A family 46 carbohydrate-binding module (BhCBM46) from B. halodurans endo-β-1,4-glucanase B (CelB) was purified and crystallized, and data were collected for the native form and a selenomethionine derivative to 2.46 and 2.3 Å resolution, respectively.
Keywords: endo-β-1,4-glucanase; carbohydrate-binding module family 46; Bacillus halodurans
Abstract
Plant cell-wall polysaccharides offer an abundant energy source utilized by many microorganisms, thus playing a central role in carbon recycling. Aerobic microorganisms secrete carbohydrate-active enzymes (CAZymes) that catabolize this composite structure, comprising cellulose, hemicellulose and lignin, into simple compounds such as glucose. Carbohydrate-binding modules (CBMs) enhance the efficacy of associated CAZYmes. They are organized into families based on primary-sequence homology. CBM family 46 contains more than 40 different members, but has yet to be fully characterized. Here, a recombinant derivative of the C-terminal family 46 CBM module (BhCBM46) of Bacillus halodurans endo-β-1,4-glucanase B (CelB) was overexpressed in Escherichia coli and purified by immobilized metal-ion affinity chromatography. Preliminary structural characterization was carried out on BhCBM46 crystallized in different conditions. The crystals of BhCBM46 belonged to the tetragonal space group I4122. Data were collected for the native form and a selenomethionine derivative to 2.46 and 2.3 Å resolution, respectively. The BhCBM46 structure was determined by a single-wavelength anomalous dispersion experiment using AutoSol from the PHENIX suite.
1. Introduction
Aerobic microorganisms secrete carbohydrate-active enzymes (CAZymes) as free-standing proteins that break down the plant cell-wall polysaccharides, comprising cellulose, hemicellulose and lignin, into simpler compounds such as ethanol and glucose that are needed to fulfil their energy requirements. CAZymes are usually modular proteins containing enzymatic catalytic domains associated with other modules such as carbohydrate-binding modules (CBMs). CBMs promote the close association of the appended catalytic domains to their targeting substrate(s), thus potentiating catalysis (Boraston et al., 2004 ▶). This proximity effect, i.e. increasing the concentration of the enzyme on the surface of the substrate, leads to a faster degradation of the polysaccharide and is pertinent to the hydrolysis of insoluble polysaccharides that characterize the majority of plant cell-wall components. CBMs also display a targeting function by directing their associated catalytic domain to their substrates within highly complex macromolecular structures such as the plant cell wall. Currently, 69 different families of CBM have been identified based on primary-sequence homology (see the CAZy website; Lombard et al., 2013 ▶). The majority of CBM families are well characterized in terms of both structure and function. However, a few, such as family 46 (CBM46), which comprises more than 40 different members, remain to be structurally and functionally characterized.
Bacillus halodurans is a rod-shaped, Gram-positive bacterium that is found in soil and water. The bacterium produces many industrially useful alkaliphilic enzymes such as proteases (protein-degrading enzymes), cellulases (cellulose-degrading enzymes) and amylases (starch-degrading enzymes). Encoded at locus BH0603 (GenBank accession No. BA000004) in the genome of B. halodurans (Takami et al., 2000 ▶) is a putative modular endo-β-1,4-glucanase (CelB) composed of an N-terminal glycoside hydrolase family 5 catalytic module (GH5) followed by an immunoglobulin-like module (Ig) and a C-terminal family 46 CBM (BhCBM46; Fig. 1 ▶). BhCBM46 binds to insoluble forms of cellulose, in particular Avicel, while the associated catalytic domain is a typical GH5 endocellulase (Wamalwa et al., 2006 ▶). BhCBM46 forms a significant part of the CelB polypeptide, and truncation studies reveal that removal of BhCBM46 results in an enzyme with a very limited overall action towards cellulosic substrates (Wamalwa et al., 2006 ▶). Thus, these data suggest that CBM46 members might display unique properties within the CBMs, and structural information on BhCBM46 will lead to a better understanding of its targeting function towards different substrates. In the present study, we describe the overproduction, purification, crystallization and preliminary X-ray analysis of both the native form and a selenomethionine derivative of BhCBM46.
