The crystal structure of the Lymphocytic choriomeningitis virus (LCMV) nucleoprotein C-terminal domain adopts the fold of an exonuclease and provides an important structural template for the study of this prototypic arenavirus.
Keywords: Lymphocytic choriomeningitis virus, nucleoprotein
Abstract
The X-ray crystal structure of the Lymphocytic choriomeningitis virus nucleoprotein C-terminal immunosuppressive domain (LCMV NPΔ340) was determined to 2.0 Å resolution. The structure indicates that LCMV NPΔ340, like the other structurally characterized arenaviral nucleoproteins, adopts the fold of an exonuclease. This structure provides a crucial three-dimensional template for functional exploration of the replication and immunosuppression of this prototypic arenavirus.
1. Introduction
Lymphocytic choriomeningitis virus (LCMV), a category A pathogen, is the prototypic member of the Arenaviridae family. The arenavirus family is composed of some of the world’s deadliest viruses, such as Junín virus and Lassa virus, which are endemic to regions of South America and Western Africa, respectively. In the most severe cases, arenaviral infection can result in viral hemorrhagic fever, multi-organ failure and death within two weeks of the initial infection (Frame et al., 1970 ▶). LCMV has a worldwide distribution and causes birth defects and neurological complications (Bonthius, 2012 ▶; Waggoner et al., 2013 ▶; Razonable, 2011 ▶). Importantly, LCMV can be studied at Biosafety Level 2 (BSL-2), unlike other members of the arenavirus family, which require BSL-4 containment. Thus, many studies to understand arenavirus replication and immunosuppression use – and will continue to use – LCMV as a model system. The study of LCMV as an experimental model system has led to seminal insights into immunobiology, viral replication and viral pathogenesis (Lapošová et al., 2013 ▶; Oldstone, 1975 ▶, 2013 ▶; Oldstone et al., 1985 ▶; Zhou et al., 2012 ▶; Borrow et al., 2010 ▶; Ng et al., 2011 ▶).
There are currently very limited therapeutic options available to combat arenavirus pathogenesis beyond ribavirin and supportive care. Thus, the Arenaviridae family poses an ongoing challenge for medical treatment, and much still remains to be understood about the molecular nature of replication and pathogenesis.
The arenaviral genome encodes only four proteins: a glycoprotein, GP; an RNA-dependent RNA polymerase, L; a matrix protein, Z; and the nucleoprotein, NP. NP is the most abundantly produced protein during viral infection and is multifunctional, with key roles in host immunosuppression, viral replication and encapsidation of the viral genome (Pythoud et al., 2012 ▶; Rodrigo et al., 2012 ▶; Ortiz-Riaño et al., 2011 ▶; Pinschewer et al., 2003 ▶; López et al., 2001 ▶). NP is divided into N-terminal and C-terminal domains, of which the C-terminal domain is immunosuppressive (Ortiz-Riaño et al., 2011 ▶; Borrow et al., 2010 ▶; Martínez-Sobrido et al., 2006 ▶, 2009 ▶; Pythoud et al., 2012 ▶; Rodrigo et al., 2012 ▶). One of several possible avenues of immunosuppression by NP is its specific 3′–5′ exoribonuclease digestion of pathogen-associated double-stranded RNA (dsRNA; Hastie et al., 2011 ▶, 2012 ▶; Jiang et al., 2013 ▶). Most available arenavirus NP structures are from BSL-4 pathogens such as Lassa virus (Jiang et al., 2013 ▶; Hastie et al., 2011 ▶, 2012 ▶; Brunotte et al., 2011 ▶; Qi et al., 2010 ▶) and Junín virus (Zhang et al., 2013 ▶). No structure is yet available for LCMV NP. Provision of an LCMV NP structure would allow targeted mutagenic analysis of the precise sequence in these viral and immune assays. In an effort to further our understanding of the structure and function of the arenavirus nucleoprotein, we determined the X-ray crystal structure of the C-terminal immunosuppressive domain of LCMV NP (composed of residues 340–558 and termed NPΔ340).
