Abstract
BioC is a recently created XML format to share text data and annotations, and an accompanying input/output library to promote interoperability of data and tools for natural language processing of biomedical text. This article reports the use of BioC to address a common challenge in processing biomedical text information—that of frequent entity name abbreviation. We selected three different abbreviation definition identification modules, and used the publicly available BioC code to convert these independent modules into BioC-compatible components that interact seamlessly with BioC-formatted data, and other BioC-compatible modules. In addition, we consider four manually annotated corpora of abbreviations in biomedical text: the Ab3P corpus of 1250 PubMed abstracts, the BIOADI corpus of 1201 PubMed abstracts, the old MEDSTRACT corpus of 199 PubMed® citations and the Schwartz and Hearst corpus of 1000 PubMed abstracts. Annotations in these corpora have been re-evaluated by four annotators and their consistency and quality levels have been improved. We converted them to BioC-format and described the representation of the annotations. These corpora are used to measure the three abbreviation-finding algorithms and the results are given. The BioC-compatible modules, when compared with their original form, have no difference in their efficiency, running time or any other comparable aspects. They can be conveniently used as a common pre-processing step for larger multi-layered text-mining endeavors.
Database URL: Code and data are available for download at the BioC site: http://bioc.sourceforge.net.
Introduction
The BioCreative (http://www.biocreative.org/) challenge evaluations—since their inception in 2003—have been a community-wide effort for evaluating text-mining information extraction systems applied to the biomedical domain. Given the emphasis on promoting scientific progress, BioCreative meetings have consistently sought to make available both suitable information extraction systems that handle life science literature and suitable ‘gold standard’ data for training and testing these systems (1–4). The BioCreative IV Interoperability track (http://www.bio creative.org/tasks/biocreative-iv/track-1-interoperability/) follows the guidelines established in previous BioCreative meetings, and specifically addresses the goal of interoperability—a major barrier for wide-scale adoption of the developed text-mining tools. As a solution, BioC (5) is an interchange format for tools for biomedical natural language processing. BioC is a simple XML format, specified by DTD (Document Type Definition), to share text documents and annotations. The BioC annotation approach allows many different annotations to be represented, including sentences, tokens, parts of speech and named entities such as genes or diseases. The Interoperability Track at BioCreative IV led to the creation of a number of tools and corpora to encourage even broader use and reuse of BioC data and tools (6).
This article reports on the contributions of our team to the BioC repository in the form of BioC-compliant modules that address the abbreviation definition detection task in biomedical text. These modules can be seamlessly coupled with other BioC code and used with any BioC-formatted corpora. We also present several BioC-formatted corpora to test the abbreviation definition detection task. These corpora can serve as gold standard data for developing better machine-learning methods for abbreviation definition recognition. They could even be used as a starting point for adding further annotations of other important biomedical entities such as genes, diseases, etc.
Abbreviation Identification in The Biomedical Domain
The past 20 years have seen a dramatic increase in the interest for automatic extraction of biological information from scientific text, and particularly from MEDLINE® abstracts. One characteristic of these documents is the frequent use of abbreviated terms. Abbreviated terms appear not only in the scientific text, but they are also frequent in user queries requesting the retrieval of those documents (7). Two related specific issues are—(i) the high rate at which new abbreviations are introduced in biomedical texts, and (ii) the ambiguity of those abbreviations. Existing databases, ontologies and dictionaries must be continually updated with new abbreviations and their definitions. To help resolve this problem, several techniques have been introduced to automatically extract abbreviations and their definitions from MEDLINE abstracts (8–11).
Abbreviation identification is the task of processing text to extract explicit occurrences of abbreviation-definition pairs. For example, in the sentence ‘Body mass index (BMI) of the dyslipidemic diabetic patients was significantly higher for females.’ ‘BMI’ is the abbreviation, and ‘Body mass index’ is its definition. This task requires both the identification of sentences that contain <Abbreviation,Definition> candidate pairs from text, and identification of exact string boundaries. An abbreviation—a ShortForm—is a shorter term that represents a longer word or phrase, which often refers to an important biomedical entity. The definition—the LongForm—is searched for in the same sentence as the ShortForm. An important clue that is shared by abbreviation-detection methods is the presence of parenthetical text and the assumption that parenthetical text signals the presence of an abbreviation definition. Two cases are distinguished:
LongForm (ShortForm), where the sentence mentions the long form first, following with the abbreviation in parenthesis, and
ShortForm (LongForm), where the sentence introduces the abbreviation first, following with the definition in parenthesis.
