New features of an antiprotozoal DNA minor-groove binding drug, which acts as a cross-linking agent, are presented. It also fills the minor groove of DNA completely and prevents the access of proteins. These features are also expected for other minor-groove binding drugs when associated with suitable DNA targets.
Keywords: CD27, minor-groove binding drug, AT-rich DNA, d(AAAATTTT)2
Abstract
The DNA of several pathogens is very rich in AT base pairs. Typical examples include the malaria parasite Plasmodium falciparum and the causative agents of trichomoniasis and trypanosomiases. This fact has prompted studies of drugs which interact with the minor groove of DNA, some of which are used in medical practice. Previous studies have been performed almost exclusively with the AATT sequence. New features should be uncovered through the study of different DNA sequences. In this paper, the crystal structure of the complex of the DNA duplex d(AAAATTTT)2 with the dicationic drug 4,4′-bis(imidazolinylamino)diphenylamine (CD27) is presented. The drug binds to the minor groove of DNA as expected, but it shows two new features that have not previously been described: (i) the drugs protrude from the DNA and interact with neighbouring molecules, so that they may act as cross-linking agents, and (ii) the drugs completely cover the whole minor groove of DNA and displace bound water. Thus, they may prevent the access to DNA of proteins such as AT-hook proteins. These features are also expected for other minor-groove binding drugs when associated with all-AT DNA. These findings allow a better understanding of this family of compounds and will help in the development of new, more effective drugs. New data on the biological interaction of CD27 with the causative agent of trichomoniasis, Trichomonas vaginalis, are also reported.
1. Introduction
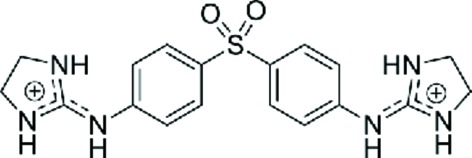
Enormous progress has been achieved in the past in the study of small-molecule ligands that have affinity for the DNA minor groove, as recently reviewed by Sheng et al. (2013 ▶). More complex types of drug binding to DNA have also been reviewed by Boer et al. (2009 ▶). Dervan and coworkers have carried out an extensive series of studies (Sheng et al., 2013 ▶; Chenoweth & Dervan, 2009 ▶) aimed at developing ligands that recognize specific DNA sequences. Some intercalating drugs also favour binding through the minor groove (Niyazi et al., 2012 ▶). The main group of studies has concentrated on the interaction of different drugs with AT-rich DNA regions, mainly with the Dickerson–Drew dodecamer d(CGCGAATTCGCG), which easily provides crystals with high resolution suitable for X-ray analysis. However, there is no evidence that AATT is the preferred sequence of interaction in vivo. In fact, little is known of the eventual DNA-sequence selectivity. We therefore decided to study the interaction of the all-AT DNA sequence d(AAAATTTT) with the dication 4,4′-bis(imidazolinylamino)diphenylamine (CD27; shown in Fig. 1 ▶).
Figure 1.
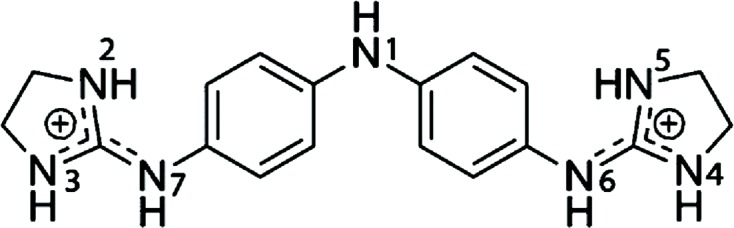
Chemical structure of CD27.
CD27 is chemically related to diamidines, a class of dicationic DNA minor-groove binders with a long history of clinical success as antiprotozoal agents (Paine et al., 2010 ▶; Soeiro et al., 2005 ▶). In the last few years, our group has discovered a set of similar related compounds (i.e. bisimidazolinium diphenyl compounds) that kill African trypanosomes, the aetiological agent of sleeping sickness, very efficiently in vitro (Dardonville & Brun, 2004 ▶). In addition, some of these compounds were also very active in vitro against the malaria parasite Plasmodium falciparum (Rodríguez et al., 2008 ▶). CD27, in particular, proved to be a potent inhibitor of Trypanosoma brucei growth in vitro (Dardonville & Brun, 2004 ▶) and in vivo. This compound was able to cure 100% of mice in the STIB900 murine model of stage 1 sleeping sickness (Dardonville et al., 2006 ▶), but was not effective in the late (CNS) stage of the illness owing to poor blood–brain barrier permeability (Nieto et al., 2011 ▶). Here, we provide additional evidence of its antiprotozoal activity by demonstrating a growth-inhibiting effect on Trichomonas vaginalis parasites, the pathogens responsible for the most common sexually transmitted infection in the world (Johnston & Mabey, 2008 ▶). T. vaginalis is a monogenetic, anaerobic, amitochondrial parasite, and as such is very different from kinetoplastid parasites such as Trypanosoma species, which are digenetic, aerobic and have functional mitochondria that perform essential functions. Despite these differences, the genomes of these parasites have in common a high content of AT base pairs. Thus, it was of interest to assess the effects of bisimidazolines on T. vaginalis and compare this with the effects on trypanosomes, apart from the inherent interest in new drug leads against this major human pathogen (Johnston & Mabey, 2008 ▶).
