The high-resolution crystal structure of a free-standing carrier protein from Acinetobacter baumannii that belongs to a larger NRPS-containing operon, encoded by the ABBFA_003406–ABBFA_003399 genes of A. baumannii strain AB307-0294, that has been implicated in A. baumannii motility, quorum sensing and biofilm formation, is presented.
Keywords: natural product biosynthesis, peptidyl carrier proteins, acyl carrier proteins, Acinetobacter baumannii, motility, biofilm formation, nonribosomal peptide synthetase
Abstract
Microorganisms produce a variety of natural products via secondary metabolic biosynthetic pathways. Two of these types of synthetic systems, the nonribosomal peptide synthetases (NRPSs) and polyketide synthases (PKSs), use large modular enzymes containing multiple catalytic domains in a single protein. These multidomain enzymes use an integrated carrier protein domain to transport the growing, covalently bound natural product to the neighboring catalytic domains for each step in the synthesis. Interestingly, some PKS and NRPS clusters contain free-standing domains that interact intermolecularly with other proteins. Being expressed outside the architecture of a multi-domain protein, these so-called type II proteins present challenges to understand the precise role they play. Additional structures of individual and multi-domain components of the NRPS enzymes will therefore provide a better understanding of the features that govern the domain interactions in these interesting enzyme systems. The high-resolution crystal structure of a free-standing carrier protein from Acinetobacter baumannii that belongs to a larger NRPS-containing operon, encoded by the ABBFA_003406–ABBFA_003399 genes of A. baumannii strain AB307-0294, that has been implicated in A. baumannii motility, quorum sensing and biofilm formation, is presented here. Comparison with the closest structural homologs of other carrier proteins identifies the requirements for a conserved glycine residue and additional important sequence and structural requirements within the regions that interact with partner proteins.
1. Introduction
Many microorganisms produce peptide natural products via novel secondary metabolic biosynthetic pathways (Gross & Loper, 2009 ▶; Li & Vederas, 2009 ▶; Meinwald, 2011 ▶). These products include the siderophore enterobactin, the biosurfactant surfactin, and antibiotics and cytostatic agents such as vancomycin and bleomycin, respectively, that have given rise to commercial therapeutics. These substances are all produced by nonribosomal peptide synthetases (NRPSs; Fischbach & Walsh, 2006 ▶). These molecular machines use a wide range of substrate amino acids to catalyze peptide synthesis independently of the ribosome.
The NRPSs use a modular catalytic strategy in which multiple protein activities that are required for the incorporation of a single amino acid into the final peptide define a single module. Typically, one module is present for each amino acid incorporated into the polypeptide. Most commonly, these multiple domains are joined in a single polypeptide, and large NRPSs that contain multiple modules and thousands of amino acids are not uncommon. During NRPS biosynthesis, the peptide intermediates are covalently attached to a peptidyl carrier protein (PCP) domain (Mercer & Burkart, 2007 ▶), which delivers the substrate to neighboring catalytic domains. The PCP domains are small ∼75-residue units that are post-translationally modified with a phosphopantetheine cofactor that binds the amino acid and peptide intermediates as a thioester (Beld et al., 2013 ▶). Unlike the more common multi-domain architecture, some NRPS systems contain catalytic and carrier domains expressed as single free-standing proteins. These so-called type II systems provide additional challenges that arise from the need for specific protein–protein interactions to govern proper biosynthesis.
A similar modular architecture is used by the polyketide synthase (PKS) machinery to incorporate malonate starter units into polyketide natural products. PKS enzymes also utilize integrated carrier proteins, described as ACPs to reflect the acyl substrates, that shuttle the polyketide intermediates between catalytic domains (Keatinge-Clay, 2012 ▶; Strieker et al., 2010 ▶). Finally, fatty-acid synthase (FAS) enzymes use ACP domains to deliver the acyl groups to catalytic domains during the iterative elongation of fatty acids.
These three types of carrier proteins have been studied functionally, and structures of carrier domains have been determined by both crystallography and NMR. These structures illustrate that the carrier proteins adopt a common fold containing four helices (Crosby & Crump, 2012 ▶; Mercer & Burkart, 2007 ▶). Helices α1, α2 and α4 are longer and are mostly parallel, while the shorter helix α3 lies nearly perpendicular to the other three. The conserved serine residue that receives the phosphopantetheine cofactor lies at the start of helix α2. The structural characterization of the holo and apo forms of the TycC3 PCP (Koglin et al., 2006 ▶) illustrates multiple states of the protein in solution that are dynamically interconverting (designated the A and H states for the unique apo and holo states, respectively, as well as a third state that is shared by both apo and holo forms of the protein and is designated the A/H state). The authors of the recent crystal structure of BlmI (Lohman et al., 2014 ▶), a type II PCP, note that carrier protein domains from X-ray crystal structures are predominantly in the A/H state and suggest that the alternate conformations observed for TycC3 may result from excising this carrier protein from the larger type I architecture.
Many human pathogens contain small NRPS clusters that are involved in the production of novel uncharacterized peptides. Acinetobacter baumannii, a Gram-negative bacterium that causes infectious outbreaks in multiple healthcare settings (Howard et al., 2012 ▶), contains a small NRPS cluster derived from eight genes. Based on the presence of two adenylation domains (within a four-domain NPRS protein and the free-standing adenylation domain), this pathway is likely to form two separate acyl adenylates and is expected to produce a dipeptide or a derivative thereof.