Of these, the first alternative is observed much more frequently in practice, and in that case, the search for the LongForm is performed on the text string between the beginning of the sentence and the open parenthesis.
Abbreviation Definition Finding Algorithms
In this study, we focus on three abbreviation definition finding algorithms and modify them to work with the BioC format. All three algorithms described here approach the abbreviation identification problem as described above, but have several differences and unique characteristics which we describe as follows:
1. The Schwartz and Hearst algorithm
The Schwartz and Hearst algorithm (8) identifies <ShortForm, LongForm> candidates using this rule: if the expression within parentheses contains three or more tokens then case (ii) is assumed, otherwise case (i) is assumed. The LongForm is always longer than the ShortForm. A ShortForm is verified to start with an alphanumeric character, to contain at least one letter character and to contain a reasonable number of characters. A LongForm candidate then is extracted from the string so that it contains at most min (|SF|+5, |SF| × 2) tokens, where |SF| is the number of characters in the ShortForm. Next, the algorithm traverses both strings right to left, matching the characters in the ShortForm to find the shortest LongForm string. With the exception of the first ShortForm character that has to match a character at the beginning of a word in the LongForm candidate, the rest of the characters can match anywhere in the LongForm string, as long as they are in sequential order. The algorithm is simple, fast, and produces good results (http://bio text.berkeley.edu/software.html).
2. The Ab3P algorithm
The Ab3P algorithm, developed by Sohn et al. (9) is another pattern-matching approach to abbreviation definition detection. This algorithm defines specific pattern-matching rules which the authors called strategies. Depending on the matching strategy and the length of the short form, they estimate an accuracy measure called pseudo-precision. Pseudo-precision provides a computed reliability estimate for an identified <ShortForm, LongForm> pair, without any human judgment. This algorithm is also very fast and provides high-precision results.
3. The NatLAb algorithm
Rule-based methods, such as the Schwartz and Hearst algorithm and Ab3P, are successful at identifying abbreviation definition pairs with high precision. However, it is challenging for such approaches to identify non-typical pairs, such as <3-D, three dimensional >, or out-of-order matches, such as <T(m), melting temperature>. Machine-learning methods have the potential of recognizing such non-trivial or irregular pairs and improving the recall, if enough training data are provided. NatLAb (Natural Learning for Abbreviations), developed by Yeganova et al. (11) is an example of such an algorithm.
NatLAb is a supervised learning approach whose features, inspired by the basic rules defined in Sohn et al., describe a mapping between a character in a potential ShortForm and a character in a potential LongForm. However, in contrast to Ab3P, Yeganova et al. do not combine these features into hand-crafted strategies. They provide the learner with all these features and feature pairs and let the training process weight them. Feature weights are then used to identify abbreviation definitions.
Converting Into BioC
A BioC-compliant module is a software module that is able to process input data in BioC format, and produce results in BioC format.
BioC-compliant abbreviation identification modules
We converted the original software tools for abbreviation definition recognition into BioC-compliant tools. Each algorithm was modified to accept data input in the BioC format and to produce results in the BioC format. The original Schwartz and Hearst software is written in Java, the original Ab3P software is written in C++ and the original NatLAb software is written in Perl. As a result, each implementation used a different BioC library; the necessary links were established so the Schwartz and Hearst algorithm could flow seamlessly with the rest of the BioC-Java code, the Ab3P algorithm with the BioC-C++ code and NatLAb with a Perl-BioC implementation (12). The BioC-compliant modules differ from the originals in these main points:
- The BioC format includes the precise locations of annotations in the original text. Because the original algorithms did not track the location of their recognized abbreviations, we modified their BioC-compliant versions to produce exact annotation offsets, as a result attaining a richer output. Retrofitting this tracking required considerable effort.
- Any text element in a BioC passage, or sentence, is processed for abbreviation definitions. The precise text offsets are produced accordingly. The original algorithms only read the original input one line at a time; whether a line was processed for abbreviations depended on the expected format of the program. In BioC, there is no concept of a ‘line’.
- The new modules produce results in BioC format and the recognized abbreviations and their definitions can be compared with the output of any other BioC-compliant abbreviation definition recognition tool which uses the same keyfile. This is demonstrated in Table 3 in the Results section, where we compare the results of these algorithms on four independent biomedical abbreviation corpora. Next we describe four abbreviation definition corpora and the representation of abbreviations in BioC format.