The DNA-binding properties of CD27 have previously been studied using different techniques such as thermal melting curves [T m = 38.5° for poly(dAdT)2; Dardonville et al., 2006 ▶)], fluorescence intercalator displacement (FID) and biosensor surface plasmon resonance (SPR; Glass et al., 2009 ▶). The crystal structure of CD27 bound to the self-complementary nucleotide d(CTTAATTCGAATTAAG)2 has previously been determined using a host–guest approach (Glass et al., 2009 ▶). The compound was found to interact with the two central AATT sequences in a similar way to that found in other drugs which interact with the Dickerson–Drew dodecamer.
In the current paper, we describe a completely different DNA interaction behaviour of CD27: the compound completely covers the minor groove of the two A-tracts of the oligonucleotide d(AAAATTTT)2. Moreover, we found that the drug may interact with a neighbouring DNA molecule. These results show the need to study the interaction of drugs with the minor groove of different AT-rich sequences. In this sense, we have recently reported striking results for the interaction of pentamidine with an alternating AT oligonucleotide (Moreno et al., 2010 ▶).
2. Materials and methods
2.1. Synthesis
The deoxyoligonucleotide d(AAAATTTT) was synthesized at the Pasteur Institute as the ammonium salt on an automatic synthesizer by the phosphoramidite method. It was purified by gel filtration and reverse-phase HPLC.
The synthesis of N 1-(4,5-dihydro-1H-imidazol-2-yl)-N 4-{4-[(4,5-dihydro-1H-imidazol-2-yl)amino]phenyl}benzene-1,4-diamine (CD27) was performed following a procedure described previously (Dardonville et al., 2000 ▶). Two equivalents of N,N′-di(tert-butoxycarbonyl)imidazolidine-2-thione were reacted with one equivalent of 4,4-diaminodiphenylamine in the presence of mercuric chloride and an excess of triethylamine in dimethylformamide to give, after purification by silica chromatography, the corresponding fully Boc-protected derivative of CD27: tetra-tert-butyl 2,2′-{[azanediylbis(4,1-phenylene)]bis(azanediyl)}bis(imidazolidine-1,3-dicarboxylate). Removal of the Boc protecting groups was carried out with an HClgas-saturated solution of dioxane at room temperature overnight. Recrystallization from methanol/diethyl ether yielded pure CD27 as the dihydrochloride salt.
2.2. Biological assays
The assays used in order to determine the effect of CD27 and related drugs on Trichomonas species are given in the Supporting Information1. In brief, compound susceptibility was tested using either the fluorescent dye resorufin as an indicator of viability (only live cells metabolize it to the nonfluorescent dihydroresorufin) or the fluorophore propidium iodide (to measure cell numbers based on the binding of propidium iodide to their DNA and RNA) (Natto et al., 2012 ▶).
2.3. Crystallization
The crystals were grown by vapour diffusion at 15°C using the hanging-drop method. We explored various divalent cations for crystallization by DLS (dynamic light scattering) and found that Mn2+ was the most appropriate. A detailed report is given in the Supporting Information. Thus, we used the following conditions: a pre-incubated DNA–CD27 complex in sodium cacodylate buffer was added to a drop with final concentrations of 0.25 mM DNA duplex, 0.75 mM CD27, 40 mM sodium cacodylate buffer pH 6.5, 8 mM MnCl2, 0.5 mM spermine and 5% MPD and equilibrated against a 30% MPD reservoir. MPD acts both as a precipitant and a cryoprotectant. After two weeks, polyhedral crystals appeared (shown in the Supporting Information).
2.4. Data collection and structure determination
The crystals were flash-cooled at −173°C. A PILATUS 6M detector on beamline BL13-XALOC at the ALBA synchrotron was used for data collection, at a wavelength of 0.979 Å, to a maximum resolution of 2 Å. A summary of crystal data and refinement statistics is given in Table 1 ▶. The data were integrated using XDS (Kabsch, 2010 ▶) and were scaled using SCALA (Evans, 2006 ▶). The space group turned out to be hexagonal, as confirmed using POINTLESS (Evans, 2006 ▶), which indicated P6122 and P6522 as possible space groups. A theoretical B-DNA model was constructed using TURBO-FRODO (http://www.afmb.univ-mrs.fr/-TURBO-), with a base-pair stacking of 3.25 Å, a uniform base-pair twist of 36° and Watson–Crick base-pair bonding. It was used as a starting search model for molecular replacement, but without success even after an exhaustive search in different hexagonal and monoclinic space groups. Since the diffraction pattern showed three orientations of stacked oligonucleotides crossed by about 60° to their neighbours, we generated a possible arrangement of the oligonucleotides in the crystal with these requirements: we built another theoretical search model formed by a column of three B-DNA duplexes which had a −26° virtual twist between terminal base pairs (Campos et al., 2006 ▶). A final solution was only obtained after using the column of three duplexes as a search model in the monoclinic space group C121 (a = 135.29, b = 78.10, c = 90.54 Å, β = 90.02°), where the asymmetric unit is formed by three columns of duplexes. In the first round of molecular replacement we could place one column using Phaser (McCoy et al., 2007 ▶). This replacement was refined with REFMAC5 (Murshudov et al., 2011 ▶). Firstly, a rigid-body refinement was performed up to 3.4 Å resolution with duplexes defined as groups. After a few cycles of maximum-likelihood isotropic restrained refinement with Watson–Crick hydrogen-bond distances restrained, the obtained model was used as a search model for molecular replacement with Phaser. Thus, it was possible to place the three columns (nine duplexes) of the full asymmetric unit in the correct position. In this model the hexagonal screw axis symmetry and its direction were clear. Thus, the correct space group is P6122 (a = b = 78.05, c = 91.66 Å). Before translating the solution to the hexagonal space group the drug was placed in the minor groove with two drugs per duplex using Coot (Emsley et al., 2010 ▶). The drug coordinates from a previous structure (PDB entry 3fsi; Glass et al., 2009 ▶) were not suitable since they contained several anomalous bonds and angles. Therefore, a stereochemical restraint dictionary was generated for CD27 with the help of the Grade web server (http://grade.globalphasing.org). The values obtained were confirmed by an ab initio calculation (at B3LYP/6-311++G*) and by comparison with related compounds in the Cambridge Crystallographic Database (http://www.ccdc.cam.ac.uk). The final model was formed by one and a half DNA duplexes and three CD27 molecules. Using this model, molecular replacement with Phaser led us to the correct placement of the final model. A stereochemical restraint dictionary was generated for CD27 with the help of the Grade web server. Several cycles of maximum-likelihood isotropic restrained refinement were performed using REFMAC5 to 2.1 Å resolution, with Watson–Crick hydrogen-bond distances restrained. Noncrystallographic symmetry (NCS) was defined between single strands of DNA, jelly body set to 0.01. The external Grade CIF dictionary was used. For the last round of refinement the NCS was turned off and the correlation factors decreased to final values of R work = 0.236 and R free = 0.251 in the resolution range 20–2.1 Å, with a completeness of 97% (a 5% set of free reflections was used as an independent cross-validation indicator of the progress of refinement). No divalent ions were detected. Solution coordinates have been deposited in the Protein Data Bank as PDB entry 4ocd. The DNA structural parameters were analyzed with the help of the 3DNA software (http://x3dna.org/). Drawings were prepared with PyMOL (http://www.pymol.org).