This operon has been implicated in bacterial motility and quorum sensing (Clemmer et al., 2011 ▶), two phenotypes that are dependent on the production of acyl-homoserine lactone signaling molecules. A random screen for mutants of the M-2 strain of A. baumannii that exhibit reduced motility identified transposon insertions into two genes within this operon (Clemmer et al., 2011 ▶). These genes were additionally noted to be upregulated in response to quorum signals. Subsequently, transcriptome analysis of strain ATCC17978 demonstrated that mRNA encoding the type II carrier protein, annotated as gene A1S_0114, was exclusively expressed in biofilms and was not detected in planktonic cells (Rumbo-Feal et al., 2013 ▶). Furthermore, the genes of this operon were overexpressed in biofilms by tenfold to 150-fold when compared with either exponential or stationary phase planktonic cell cultures. When the A1S_0114 gene was disrupted, there was an eightfold reduction in biofilm formation compared with the wild-type strain. Taken together, this evidence suggests that the carrier protein and this natural product operon are important in motility, quorum sensing and biofilm formation, phenotypes that are closely associated with bacterial virulence.
As an initial step towards understanding this uncharacterized NRPS pathway, we have subjected the core domains to structural investigation. Here, we report the structure of the free-standing carrier protein domain from this operon from A. baumannii strain AB307-0294 (Adams et al., 2008 ▶). The biological function of this carrier protein is currently unknown; however, the co-translational expression of the operon suggests that this carrier domain may contribute a substrate to the NRPS system. Within the NRPS machinery, a type II PCP is a relatively rare occurrence. Du & Shen (1999 ▶) identified and characterized BlmI from the NRPS pathway involved in the synthesis of bleomycin, a protein that has recently been structurally characterized (Lohman et al., 2014 ▶). Here, we present the crystal structure of this carrier protein and compare it with other carrier protein domains. Using features that have been described to distinguish the three types of carrier protein domains, we note that the A. baumannii carrier protein is more similar to the carrier proteins of natural product biosynthetic operons than to the ACPs of fatty-acid biosynthesis. We further characterize several conserved sequence motifs and compare the regions of the proteins that interact with biochemical partners.
2. Materials and methods
2.1. Cloning, expression and purification of A3404
For the overexpression of A34041 in Escherichia coli, we PCR-amplified the gene encoding A3404 (NCBI accession YP_002327276) from AB307-0294 genomic DNA and ligated the gene into the pET-15b-TEV expression vector (Kapust et al., 2001 ▶). This yielded a construct that produced A3404 with a pentahistidine tag at the N-terminus that was cleaved by Tobacco etch virus (TEV) protease. The a3404 gene was cloned from A. baumannii strain AB307 genomic DNA (a gift from Dr Thomas Russo, University at Buffalo) by PCR. The primers used were 5′-ATT TTC AGG GCC ATA TGA ATA AAG ATA AAG CTT ACT GGA G-3′ and 5′-GTT AGC AGC CGG ATC CTC CTC ATG AAG CAA CTC CCT GC-3′. The 261-nucleotide gene was cloned using NdeI and BamHI restriction sites (bold) into a modified pET-15b plasmid that contained a TEV protease site, and the sequence was confirmed by DNA sequencing. The resultant plasmid was used for expression in E. coli BL21(DE3) cells. Following inoculation with a small-scale overnight culture, a 1 l culture of cells was grown to an OD600 of ∼0.6 at 37°C and induced with 0.5 mM IPTG for 3 h. The cells were then pelleted by centrifugation and were either used immediately for protein purification or were flash-frozen in liquid nitrogen and stored at −80°C.
Cells were lysed by sonication in a buffer consisting of 50 mM HEPES (pH 7.5 at 4°C), 150 mM NaCl, 10 mM imidazole. The protein was purified by nickel ion-immobilized metal-affinity chromatography (IMAC). Following protein adsorption, the column was washed with 40 mM imidazole followed by elution of tagged A3404 with lysis buffer containing 300 mM imidazole. The purified protein was dialyzed overnight at 4°C against 1 l cleavage buffer consisting of 50 mM HEPES (pH 8.0 at 4°C), 150 mM NaCl, 0.5 mM EDTA with TEV protease included in the dialysis bag with the His5-tagged protein and allowed to react overnight at 4°C during the dialysis step. The cleaved protein was passed over the same IMAC column and the untagged protein was collected in the flowthrough. The final protein, containing an N-terminal Gly-His sequence remaining after TEV cleavage, was dialyzed against 10 mM HEPES (pH 7.5 at 4°C), 50 mM NaCl. From 1 l of cells, ∼10 mg protein was obtained. The protein stock was frozen by pipetting directly into liquid nitrogen for storage at −80°C (Deng et al., 2004 ▶).
2.2. Crystallization of A3404 and structure determination
Crystallization of apo A3404 was achieved via an initial screen with sparse-matrix conditions that utilized a broad array of PEG-based and salt-based precipitants (Carter & Carter, 1979 ▶; Jancarik & Kim, 1991 ▶). The final crystals of A3404 were grown at 4°C by hanging-drop vapor diffusion with a precipitant consisting of 25%(v/v) PEG 400, 5%(v/v) MPD, 0.2 mM TCEP, 50 mM CHES pH 9.0. Despite numerous attempts, we could not obtain crystals of the holo protein. Two sets of diffraction data were collected, with the first being collected at 100 K using a Rigaku MicroMax-007 microfocus X-ray generator, Osmic Max-Flux confocal focusing mirrors and a Saturn 944+ CCD detector. A higher resolution data set was subsequently collected using a wavelength of 0.9795 Å on SSRL beamline 9-2 equipped with a Si(111) double-crystal monochromator and a 325 mm MAR Mosaic CCD detector. Diffraction images were processed and scaled with the HKL-2000 suite (Otwinowski & Minor, 1997 ▶) and were converted to structure factors with TRUNCATE from the CCP4 software suite (Winn et al., 2011 ▶).