Table 3.
Results of BioC-compliant abbreviation detection modules when tested on BioC-formatted abbreviation corpora
| Corpus/Shwartz&Hearst results | Ab3P | BIOADI | MEDSTRACT | Schwartz&Hearst |
|---|---|---|---|---|
| Shwartz&Hearst results | ||||
| Precision | 0.950 | 0.943 | 0.986 | 0.928 |
| Recall | 0.788 | 0.765 | 0.893 | 0.763 |
| F-score | 0.861 | 0.844 | 0.937 | 0.837 |
| Ab3P results | ||||
| Precision | 0.971 | 0.952 | 0.993 | 0.929 |
| Recall | 0.836 | 0.770 | 0.906 | 0.770 |
| F-score | 0.898 | 0.851 | 0.947 | 0.842 |
| NatLAb results | ||||
| Precision | 0.927 | 0.885 | 0.924 | 0.856 |
| Recall | 0.879 | 0.833 | 0.918 | 0.824 |
| F-score | 0.903 | 0.858 | 0.921 | 0.840 |
BioC-formatted corpora
BioC Representation of Abbreviations. The first step in converting a given corpus into BioC format is deciding how to represent the information present in the corpus. This representation is what is contained in the keyfile that must accompany a BioC corpus. Figures 1 and 2 illustrate the BioC format representation for abbreviation annotations that we used in the corpora used for this study. The original abbreviation annotations in these corpora consisted of annotations in the form of <ShortForm, LongForm> pairs. To capture this, and to make the corpora versatile for other possible biomedical information retrieval studies, we use this markup:
- For annotation elements, the infon key=type, with value ABBR, semantically identifies the annotations as abbreviations. This allows the possibility of having multiple layers of annotations on the same textual data, even including annotations on other entity types that may also overlap. All such annotations can be added without confusion.
- Each part of an abbreviation is identified by an additional infon element, key=ABBR, with value ShortForm or LongForm, thus preserving the original text’s role in an abbreviation definition <ShortForm, LongForm> pair.
- Finally, a relation element reflects the pairing between a ShortForm and a corresponding LongForm, as indicated by the role attribute. The same infon, key=type, with value ABBR, is repeated for the relation to make it easier at a semantic level to distinguish what is being annotated and how they relate together.
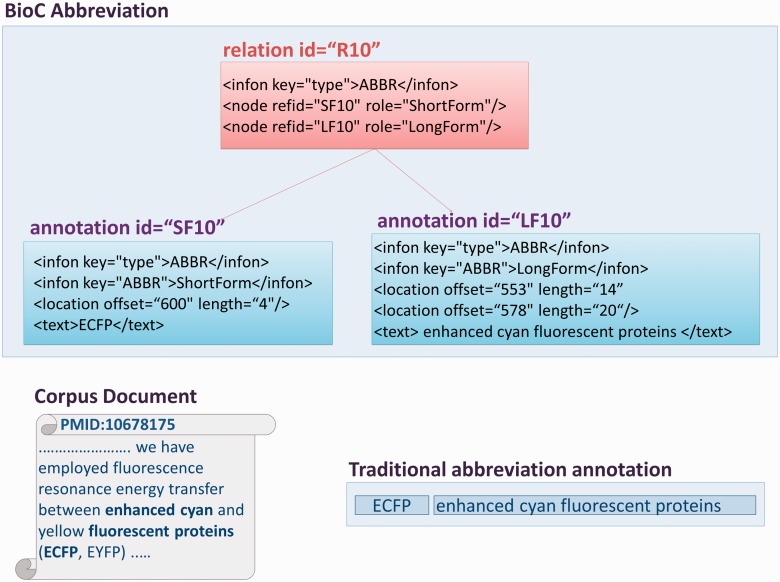
Figure 1.

Illustration of abbreviation annotation in BioC format.
Figure 2.
A graphical representation of abbreviation annotations in BioC format. The excerpt from one of the corpus documents contains multi-segmented abbreviation long forms. The traditional ShortForm, LongForm pairing is shown in the figure, as well as the infons detailing BioC annotations for an abbreviation, and the relation between them, with the corresponding precise text offsets.
Documents may contain multiple abbreviation definitions for the same short form, as we also discovered during our annotation effort. Moreover, an annotated mention may be composed of several non-consecutive strings. One such example is shown in Figure 2. However, these issues are easily addressed in BioC, via the location element. The location element links the annotation definition to the exact textual coordinates where it is mentioned, and also allows for defining mentions composed of multiple non-consecutive substrings. Note that, the location information was not present in the original annotations of these corpora, so this is an important enrichment over the original versions.