Table 1. Data-collection and refinement statistics.
Values in parentheses are for the highest resolution shell.
| Data collection | |
| Beamline | BL13-XALOC, ALBA |
| Wavelength (Å) | 0.97949 |
| Resolution range (Å) | 39–2.1 (2.21–2.10) |
| Space group | P6122 |
| Unit-cell parameters (Å, °) | a = b = 78.01, c = 91.55, α = β = 90, γ = 120 |
| Total reflections | 114934 (9823) |
| Unique reflections | 10091 (1433) |
| Multiplicity | 11.4 (6.9) |
| Completeness (%) | 99.8 (99.8) |
| 〈I/σ(I)〉 | 30.2 (3.0) |
| Wilson B factor (Å2) | 59.64 |
| R merge † | 0.036 (0.578) |
| Refinement | |
| R work ‡/R free § | 0.2361/0.2510 |
| No. of reflections | 9432 |
| No. of non-H atoms | 622 |
| DNA duplexes per asymmetric unit | 1.5 |
| Ligands | 3 CD27 |
| Waters | 64 |
| R.m.s.d., bond lengths (Å) | 0.0238 |
| R.m.s.d., angles (°) | 1.6525 |
| Average B factor (Å2) | 60.32 |
R
merge = 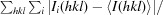
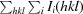 .
.
R
work and R
free were calculated as R = 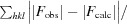
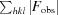 .
.
R free is the R factor evaluated for the reflections (5%) used for cross-validation during refinement.
3. Results
3.1. Effects of bisimidazolines on T. vaginalis
Compound CD27 and two closely related analogues were tested in vitro against the human pathogen T. vaginalis using two different protocols: the resorufin and the propidium iodide (PI) assays, respectively (Natto et al., 2012 ▶). In general, these compounds showed weak or no activity against T. vaginalis. However, CD27 displayed the lowest EC50 of the series, whereas its guanidine analogue CD25 was approximately twofold less active against this pathogen. The opposite results were obtained against Trypanosoma brucei rhodesiense, as shown in Table 2 ▶. Taken together, the high anti-T. brucei activity and the weak anti-trichomonal activity of these compounds are completely consistent with kinetoplastid DNA targeting (or at least a mitochondrial target) being more important than nuclear DNA. The AT-rich minicircles in kinetoplasts (Jensen & Englund, 2012 ▶) appear to be the target for drug interaction, given their unique structural features. This is in part driven by the strong accumulation of cations in the mitochondria of trypanosomes because of the mitochondrial membrane potential (Ibrahim et al., 2011 ▶).
Table 2. In vitro activity of CD27 and related analogues against T. vaginalis .
| T. vaginalis † | T. brucei rhodesiense ‡ | ||||
|---|---|---|---|---|---|
| EC50 (µM) | EC50 (µM) | ||||
| Compound | Structure | Resorufin assay | Propidium iodide assay | Alamar Blue assay | T m § (°C) |
| CD27 |
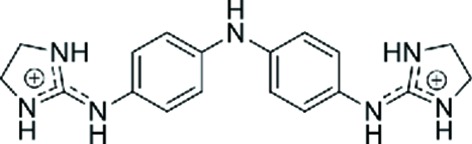
|
36.8 ± 6.9 | 25.1 ± 6.5 | 0.069 | 38.5 |
| CD25 |
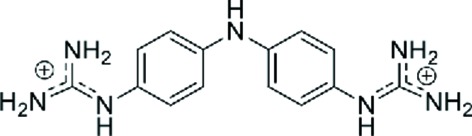
|
65.9 ± 9.7 | 48.7 ± 3.9 | 0.022 | 29.6 |
| CD29 |
|
>100 | 71.7 ± 6.1 | 32.4 | 12.1 |
| Reference drug | Metronidazole | 0.66 ± 0.20 | 0.20 ± 0.03 | — | |
Trichomonas vaginalis trophozoites of the metronidazole-susceptible strain G3.
