Abstract
Castration-resistant prostate cancer (CRPC) continues to pose a significant clinical challenge with new generation second line hormonal therapies affording limited improvement in disease outcome. As the androgen receptor (AR) remains a critical driver in CRPC, understanding the determinants of its transcriptional activity is important for developing new AR targeted therapies. FOXA1 is a key component of the AR transcriptional complex yet its role in prostate cancer progression and the relationship between AR and FOXA1 are not completely resolved. It is well established that FOXA1 levels are elevated in advanced prostate cancer and metastases. We mimicked these conditions by over-expressing FOXA1 in the androgen-responsive LNCaP prostate cancer cell line and observed a significant increase in AR genomic binding at novel regions that possess increased chromatin accessibility. High levels of FOXA1 resulted in increased proliferation at both sub-optimal and high 5α-dihydrotestosterone (DHT) concentrations. Immunohistochemical staining for FOXA1 in a clinical prostate cancer cohort revealed that high FOXA1 expression is associated with shorter time to biochemical recurrence after radical prostatectomy (HR 5.0, 95% CI 1.2-21.1, p=0.028), positive surgical margins and higher stage disease at diagnosis. The gene expression program that results from FOXA1 over-expression is enriched for PTEN, Wnt and other pathways typically represented in CRPC gene signatures. Together these results suggest that in an androgen-depleted state, elevated levels of FOXA1 enhance AR binding at genomic regions not normally occupied by AR, which in turn facilitates prostate cancer cell growth.
Keywords: Androgen receptor, FOXA1, prostate cancer, CRPC, genomics
Prostate cancer is the most common non-cutaneous cancer in men accounting for almost one third of all newly diagnosed cancers (1). The standard first line of treatment for metastatic prostate cancer is androgen deprivation therapy (ADT), which typically induces regression of the tumor. Despite a high initial response rate in nearly all cases, resistance to ADT occurs resulting in tumor regrowth which is termed castration-resistant prostate cancer (CRPC) (2). Tumors that become resistant to ADT pose a significant clinical challenge. The development of new hormonal therapies that target androgen biosynthesis (e.g. Abiraterone) or the androgen receptor directly (e.g. Enzalutamide) has produced improved outcomes for patients with CRPC, but they are not universally effective and responses are not durable (3, 4). The androgen receptor (AR) remains active and critical for tumor growth in CRPC despite low circulating levels of androgens, but typically there are alterations in the receptor structure or function that allow it to retain activity despite ADT. Documented methods for circumvention of ADT include AR gene amplification, acquisition of mutations, genomic rearrangements and alternative splicing of AR (2, 5-8). An additional mechanism is altered interaction of AR with key transcriptional cofactors (9).
The forkhead transcription factor FOXA1 is a key member of the AR transcriptional complex, which has been shown to interact directly with AR through the hinge domain (10). FOXA1 functions primarily as a pioneer factor, binding to closed chromatin regions through its winged helix domain, which has a structure akin to that of linker histones. FOXA1 association with chromatin contributes to changes in chromatin accessibility, rendering these regions more accessible to nuclear receptors such as AR (11), ER (12) and PR (13) . As such, FOXA1 plays a key role in demarcating the tissue specific binding sites of these nuclear receptors (14). FOXA1, expressed in the peripheral zone of the human prostate (15), is essential for hormone induced ductal branching and epithelial cell maturation of the prostate gland during puberty (16). Furthermore, FOXA1 plays a central role in AR driven gene expression in both the normal prostate and prostate cancers (11, 17, 18).
There are a number of conflicting reports regarding the role that FOXA1 plays in the progression of prostate cancer to castrate resistant disease and its expression has been associated with both good and poor prostate cancer patient outcome. One study suggested that FOXA1 mRNA levels are moderately up-regulated in primary cancer when compared to benign disease, but unexpectedly it was down-regulated in metastasis (19). In contrast to this observation, a number of immunohistochemical studies have shown that FOXA1 is a marker of poor outcome in prostate cancer, with high FOXA1 levels being associated with a shorter time to biochemical recurrence (15, 20) or prostate cancer-specific death (21). High expression at the protein level has also been noted in the majority (82-89%) of metastatic and CRPC samples (15, 22).