2. Materials and methods
2.1. Protein expression and purification
The LCMV NPΔ340 gene was subcloned into the pET-46 Ek/LIC vector (Novagen) with an N-terminal His6 affinity tag. The protein was overexpressed in Escherichia coli in LB medium (supplemented with ZnCl and MgCl2 to 100 µM each) and purified by Ni–NTA affinity chromatography (50 mM Tris pH 8.0, 300 mM NaCl, 20 mM imidazole and eluted with the same supplemented to 500 mM imidazole). The protein was further purified by size-exclusion chromatography on a GE Superdex 75 column (10 mM Tris pH 8.0, 300 mM NaCl).
2.2. Crystallization and data collection
Two structures of LCMV NPΔ340 were determined from crystals that belonged to different crystallographic space groups. The first structure was solved in space group P21 from crystals obtained by setting up 4 µl vapour-diffusion sitting drops with 2 µl 10.5 mg ml−1 NPΔ340 plus 2 µl reservoir solution (0.35 M potassium phosphate, 20% PEG 3350). Crystals were cryoprotected with mother liquor supplemented with 20% glycerol. Native data for the P21 space-group crystals were collected on beamline 8.2.2 at the Advanced Light Source (ALS) in Berkeley, California, USA. The second structure was solved in space group P65 from crystals that grew after incubation of the post-Superdex 75 sample at 1.15 mg ml−1 in an Eppendorf tube for several weeks at 4°C. These were cryoprotected with the Superdex 75 buffer (10 mM Tris pH 8.0, 300 mM NaCl) supplemented with 40% glycerol. Native data for the P65 space-group crystals were collected on beamline 23-ID at the Advanced Photon Source (APS) in Argonne, Illinois, USA.
2.3. Structure solution and refinement
All data were indexed using d*TREK (Pflugrath, 1999 ▶). The structures were determined by molecular replacement using the Cα chain of Lassa virus (LASV) NPΔ340 (PDB entry 4fvu; Hastie et al., 2012 ▶) as a search model, built in Coot (Emsley & Cowtan, 2004 ▶) and refined with PHENIX (Adams et al., 2010 ▶) (Table 1 ▶). The atomic coordinates and diffraction data for the LCMV NPΔ340 structures have been deposited in the Protein Data Bank with accession codes 4o6h and 4o6i.
Table 1. Data-collection and refinement statistics for LCMV NP340.
| Crystal | Form A | Form B |
|---|---|---|
| PDB code | 4o6h | 4o6i |
| Data collection | ||
| Space group | P21 | P65 |
| Unit-cell parameters | ||
| a () | 92.86 | 89.96 |
| b () | 94.40 | 89.96 |
| c () | 145.12 | 145.94 |
| () | 90.00 | 90.00 |
| () | 102.30 | 90.00 |
| () | 90.00 | 120.00 |
| Resolution () | 47.32.8 (2.92.8) | 39.02.0 (2.12.0) |
| R merge | 0.1 (0.5) | 0.1 (0.7) |
| I/(I) | 6.2 (1.6) | 7.7 (1.9) |
| Completeness (%) | 97.2 (92.9) | 100 (100) |
| Multiplicity | 3.4 (3.4) | 10.6 (10.8) |
| Wavelength () | 0.9793 | 1.0332 |
| Refinement | ||
| Resolution () | 2.8 | 2.0 |
| No. of reflections | 58766 | 45034 |
| R work/R free | 0.227/0.261 | 0.209/0.228 |
| No. of atoms | ||
| Protein | 12463 | 3198 |
| Ligand/ion | 36 | 54 |
| Water | 0 | 142 |
| B factors (2) | ||
| Wilson (average) | 83 | 64 |
| Protein | 76 | 66 |
| Ligand/ion | 76 | 83 |
| Water | N/A | 64 |
| R.m.s. deviations | ||
| Bond lengths () | 0.003 | 0.003 |
| Bond angles () | 0.68 | 0.64 |
| Ramachandran plot (%) | ||
| Favored region | 96.7 | 97.0 |
| Allowed region | 3.3 | 3.0 |
2.4. Alignment of LCMV NPΔ340
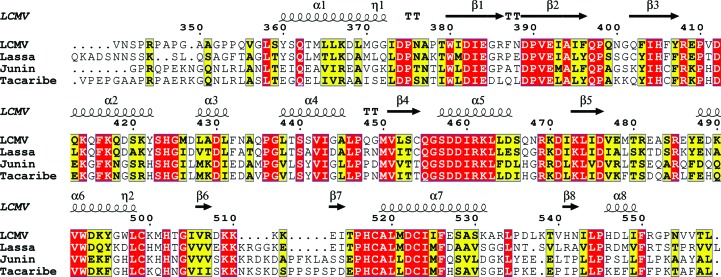
Sequence alignment was performed using the Clustal Omega web server (Sievers et al., 2011 ▶; http://www.ebi.ac.uk/Tools/msa/clustalo/) to align the NP C-terminal domains of LCMV, Lassa virus, Junín virus and Tacaribe virus (UniProt sequences P09992, P13699, P14239 and P18140, respectively) retrieved from http://www.uniprot.org. The alignment was visualized via the ESPript web server (http://espript.ibcp.fr/ESPript/ESPript/) and displayed using a Risler similarity calculation with a global similarity score of 70% (yellow residues) and with identical residues shown in red.
