 ▶).
Here, we show that the replacement of the His114 [2Fe–2S] cluster-coordinating side chain in NAF-1 with a Cys residue (H114C) significantly affects the NAF-1 cluster properties. The [2Fe–2S] cluster of NAF-1 H114C is approximately 25 times more stable than the native NAF-1 cluster, and its reduction potential (E m) is ∼300 mV more negative at pH 7.0. Strikingly, the NAF-1 H114C protein is incapable of acting as a cluster donor protein. In this study we report the crystal structure of NAF-1 H114C. Overall, the changes in the structure of NAF-1 H114C are localized to the [2Fe–2S] region, and the effects of the NAF-1 H114C mutation on the properties of the cluster are quite dramatic. Taken together, our results suggest that the H114C mutation is an ideal experimental tool to elucidate the importance of the redox potential and/or [2Fe–2S] cluster donor capabilities of NAF-1 in cellular studies.
2. Materials and methods
2.1. Expression and purification of NAF-1 proteins and ferredoxin
Expression and purification of NAF-1 and NAF-1 H114C was performed as described previously (Conlan et al., 2009 ▶). For crystallization purposes, where a higher grade of purity is required, an additional cation-exchange chromatography (HiTrap, GE Healthcare) step was conducted as described previously (Paddock et al., 2007 ▶). Expression and purification of ferredoxin (Fd) in both holo and apo forms was performed as described previously (Fish et al., 2005 ▶).
2.2. Optical spectroscopy
All UV–visible absorption spectra were measured from the near-UV to the near-IR (250–700 nm) on a Cary 300 Bio UV–Visible spectrophotometer (Varian, Palo Alto, California, USA) at room temperature. The protein samples analyzed, NAF-1, NAF-1 H114C and ferredoxin (Fd), were at concentrations of 20 µM protein in 20 mM Tris–HCl pH 8.0, 100 mM NaCl buffer. The stabilities of the [2Fe–2S] clusters were determined from monitoring their characteristic absorbance at 458 nm as a function of time with ∼20 µM NAF-1 and NAF-1 H114C proteins in 20 mM Tris–HCl pH 5.5, 100 mM NaCl on a Synergy 2 microplate reader (BioTek Instruments).
2.3. Determination of the E m of NAF-1 and NAF-1 H114C proteins
The oxidized states of the [2Fe–2S] clusters were monitored by their absorbance at 458 nm for E m determination as described previously (Zuris et al., 2011 ▶). 100 µM NAF-1 (native or H114C mutant) in 25 mM Tris–HCl pH 7.0, 100 mM NaCl was used. The ambient redox potential was adjusted by adding dithionite and was measured using an Ag/AgCl dual reference and working electrode (Microelectrodes, Bedford, New Hampshire, USA). The reduction potentials were adjusted to SHE for presentation. An elixir of redox mediators was used to ensure equilibration and redox stability. All mediators were purchased from Sigma–Aldrich. The mediators employed and their respective concentrations were 1,4-benzoquinone (50 µM), methylene blue (25 µM), menadione (50 µM), 1,4-naphthoquinone (25 µM), anthraquinone-2-sulfonate (25 µM), dithiothreitol (50 µM) and methyl viologen (2 µM). The standard midpoint (redox) potential was determined from a fit of the Nernst equation to the data as described previously (Dutton, 1978 ▶).
2.4. Native PAGE for detecting [2Fe–2S] cluster transfer from NAF-1 and NAF-1 H114C proteins to apo-ferredoxin (apo-Fd)
Native NAF-1 and the H114C mutant were incubated with apo-Fd. The NAF-1 and apo-Fd concentrations were 200 and 400 µM, respectively, so that their bands (and also absorption spectra) could be clearly visualized. NAF-1 and apo-Fd were incubated under vigorous aeration in the presence of 2% β-mercaptoethanol for 20 min. Transfer of the [2Fe–2S] cluster from NAF-1 to apo-Fd was then analyzed by native PAGE (Zuris et al., 2011 ▶; Tamir et al., 2013 ▶) stained with Coomassie Blue to verify the constant presence of both proteins during the cluster-transfer reaction. The red band visualized on the native gels results from the iron of the [2Fe–2S] cluster of the proteins. In order to verify whether changes in electrostatic or hydrophobic environments affect the cluster transfer from native NAF-1 and its H114C mutant to apo-Fd, the cluster-transfer assays were performed in the absence and presence of 20 mM MgCl2 and 20% glycerol. In order to test the possibility of cluster transfer in the reverse direction, 4-Cys-coordinated holo-Fd was used as a donor to the 3-Cys-1-His apo NAF-1 acceptor in the presence of cysteine reducing agent.