Figure 1.
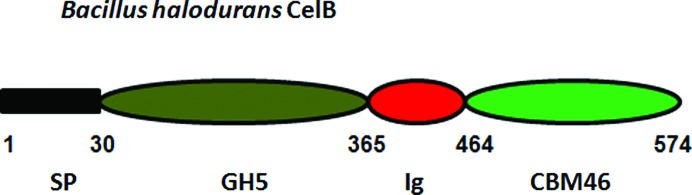
Schematic showing the modular architecture of full-length B. halodurans endo-β-1,4-glucanase (CelB). SP is the N-terminal signal peptide, GH5 is the catalytic module belonging to glycoside hydrolase family 5, Ig is the immunoglobulin-like module and CBM46 is the family 46 carbohydrate-binding module. The BhCBM46 construct used in this study covers the residue range 457–563. The boundary between Ig and BhCBM46 cannot be accurately predicted by sequence alignment and the C-terminus is expected to be flexible.
2. Materials and methods
2.1. Protein production and purification
B. halodurans genomic DNA was used and the gene encoding BhCBM46 was amplified and inserted into the pET-28a vector (Novagen) so that the encoded recombinant protein contained an N-terminal His6 tag (Wamalwa et al., 2006 ▶). The resulting plasmid was termed pCBM46_28a. Escherichia coli BL21 cells harbouring pCBM46_28a were cultured in Luria–Bertani broth at 310 K to mid-exponential phase (A 600 nm = 0.6) and recombinant protein overproduction was induced by the addition of 1 mM isopropyl β-d-1-thiogalactopyranoside and incubation for a further 16 h at 292 K. The His6-tagged recombinant protein was purified from cell-free extracts by immobilized metal-ion affinity chromatography (IMAC) as described previously (Najmudin et al., 2010 ▶). Purified BhCBM46 was buffer-exchanged into 50 mM HEPES–HCl pH 7.5, 200 mM NaCl, 5 mM CaCl2 but was not subjected to gel filtration.
Preparation of E. coli to generate selenomethionylated BhCBM46 (SeMet-BhCBM46) was performed as described in Najmudin et al. (2006 ▶) and the protein was purified using the same procedures as employed for the native BhCBM46. Purified BhCBM46 was concentrated using an Amicon 10 kDa molecular-mass centrifugal concentrator and washed three times with 5 mM DTT (for the SeMet protein) or water (for native BhCBM46). The recombinant BhCBM46 contains an N-terminal His6 tag (MGSSHHHHHHSSGLVPRGSHMAS) and amino-acid residues 457–563 of CelB, giving a total of 130 amino-acid residues (including two internal methionines) with an approximate molecular mass of 14.3 kDa, as also analyzed by SDS–PAGE (Fig. 2 ▶).
Figure 2.