3. Results
3.1. Overall structure of LCMV NPΔ340: conservation of the exonuclease fold
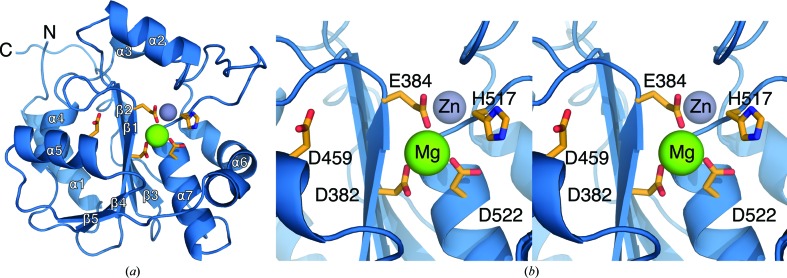
The structure of the LCMV NP immunosuppressive C-terminal domain was solved to 2.0 Å resolution and provides a continuous atomic map from residue 355 to the ultimate C-terminal residue 558 (Fig. 1 ▶ a). As expected, the structure is similar in overall fold to other structurally characterized arenavirus NPs. All C-terminal domain crystal structures of arenaviral NPs adopt the fold of the DEDDh superfamily of exonucleases (Qi et al., 2010 ▶; Hastie et al., 2011 ▶, 2012 ▶; Jiang et al., 2013 ▶; Zhang et al., 2013 ▶; Brunotte et al., 2011 ▶). Accordingly, the arenavirus NP also functions as a dsRNA-specific exonuclease (Hastie et al., 2011 ▶, 2012 ▶; Jiang et al., 2013 ▶). LCMV NP is 61% identical in sequence to LASV NP (Fig. 2 ▶), and the r.m.s.d. between the LCMV and LASV structures is only 0.48 Å (Supplementary Fig. S1 and Supplementary Table S11). Further, LCMV NP coordinates a structural Zn metal using the residues Glu392, Cys499, His502 and Cys518 that are highly conserved across the arenavirus family (Jiang et al., 2013 ▶; Hastie et al., 2011 ▶, 2012 ▶; Brunotte et al., 2011 ▶; Qi et al., 2010 ▶). LCMV NPΔ340 also coordinates a metal ion that is observable in the electron-density map and has been modeled as Mg (supplemented to the expression media), which clearly interacts with the catalytic residues of the similarly conserved DEDDh exonuclease motif (Asp382, Glu384, Asp459, Asp522 and His517; Fig. 1 ▶ b). The metal is directly coordinated by the residues Asp382, Glu384 and Asp522, while the other residues of the DEDDh motif are involved in the coordination of a second catalytic metal ion and the associated exonuclease catalytic mechanism (Jiang et al., 2013 ▶). Previous studies have shown the exonuclease activity of Lassa virus NP to be dependent upon the coordination of a divalent cation cofactor, with a preference for Mn2+, Co2+ and Mg2+ (Hastie et al., 2011 ▶; Qi et al., 2010 ▶).
Figure 1.
Structure of LCMV NPΔ340. (a) Overall structure of LCMV NPΔ340 shown as a cartoon with the exonuclease active-site residues illustrated as orange sticks and the coordinated Zn and Mg metals shown as gray and green spheres, respectively. (b) Enlarged stereoview of the LCMV NPΔ340 DEDDh exonuclease active-site residues labeled and shown as orange sticks. The active-site metal cofactor has been modeled as an Mg and is illustrated here as a green sphere.
Figure 2.