2.5. Crystallization, data collection and structure solution of NAF-1 and its H114C mutant
Crystallization conditions for the native NAF-1 were based on previously published conditions (Conlan et al., 2011 ▶) and were further refined using the sitting-drop vapor-diffusion method at 20°C. Crystals of native NAF-1 were obtained from a 4 µl drop consisting of equal amounts of protein solution (80 mg ml−1) and reservoir solution (35% PEG 3350, 0.1 M Tris–HCl pH 8.0, 50 mM NaCl). Rod-shaped crystals of NAF-1 appeared within 24 h and reached their final size within several days. Crystals of NAF-1 H114C were obtained from two different conditions by streak-seeding (Stura & Wilson, 1991 ▶) after 6 h of pre-equilibration, using the native crystals as the source of microseeds. In this regard, the 4 µl drop consisted of equal amounts of protein solution (50 mg ml−1) and reservoir solution consisting of 35% PEG 2000, 100 mM Tris–HCl pH 8.0, 100–200 mM NaCl or 35% PEG 3500, 100 mM Tris–HCl pH 8.0, 100–200 mM NaCl. Rod-shaped crystals similar to those of the wild type appeared within 24 h. In addition, a different form of thin plates appeared within 8 d in the same conditions. Prior to data collection, crystals were cryoprotected in a solution consisting of 17–20% glycerol and the reservoir solution and were immediately flash-cooled in liquid nitrogen. Crystallographic data for all crystals were collected at the European Synchrotron Radiation Facility (ESRF), Grenoble, France.
Crystallographic data for the native NAF-1 crystal were collected on the ID23-1 beamline at the ESRF at a temperature of 100 K and a wavelength of 1.0 Å. Crystals diffracted to a maximal resolution of 1.65 Å and the data were integrated and scaled using the HKL-2000 suite (Otwinowski & Minor, 1997 ▶). The crystal belonged to the orthorhombic space group P212121, with unit-cell parameters a = 41.0, b = 48.7, c = 73.7 Å and with two NAF-1 molecules in the asymmetric unit. Although the unit-cell parameters are highly similar to those of the previously published NAF-1 structure (Conlan et al., 2011 ▶), we elected to solve the structure via molecular replacement using AutoMR (Adams et al., 2010 ▶) in the resolution range 35.0–4.5 Å with the atomic coordinates of human Miner1 (PDB entry 3fnv, now designated NAF-1; Conlan et al., 2009 ▶) as a search model after removing all solvent molecules. The structure was then refined by seven cycles of restrained refinement using PHENIX (Echols et al., 2012 ▶). The structure was fitted into electron-density maps using the graphics program Coot (Emsley & Cowtan, 2004 ▶). The structure was further refined using REFMAC (Murshudov et al., 2011 ▶) restrained refinement with the maximum-likelihood option, and solvent molecules were added utilizing ARP/wARP (Morris et al., 2003 ▶).
The rod-shaped NAF-1 H114C crystals, which were highly similar in shape to those of native NAF-1, showed extremely poor diffraction characteristics, with a maximal resolution of 3.6 Å and a mosaicity of 2.5°. However, the plate-like crystals diffracted to a maximal resolution of 1.58 Å. Data were collected on the BM14 beamline at the ESRF at 100 K and were integrated and scaled using the HKL-2000 suite (Otwinowski & Minor, 1997 ▶). The crystals belonged to the orthorhombic space group P212121, with unit-cell parameters a = 48.4, b = 61.0, c = 135.2 Å and with six NAF-1 H114C molecules in the asymmetric unit. The structure of NAF-1 H114C was solved via molecular replacement using Phaser (McCoy et al., 2007 ▶) as implemented in CCP4i (Potterton et al., 2003 ▶) in the resolution range 35–3.5 Å using the coordinates of the refined NAF-1 as the search model after removing solvent molecules. The model was then refined using the REFMAC restrained option in the resolution range 30.5–1.58 Å, and solvent molecules were added utilizing ARP/wARP (Morris et al., 2003 ▶). The structure was fitted into the electron-density maps using Coot (Emsley & Cowtan, 2004 ▶) with His114 mutated to a Cys.
3. Results
3.1. The structures of NAF-1 and NAF-1 H114C proteins
The structures of NAF-1 and NAF-1 H114C were both refined to better than 1.6 Å resolution (Table 1 ▶). The final refined NAF-1 structure consisted of 1034 protein and 87 solvent atoms and two [2Fe–2S] clusters (Table 1 ▶). R cryst and R free converged to values of 13.5% and 15.0%, respectively (Table 1 ▶). The final NAF-1 H114C structure consisted of 3082 protein and 264 solvent atoms with six [2Fe–2S] clusters. R cryst and R free converged to values of 16.0% and 19.6%, respectively (Table 1 ▶).
Table 1. Crystallographic data-collection and refinement statistics.