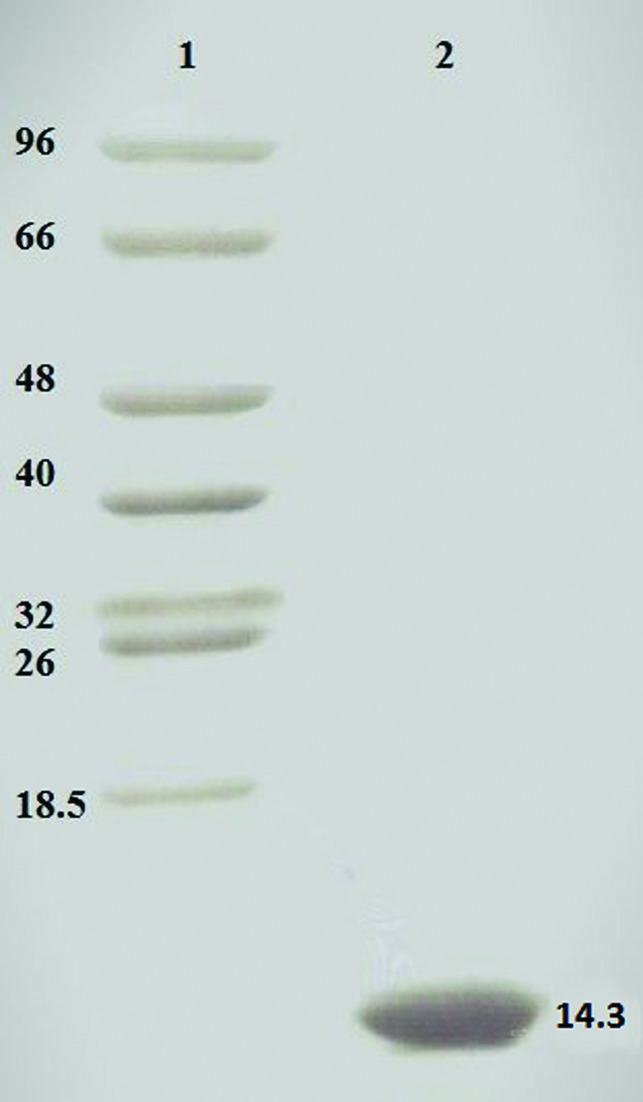
A Coomassie Brilliant Blue-stained 16% PAGE gel evaluation of protein purity. Lane 1, molecular-mass markers (labelled in kDa); lane 2, native BhCBM46. Similar purity was obtained for the SeMet-BhCBM46.
2.2. Crystallization
Crystallization conditions were screened by the sitting-drop vapour-phase-diffusion method using the commercial kits Crystal Screen, Crystal Screen 2, PEG/Ion and PEG/Ion 2 from Hampton Research (California, USA) and JBScreen 6–10 from Jena Bioscience using the Oryx8 robotic nanodrop dispensing system (Douglas Instruments). Two drops per well containing 30 µl reservoir solution were prepared: one consisting of 0.7 µl 28 mg ml−1 BhCBM46 and 0.7 µl reservoir solution, and one consisting of 0.7 µl 14 mg ml−1 BhCBM46 with 10 mM 1,4-β-d-cellohexaose (C6) and 0.7 µl reservoir solution. Crystals grew after four months in the following condition for both setups: 0.4 M potassium sodium tartrate tetrahydrate (Fig. 3 ▶ a).
Figure 3.
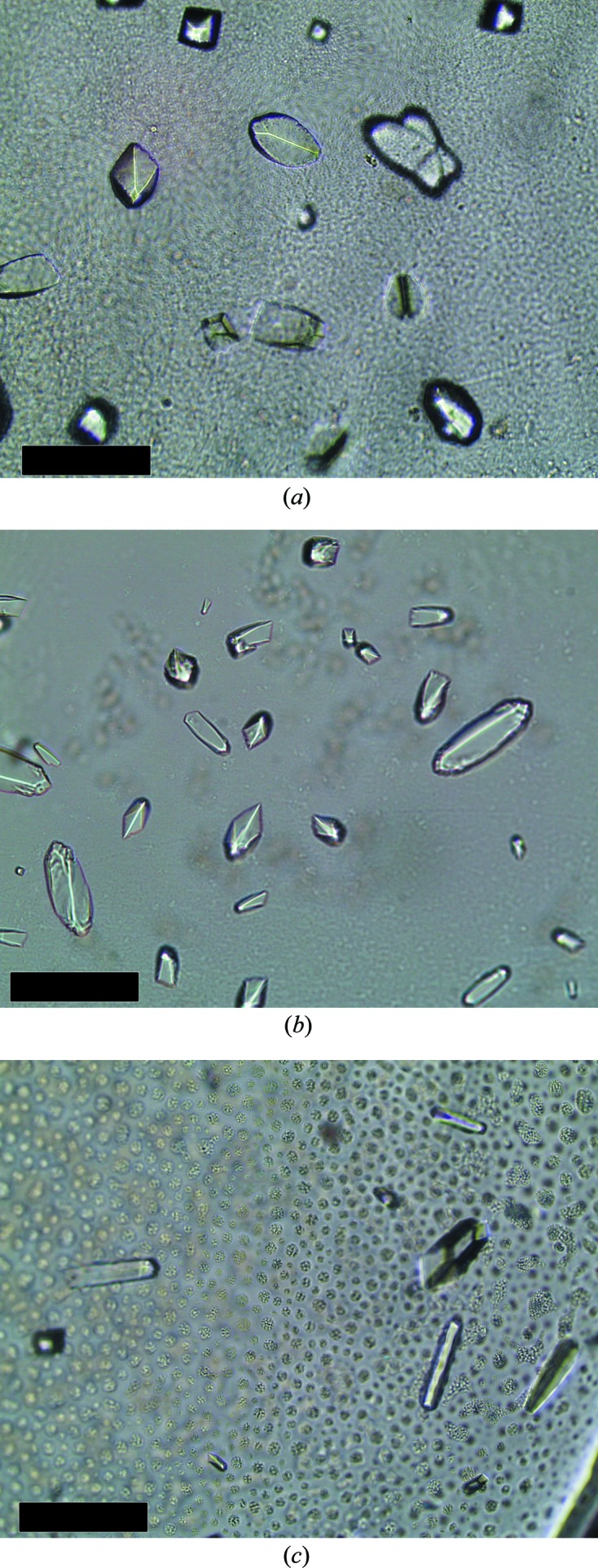
Crystals of BhCBM46 (with 10 mM 1,4-β-d-cellohexaose) and SeMet-BhCBM46 obtained by both sitting-drop and hanging-drop vapour-diffusion methods. (a) 0.4 M potassium sodium tartrate tetrahydrate, (b) 0.75 M potassium sodium tartrate tetrahydrate, (c) 18 mg ml−1 SeMet-BhCBM46 in 0.2 M ammonium sulfate, 30%(w/v) polyethylene glycol 4000. The longest dimension did not exceed 0.1 mm for any of the crystals. The black scale bar represents 0.1 mm.