Abbreviation definition Corpora in BioC Format. We converted four abbreviation definition recognition corpora to BioC format. These corpora are—the Ab3P corpus (9), the BIOADI corpus (10), an earlier version of the MEDSTRACT corpus (13) and the Schwartz and Hearst corpus (8). Statistics on abbreviation occurrences in the different corpora are contained in Table 1.
Table 1.
Characteristics of abbreviation definition corpora in biomedical literature
| Corpora | Ab3P | BIOADI | MEDSTRACT | Schwartz&Hearst |
|---|---|---|---|---|
| Number of abstracts | 1250 | 1201 | 199 | 1000 |
| Number of abbreviation definitions | 1223 | 1720 | 159 | 979 |
| Number of unique abbreviation definitions (across the whole corpus) | 1113 | 1421 | 152 | 842 |
| Number of unique abbreviations (ShortForms) | 998 | 1330 | 146 | 724 |
The original versions of these corpora resembled each other in that they all consisted of text files where documents were separated by blank lines. For each document we were given a passage of text. In the Ab3P corpus, the text for each document is divided into PubMed title and PubMed abstract lines, and in the BIOADI corpus, the title and abstract lines are concatenated together. In the Schwartz and Hearst corpus we were additionally given author list, affiliation field as well as publication type and venue. Finally, for the MEDSTRACT corpus (The MEDSTRACT corpus used for this study was downloaded from www.medstract.org in 2008 for an earlier study on abbreviations.), we were given bibliographic information in addition to the title and abstract of the journal articles, but PubMed document IDs (PMIDs) were missing. We used the journal citation information and author list to find the corresponding PMIDs for the MEDSTRACT corpus documents.
The original versions of Ab3P, BIOADI and MEDSTRACT contained each document followed by its list of <ShortForm, LongForm> pairs of abbreviations mentioned in the title or abstract, while the Schwartz and Hearst corpus contained an XML-like format where each definition was notated with in-text tags that were connected via an identification number, for example <Long id =N>, <Short id=N>. This format caused some difficulties. For example, corrections were required for: definitions with tags containing mismatched identifiers, malformed tags (for both long and short forms), repeated use of the same identifier for more than one abbreviation definition and incorrect tags.
In converting these corpora to BioC format, first, we kept all relevant text for abbreviation definition detection: title, abstract (and affiliation for the Schwartz and Hearst corpus). We left out author names and publication venue information for the MEDSTRACT and Schwartz and Hearst corpora, but these may be easily retrieved using the PMIDs. The major work in performing the conversion of these corpora to BioC format involved identifying the correct offsets for each defined abbreviation in the corresponding text. This step, in some cases, included multiple occurrences of a definition within the same passage, as well as correct identification of multiple offsets for multi-substring LongForm definitions.
During the BioC conversion process, we reviewed all the original annotations; and when searching for their offset locations in the text, new potential abbreviation pairs were found. All potential abbreviation cases and borderline cases were considered in detail. These were discussed by all the authors for inclusion or exclusion. The review process included searches on PubMed to find other publications that used the same abbreviation definition. This detailed analysis resulted in a handful of abbreviation pairs which were excluded from the corpora, and a few valid additional abbreviations which were added to the data. The final numbers for each corpus are shown in Table 1. As a result of these changes, the final numbers reflect a difference in the total number of annotations per corpus, when compared to the original publications. Problems of consistency are not surprising in corpora annotated by a single annotator. Different people reading the same document raise different points and catch missed definitions, thus ensuring a better quality for the final product.
Finally, we compared the four corpora for overlapping documents. We found that there was minimal to no overlap between the four corpora, as shown in Table 2. Continuing on this line of comparison, we extracted all MeSH terms assigned to all documents in each corpus, and performed a comparison between them. Figure 2 shows the relative importance of these terms for each corpus. The bare minimum overlap in document count, as shown in Table 2, as well as the collective MeSH terms of these corpora, as illustrated in Figure 2, demonstrates that these corpora can safely be combined into a larger abbreviation definition resource. This may help build more sophisticated abbreviation definition recognition models (Figure 3).
Table 2.