Values in parentheses are for the last shell.
| NAF-1 H114C | Native NAF-1 | |
|---|---|---|
| ESRF beamline | BM14 | ID23-1 |
| Wavelength () | 0.98 | 1.0 |
| Space group | P212121 | P212121 |
| Unit-cell parameters () | a = 48.40, b = 60.99, c = 135.24 | a = 41.01, b = 48.65, c = 73.73 |
| Resolution range () | 50.01.58 (1.611.58) | 50.01.65 (1.681.65) |
| Unique reflections | 50138 | 16326 |
| Multiplicity | 3.8 (3.5) | 4.2 (4.0) |
| R merge(I) | 0.076 (0.601) | 0.058 (0.614) |
| CC1/2 (last shell) | 0.737 | 0.802 |
| Completeness (%) | 95.11 (70.46) | 93.88 (61.15) |
| I/(I) | 16.9 (1.9) | 19.19 (2.89) |
| No. of protein atoms | 3082 | 1034 |
| No. of ligand atoms | 24 | 8 |
| No. of solvent atoms | 264 | 87 |
| R factor (%) | 15.97 | 13.48 |
| R free (%) | 19.60 | 15.04 |
| Average B factor (2) | ||
| Protein | 20.39 | 20.93 |
| Ligand | 12.01 | 10.65 |
| Solvent | 30.13 | 33.30 |
| R.m.s. deviation from ideality | ||
| Bond lengths () | 0.03 | 0.01 |
| Bond angles () | 1.89 | 1.84 |
| Ramachandran plot | ||
| Favored (%) | 97.9 | 99.2 |
| Allowed (%) | 2.1 | 0.8 |
| Outliers (%) | 0 | 0 |
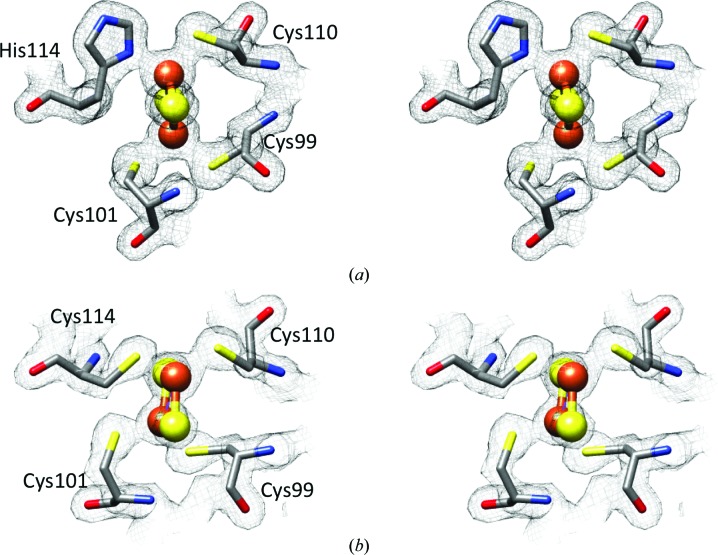
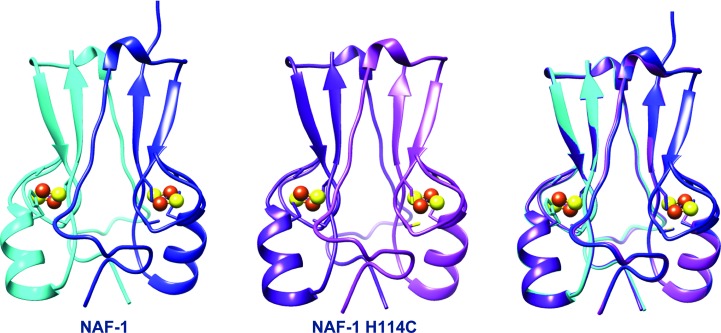
The overall structures of NAF-1 and NAF-1 H114C are highly similar, with a β-rich cap domain and helical regions, where the latter comprises the cluster binding domain (Fig. 1 ▶). In this regard, the structure consists of a functional dimer that interlaces the N-terminal part including the first β-strand of the other monomer. The functional dimer binds two [2Fe–2S] clusters coordinated by three cysteines and one histidine side chain (Fig. 2 ▶). The mutation of His114 to Cys causes no significant changes in the cluster binding region. In fact, the three unchanged cysteine ligands (Cys99, Cys101 and Cys110) superimpose in the native and H114C mutant NAF-1 structures. The only significant difference is at the site of mutation, where Cys114 displays a somewhat larger bond length by 0.2 Å on average, probably owing to the overall size of the respective residues (Table 2 ▶). A similar increase in binding distance upon His-to-Cys (H87C) mutation was observed in the structures of mitoNEET (Conlan et al., 2011 ▶; Table 2 ▶). The overall structures of the NAF-1 dimer (native and H114C mutant) are almost identical. Because the native structure includes only one dimer, whereas NAF-1 H114C contains three dimers in the asymmetric unit, we have a comparative tool to examine the changes and structural deviations. In this context, the superposition of the dimers exhibited low r.m.s.d. values ranging between 0.56 and 0.26 Å (Table 3 ▶). The high structural similarity between native NAF-1 and NAF-1 H114C encouraged us to test the biophysical, biochemical and functional effects of the mutation. Since the function of proteins largely depends on their cluster, the effect of the mutation on the properties of the NAF-1 [2Fe–2S] cluster were investigated.