Initially, crystals of SeMet-BhCBM46 were obtained by the hanging-drop vapour-diffusion method with equal volumes (1 µl) of protein solution (15.5 mg ml−1 in 5 mM DTT) with 10 mM of C6 and reservoir solution per well (containing 1 ml reservoir solution) from a fine screen based around the successful condition for the native crystals. Crystals grew after four months in the following condition: 0.75 M potassium sodium tartrate tetrahydrate (Fig. 3 ▶ b). However, these crystals gave poor diffraction. Subsequently, crystals of SeMet-BhCBM46 were obtained by the sitting-drop vapour-phase-diffusion method using the commercial kits Crystal Screen and Crystal Screen 2 from Hampton Research (California, USA) using the Oryx8 robotic nanodrop dispensing system (Douglas Instruments). A single drop consisting of 0.7 µl 18 mg ml−1 SeMet-BhCBM46 and 0.7 µl reservoir solution per condition was set up. Crystals grew after six months in the following condition: 0.2 M ammonium sulfate, 30%(w/v) polyethylene glycol 4000 (Fig. 3 ▶ c). All crystallization trials were carried out at 292 K. The crystals were cryocooled in liquid nitrogen after soaking in the cryoprotectant [30%(v/v) glycerol added to the crystallization buffer] for a few seconds.