Bloodstream-form trypomastigotes of Trypanosoma brucei rhodesiense strain STIB900. These data have previously been reported (Dardonville & Brun, 2004 ▶; Rodríguez et al., 2008 ▶) and are given here for comparative purposes.
Thermal melting for complexes with the poly(dAdT) oligonucleotide (Rodríguez et al., 2008 ▶).
3.2. Structure of the complex
The drug–DNA complex crystallized in a P6122 unit cell with an asymmetric unit which contained three drugs plus one and a half duplexes, as shown in Fig. 2 ▶(a). Views of the unit cell are given in the Supporting Information.
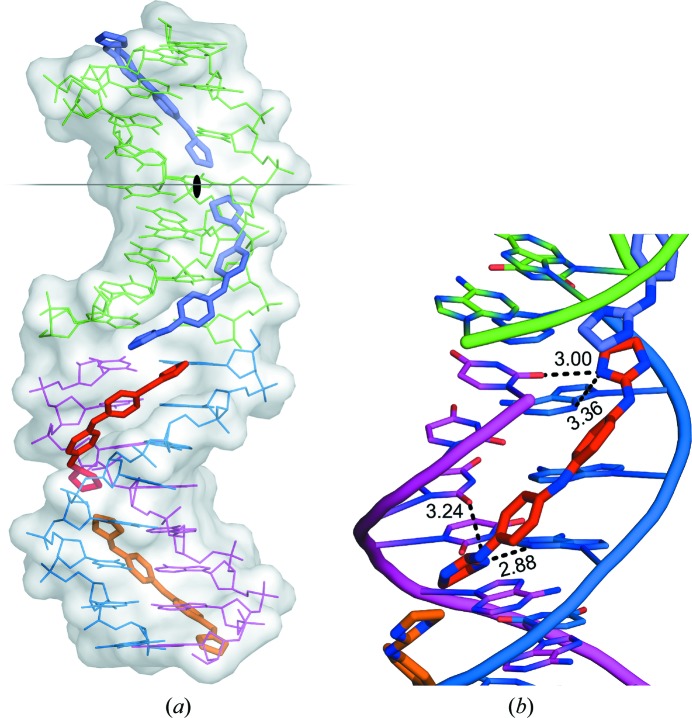
Figure 2.
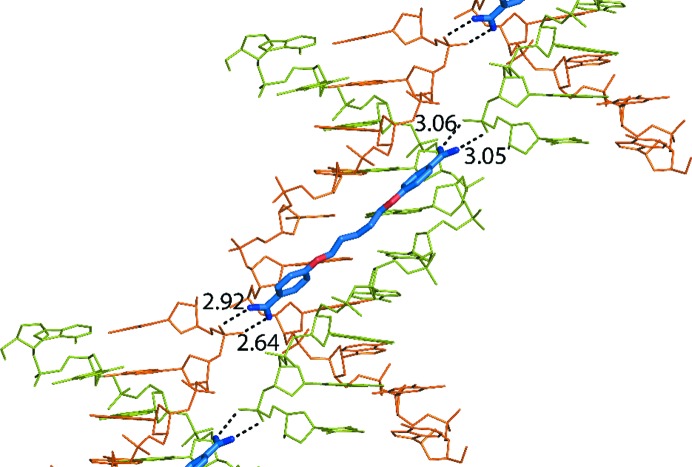
(a) View of the different crystallographic units of the complex. The black lozenge indicates one of the dyad axes. There are three independent oligonucleotide chains. One of them (green) forms a duplex with an identical symmetric chain. The other two (blue and magenta) form another DNA duplex. Three crystallographically independent drug molecules are indicated in different colours. (b) Hydrogen bonds formed by one CD27 drug with minor-groove atoms of the DNA duplex. N atoms of the drug are shown in dark blue. All three drugs show similar interactions.
The duplexes are stacked and organized as infinite continuous columns which cross in space at 60°. The drug molecules completely fill the minor groove of the DNA duplexes. No water molecules remain in the minor groove. Thus, the complex appears as a pseudo-continuous triple helix with one drug and two phosphodiester strands.
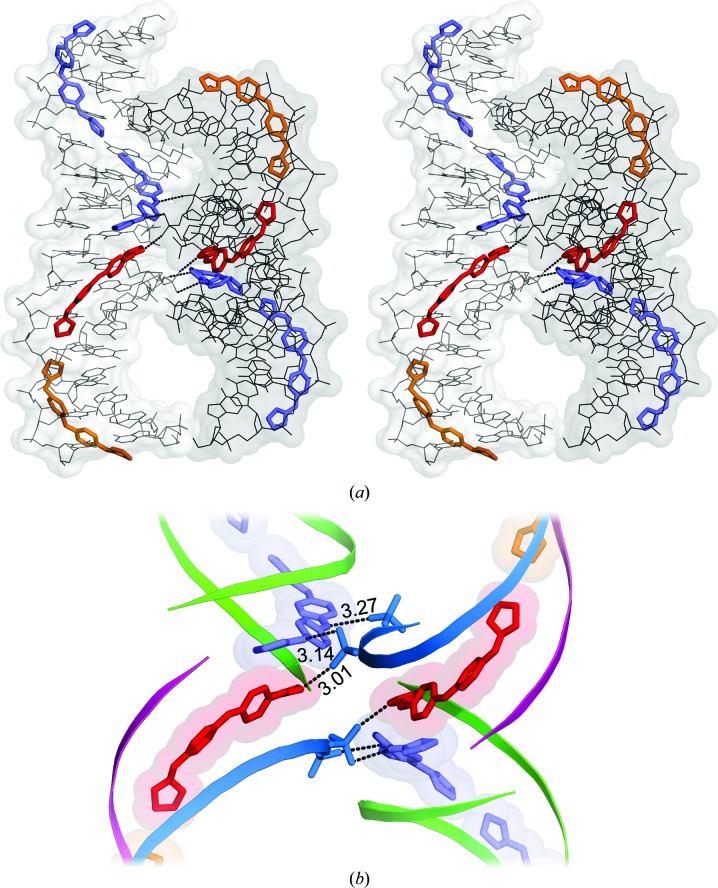
As shown in the Supporting Information, the DNA–drug columns cross in space and are surrounded by large solvent channels. Crossings are stabilized in part by the interaction of the drug molecules with neighbouring DNA phosphates. Such interactions are shown in Fig. 3 ▶. A network of associated water molecules is also present in this region (not shown) and contributes to the stability of the crossings of DNA duplexes through phosphate–water–phosphate bridges. Drug F (orange) and DNA chain C (magenta) do not participate in the interactions: they face a solvent space in the crystal.