Figure 1.
The 2F o − F c electron-density map at 1.5σ depicting the [2Fe–2S] cluster and its interactions with NAF-1 for (a) NAF-1 and (b) NAF-1 H114C.
Figure 2.
The crystal structure of NAF-1 consists of an interlaced dimer (left; shown in blue and cyan). The structure of NAF-1 H114C exhibits a highly similar topology (center), as can also be observed in the superposition (right).
Table 2. Distances between His/Cys and Fe ().
Wild-type NAF-1.
| Fe1Cys99 | Fe1Cys101 | Fe2Cys110 | Fe2His114 | |
|---|---|---|---|---|
| Monomer A | 2.34 | 2.29 | 2.29 | 2.16 |
| Monomer B | 2.32 | 2.29 | 2.35 | 2.19 |
| Average | 2.33 | 2.29 | 2.32 | 2.18 |
NAF-1 H114C.
| Fe1Cys99 | Fe1Cys101 | Fe2Cys110 | Fe2Cys114 | |
|---|---|---|---|---|
| Monomer A | 2.13 | 2.11 | 2.27 | 2.34 |
| Monomer B | 2.26 | 2.23 | 2.28 | 2.33 |
| Monomer C | 2.19 | 2.08 | 2.27 | 2.32 |
| Monomer D | 2.19 | 2.15 | 2.30 | 2.32 |
| Monomer E | 2.18 | 2.11 | 2.28 | 2.31 |
| Monomer F | 2.25 | 2.12 | 2.26 | 2.34 |
| Average | 2.20 | 2.13 | 2.28 | 2.33 |
Wild-type mitoNEET (PDB entry 2qh7).
| Fe1Cys72 | Fe1Cys74 | Fe2Cys83 | Fe2Cys87 | |
|---|---|---|---|---|
| Monomer A | 2.33 | 2.15 | 2.32 | 2.19 |
| Monomer B | 2.32 | 2.24 | 2.30 | 2.14 |
| Average | 2.33 | 2.20 | 2.31 | 2.17 |
MitoNEET H87C (PDB entry 3plq).
| Fe1Cys72 | Fe1Cys74 | Fe2Cys83 | Fe2Cys87 | |
|---|---|---|---|---|
| Monomer A | 2.36 | 2.17 | 2.31 | 2.39/2.19 |
| Monomer B | 2.34 | 2.17 | 2.33 | 2.33/3.17 |
| Average | 2.35 | 2.17 | 2.32 |
Table 3. Superposition of native (WT) NAF-1 on the three available dimers of NAF-1 H114C.
| WT NAF-1 | NAF-1 H114C | R.m.s.d. () |
|---|---|---|
| Dimer 1 | Dimer 1 | 0.45 |
| Dimer 1 | Dimer 2 | 0.26 |
| Dimer 1 | Dimer 3 | 0.56 |
3.2. Increased stability of the [2Fe–2S] clusters of NAF-1 H114C
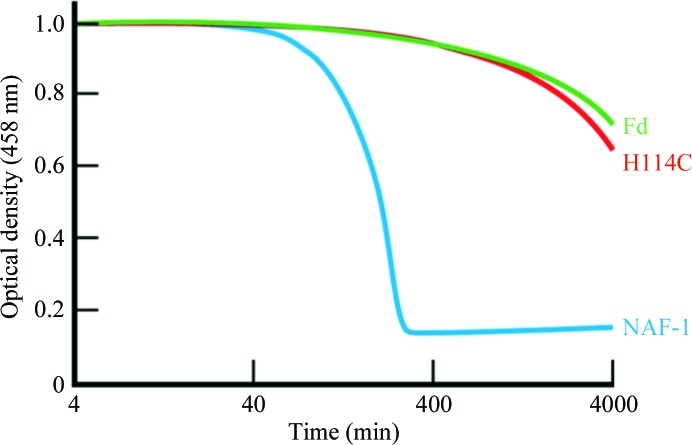
The stability of native NAF-1 is strongly pH-dependent (Conlan et al., 2009 ▶). This observation led to the suggestion that the strong pH dependence of the stability of native NAF-1 was associated with titration of His114. For this reason, we pursued a more detailed comparative study of the stability of the [2Fe–2S] clusters of both NAF-1 and NAF-1 H114C in acidic pH (pH 5.5) over time. The cluster stability was determined by monitoring the change in the characteristic absorption of the iron–sulfur cluster at 458 nm as a function of time at 37°C. NAF-1 H114C displayed a ∼25-fold increased stability over that observed for the NAF-1 protein (Fig. 3 ▶). This high stability is very similar to that of the [2Fe–2S] cluster of ferredoxin (Fd), an iron–sulfur protein, in which the [2Fe–2S] cluster is coordinated by the classical 4-Cys ligation geometry.