2.3. Data collection and processing
Data sets were collected from various BhCBM46 crystals at the Diamond Light Source (DLS; Harwell, England) using a Quantum 315r charge-coupled device detector (ADSC) on beamline I04-1 and a PILATUS 6M detector (Dectris) on beamline I04, with the crystals cooled to 100 K using a Cryostream (Oxford Cryosystems). X-ray radiation of 0.97976 Å wavelength as determined by an energy scan was used to carry out data collection at the Se peak for a single-wavelength anomalous diffraction experiment (f′ = 6.12 and f′′ = −8.41 for the best data). The data were processed using iMosflm (Battye et al., 2011 ▶) and AIMLESS (Evans, 2011 ▶) from the CCP4 suite (Winn et al., 2011 ▶). The crystals were fragile and suffered radiation damage during or by the end of data collection, as was evident by a loss of resolution, an increase in mosaicity and a change in the unit-cell parameters during processing. All the diffracting BhCBM46 crystals belonged to the tetragonal space group (I4122), with two molecules in the asymmetric unit, a solvent content of ∼50% and a Matthews coefficient of ∼2.5 Å3 Da−1 (Matthews, 1968 ▶). The data collected for the SeMet-BhCBM46 were from the best diffracting crystal (Fig. 3 ▶ c) and were used to solve the BhCBM46 structure. 360° of data were collected with a Δϕ of 0.2°. The crystal diffracted to a resolution of 2.3 Å. Attempts were made to collect data at the inflection-point and remote wavelengths, but the crystals did not survive. The native crystals (Fig. 3 ▶ b) were even more fragile and suffered radiation damage before the end of data collection, as evident from the high mosaicity and R merge values at this resolution. 140° of data were collected with a Δϕ of 0.2°. Data-collection statistics are presented in Table 1 ▶. The SeMet-BhCBM46 structure was determined by a single-wavelength anomalous dispersion experiment using AutoSol (Terwilliger et al., 2009 ▶) from the PHENIX suite (Adams et al., 2010 ▶). Seven heavy-atom sites were identified with a figure of merit of 0.454 and an overall score of 49.85. Four of these corresponded to the four well defined internal SeMet residues expected in the BhCBM46 dimer. The final model after AutoBuild (Terwilliger et al., 2008 ▶) placed 126 amino-acid residues out of a potential 260 in 20 fragments with an R work and R free of 0.472 and 0.515, respectively. The three-dimensional structure for the native BhCBM46 was solved by molecular replacement using Phaser (McCoy et al., 2007 ▶) with the SeMet-derivative model as a search model, giving a TFZ of 22.0 and an LLG of 2404. Further structure refinement and analysis are ongoing.
Table 1. Data-collection statistics.
Values in parentheses are for the highest resolution shell.
| Data set | SeMet peak | Native |
|---|---|---|
| Beamline | I04, DLS | I04-1, DLS |
| Space group | I4122 | I4122 |
| Wavelength (Å) | 0.97976 | 0.9200 |
| Unit-cell parameters | ||
| a (Å) | 120.85 | 121.19 |
| b (Å) | 120.85 | 121.19 |
| c (Å) | 76.38 | 77.28 |
| Resolution limits (Å) | 85.45–2.30 (2.38–2.30) | 44.37–2.46 (2.56–2.46) |
| Average mosaicity (°) | 0.23 | 1.53 |
| No. of observations | 246067 (9270) | 87997 (9188) |
| No. of unique observations | 12870 (1231) | 10730 (1191) |
| Multiplicity | 19.1 (7.5) | 8.2 (7.7) |
| Completeness (%) | 100 (99.6) | 99.8 (100.0) |
| 〈I/σ(I)〉 | 17.1 (3.8) | 9.7 (1.3) |
| CC1/2 † | 0.99 (0.89) | 0.998 (0.613) |
| R merge ‡ (%) | 12.4 (79.7) | 12.6 (163.5) |
| R p.i.m. § (%) | 2.9 (30.4) | 4.6 (61.5) |
CC1/2 is the half-data-set correlation coefficient (Diederichs & Karplus, 2013 ▶).
R
merge = 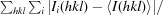
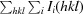 , where Ii(hkl) is the ith intensity measurement of reflection hkl, including symmetry-related reflections, and 〈I(hkl)〉 is its average.
, where Ii(hkl) is the ith intensity measurement of reflection hkl, including symmetry-related reflections, and 〈I(hkl)〉 is its average.
R
p.i.m. = 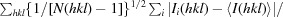
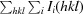 , where 〈I(hkl)〉 is the average of symmetry-related observations of a unique reflection.
, where 〈I(hkl)〉 is the average of symmetry-related observations of a unique reflection.