The A3404 structure was initially determined by molecular replacement by using the first model (converted to polyalanine) of the Asl1650 protein ensemble (Johnson et al., 2006 ▶; PDB entry 2afd) as a search model against a data set collected on the home source. EPMR (Kissinger et al., 2001 ▶) was used to identify the locations of the two molecules in the asymmetric unit. More than 20 models of ACP and PCP proteins were probed using multiple molecular-replacement programs before the successful search model was found using a polyalanine chain derived from model 1 of PDB entry 2afd, an NMR structure with 29% identical (45% similar) residues to A3404. Following data collection to higher resolution, the low-resolution model was used with Phaser as utilized by the PHENIX (Adams et al., 2010 ▶) molecular-replacement GUI. Refinement of the initial solution with PHENIX resulted in a model with an R cryst of 14.9% (R free of 16.8%).
The final model contains 1294 protein atoms and 311 solvent molecules. Additionally, there were two molecules of MPD bound near the pantetheine-binding motif in both subunits, a single molecule of ethylene glycol and a partial molecule of polyethylene glycol. Statistics for the data collection and refinement are presented in Table 1 ▶. The structure-based sequence alignment was generated with the DALI server (Holm & Rosenström, 2010 ▶); the structures were aligned with CHIMERA (Pettersen et al., 2004 ▶).
Table 1. Crystallographic and refinement data.
Values in parentheses are for the highest resolution shell. Because of the high noncrystallographic symmetry, the R free reflections were generated in thin shells. The high-resolution R free value is reported for data from 1.32 to 1.30 resolution.
| Data collection | |
| Source | BL9-2, SSRL |
| Resolution () | 31.221.30 |
| Space group | P65 |
| Unit-cell parameters (, ) | a = b = 61.81, c = 76.85, = = 90, = 120 |
| R merge (%) | 6.0 (52.6) |
| Completeness (%) | 99.1 (97.2) |
| I/(I) | 19.3 (5.7) |
| Multiplicity | 10.3 (8.0) |
| Total reflections | 415971 |
| Unique reflections | 40546 |
| Refinement | |
| R cryst (%) | 14.9 (16.1) |
| R free (%) | 16.8 (16.6) |
| Wilson B factor (2) | 11.90 |
| Average B factors (2) | |
| Overall | 16.7 |
| Macromolecules | 11.7 |
| Solvent | 27.0 |
| R.m.s.d., bond lengths () | 0.005 |
| R.m.s.d., angles () | 1.00 |
| No. of atoms | |
| Total | 1624 |
| Macromolecules | 1294 |
| Ligands | 16 |
| Water | 311 |
| Ramachandran favored (%) | 98 |
| Ramachandran outliers (%) | 0 |
| Clashscore† | 2.24 |
| PDB code | 4hkg |
The MolProbity clashscore placed this structure in the 99th percentile.
3. Results and discussion
3.1. Sequence analysis of the A. baumannii NRPS cluster
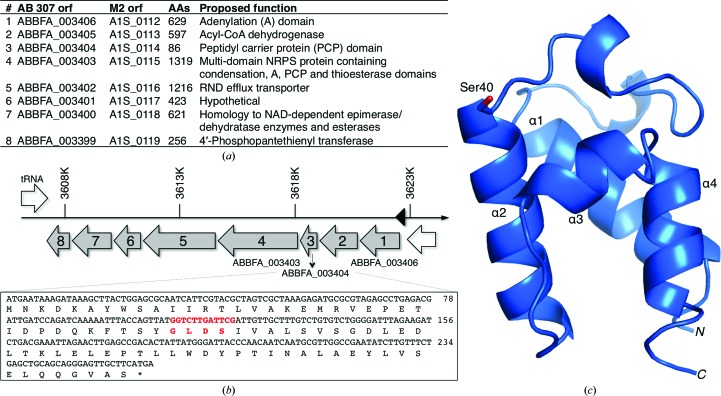
The novel synthetic pathway encoded by A. baumannii under investigation in this work is approximately 15 kb of DNA in length and contains eight open reading frames (Fig. 1 ▶). This cluster has been identified in all available genomic sequences of A. baumannii (Adams et al., 2008 ▶; Smith et al., 2007 ▶; Vallenet et al., 2008 ▶) and is not present in the nonpathogenic SDF strain or the related species A. baylyi. This predicted operon lies downstream of a transcriptional regulatory protein, ABBFA_003407, with homology to the PhzR and LuxR regulators as well as the acyl-homoserine lactone synthase CepI at ABBFA_003409.
Figure 1.