Alignment of the LCMV NPΔ340 primary structure. The primary structure of LCMV NPΔ340 was aligned with those of other structurally characterized arenaviruses. The secondary structure of LCMV NPΔ340 is shown along the top of each alignment row. Highly conserved residues are shown in red, while residues that share >70% sequence similarity are shown in yellow.
The ‘basic loop’ region outside the active site (residues 507–515 in LCMV NP) is clearly defined in this structure. The precise role of this loop is unclear, although mutation of the basic residues in this region has been shown to decrease the exonuclease activity (Hastie et al., 2011 ▶). The crystal structure of the Lassa virus NP exonuclease domain in complex with dsRNA indicates that the complementary strand extends up along this basic loop as it is split from the substrate strand feeding into the enzymatic active site (Hastie et al., 2012 ▶). Mutagenic loss of the basic charge in this loop may decrease exonuclease function via loss of stabilizing binding of the complementary dsRNA strand.
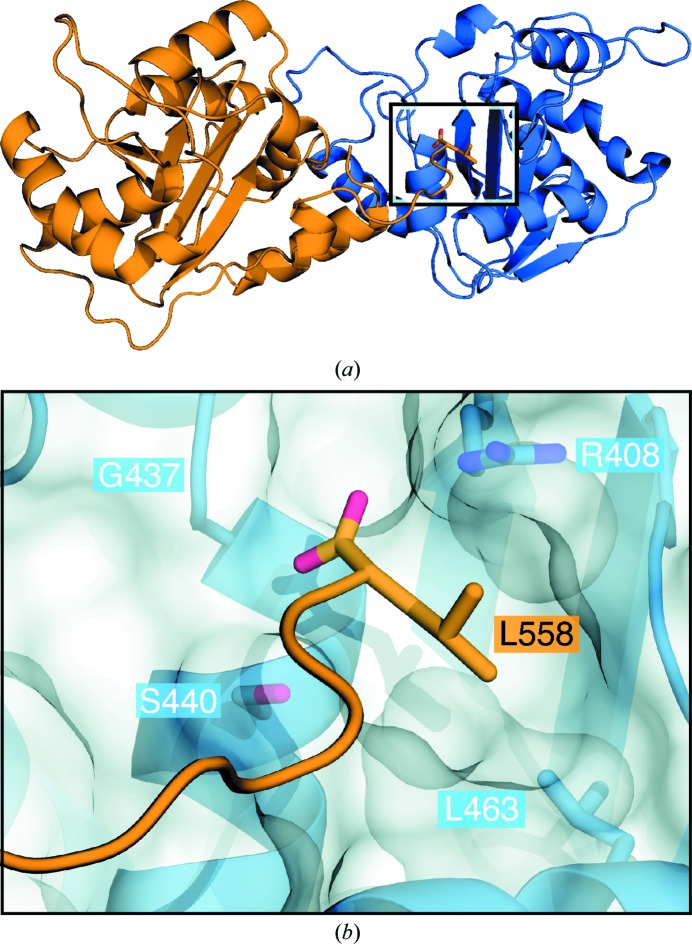
3.2. Crystal packing
The asymmetric unit of each crystal is composed of NPΔ340 molecules interacting through a dimeric interface (Fig. 3 ▶ a). The P21 crystal contains eight NPΔ340 molecules in the asymmetric unit, whereas the P65 crystal contains two NPΔ340 molecules in the asymmetric unit. Each of the ten independent copies of NP from either of the asymmetric units can be aligned onto any other with an average r.m.s.d. of 0.31 Å. The two molecules of the P65 asymmetric unit are related by noncrystallographic symmetry through a twofold rotational axis that forms a dimeric interface between the two molecules. Interestingly, this dimer of NPΔ340 in the P65 crystal can be used to build up the crystal packing in the P21 crystal as well. Of the eight molecules present in the asymmetric unit of the P21 crystal, four are paired as dimers identical to that observed in the P65 crystal packing, while the remaining four molecules are positioned towards the edge of the asymmetric unit, enabling them to form a complete dimer within the crystal upon translation of the asymmetric unit. Alignment of the different dimers from each crystal results in an average r.m.s.d. of 1.88 Å. The dimer interfaces are nearly identical, with slight relative motions toward the outer edge of the dimer. Thus each crystal, although belonging to a different space group and incorporating a different number of molecules into its asymmetric unit, is constructed from the same dimeric structure of LCMV NPΔ340 (Supplementary Fig. 2 ▶).