The overlap between corpora identified as number of documents that they have in common
| Corpora | Ab3P | BioADI | Medstract | Schwartz&Hearst |
|---|---|---|---|---|
| Ab3P | 1250 | 0 | 0 | 1 |
| BioADI | 1201 | 0 | 6 | |
| Medstract | 199 | 2 | ||
| Schwartz&Hearst | 1000 |
Figure 3.
Word cloud (http://www.wordle.net/create) representations of MeSH terms found in each corpus: Ab3P (top left), BIOADI (top right), MEDSTRACT (bottom left) and Schwartz and Hearst (bottom right). The MeSH terms confirm each corpus’ original intent: Ab3P was intended as a representation of all biomedical literature in PubMed, BIOADI is the corpus used in the BioCreative II gene normalization challenge, half of MEDSTRACT documents were a result of the search term ‘gene’ on MEDLINE restricted to a small group of biomedical journals and Schwartz and Hearst was a selection of documents returned as a result of the search term ‘yeast’ applied to PubMed.
Results
We tested the three abbreviation identifying modules on the Ab3P, BIOADI, MEDSTRACT and Schwartz and Hearst corpora as shown in Table 3. Results are based on the new gold standard annotations in the four abbreviation corpora. When compared to the outputs of the algorithms’ original versions, the BioC-compliant modules produced the same results. The BioC versions, however, have the advantage of being easily combined with other BioC-compliant tools to produce a more complex biomedical text processing system.
Conclusions
We present three easy-to-use, BioC-compatible abbreviation definition recognizing modules for biomedical text. The original tools corresponding to Ab3P, Schwartz and Hearst and NatLAb algorithms, have only been altered to read and produce the enriched BioC format. The new BioC-compatible modules faithfully preserve their original efficiency, running-time power and other complexity-related aspects, so they can be confidently used as a common pre-processing step for larger multi-layered text-mining endeavors.
We also present four BioC-formatted abbreviation definition recognition corpora that can be used to test the above modules, as well as to study new natural language processing tools. The new versions of the modules, as well as the accompanying corpora, are freely available to the community, through the BioC website: http://bioc.source forge.net.
Funding
This research was supported by the Intramural Research Program of the NIH, National Library of Medicine.
Conflict of interest. None declared.
References
- 1.Hirschman L., Yeh A., Blaschke C., et al. (2005) Overview of BioCreAtIvE: critical assessment of information extraction for biology. BMC Bioinformatics, 6, S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Krallinger M., Morgan A., Smith L., et al. (2008) Evaluation of text-mining systems for biology: overview of the Second BioCreative community challenge. Genome Biol., 9, S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arighi C.N., Lu Z., Krallinger M., et al. (2011) Overview of the BioCreative III Workshop. BMC Bioinformatics, 12, S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu C.H., Arighi C.N., Cohen K.B., et al. (2012) BioCreative-2012 virtual issue. Database, 2012, bas049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Comeau D.C., Islamaj Doğan R., Ciccarese P., et al. (2013) BioC: a minimalist approach to interoperability for biomedical text processing. Database, 2013, bat064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Comeau D.C., Batista-Navarro R.T., Dai H.-J., et al. (2014) BioC interoperability track overview. Database, 2014, bau053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Islamaj Dogan R., Murray G.C., Neveol A., et al. (2009) Understanding PubMed user search behavior through log analysis. Database, 2009, bap018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schwartz A.S., Hearst M.A. (2003) A simple algorithm for identifying abbreviation definitions in biomedical text. Pac. Symp. Biocomput., 451–462 [PubMed] [Google Scholar]
- 9.Sohn S., Comeau D.C., Kim W., et al. (2008) Abbreviation definition identification based on automatic precision estimates. BMC Bioinformatics, 9, 402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kuo C.J., Ling M.H., Lin K.T., et al. (2009) BIOADI: a machine learning approach to identifying abbreviations and definitions in biological literature. BMC Bioinformatics, 10, S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yeganova L., Comeau D.C., Wilbur W.J. (2011) Machine learning with naturally labeled data for identifying abbreviation definitions. BMC Bioinformatics, 12, S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu W., Comeau D.C., Islamaj D.R., et al. (2013) Extending BioC implementation to more languages. Proceedings of the Fourth BioCreative Challenge Evaluation Workshop, 1, 31–37 [Google Scholar]
- 13.Pustejovsky J., Castano J., Cochran B., et al. (2001) Automatic extraction of acronym-meaning pairs from MEDLINE databases. Stud. Health Technol. Inform., 84, 371–375 [PubMed] [Google Scholar]