Figure 3.
(a) Stereoview of crossed oligonucleotide duplexes, showing the interaction of two drugs with neighbouring phosphates of a symmetrical DNA chain. (b) Enlarged view of the interaction of two drugs with the neighbouring phosphates of a symmetrical DNA chain. A dyad axis runs through the centre of the figure.
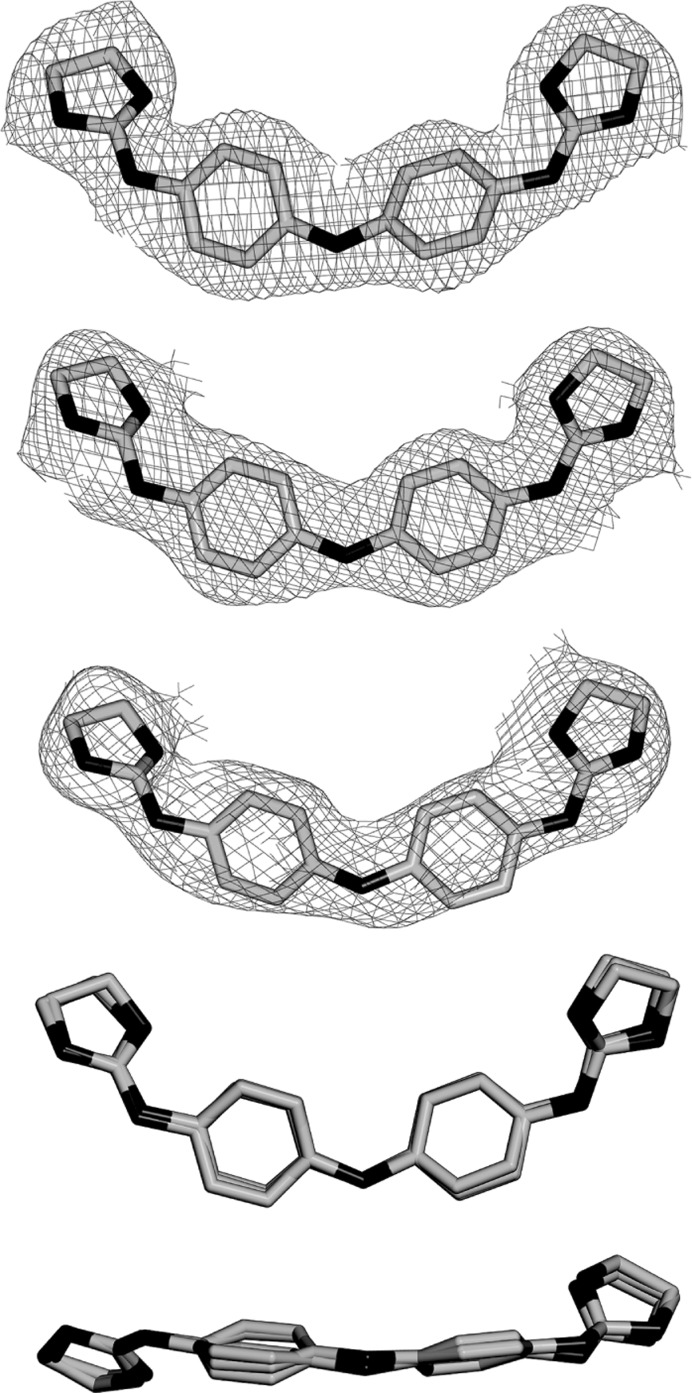
3.3. Drug conformation and interactions
The three crystallographically independent CD27 molecules have very similar conformations, as shown in Fig. 4 ▶, with maximum r.m.s. differences of 0.17 Å. Interestingly, they interact with the A-tracts and not with the central AATT sequence. In most previous studies (Sheng et al., 2013 ▶), minor-groove binding drugs were found in association with the GAATTC sequence. All three CD27 drugs form tight van der Waals interactions with the minor groove of DNA along the whole molecule. They have clearly different ends, in spite of the fact that CD27 has a symmetrical chemical structure (Fig. 1 ▶). On the one hand, the imidazoline rings placed in the centre of the duplexes show van der Waals interactions through their coplanar edges, with distances in the range 3.6–4.0 Å. On the other hand, the imidazoline rings at the other end of the molecule are placed in the terminal region of each DNA duplex: they are π-stacked with the terminal imidazoline of the CD27 neighbour. The charged terminal imidazoline groups also interact with different bases in the minor groove, as shown in Fig. 2 ▶(b) and Table 3 ▶. They form bifurcated hydrogen bonds with thymine and adenine atoms in opposite DNA strands.
Figure 4.