Figure 3.
LCMV NPΔ340 asymmetric unit dimeric interface. (a) Side view of the dimeric interface involving the N-terminal helix and the insertion of the ultimate C-terminal residue, shown as sticks, into a hydrophobic pocket (boxed region). The monomers of the asymmetric unit are shown as blue and orange cartoons. (b) Enlarged view of the boxed region showing the highly conserved residues involved in the formation of the hydrophobic pocket and the interaction with the C-terminal residue Leu558.
Analysis of this interface was performed using the Protein Interfaces, Surfaces and Assemblies service (PISA) at the European Bioinformatics Institute (http://www.ebi.ac.uk/msd-srv/prot_int/pistart.html; Krissinel & Henrick, 2007 ▶). The analysis confirms a dimerization interface of 1032.1 Å2 for each subunit, which is equivalent to 8.9% of the total accessible solvent area of each monomer.
The dimeric interface involves an interesting NP–NP interaction, observed in both the P21 and the P65 structures, in which the ultimate C-terminal residue of LCMV NP inserts into a hydrophobic pocket of a neighboring NP monomer (Fig. 3 ▶ b). Both the C-terminus and the recipient hydrophobic pocket are conserved across arenaviruses. The C-terminal residue of all classified arenavirus NPs is hydrophobic (either Leu, Met or Ile), with leucine as the terminal residue in 75% of the viral species. The recipient hydrophobic pocket in LCMV is composed of residues Arg408, Gly437, Ser440 and Leu463 that are 100% conserved across arenaviruses.
Notably, a similar interaction between the C-terminus and the equivalent hydrophobic binding pocket is also formed in the crystal packing of all structures of the LASV NP C-terminal domain (NPΔ340; Jiang et al., 2013 ▶; Hastie et al., 2011 ▶, 2012 ▶). In LASV NPΔ340, the C-terminus extends from one monomer to another, forming a crystal-packing polymer tethered by the C-terminus (Supplementary Fig. 3 ▶ a). By contrast, in the structure of LCMV NPΔ340 the interaction is reciprocal between two monomers and, in conjunction with hydrophobic interactions between the N-terminal α-helices (Supplementary Fig. S4), results in the formation of discrete dimers. Investigation is currently under way to determine whether this dimeric interface is of biological relevance.
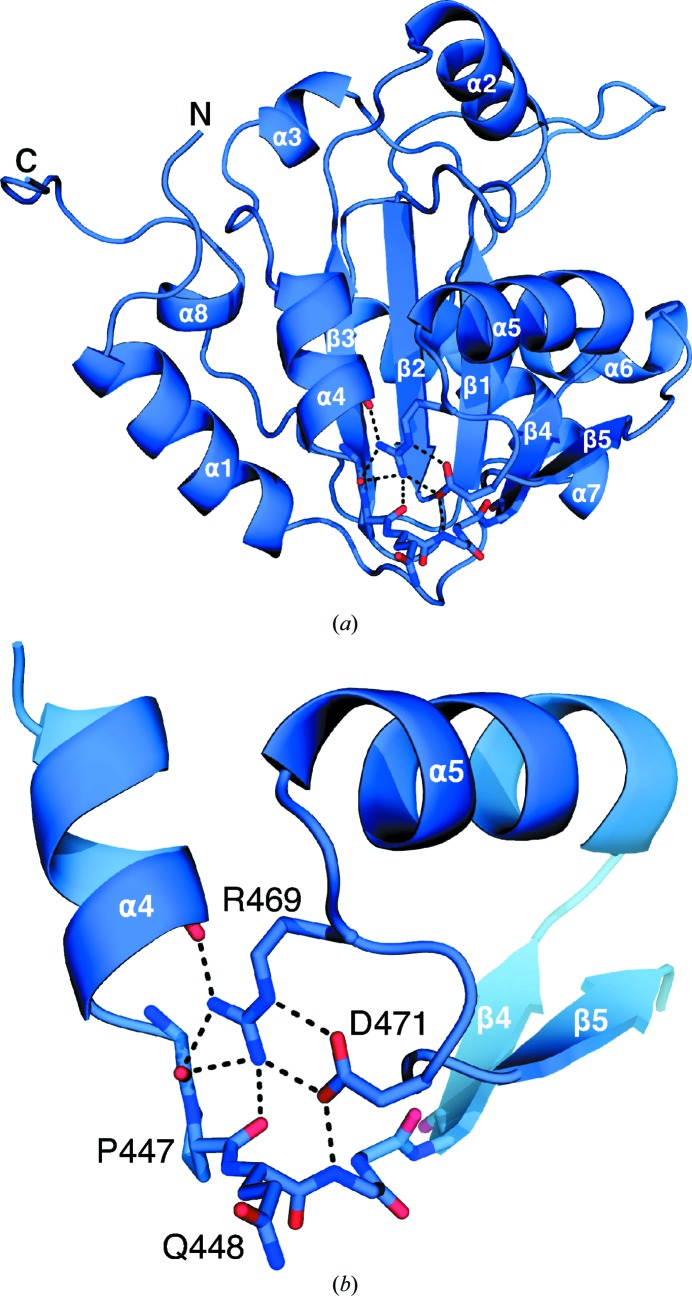
3.3. Role of residue Asp471
Ortiz-Riano and coworkers observed that LCMV residue Asp471 is essential for NP oligomerization and viral replication. However, the exact role of Asp471 in NP self-association is unknown (Ortiz-Riaño et al., 2012 ▶). This structure confirms that Asp471 plays a key role in the stabilization of the α5 helix region through polar interactions with Arg469 and the backbone N atom of Gly449 (Fig. 4 ▶).
Figure 4.