Acknowledgments
The authors acknowledge financial support from Fundacão para a Ciência e a Tecnologia, Portugal through projects PTDC/BIA-PRO/103980/2008 and PTDC/BIAPRO/100359/2008. This work was supported by the European Union Seventh Framework Programme (FP7 2007–2013) under the WallTraC project (grant agreement No. 263916). This paper reflects the authors’ views only. The European Community is not liable for any use that may be made of the information contained herein. We acknowledge Diamond Light Source, Oxfordshire, England for provision of synchrotron-radiation facilities (beamlines I04 and I04-1). We would also like to thank Cecília Bonifácio for help with crystallization, Drs Juan Sanchez-Weatherby, Carina Lobley, Pedro Bule and Joana L. A. Brás for help with data collection and the European Community’s Seventh Framework Programme (FP7/2007–2013) under BioStruct-X (grant agreement No. 283570, proposal No. Biostruct-X_4399) for funding.
References
- Adams, P. D. et al. (2010). Acta Cryst. D66, 213–221.
- Battye, T. G. G., Kontogiannis, L., Johnson, O., Powell, H. R. & Leslie, A. G. W. (2011). Acta Cryst. D67, 271–281. [DOI] [PMC free article] [PubMed]
- Boraston, A. B., Bolam, D. N., Gilbert, H. J. & Davies, G. J. (2004). Biochem. J. 382, 769–781. [DOI] [PMC free article] [PubMed]
- Diederichs, K. & Karplus, P. A. (2013). Acta Cryst. D69, 1215–1222. [DOI] [PMC free article] [PubMed]
- Evans, P. R. (2011). Acta Cryst. D67, 282–292. [DOI] [PMC free article] [PubMed]
- Lombard, V., Golaconda Ramulu, H., Drula, E., Coutinho, P. M. & Henrissat, B. (2013). Nucleic Acids Res. 42, D490–D495. [DOI] [PMC free article] [PubMed]
- Matthews, B. W. (1968). J. Mol. Biol. 33, 491–497. [DOI] [PubMed]
- McCoy, A. J., Grosse-Kunstleve, R. W., Adams, P. D., Winn, M. D., Storoni, L. C. & Read, R. J. (2007). J. Appl. Cryst. 40, 658–674. [DOI] [PMC free article] [PubMed]
- Najmudin, S., Guerreiro, C. I. P. D., Carvalho, A. L., Prates, J. A. M., Correia, M. A. S., Alves, V. D., Ferreira, L. M. A., Romão, M. J., Gilbert, H. J., Bolam, D. N. & Fontes, C. M. G. A. (2006). J. Biol. Chem. 281, 8815–8828. [DOI] [PubMed]
- Najmudin, S., Pinheiro, B. A., Prates, J. A. M., Gilbert, H. J., Romão, M. J. & Fontes, C. M. G. A. (2010). J. Struct. Biol. 172, 353–362. [DOI] [PubMed]
- Takami, H., Nakasone, K., Takaki, Y., Maeno, G., Sasaki, R., Masui, N., Fuji, F., Hirama, C., Nakamura, Y., Ogasawara, N., Kuhara, S. & Horikoshi, K. (2000). Nucleic Acids Res. 28, 4317–4331. [DOI] [PMC free article] [PubMed]
- Terwilliger, T. C., Adams, P. D., Read, R. J., McCoy, A. J., Moriarty, N. W., Grosse-Kunstleve, R. W., Afonine, P. V., Zwart, P. H. & Hung, L.-W. (2009). Acta Cryst. D65, 582–601. [DOI] [PMC free article] [PubMed]
- Terwilliger, T. C., Grosse-Kunstleve, R. W., Afonine, P. V., Moriarty, N. W., Zwart, P. H., Hung, L.-W., Read, R. J. & Adams, P. D. (2008). Acta Cryst. D64, 61–69. [DOI] [PMC free article] [PubMed]
- Wamalwa, B. M., Sakka, M., Shiundu, P. M., Ohmiya, K., Kimura, T. & Sakka, K. (2006). Appl. Environ. Microbiol. 72, 6851–6853. [DOI] [PMC free article] [PubMed]
- Winn, M. D. et al. (2011). Acta Cryst. D67, 235–242.