(a) The NRPS cluster from A. baumannii (with the gene nomenclature from both the AB307-0294 and ATCC17978/M2 strains). Protein sizes and proposed functions are included. (b) The genes are organized in a polycistronic operon containing eight genes (grey) preceded by a transcriptional regulatory protein (white). The sequences of the a3404 gene and protein are shown, with the carrier protein phosphopantetheinylation motif in red. (c) A ribbon diagram of A3404 highlights the four primary helices, α1–α4, and the long turn between the first two helices that contains two single-turn 310-helices. Ser40, the site of phosphopantetheinylation, is shown in a stick representation.
The operon has all of the hallmarks of a pathway that produces a novel natural product. Gene a3399 encodes a phosphopantetheinyl transferase, or holo ACP synthase, that converts the carrier proteins from the apo to the holo state (Beld et al., 2013 ▶). The operon encodes three NRPS proteins, an adenylation domain at a3406, a free-standing (type II) carrier protein at a3404 and a four-domain NRPS at a3403, which is composed of condensation, adenylation, carrier protein and thioesterase domains. A3404 is a free-standing carrier protein that could deliver a substrate to the NRPS system. Also encoded by the operon are two proteins, at a3400 and a3405, that show homology to NAD-dependent enzymes, as well as a hypothetical protein at a3401 that has no homology to characterized proteins but could function upstream or downstream to either generate alternate substrates or to modify the released product. Finally, the operon also encodes A3402, an efflux transporter that would be available to transport the final product outside of the cell.
The a3404 gene encodes a protein of 88 amino acids with a calculated pI of 4.22 and a molecular weight of 9643 Da. Similar to the carrier protein domains of NRPSs and the ACP domains of PKSs and FASs, A3404 has the highly conserved 4′-phosphopantetheinylation signature motif of GLDS. We examined this motif in the sequences of the more than 40 000 members of the Pfam family of carrier proteins, designated as PF00550, phosphopantetheine-attachment site. In this motif, the initial glycine three residues in front of the modified serine is observed in 91% of the family members. Two residues in front of the serine is a glycine (40%), leucine (19%), alanine (14%) or another hydrophobic residue such as valine or isoleucine (24% in total). The residue immediately in front of the serine is most commonly an aspartic acid (67%), histidine (17%) or asparagine (6%). This motif is thus best described as G-(G/L/A)-(D/H/N)-S.
3.2. Structure determination of A3404
The single-domain A3404 protein structure was initially determined using a data set collected on a home-source X-ray generator by molecular replacement with PDB entry 2afd as a search model. Following higher resolution data collection, the structure in progress was used as a model for molecular replacement. The asymmetric unit contains two A3404 monomers. The final model of A3404 contains 81 residues for each chain; both monomers are missing Gly−1 and His0, remnants from the N-terminal purification tag that remain after TEV cleavage, and Met1, as well as five C-terminal residues (Gln82–Ser86). Crystallographic and refinement data statistics are shown in Table 1 ▶.
The domain structure of A3404 is the archetypal carrier protein consisting of four α-helices. The conserved serine that is the site of the 4′-phosphopantetheinylation modification, Ser40, is positioned at the N-terminal end of the second helix (Fig. 1 ▶). Multiple carrier protein structures from NRPS clusters and ACPs from PKS and FAS systems have been determined by X-ray crystallography and NMR (Crosby & Crump, 2012 ▶; Mercer & Burkart, 2007 ▶). Although there is significant structural variation among the previous structures, nearly all structures retain the four main helices. An extended loop joins helices α1 and α2. In the case of A3404, this loop contains two single-turn 310-helices. While helices α1, α2 and α4 are of similar lengths and are roughly parallel, helix α3 is shorter and is nearly perpendicular to helices α2 and α4. Using the DALI alignment server (Holm & Rosenström, 2010 ▶), the 14 closest structural homologs to A3404 were determined (all resulting in Z-scores greater than 10; Table 2 ▶). Within these structures, the sequence similarity ranged from 13% for an inhibitor-bound adenylation-PCP domain module from Pseudomonas aeruginosa (Mitchell et al., 2012 ▶) to 32% for the 2afd structure (Johnson et al., 2006 ▶) that was used as a model for molecular replacement. This sampling of structures, which included a wide array of representative carrier proteins, all had less than 2.5 Å root-mean-square displacement of Cα positions. Interestingly, the list of the proteins that are the closest homologs contains equal numbers of structures determined by X-ray crystallography and by NMR spectroscopy.
Table 2. Top unique homologs of A3404 from the DALI server (Z score > 10).