Stabilizing interactions of Asp471. (a) Cartoon representation of LCVM NPΔ340 showing the region surrounding Arg469 and Asp471 (shown as sticks). (b) Enlarged view of the region around residue Asp471 showing electrostatic interactions between Asp471, Arg469 and the main chain of Pro447 and Gln448 indicated as black dashed lines.
4. Discussion
In this work, we provide two crystal structures of the immunosuppressive C-terminal domain of LCMV NP at 2.0 and 2.8 Å resolution. The crystal structures reveal that the DEDDh exonuclease fold and both bound metal ions are conserved in LCMV NP.
Further, a conserved dimeric interface is observed in both crystal structures presented here (in the P21 and the P65 packing). This conserved interface is especially interesting because it involves the insertion of the C-terminal Leu558 residue into the hydrophobic pocket of a neighboring NPΔ340 molecule, an insertional interface that is nearly identical to that observed in all of the current LASV NPΔ340 structures (Jiang et al., 2013 ▶; Hastie et al., 2011 ▶, 2012 ▶). It is possible that this insertion serves as a tether to link together NP monomers in the functional ribonucleoprotein complex while allowing a great degree of flexibility between tethered subunits. Future experiments should explore the biological relevance of the NP C-terminus.
This structure illustrates that residue Asp471 of LCMV NP forms a salt bridge with Arg469 to stabilize the region around the α4 and α5 helices. A similar salt bridge is also observed between equivalent Asp and Arg residues of the Lassa virus, Tacaribe virus and Junín virus NP structures (Jiang et al., 2013 ▶; Hastie et al., 2011 ▶, 2012 ▶; Brunotte et al., 2011 ▶; Qi et al., 2010 ▶; Zhang et al., 2013 ▶). Mutation of Asp471 to Ala in LCMV NP has been shown to abrogate NP–NP oligomerization (Ortiz-Riaño et al., 2012 ▶). In the structure of full-length trimeric Lassa virus NP containing both the N-terminal and C-terminal domains, residue Asp478 (equivalent to LCMV NP Asp471) is too far from the oligomeric interfaces to be directly involved in trimerization and is too far from the N–C interface within a single monomer to influence interdomain association (Supplementary Fig. S5; Brunotte et al., 2011 ▶; Qi et al., 2010 ▶). Instead, we propose that the role of Asp471 is to interact with Arg469. Together, this pair of residues located on the loop between helix α5 and β-sheet β5 forms multiple hydrogen bonds to the backbone and side chains of the loop between helix α4 and strand β4 (Fig. 4 ▶). Stabilization of the region around the α4 and α5 helices by Asp471/Arg469 may be important for NP–NP self-association either by contributing to the structural stability of the fold of the monomer itself or through an as-yet-unrevealed interface of the ribonucleoprotein assembly, which could be different from the trimer. Additional studies will be required to elucidate the precise function of this residue in NP–NP self-association.