OMIT 2F o − F c electron-density map of the three drugs in the complex at the 1σ level. D is at the top, followed by E and F below. The bottom two frames show a superposition of the three drugs in two perpendicular views.
Table 3. Hydrogen bonds formed by CD27 in the minor groove of d(AAAATTTT)2 and external interactions with neighbouring phosphates.
All values are given in Å. A spatial representation of drugs D and E is shown in Figs. 2 ▶(b) and 3 ▶(b). The hydrogen bonds are ordered from the centre to the end of the duplex.
| Atoms involved | Drug D | Drug E | Drug F |
|---|---|---|---|
| N3(A4)–N4(CD27) | 3.06 | 2.88 | 2.77 |
| O2(T6′)–N4(CD27) | 3.08 | 3.24 | 3.26 |
| N3(A2)–N3(CD27) | 3.28 | 3.36 | 3.34 |
| O2(T8′)–N3(CD27) | 2.90 | 3.00 | 2.80 |
| N1(CD27)–OP1(A2 oligo B′) | 3.27 | — | — |
| N7(CD27)–OP2(A3 oligo B′) | 3.14 | — | — |
| N7(CD27)–OP1(A3 oligo B′) | 3.24 | — | — |
| N2(CD27)–OP1(A3 oligo B′) | — | 3.01 | — |
The central amino group of the drug always faces away from the DNA. In molecule D it interacts with a phosphate from a neighbouring DNA molecule, as shown in Fig. 3 ▶. In the case of molecule E, it is associated with a water molecule in the solvent.
As we have just shown, the three independent CD27 molecules present very similar features. However their interactions with neighbouring phosphates are different. Drug D interacts with two phosphates, whereas drug E interacts with only one. Drug F has no external interactions. As a result the latter has higher B factors and is more disordered, as can be appreciated in Fig. 4 ▶. These differences are mainly owing to their different positions in the crystal. Molecule F faces a solvent region and no external interactions are possible.
3.4. DNA structure
The two crystallographically independent duplexes in the structure are very similar, with an r.m.s. difference of 0.49 Å between the two duplexes (A–A and B–C). The duplexes show the standard features of A·T base pairs, with an average propeller twist of −13.6°. The duplexes are rather straight; the roll angles of individual steps have values of below 5°. This was confirmed using the CURVES program (Lavery et al., 2009 ▶), which shows only a slight curvature in the case of the B–C duplex. The strong bending found in solution for this sequence (Stefl et al., 2004 ▶) is absent in our crystal structure. The presence of the drug in the minor groove probably restricts the bending of the duplex.
An unexpected feature of the DNA structure is the high twist value in the AA/TT base steps, which have an average value of 38.5°. In previous structures an average of 35° was reported (Gorin et al., 1995 ▶; Subirana & Faria, 1997 ▶). The difference is probably owing to the fact that the latter average included mainly AATT sequences, which have low twist values. High values are also observed in other structures that have AAA sequences (Edwards et al., 1992 ▶; Valls et al., 2005 ▶). Thus, we can conclude that long adenine stretches will have a high value of twist, which may explain some of the anomalous features observed in A tracts.
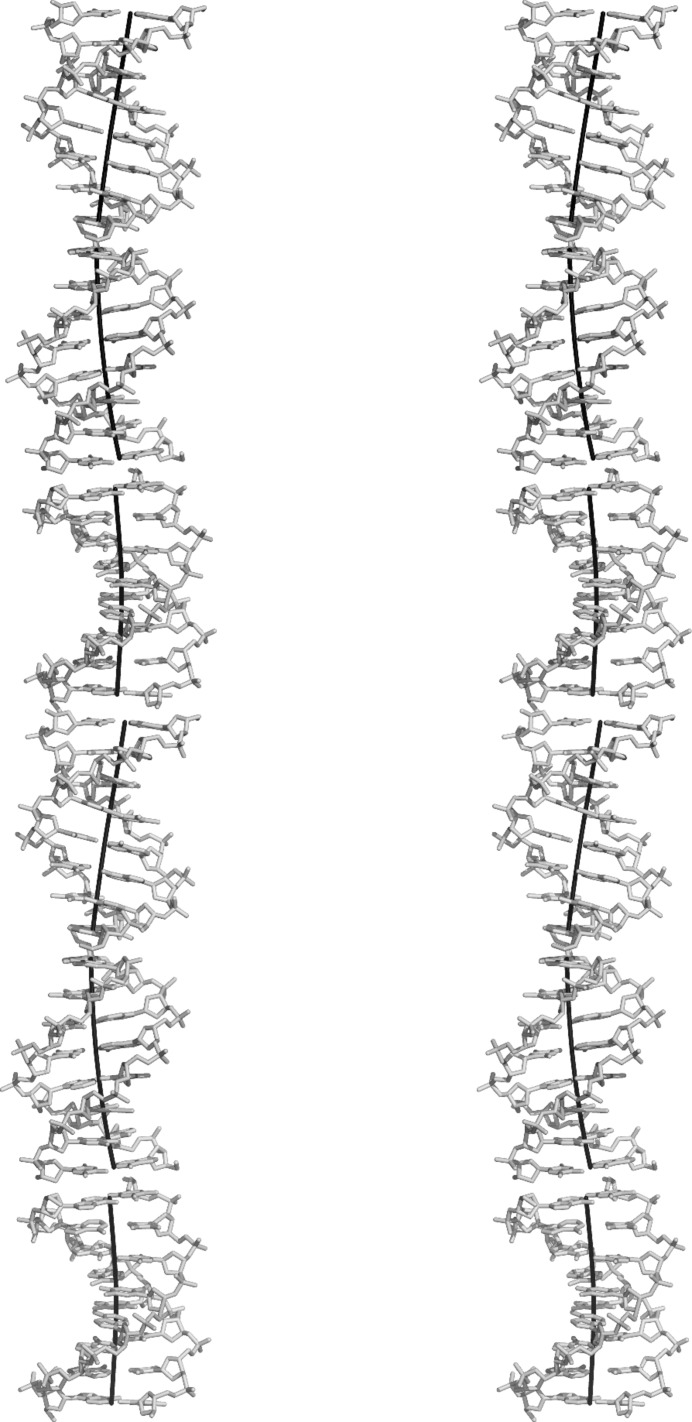
The duplexes in our structure are organized as infinite columns, as shown in Fig. 5 ▶. They are similar to those described for other all-AT octamer duplexes (Valls et al., 2005 ▶; Campos et al., 2006 ▶). Thus, the value of twist in the base step TA between terminal bases of neighbouring duplexes is negative (−26°). In our case there are three duplexes in the repeating unit, so that the average rotation angle Ω between neighbouring duplexes is 240°. In other octamers (Valls et al., 2005 ▶; Campos et al., 2006 ▶) it is smaller at close to 230°. Another feature of these columns is that the angle of the axis of each duplex with respect to the overall axis of the column is 9°. Thus, the duplexes are organized as a smooth coiled coil (De Luchi et al., 2011 ▶), as shown in Fig. 5 ▶.