| Molecule | PDB code | Type | Z † | R.m.s.d. () | Res.‡ | Align.§ | Identity (%) | Method | Reference |
|---|---|---|---|---|---|---|---|---|---|
| CurA ACPI | 2liu | PKS ACP | 14.0 | 1.0 | 99 | 80 | 30 | NMR | Busche et al. (2011 ▶) |
| BlmI | 4i4d | Type II NRPS PCP | 13.9 | 1.6 | 83 | 78 | 19 | X-ray | Lohman et al. (2014 ▶) |
| Protein ASL1650 | 2afd | PKS/NRPS carrier protein | 13.0 | 1.4 | 88 | 79 | 32 | NMR | Johnson et al. (2006 ▶) |
| Erythronolide synthase | 2ju2 | PKS ACP | 12.0 | 1.6 | 95 | 79 | 29 | NMR | Alekseyev et al. (2007 ▶) |
| EntF | 3tej | NRPS PCP | 11.5 | 2.0 | 320 | 72 | 19 | X-ray | Liu et al. (2011 ▶) |
| TtACP | 1x3o | Thermus thermophilus ACP | 11.2 | 1.8 | 78 | 80 | 23 | X-ray | RIKEN Structural Genomics/Proteomics Initiative (unpublished work) |
| Tyrocidine synthetase 3 | 2jgp | Type II NRPS PCP | 11.0 | 1.9 | 520 | 74 | 19 | X-ray | Samel et al. (2007 ▶) |
| SaACP | 4dxe | Staphylococcus aureus ACP | 10.7 | 1.8 | 75 | 77 | 16 | X-ray | Center for Structural Genomics of Infectious Diseases (unpublished work) |
| Mupircin ACP | 2l22 | Tandem PKS ACP | 10.6 | 1.8 | 76 | 183 | 22 | NMR | Haines et al. (2013 ▶) |
| ScACP | 2koq | Streptomyces coelicolor ACP | 10.4 | 2.1 | 79 | 81 | 14 | NMR | Posko et al. (2010 ▶) |
| RcACP | 2xz1 | Rice ACP | 10.4 | 2.2 | 76 | 82 | 18 | X-ray | Guy et al. (2011 ▶) |
| PfACP | 3gzm | Plasmodium falciparum ACP | 10.2 | 2.0 | 77 | 81 | 18 | X-ray | Gallagher Prigge (2010 ▶) |
| RpACP | 2kw2 | Rhodopseudomonas palustris ACP | 10.1 | 1.9 | 74 | 101 | 19 | NMR | Ramelot et al. (2012 ▶) |
| SoACP | 2fve | Spinach ACP | 10.0 | 2.1 | 77 | 82 | 18 | NMR | Zornetzer et al. (2006 ▶) |
The Z-score is a pairwise comparison score to allow ranking of the results.
The number of total residues in a given structure.
The number of residues that were aligned with the query sequence (A3404).
3.3. Comparison of the structure of A3404 to other carrier proteins
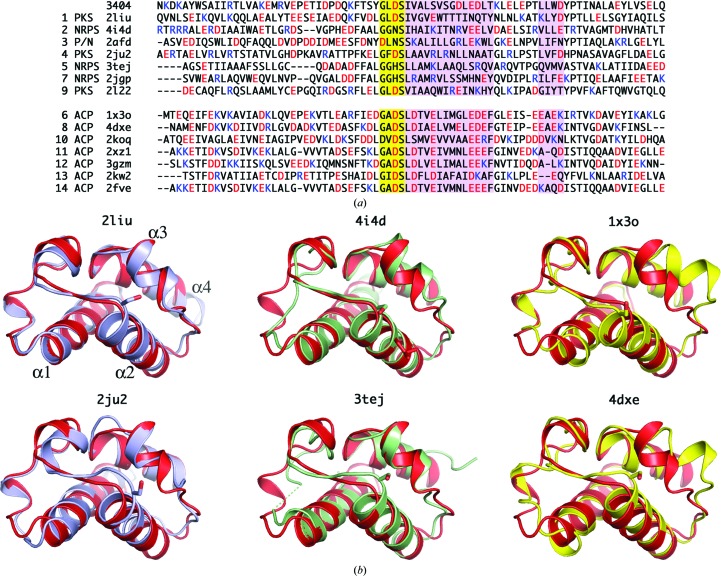
The classification of carrier proteins into one of the three classes based on sequence or structure alone can be difficult and, indeed, genomic context is another important tool that should be considered. The co-expression of A3404 with the adenylation domain of A3406 and the four-domain NRPS protein at A3403 suggests that this protein will serve in a natural product pathway. We therefore examined the 14 protein structures that were most closely related as predicted by the DALI server more closely. Interestingly, the 14 proteins contain seven ACPs from fatty-acid synthesis and transport and seven proteins from natural product (NRPS or PKS) pathways. The top five proteins as scored by DALI, and six of the top seven, are all from natural product systems. One protein, a carrier protein from Anabaena, is of unclear function; however, the authors considered it to be a carrier protein for either an NRPS or PKS (or hybrid) cluster (Johnson et al., 2006 ▶).
The regions of the carrier protein that are most important for distinguishing among the different types are the loop between helix α1 and α2, the α2 helix itself and the α3 helix (Crosby & Crump, 2012 ▶; Lai et al., 2006 ▶; Lohman et al., 2014 ▶; Mercer & Burkart, 2007 ▶). Not surprisingly, these are the regions of the proteins that interact with partner proteins, largely owing to the proximity to the site of loading at the start of the α2 helix. We examined the multiple sequence alignment generated from DALI and additionally examined the structures of each protein compared with A3404 (Fig. 2 ▶). This limited alignment of closely related structures provides some insight into the comparison between the three types of carrier protein. Firstly, we examined the sequence of the pantetheine-binding motif. Of interest, all ACPs, whether from FAS or PKS systems, contained an aspartic acid immediately preceding the serine residue. This trend is consistent with larger alignments presented by others (Crosby & Crump, 2012 ▶; Lohman et al., 2014 ▶); however, exceptions are clearly present. The acidic nature of the hydrophilic face of helix α2 is also quite striking. All FAS ACPs except for PDB entry 2koq contain acidic residues at the second and fifth residues following the pantetheinylated serine and are much more highly acidic at the C-terminal end of the helix. Similarly, the amino acids that immediately precede helix α3 also are much more highly acidic in the FAS ACP sequences. These two features are also similar to the observations used to characterize the Asl1650 carrier protein (Johnson et al., 2006 ▶).