The structural zinc ion observed in LCMV NPΔ340 and all other arenavirus NP structures except for the recently solved Junín virus NP C-terminal domain (Zhang et al., 2013 ▶) is likely to play a key role in the structural stability of the protein. In the Junín virus structure (PDB entry 4k7e) residues 407–431 and 499–521 are not visualized. These regions are proximal to the zinc-coordination site and may be stabilized upon zinc binding. It is possible that the absence of zinc has rendered them disordered and functionally inactive. Indeed, the exonuclease function was not catalytically active in this preparation of NP (Zhang et al., 2013 ▶). The zinc ion is not the ion incorporated directly into the DEDDh exonuclease active site, but may play a role in structural stability of the domain as a whole and may therefore be necessary for correct folding and positioning of the active site. Notably, the presence of a zinc metal ligand in Junín virus NP has previously been reported (Tortorici et al., 2001 ▶). The fact that the Junín virus NP used in crystallization lacked zinc and was catalytically inactive as an exonuclease, contrary to the observations for Lassa virus and Tacaribe virus NP (Jiang et al., 2013 ▶; Hastie et al., 2011 ▶, 2012 ▶; Qi et al., 2010 ▶) and both structures of LCMV NPΔ340 presented here, suggests that it may be possible to achieve an alternate Junín virus NP structure that is zinc-bound and catalytically active
These structures of LCMV NPΔ340 enable the mapping of characterized mutant phenotypes in viruses and infected cells directly to the X-ray structure of NP of the infecting virus. We may now use these LCMV crystal structures to design mutants and directly understand structure–function relationships using sequence-matched structural templates for mutagenesis.
Supplementary Material
Supporting Information. DOI: 10.1107/S1399004714007883/mn5054sup1.pdf
PDB reference: LCMV NPΔ340, 4o6h
PDB reference: 4o6i
Acknowledgments
We thank the staff of ALS beamline 8.2.2 and APS beamline 23-ID for assistance with data collection and Dr Juan Carlos de la Torre for the gift of LCMV NP cDNA and helpful discussions. We acknowledge NIH grant HHSN27220090049C, an Investigators in the Pathogenesis of Infectious Disease award and the Skaggs Institute of Chemical Biology for support. This is manuscript #26036 from The Scripps Research Institute.
Footnotes
Supporting information has been deposited in the IUCr electronic archive (Reference: MN5054).
References
- Adams, P. D. et al. (2010). Acta Cryst. D66, 213–221.
- Bonthius, D. J. (2012). Semin. Pediatr. Neurol. 19, 89–95. [DOI] [PMC free article] [PubMed]
- Borrow, P., Martínez-Sobrido, L. & de la Torre, J. C. (2010). Viruses, 2, 2443–2480. [DOI] [PMC free article] [PubMed]
- Brunotte, L., Kerber, R., Shang, W., Hauer, F., Hass, M., Gabriel, M., Lelke, M., Busch, C., Stark, H., Svergun, D. I., Betzel, C., Perbandt, M. & Günther, S. (2011). J. Biol. Chem. 286, 38748–38756. [DOI] [PMC free article] [PubMed]
- Emsley, P. & Cowtan, K. (2004). Acta Cryst. D60, 2126–2132. [DOI] [PubMed]
- Frame, J. D., Baldwin, J. M. Jr, Gocke, D. J. & Troup, J. M. (1970). Am. J. Trop. Med. Hyg. 19, 670–676. [DOI] [PubMed]
- Hastie, K. M., Kimberlin, C. R., Zandonatti, M. A., MacRae, I. J. & Saphire, E. O. (2011). Proc. Natl Acad. Sci. USA, 108, 2396–2401. [DOI] [PMC free article] [PubMed]