Figure 3.
The stability of the [2Fe–2S] cluster of NAF-1 H114C is greatly increased. The cluster stabilities of NAF-1 (blue), NAF-1 H114C (red) and Fd (green) were monitored over time by optical spectroscopy at 458 nm, which is attributed to cluster absorbance at 37°C. The absorbance decay corresponds to loss of the [2Fe–2S] cluster. The cluster is much more stable in NAF-1 H114C compared with native NAF-1 protein and its stability is almost equal to that in Fd.
3.3. A dramatic shift in the redox potential of the [2Fe–2S] clusters in NAF-1 H114C
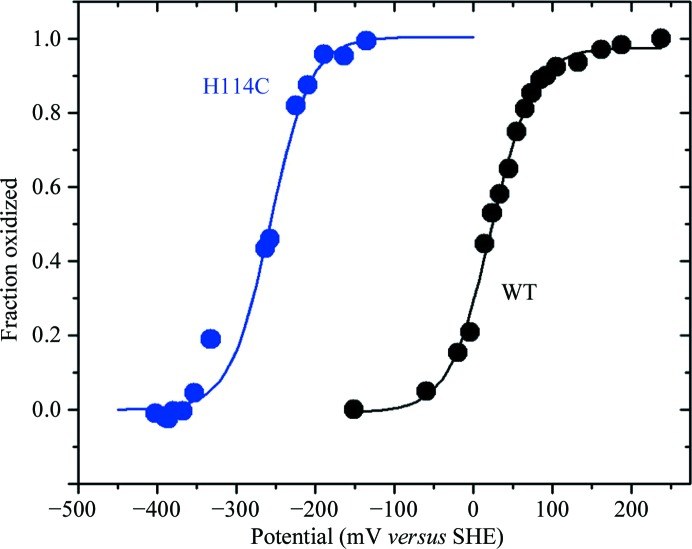
The [2Fe–2S] cluster of NAF-1 is redox-active (Conlan et al., 2009 ▶). The reduction potential (E m) of NAF-1 and NAF-1 H114C was measured to characterize the effect of the single substitution of His114 by Cys on this important characteristic of the cluster. The redox potential (E m) of the [2Fe–2S] centers, i.e. [2Fe–2S]2+/[2Fe–2S]+, of the [2Fe–2S] cluster of the NAF-1 or NAF-1 H114C protein was measured spectroscopically by monitoring the change in absorbance at 458 nm as a function of the measured redox potential (Fig. 4 ▶). The data were fitted by the Nernst equation as described by Zuris et al. (2010 ▶). At pH 7.0, the E m value of the native NAF-1 is +24 mV (± 5 mV) (Fig. 4 ▶, black trace). The E m value of the [2Fe–2S] cluster of the NAF-1 H114C protein at pH 7.0 is −280 mV (± 10 mV) (Fig. 4 ▶, blue trace), a difference of ∼−300 mV from that measured for native NAF-1 (Fig. 4 ▶, black trace). This dramatic shift is in the range observed for a 4-Cys-coordinated ferrodoxin-type [2Fe–2S] cluster protein (Jung et al., 1999 ▶).
Figure 4.
Substitution of a single residue, His114, by Cys in NAF-1 alters the redox potential of the [2Fe–2S] cluster (E m) of NAF-1. E m of the NAF-1 H114C [2Fe–2S] cluster is shifted by >−300 mV compared with native NAF-1. Native NAF-1 (black) has a redox potential of approximately 24 mV (±5 mV). The point mutant H114C (blue) has a substantially shifted redox potential of −280 mV (±10 mV).
3.4. The point mutation of His114 of NAF-1 to Cys abolishes the protein cluster donation function
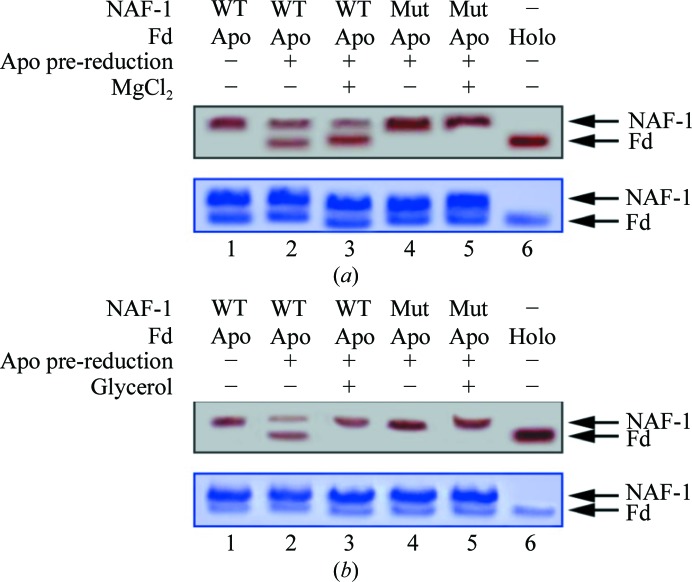
In our previous studies, we demonstrated that NAF-1 is a cluster-transfer protein (Tamir et al., 2013 ▶). To test whether changes in the electrostatic or hydrophobic environment of the cluster alter its transfer from NAF-1 or NAF-1 H114C to apo-Fd, we performed cluster-transfer assays in the absence or presence of 20 mM MgCl2 and 20% glycerol. Cluster transfer from NAF-1 to the apo-Fd acceptor was slightly accelerated in the presence of MgCl2 (Fig. 5 ▶ a). Interestingly, the presence of 20% glycerol resulted in the abolition of cluster transfer from native NAF-1 to apo-Fd (Fig. 5 ▶ b), indicating that hydrophobic interactions are probably involved in protein–protein interactions. Cluster transfer was not observed for the NAF-1 H114C protein to apo-Fd under any conditions (Fig. 5 ▶). In addition, cluster transfer was unidirectional from NAF-1 to apo-Fd only.