Figure 2.
The 14 structures of closest homologs as identified by the DALI server were compared with A3404. (a) Sequence alignment of the homologous proteins. The first three columns represent the rank in the DALI scoring, the type of protein and the PDB code. Proteins in the top half of the alignment are from NPRS or PKS clusters, while proteins in the bottom half are ACPs from fatty-acid synthesis and transport. The pantetheinylation motif is highlighted in yellow; helices α2 and α3 are shaded in pink. In the alignment, acidic amino acids are red, while basic residues are blue. (b) A ribbon diagram of A3404 (red) is superimposed on the top two closest homologs of each of the three carrier protein types. The same orientation is used in all panels and the helix designations are shown in the top left panel. The two PKS acyl carrier proteins 2liu and 2ju2 are shown in light blue. Two NRPS PCP domains, the type II 4i4d and the type I 3tej, are shown in green. Two acyl carrier proteins (1x3o and 4dxe) are shown in yellow. In all structures, the serine residue at the start of helix α2 is shown in a stick representation.
The structure of A3404 was compared with all 14 of the most closely related ACP structures, and structural alignments for the two most similar structures of each class are shown in Fig. 2 ▶(b). The structure of the ACP from the curamicin PKS (PDB entry 2liu) illustrates the best alignment with A3404, which is also reflected by the lowest r.m.s. displacement of Cα positions (Table 2 ▶). In particular, the path traced by the main chain in the divergent loop between helices α1 and α2 and the α3 helix are very similar. The positional conservation with a second PKS ACP is also quite good, although differences in the position of the second 310-helix of the loop that precedes helix α2 are more pronounced. The comparison with the two NRPS PCP structures, BlmI and EntF, show comparable overall similarities. A noteworthy difference is the lack of the first 310-helix in the two PCP structures. Finally, comparison of the A3404 structure with the FAS ACP structures shows larger differences in the loop between helices α1 and α2, and, most strikingly, the orientation of the α3 helix. This potentially reflects the predominantly acidic nature of the loop immediately before this helix.
Fig. 2 ▶(b) presents the structures of carrier protein domains from the perspective of the partner protein. The right half of the molecules represents helices α2 and α3 and the loop that joins them. What is striking from the sequence alignment is the number of negatively charged residues in this region of the ACPs of FAS systems. Of the seven sequences shown, there are an average of more than seven aspartic or glutamic acid residues within this 25-residue stretch. In contrast, the carrier proteins from PKS or NRPS systems show only an average of less than two anionic residues. The A3404 protein has six glutamic acid residues. It seems, however, that this does not imply that A3404 is an ACP from fatty acid metabolism. Rather, this appears to be a function of the type II nature of this protein. BlmI, the recently characterized type II PCP (Lohman et al., 2014 ▶), has five acidic residues, as do SgcC2 and MdpC2, two additional type II PCPs from hybrid NRPS/PKS systems (Van Lanen et al., 2006 ▶, 2007 ▶). It is possible, then, that the highly acidic nature of this region of the protein does not reflect the specific function of the protein as an ACP or PCP but is rather a requirement of the type II carrier proteins. These three proteins, along with A3404, contain the cluster of negatively charged residues at the C-terminal end of helix α2; however, none of them contains the anionic residues at the N-terminal portion of this helix.
The first glycine of the pantetheine attachment motif is the most highly conserved residue of the PF00550 family, not including the serine that serves as the necessary pantetheine attachment site. The ϕ and ψ angles of this residue in A3404 are 94.7 and 2.6°, respectively, in chain A, and 97.8 and 0.7°, respectively, in chain B. These angles place this residue in a region of the Ramachandran plot that is not allowed for all side-chain-bearing residues. These angles are highly conserved in the closest homologous carrier proteins, including BlmI (85.4 and 12.3°), CurA (142.6 and 5.5°) and the DEBS synthase (112.0 and 56°). This glycine residue is positioned at the end of the 310-helix and allows the chain to adopt a uniform path to the start of the α2 helix. In the 12 structures in Table 2 ▶ that contain a glycine at this position, the ϕ angles range from 63 to 142° and the ψ angles range from −6 to 88°. It appears that this is a structurally conserved configuration that is consistent in a wide variety of carrier protein structures either in isolation or interacting with catalytic domains. The two proteins structures that do not contain a glycine here show main-chain torsion angles ϕ, ψ of −50, −26° (PDB entry 2afd, residue Asp44) and 74, 85° (PDB entry 2koq, residue Asp37). This highly strained position in the Streptomyces coelicolor ACP structure lies just outside the allowable region of the Ramachandran plot for a nonglycine residue.
4. Conclusion
Our laboratory is interested in the production of novel natural products, and in particular we have focused our attention on the NRPS enzymes that are responsible for the production of peptide siderophores. Using the enterobactin and pyoverdine systems of E. coli and P. aeruginosa, respectively, we have determined the structures of several NRPS domains and associated enzymes. To expand these efforts, we have begun to pursue a novel cluster from the human pathogen A. baumannii. The A3404 protein is part of an operon derived from the ABBFA_003406–ABBFA_003399 genes of A. baumannii strain AB307-0294 (Adams et al., 2008 ▶). Recent studies have demonstrated that genetic disruptions of this operon result in reductions in bacterial motility (Clemmer et al., 2011 ▶) and biofilm formation (Rumbo-Feal et al., 2013 ▶). This report represents our initial structural characterization of a protein within this pathway. Structural, biochemical and biological experiments are under way to isolate and identify the product of this novel pathway.