Figure 5.
Stereoview of the helical organization of the duplex columns in the crystal. The axis of each individual duplex is also indicated (calculated with CURVES). The drug is not shown.
4. Discussion
In the present study, we have found that the CD27 molecule completely covers the entire minor groove of the DNA duplexes. Since the duplexes are stacked in columns, the complex appears as a continuous triple helix formed by two single strands of DNA and one strand of CD27 molecules arranged end to end (Fig. 2 ▶ a). To our knowledge, complete coverage of the minor groove has not been described previously, with the exception of the complex of the unusual duplex d(CCCCCIIIII)2 with netropsin (Chen et al., 1998 ▶).
Another unique feature of the complex is the interaction of CD27 with the phosphates of neighbouring molecules in the crystal, as shown in detail in Fig. 3 ▶. Interactions are found both in the terminal charged groups of CD27 and in its central N1 atom. Similar features have been observed (Moreno et al., 2010 ▶) in the complex formed by pentamidine and the alternating duplex d(ATATATATAT)2. A scheme of the interactions is presented in Fig. 6 ▶. Such interactions are allowed by crystal packing. In contrast, in the studies performed with the conventional Dickerson dodecamer d(CGCGAATTCGCG)2 (Sheng et al., 2013 ▶) no external interactions are found since the drug is always completely buried inside the minor groove. In this case, crystal packing also prevents interaction of the drugs with neighbouring molecules. The interactions with neighbouring phosphates that we have described are certainly a feature of this sequence, and demonstrate that minor-groove binding drugs may interact with neighbouring molecules, including other DNA duplexes. It is likely that other drugs might show similar interactions when bound to appropriate DNA sequences. The formation of cross-links may be a feature related to their biological action.
Figure 6.
Interaction of pentamidine with neighbouring molecules in a complex with DNA (Moreno et al., 2010 ▶). Hydrogen bonds between the terminal N atoms of pentamidine and phosphates are indicated.
5. Conclusions
Our studies show two new features of DNA complexes with minor-groove binding drugs: (i) the drugs completely fill the minor groove and displace water in the AT-rich minor groove of DNA and (ii) the drugs protrude from the DNA and interact with neighbouring molecules. These findings demonstrate that further studies of oligonucleotides with different sequences are required in order to fully understand the structural features of the interaction of DNA with drugs.
Supplementary Material
PDB reference: d(AAAATTTT)2–CD27, 4ocd
Biological assays, DLS and supplementary figures.. DOI: 10.1107/S139900471400697X/dz5324sup1.pdf
Acknowledgments
This work was supported through research fellowships BFU-2009-10380 and SAF-2009-10399 from MICINN, Spain and AQU-2009-SGR-1208 from the Catalan Government, grant No. S8306 from the Government of Saudi Arabia and a Doctoral Fellowship to FAR (CONACYT: 212993). The authors are also grateful to Dr Anthonius Eze for technical assistance and to J. Juanhuix and F. Gil at the ALBA Synchrotron BL13-XALOC beamline. We also thank Professor Isabel Rozas for providing the optimized structure of CD27 and Professor G. Murshudov, Drs F. Long and J. Pous for advice during refinement.
Footnotes
Supporting information has been deposited in the IUCr electronic archive (Reference: DZ5324).
References
- Boer, D. R., Canals, A. & Coll, M. (2009). Dalton Trans., pp. 399–414. [DOI] [PubMed]
- Campos, L., Valls, N., Urpí, L., Gouyette, C., Sanmartín, T., Richter, M., Alechaga, E., Santaolalla, A., Baldini, R., Creixell, M., Ciurans, R., Skokan, P., Pous, J. & Subirana, J. A. (2006). Biophys. J. 91, 892–903. [DOI] [PMC free article] [PubMed]
- Chen, X., Mitra, S. N., Rao, S. T., Sekar, K. & Sundaralingam, M. (1998). Nucleic Acids Res. 26, 5464–5471. [DOI] [PMC free article] [PubMed]
- Chenoweth, D. M. & Dervan, P. B. (2009). Proc. Natl Acad. Sci. USA, 106, 13175–13179. [DOI] [PMC free article] [PubMed]
- Dardonville, C., Barrett, M. P., Brun, R., Kaiser, M., Tanious, F. & Wilson, W. D. (2006). J. Med. Chem. 49, 3748–3752. [DOI] [PubMed]
- Dardonville, C. & Brun, R. (2004). J. Med. Chem. 47, 2296–2307. [DOI] [PubMed]
- Dardonville, C., Goya, P., Rozas, I., Alsasua, A., Martín, M. I. & Borrego, M. J. (2000). Bioorg. Med. Chem. 8, 1567–1577. [DOI] [PubMed]
- De Luchi, D., Urpí, L., Subirana, J. A. & Campos, L. (2011). Ind. Eng. Chem. Res. 50, 5218–5224.