Figure 5.
The NAF-1 [2Fe–2S] cluster-transfer function is abrogated in NAF-1 H114C. NAF-1 was incubated at 37°C for 20 min with β-mercaptoethanol pre-reduced apo-Fd and run on a native gel. In the upper native gels in each panel the red-colored bands are indicative of the [2Fe–2S] cluster in the two proteins; the upper red band represents NAF-1 (labeled NAF-1 on the right side of the gel) and the lower red band represents holo-Fd (labeled Fd). (a) Cluster transfer from NAF-1 to the pre-reduced apo-Fd in the absence and presence of MgCl2. The native (WT) NAF-1 (lane 1) indicated by the red-colored upper band decreases in intensity upon incubation with pre-reduced apo-Fd, with a concomitant increase in the intensity of the lower red band owing to the formation of holo-Fd (lane 2). In the presence of MgCl2, the cluster transfer from NAF-1 to apo-Fd is slightly greater (lane 3). NAF-1 H114C completely lost the cluster-transfer function in the absence (lane 4) or presence (lane 5) of MgCl2. Lane 6 contains holo-Fd for reference. The lower gel was stained with Coomassie Blue to confirm that the protein levels were the same in all experiments. (b) Cluster transfer in the presence of glycerol. The native NAF-1 (WT; lane 1), when incubated with pre-reduced apo-Fd, transfers its cluster to the latter to form holo-Fd (lower red band, lane 2). The presence of glycerol prevents cluster transfer from NAF-1 to pre-reduced apo-Fd (lane 3). NAF-1 H114C does not function as a cluster-donor protein in the absence (lane 4) or the presence of glycerol (lane 5). Lane 6 contains the control holo-Fd.
4. Discussion
The aim of this study was to determine the structural and biophysical characteristics that result from the substitution of the single noncysteine ligand of the NAF-1 [2Fe–2S] cluster, His114, with Cys. The single His coordination marks one of the unique structural features of the NEET protein family. Here, we report that a single H114C substitution in NAF-1 leads to a major increase in the stability of the mutant [2Fe–2S] cluster and to a dramatic shift in the mutant cluster redox potential, E m, compared with the native protein. Moreover, the His-to-Cys substitution, and its induced biophysical changes, eliminates the cluster-transfer property of NAF-1. The biophysical and biochemical characteristics are supported by a high-resolution structure determination to 1.58 Å resolution (R factor = 16.0%) of NAF-1 H114C in comparison to the 1.65 Å resolution (R factor = 13.5%) structure of the native NAF-1 protein. As shown in Fig. 2 ▶, the NAF-1 H114C protein backbone structure is nearly identical to that of native NAF-1, with major structural consequences only being found at the site of the mutation, i.e. the cluster binding region (Fig. 1 ▶). Although the structural differences are localized, the consequences for the cluster properties are pronounced.
The 4-Cys-coordinated cluster of NAF-1 H114C has a 25-fold increased stability compared with the cluster of the native protein. As the structural changes are localized to the mutation site, the increased stability is likely to be owing to the increased anionic environment and the loss of the properties of His114 (Fig. 3 ▶). Another consequence of the mutation is a significant shift in the E m of the NAF-1 H114C cluster by −300 mV. Since the conformational changes are localized to the site of the mutation, the altered E m is a consequence of the more anionic properties of the cysteine ligand in comparison with the neutral histidine in the electrostatic environment of the cluster. Replacement of the single histidine ligand with cysteine results in an E m that falls into the redox range of other 4-Cys-coordinated iron–sulfur proteins such as ferredoxin (Meyer, 2008 ▶). The third consequence of the mutation is the abolition of the ability of the NAF-1 mutant to donate its iron–sulfur cluster to apo acceptor proteins. Thus, the single histidine ligand of the cluster is key to the cluster-donor functionality of NAF-1. The cluster coordinated solely by anionic cysteines in NAF-1 H114C is more stable. As such, cluster donation may be inhibited, at least in part, by this increased stability. The His114 ligand appears to play a crucial role in iron transfer, iron metabolism or homeostasis, in which NAF-1 participates in vivo. Similar results have also been reported for the H87C mutant of the paralog mitoNEET. As for NAF1-H114C, the mutation from histidine to cysteine resulted in increased stability of the cluster and a decrease in the redox potential, yet only minor structural changes were observed (Conlan et al., 2011 ▶).