The current study presents the structure of a novel carrier protein that is encoded within this biosynthetic operon. Here, we have presented the three-dimensional structure of this protein and compared the sequence and structural features with those of related carrier protein domains from both primary (fatty-acid biosynthesis) and secondary (polyketide and nonribosomal peptide) pathways. The proteins of these processes share many structural features, owing to their shared function, namely the delivery of covalently attached substrates to a variety of interacting catalytic domains. The expression of type II carrier proteins as isolated proteins may pose different demands on their sequence and structure. For example, the free-standing proteins need to bind to their partners intermolecularly in the crowded cellular environment and may therefore require a higher affinity for their partners. Additionally, the solubility requirements for a small isolated protein may result in different global properties than for a domain that is integrated into a larger type I system.
From the structure of A3404, we have identified the features that it shares with carrier proteins of other natural product (NRPS and PKS) systems. The low pI of carrier proteins has been noted previously (Crosby & Crump, 2012 ▶; Mercer & Burkart, 2007 ▶); however, we have identified potential regions that may be required by free-standing carrier proteins from these different systems. Understanding the structural features of carrier proteins and the interfaces that they form with partner catalytic domains is a valuable step toward characterizing the potential interactions between different proteins of the NRPS and PKS pathways. Similarly, the modular nature of NRPS clusters has raised the potential for engineering these pathways to produce novel peptide products. Clearly, an improved understanding of the key elements that allow functional interactions between the carrier and catalytic domains is necessary for these efforts to succeed.
Supplementary Material
PDB reference: NRPS PCP domain, 4hkg
Acknowledgments
This work was funded in part by NIH Grant GM-068440 and a grant awarded from and administered by the Telemedicine and Advanced Technology Research Center (TATRC) of the US Army Medical Research and Materiel Command (USAMRMC), Award No. W81XWH-11-2-0218. Diffraction data were collected at the Stanford Synchrotron Radiation Laboratory, a national user facility operated by Stanford University on behalf of the US Department of Energy, Office of Basic Energy Sciences. The SSRL Structural Molecular Biology Program is supported by the Department of Energy, Office of Biological and Environmental Research, and by the National Institutes of Health National Center for Research Resources, Biomedical Technology Program and the National Institute of General Medical Sciences. We also thank Anyango Kamina for assistance with protein production and crystallization.
Footnotes
The genes from strain AB307-0294 are annotated ABBFA_00####. For simplification, we will describe the genes as a####; the encoded proteins will be designated A####. Within the ATCC17978 and M-2 strains used in the genetic studies described in §1, the genes are annotated A1S_####. Both naming conventions are included in Fig. 1 ▶.
References
- Adams, M. D., Goglin, K., Molyneaux, N., Hujer, K. M., Lavender, H., Jamison, J. J., MacDonald, I. J., Martin, K. M., Russo, T., Campagnari, A. A., Hujer, A. M., Bonomo, R. A. & Gill, S. R. (2008). J. Bacteriol. 190, 8053–8064. [DOI] [PMC free article] [PubMed]
- Adams, P. D. et al. (2010). Acta Cryst. D66, 213–221.
- Alekseyev, V. Y., Liu, C. W., Cane, D. E., Puglisi, J. D. & Khosla, C. (2007). Protein Sci. 16, 2093–2107. [DOI] [PMC free article] [PubMed]
- Beld, J., Sonnenschein, E. C., Vickery, C. R., Noel, J. P. & Burkart, M. D. (2014). Nat. Prod. Rep. 31, 61–108. [DOI] [PMC free article] [PubMed]
- Busche, A., Gottstein, D., Hein, C., Ripin, N., Pader, I., Tufar, P., Eisman, E. B., Gu, L., Walsh, C. T., Sherman, D. H., Löhr, F., Güntert, P. & Dötsch, V. (2011). ACS Chem. Biol. 7, 378–386. [DOI] [PMC free article] [PubMed]
- Carter, C. W. Jr & Carter, C. W. (1979). J. Biol. Chem. 254, 12219–12223. [PubMed]
- Clemmer, K. M., Bonomo, R. A. & Rather, P. N. (2011). Microbiology, 157, 2534–2544. [DOI] [PMC free article] [PubMed]
- Crosby, J. & Crump, M. P. (2012). Nat. Prod. Rep. 29, 1111–1137. [DOI] [PubMed]
- Deng, J., Davies, D. R., Wisedchaisri, G., Wu, M., Hol, W. G. J. & Mehlin, C. (2004). Acta Cryst. D60, 203–204. [DOI] [PubMed]
- Du, L. & Shen, B. (1999). Chem. Biol. 6, 507–517. [DOI] [PubMed]
- Fischbach, M. A. & Walsh, C. T. (2006). Chem. Rev. 106, 3468–3496. [DOI] [PubMed]
- Gallagher, J. R. & Prigge, S. T. (2010). Proteins, 78, 575–588. [DOI] [PMC free article] [PubMed]
- Gross, H. & Loper, J. E. (2009). Nat. Prod. Rep. 26, 1408–1446. [DOI] [PubMed]
- Guy, J. E., Whittle, E., Moche, M., Lengqvist, J., Lindqvist, Y. & Shanklin, J. (2011). Proc. Natl Acad. Sci. USA, 108, 16594–16599. [DOI] [PMC free article] [PubMed]
- Haines, A. S. et al. (2013). Nature Chem. Biol. 9, 685–692. [DOI] [PMC free article] [PubMed]
- Holm, L. & Rosenström, P. (2010). Nucleic Acids Res. 38, W545–W549. [DOI] [PMC free article] [PubMed]
- Howard, A., O’Donoghue, M., Feeney, A. & Sleator, R. D. (2012). Virulence, 3, 243–250. [DOI] [PMC free article] [PubMed]
- Jancarik, J. & Kim, S.-H. (1991). J. Appl. Cryst. 24, 409–411.