- Edwards, K. J., Brown, D. G., Spink, N., Skelly, J. V. & Neidle, S. (1992). J. Mol. Biol. 226, 1161–1173. [DOI] [PubMed]
- Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. (2010). Acta Cryst. D66, 486–501. [DOI] [PMC free article] [PubMed]
- Evans, P. (2006). Acta Cryst. D62, 72–82. [DOI] [PubMed]
- Glass, L. S., Nguyen, B., Goodwin, K. D., Dardonville, C., Wilson, W. D., Long, E. C. & Georgiadis, M. M. (2009). Biochemistry, 48, 5943–5952. [DOI] [PMC free article] [PubMed]
- Gorin, A. A., Zhurkin, V. B. & Olson, W. K. (1995). J. Mol. Biol. 247, 34–48. [DOI] [PubMed]
- Ibrahim, H. M. S., Al-Salabi, M. I., El Sabbagh, N., Quashie, N. B., Alkhaldi, A. A. M., Escale, R., Smith, T. K., Vial, H. J. & de Koning, H. P. (2011). J. Antimicrob. Chemother. 66, 111–125. [DOI] [PMC free article] [PubMed]
- Jensen, R. E. & Englund, P. T. (2012). Annu. Rev. Microbiol. 66, 473–491. [DOI] [PubMed]
- Johnston, V. J. & Mabey, D. C. (2008). Curr. Opin. Infect. Dis. 21, 56–64. [DOI] [PubMed]
- Kabsch, W. (2010). Acta Cryst. D66, 125–132. [DOI] [PMC free article] [PubMed]
- Lavery, R., Moakher, M., Maddocks, J. H., Petkeviciute, D. & Zakrzewska, K. (2009). Nucleic Acids Res. 37, 5917–5929. [DOI] [PMC free article] [PubMed]
- McCoy, A. J., Grosse-Kunstleve, R. W., Adams, P. D., Winn, M. D., Storoni, L. C. & Read, R. J. (2007). J. Appl. Cryst. 40, 658–674. [DOI] [PMC free article] [PubMed]
- Moreno, T., Pous, J., Subirana, J. A. & Campos, J. L. (2010). Acta Cryst. D66, 251–257. [DOI] [PubMed]
- Murshudov, G. N., Skubák, P., Lebedev, A. A., Pannu, N. S., Steiner, R. A., Nicholls, R. A., Winn, M. D., Long, F. & Vagin, A. A. (2011). Acta Cryst. D67, 355–367. [DOI] [PMC free article] [PubMed]
- Natto, M. J., Savioli, F., Quashie, N. B., Dardonville, C., Rodenko, B. & de Koning, H. P. (2012). J. Antimicrob. Chemother. 67, 933–943. [DOI] [PubMed]
- Nieto, L., Mascaraque, A., Miller, F., Glacial, F., Ríos Martínez, C., Kaiser, M., Brun, R. & Dardonville, C. (2011). J. Med. Chem. 54, 485–494. [DOI] [PubMed]
- Niyazi, H., Hall, J. P., O’Sullivan, K., Winter, G., Sorensen, T., Kelly, J. M. & Cardin, C. J. (2012). Nature Chem. 4, 621–628. [DOI] [PubMed]
- Paine, M. F., Wang, M. Z., Generaux, C. N., Boykin, D. W., Wilson, W. D., De Koning, H. P., Olson, C. A., Pohlig, G., Burri, C., Brun, R., Murilla, G. A., Thuita, J. K., Barrett, M. P. & Tidwell, R. R. (2010). Curr. Opin. Investig. Drugs, 11, 876–883. [PubMed]
- Rodríguez, F., Rozas, I., Kaiser, M., Brun, R., Nguyen, B., Wilson, W. D., García, R. N. & Dardonville, C. (2008). J. Med. Chem. 51, 909–923. [DOI] [PubMed]
- Sheng, J., Gan, J. & Huang, Z. (2013). Med. Res. Rev. 33, 1119–1173. [DOI] [PMC free article] [PubMed]
- Soeiro, M. N. C., De Souza, E. M., Stephens, C. E. & Boykin, D. W. (2005). Expert Opin. Investig. Drugs, 14, 957–972. [DOI] [PubMed]
- Stefl, R., Wu, H., Ravindranathan, S., Sklenár, V. & Feigon, J. (2004). Proc. Natl Acad. Sci. USA, 101, 1177–1182. [DOI] [PMC free article] [PubMed]
- Subirana, J. A. & Faria, T. (1997). Biophys. J. 73, 333–338. [DOI] [PMC free article] [PubMed]
- Valls, N., Richter, M. & Subirana, J. A. (2005). Acta Cryst. D61, 1587–1593. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: d(AAAATTTT)2–CD27, 4ocd
Biological assays, DLS and supplementary figures.. DOI: 10.1107/S139900471400697X/dz5324sup1.pdf