5. Conclusions
The single His ligand, His114, of the [2Fe–2S] cluster of NAF-1 confers several key properties to the cluster: (i) it mediates the cluster stability, (ii) it tunes the redox potential to be close to neutral at pH 7.0 and (iii) it confers the ability to transfer its [2Fe–2S] cluster to apo-Fd. Importantly, NAF-1 H114C alters these biophysical properties without significant changes to the global structure or to the cluster binding region, thereby indicating that the biophysical properties are influenced mainly by the properties of the side chain. The fact that the overall structure did not change and only subtle alterations near the cluster were observed indicates that the mutant protein, when introduced into cells, could reveal the underlying biological importance of NAF-1 in in vivo experiments aimed at elucidating the role that the 3-Cys-1-His [2Fe–2S] cluster plays in the function of the protein. The unique 3-Cys-1-His cluster coordination in NEET proteins is likely to regulate key functional properties of these important proteins and this could be the reason why it is highly conserved in nature.
Supplementary Material
PDB reference: NAF-1, wild type, 4oo7
PDB reference: H114C mutant, 4ooa
Acknowledgments
This work was supported by The Israeli Science Foundation (ISF863/09 and ISF865/13 to RN) and the National Institutes of Health (Grants GM54038 and GM101467 to PAJ). RN and OL are members of The MINERVA Center for Bio-hybrid Complex Systems and acknowledge funds received from the Center. We would like to thank the staff of the ESRF and EMBL Grenoble for their assistance and support in using beamlines BM-14 and ID23-1.
References
- Adams, P. D. et al. (2010). Acta Cryst. D66, 213–221.
- Amr, S., Heisey, C., Zhang, M., Xia, X.-J., Shows, K. H., Ajlouni, K., Pandya, A., Satin, L. S., El-Shanti, H. & Shiang, R. (2007). Am. J. Hum. Genet. 81, 673–683. [DOI] [PMC free article] [PubMed]
- Boucquey, M., De Plaen, E., Locker, M., Poliard, A., Mouillet-Richard, S., Boon, T. & Kellermann, O. (2006). J. Neurochem. 99, 657–669. [DOI] [PubMed]
- Chang, N. C., Nguyen, M., Bourdon, J., Risse, P.-A., Martin, J., Danialou, G., Rizzuto, R., Petrof, B. J. & Shore, G. C. (2012). Hum. Mol. Genet. 21, 2277–2287. [DOI] [PubMed]
- Chen, Y.-F., Kao, C.-H., Chen, Y.-T., Wang, C.-H., Wu, C.-Y., Tsai, C.-Y., Liu, F.-C., Yang, C.-W., Wei, Y.-H., Hsu, M.-T., Tsai, S.-F. & Tsai, T.-F. (2009). Genes Dev. 23, 1183–1194. [DOI] [PMC free article] [PubMed]
- Chen, Y.-F., Wu, C.-Y., Kirby, R., Kao, C.-H. & Tsai, T.-F. (2010). Ann. N. Y. Acad. Sci. 1201, 58–64. [DOI] [PubMed]
- Conlan, A. R., Axelrod, H. L., Cohen, A. E., Abresch, E. C., Zuris, J., Yee, D., Nechushtai, R., Jennings, P. A. & Paddock, M. L. (2009). J. Mol. Biol. 392, 143–153. [DOI] [PMC free article] [PubMed]
- Conlan, A. R., Paddock, M. L., Homer, C., Axelrod, H. L., Cohen, A. E., Abresch, E. C., Zuris, J. A., Nechushtai, R. & Jennings, P. A. (2011). Acta Cryst. D67, 516–523. [DOI] [PMC free article] [PubMed]
- Cortes, D. F., Sha, W., Hower, V., Blekherman, G., Laubenbacher, R., Akman, S., Torti, S. V. & Shulaev, V. (2011). Free Radic. Biol. Med. 50, 1565–1574. [DOI] [PMC free article] [PubMed]
- Dutton, P. L. (1978). Methods Enzymol. 54, 411–435. [DOI] [PubMed]
- Echols, N., Grosse-Kunstleve, R. W., Afonine, P. V., Bunkóczi, G., Chen, V. B., Headd, J. J., McCoy, A. J., Moriarty, N. W., Read, R. J., Richardson, D. C., Richardson, J. S., Terwilliger, T. C. & Adams, P. D. (2012). J. Appl. Cryst. 45, 581–586. [DOI] [PMC free article] [PubMed]