- Johnson, M. A., Peti, W., Herrmann, T., Wilson, I. A. & Wüthrich, K. (2006). Protein Sci. 15, 1030–1041. [DOI] [PMC free article] [PubMed]
- Kapust, R. B., Tözsér, J., Fox, J. D., Anderson, D. E., Cherry, S., Copeland, T. D. & Waugh, D. S. (2001). Protein Eng. 14, 993–1000. [DOI] [PubMed]
- Keatinge-Clay, A. T. (2012). Nat. Prod. Rep. 29, 1050–1073. [DOI] [PubMed]
- Kissinger, C. R., Gehlhaar, D. K., Smith, B. A. & Bouzida, D. (2001). Acta Cryst. D57, 1474–1479. [DOI] [PubMed]
- Koglin, A., Mofid, M. R., Löhr, F., Schäfer, B., Rogov, V. V., Blum, M. M., Mittag, T., Marahiel, M. A., Bernhard, F. & Dötsch, V. (2006). Science, 312, 273–276. [DOI] [PubMed]
- Lai, J. R., Koglin, A. & Walsh, C. T. (2006). Biochemistry, 45, 14869–14879. [DOI] [PubMed]
- Li, J. W.-H. & Vederas, J. C. (2009). Science, 325, 161–165. [DOI] [PubMed]
- Liu, Y., Zheng, T. & Bruner, S. D. (2011). Chem. Biol. 18, 1482–1488. [DOI] [PMC free article] [PubMed]
- Lohman, J. R., Ma, M., Cuff, M. E., Bigelow, L., Bearden, J., Babnigg, G., Joachimiak, A., Phillips, G. N. Jr & Shen, B. (2014). Proteins, 10.1002/prot.24485. [DOI] [PMC free article] [PubMed]
- Meinwald, J. (2011). J. Nat. Prod. 74, 305–309. [DOI] [PMC free article] [PubMed]
- Mercer, A. C. & Burkart, M. D. (2007). Nat. Prod. Rep. 24, 750–773. [DOI] [PubMed]
- Mitchell, C. A., Shi, C., Aldrich, C. C. & Gulick, A. M. (2012). Biochemistry, 51, 3252–3263. [DOI] [PMC free article] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol. 276, 307–326. [DOI] [PubMed]
- Pettersen, E. F., Goddard, T. D., Huang, C. C., Couch, G. S., Greenblatt, D. M., Meng, E. C. & Ferrin, T. E. (2004). J. Comput. Chem. 25, 1605–1612. [DOI] [PubMed]
- Płoskoń, E., Arthur, C. J., Kanari, A. L., Wattana-amorn, P., Williams, C., Crosby, J., Simpson, T. J., Willis, C. L. & Crump, M. P. (2010). Chem. Biol. 17, 776–785. [DOI] [PubMed]
- Ramelot, T. A. et al. (2012). Biochemistry, 51, 7239–7249. [DOI] [PMC free article] [PubMed]
- Rumbo-Feal, S., Gómez, M. J., Gayoso, C., Álvarez-Fraga, L., Cabral, M. P., Aransay, A. M., Rodríguez-Ezpeleta, N., Fullaondo, A., Valle, J., Tomás, M., Bou, G. & Poza, M. (2013). PLoS One, 8, e72968. [DOI] [PMC free article] [PubMed]
- Samel, S. A., Schoenafinger, G., Knappe, T. A., Marahiel, M. A. & Essen, L. O. (2007). Structure, 15, 781–792. [DOI] [PubMed]
- Smith, M. G., Gianoulis, T. A., Pukatzki, S., Mekalanos, J. J., Ornston, L. N., Gerstein, M. & Snyder, M. (2007). Genes Dev. 21, 601–614. [DOI] [PMC free article] [PubMed]
- Strieker, M., Tanović, A. & Marahiel, M. A. (2010). Curr. Opin. Struct. Biol. 20, 234–240. [DOI] [PubMed]
- Vallenet, D. et al. (2008). PLoS One, 3, e1805. [DOI] [PMC free article] [PubMed]
- Van Lanen, S. G., Lin, S., Dorrestein, P. C., Kelleher, N. L. & Shen, B. (2006). J. Biol. Chem. 281, 29633–29640. [DOI] [PubMed]
- Van Lanen, S. G., Oh, T.-J., Liu, W., Wendt-Pienkowski, E. & Shen, B. (2007). J. Am. Chem. Soc. 129, 13082–13094. [DOI] [PMC free article] [PubMed]
- Winn, M. D. et al. (2011). Acta Cryst. D67, 235–242.
- Zornetzer, G. A., Fox, B. G. & Markley, J. L. (2006). Biochemistry, 45, 5217–5227. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: NRPS PCP domain, 4hkg