- Hastie, K. M., King, L. B., Zandonatti, M. A. & Saphire, E. O. (2012). PLoS One, 7, e44211. [DOI] [PMC free article] [PubMed]
- Jiang, X., Huang, Q., Wang, W., Dong, H., Ly, H., Liang, Y. & Dong, C. (2013). J. Biol. Chem. 288, 16949–16959. [DOI] [PMC free article] [PubMed]
- Krissinel, E. & Henrick, K. (2007). J. Mol. Biol. 372, 774–797. [DOI] [PubMed]
- Lapošová, K., Pastoreková, S. & Tomášková, J. (2013). Acta Virol. 57, 160–170. [DOI] [PubMed]
- López, N., Jácamo, R. & Franze-Fernández, M. T. (2001). J. Virol. 75, 12241–12251. [DOI] [PMC free article] [PubMed]
- Martínez-Sobrido, L., Emonet, S., Giannakas, P., Cubitt, B., García-Sastre, A. & de la Torre, J. C. (2009). J. Virol. 83, 11330–11340. [DOI] [PMC free article] [PubMed]
- Martínez-Sobrido, L., Zúñiga, E. I., Rosario, D., García-Sastre, A. & de la Torre, J. C. (2006). J. Virol. 80, 9192–9199. [DOI] [PMC free article] [PubMed]
- Ng, C. T., Sullivan, B. M. & Oldstone, M. B. (2011). Curr. Opin. Virol. 1, 160–166. [DOI] [PMC free article] [PubMed]
- Oldstone, M. B. (1975). Prog. Med. Virol. 19, 84–119. [PubMed]
- Oldstone, M. B. (2013). Proc. Natl Acad. Sci. USA, 110, 4180–4183. [DOI] [PMC free article] [PubMed]
- Oldstone, M. B., Ahmed, R., Byrne, J., Buchmeier, M. J., Riviere, Y. & Southern, P. (1985). Br. Med. Bull. 41, 70–74. [DOI] [PubMed]
- Ortiz-Riaño, E., Cheng, B. Y. H., de la Torre, J. C. & Martínez-Sobrido, L. (2011). J. Virol. 85, 13038–13048. [DOI] [PMC free article] [PubMed]
- Ortiz-Riaño, E., Cheng, B. Y. H., de la Torre, J. C. & Martínez-Sobrido, L. (2012). Viruses, 4, 2137–2161. [DOI] [PMC free article] [PubMed]
- Pflugrath, J. W. (1999). Acta Cryst. D55, 1718–1725. [DOI] [PubMed]
- Pinschewer, D. D., Perez, M. & de la Torre, J. C. (2003). J. Virol. 77, 3882–3887. [DOI] [PMC free article] [PubMed]
- Pythoud, C., Rodrigo, W. W., Pasqual, G., Rothenberger, S., Martínez-Sobrido, L., de la Torre, J. C. & Kunz, S. (2012). J. Virol. 86, 7728–7738. [DOI] [PMC free article] [PubMed]
- Qi, X., Lan, S., Wang, W., Schelde, L. M., Dong, H., Wallat, G. D., Ly, H., Liang, Y. & Dong, C. (2010). Nature (London), 468, 779–783. [DOI] [PMC free article] [PubMed]
- Razonable, R. R. (2011). Curr. Opin. Organ Transplant. 16, 580–587. [DOI] [PubMed]
- Rodrigo, W. W., Ortiz-Riaño, E., Pythoud, C., Kunz, S., de la Torre, J. C. & Martínez-Sobrido, L. (2012). J. Virol. 86, 8185–8197. [DOI] [PMC free article] [PubMed]
- Sievers, F., Wilm, A., Dineen, D., Gibson, T. J., Karplus, K., Li, W., Lopez, R., McWilliam, H., Remmert, M., Söding, J., Thompson, J. D. & Higgins, D. G. (2011). Mol. Syst. Biol. 7, 539. [DOI] [PMC free article] [PubMed]
- Tortorici, M. A., Ghiringhelli, P. D., Lozano, M. E., Albariño, C. G. & Romanowski, V. (2001). J. Gen. Virol. 82, 121–128. [DOI] [PubMed]
- Waggoner, J. J., Soda, E. A. & Deresinski, S. (2013). Clin. Infect. Dis. 57, 1182–1188. [DOI] [PMC free article] [PubMed]
- Zhang, Y., Li, L., Liu, X., Dong, S., Wang, W., Huo, T., Guo, Y., Rao, Z. & Yang, C. (2013). J. Gen. Virol. 94, 2175–2183. [DOI] [PubMed]
- Zhou, X., Ramachandran, S., Mann, M. & Popkin, D. L. (2012). Viruses, 4, 2650–2669. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information. DOI: 10.1107/S1399004714007883/mn5054sup1.pdf
PDB reference: LCMV NPΔ340, 4o6h
PDB reference: 4o6i