- Emsley, P. & Cowtan, K. (2004). Acta Cryst. D60, 2126–2132. [DOI] [PubMed]
- Fish, A., Danieli, T., Ohad, I., Nechushtai, R. & Livnah, O. (2005). J. Mol. Biol. 350, 599–608. [DOI] [PubMed]
- Hou, X., Liu, R., Ross, S., Smart, E. J., Zhu, H. & Gong, W. (2007). J. Biol. Chem. 282, 33242–33246. [DOI] [PubMed]
- Jung, Y. S., Gao-Sheridan, H. S., Christiansen, J., Dean, D. R. & Burgess, B. K. (1999). J. Biol. Chem. 274, 32402–32410. [DOI] [PubMed]
- Lin, J., Zhang, L., Lai, S. & Ye, K. (2011). PLoS One, 6, e24790. [DOI] [PMC free article] [PubMed]
- Lin, J., Zhou, T., Ye, K. & Wang, J. (2007). Proc. Natl Acad. Sci. USA, 104, 14640–14645. [DOI] [PMC free article] [PubMed]
- McCoy, A. J., Grosse-Kunstleve, R. W., Adams, P. D., Winn, M. D., Storoni, L. C. & Read, R. J. (2007). J. Appl. Cryst. 40, 658–674. [DOI] [PMC free article] [PubMed]
- Meyer, J. (2008). J. Biol. Inorg. Chem. 13, 157–170. [DOI] [PubMed]
- Morris, R. J., Perrakis, A. & Lamzin, V. S. (2003). Methods Enzymol. 374, 229–244. [DOI] [PubMed]
- Murshudov, G. N., Skubák, P., Lebedev, A. A., Pannu, N. S., Steiner, R. A., Nicholls, R. A., Winn, M. D., Long, F. & Vagin, A. A. (2011). Acta Cryst. D67, 355–367. [DOI] [PMC free article] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol. 276, 307–326. [DOI] [PubMed]
- Paddock, M. L., Wiley, S. E., Axelrod, H. L., Cohen, A. E., Roy, M., Abresch, E. C., Capraro, D., Murphy, A. N., Nechushtai, R., Dixon, J. E. & Jennings, P. A. (2007). Proc. Natl Acad. Sci. USA, 104, 14342–14347. [DOI] [PMC free article] [PubMed]
- Potterton, E., Briggs, P., Turkenburg, M. & Dodson, E. (2003). Acta Cryst. D59, 1131–1137. [DOI] [PubMed]
- Sohn, Y.-S. et al. (2013). Proc. Natl Acad. Sci. USA, 110, 14676–14681.
- Stelzer, G. et al. (2011). Hum. Genomics, 5, 709–717. [DOI] [PMC free article] [PubMed]
- Stura, E. A. & Wilson, I. A. (1991). J. Cryst. Growth, 110, 270–282.
- Tamir, S., Rotem-Bamberger, S., Katz, C., Morcos, F., Hailey, K., Zuris, J., Wang, C., Conlan, A., Lipper, A., Paddock, M., Mittler, R., Onuchic, J., Jennings, P., Friedler, A. & Nechushtai, R. (2014). Proc. Natl Acad. Sci. USA, 111, 5177–5182. [DOI] [PMC free article] [PubMed]
- Tamir, S., Zuris, J. A., Agranat, L., Lipper, C. H., Conlan, A. R., Michaeli, D., Harir, Y., Paddock, M. L., Mittler, R., Cabantchik, Z. I., Jennings, P. A. & Nechushtai, R. (2013). PLoS One, 8, e61202. [DOI] [PMC free article] [PubMed]
- Wiley, S. E., Andreyev, A. Y., Divakaruni, A. S., Karisch, R., Perkins, G., Wall, E. A., van der Geer, P., Chen, Y.-F., Tsai, T.-F., Simon, M. I., Neel, B. G., Dixon, J. E. & Murphy, A. N. (2013). EMBO Mol. Med. 5, 904–918. [DOI] [PMC free article] [PubMed]
- Wiley, S. E., Murphy, A. N., Ross, S. A., van der Geer, P. & Dixon, J. E. (2007). Proc. Natl Acad. Sci. USA, 104, 5318–5323. [DOI] [PMC free article] [PubMed]
- Zuris, J. A., Halim, D. A., Conlan, A. R., Abresch, E. C., Nechushtai, R., Paddock, M. L. & Jennings, P. A. (2010). J. Am. Chem. Soc. 132, 13120–13122. [DOI] [PMC free article] [PubMed]
- Zuris, J. A., Harir, Y., Conlan, A. R., Shvartsman, M., Michaeli, D., Tamir, S., Paddock, M. L., Onuchic, J. N., Mittler, R., Cabantchik, Z. I., Jennings, P. A. & Nechushtai, R. (2011). Proc. Natl Acad. Sci. USA, 108, 13047–13052. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: NAF-1, wild type, 4oo7
PDB reference: H114C mutant, 4ooa