At the molecular level, it remains to be determined what the exact consequences of higher FOXA1 levels are for prostate cancer progression. Examination of AR binding events in CRPC cell line models and primary tissues suggests that the unique CRPC AR sites are not reliant upon FOXA1 (23, 24) and similarly, FOXA1 may have AR independent functions in CRPC (25). Despite this, FOXA1 over-expressing LNCaP prostate cancer cells exhibit increased migration and produce larger tumors in xenograft models (15, 26). Thus it is imperative to assess the consequences of elevated FOXA1 expression and the potential impact on AR signalling during prostate cancer progression.
In this study, we assessed the downstream effects of elevated FOXA1 levels on AR chromatin binding, gene expression and prostate cancer cell proliferation. We found that in cells expressing higher levels of FOXA1, AR makes novel chromatin associations and is able to drive tumor cell growth even at reduced levels of androgen, suggesting that tumors with higher FOXA1 levels may have a growth advantage following androgen deprivation.
Results
Higher levels of FOXA1 result in increased AR binding
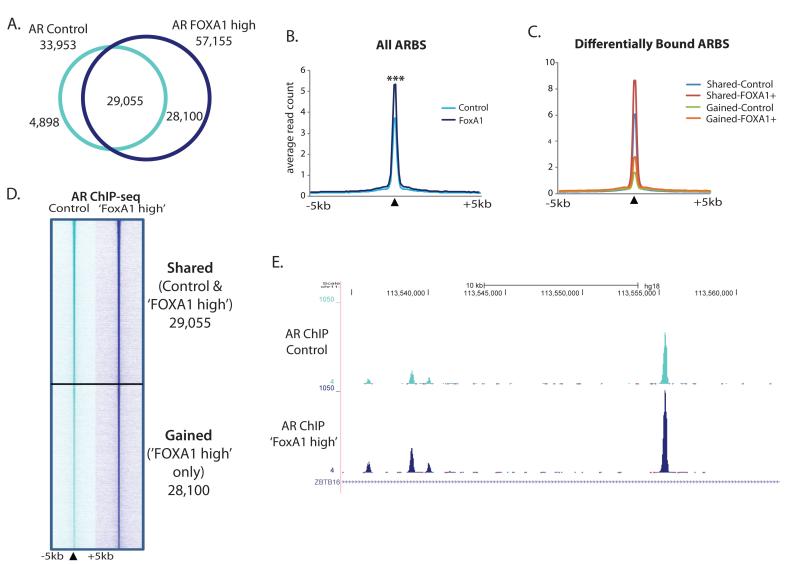
Given the disputed role that elevated levels of FOXA1 play in prostate cancer, we aimed to examine the effects of over-expression of FOXA1 in prostate cancer cells. The LNCaP prostate cancer cell line was transiently transfected with a control vector or a FOXA1 construct, which we subsequently termed ‘FOXA1 high’ cells. Western blot analysis confirmed increased FOXA1 levels compared to GFP control transfected cells (Supplementary Figure 1A). Since AR is central to both primary and castrate resistant prostate cancer, we initially focused on the effect of increased levels of FOXA1 upon AR binding. Genome wide AR binding regions were mapped in asynchronous ‘FOXA1 high’ versus GFP control cells and we found an almost two-fold increase in reproducible AR binding sites (ARBS) in ‘FOXA1 high’ cells (33,953 ARBS in control cells versus 57,155 ARBS in ‘FOXA1 high’ cells, Supplementary Table 1). The majority (86%) of the ARBS in control cells were also seen in FOXA1 high cells. However, in elevated FOXA1 conditions, an additional 28,100 ARBS were observed (Figure 1A).
Figure 1. AR binding is enhanced in the presence of high levels of FOXA1.
A. AR binding was mapped in FOXA1 over-expressing, ‘FOXA1 high’, versus Control transfected LNCaP cells using ChIP-seq. Venn diagram shows the overlap between the two datasets. B. Average binding intensity plot for AR ChIP-seq signal in control or ‘FOXA1 high’ cells at all AR binding events, p value <0.0001. C. Average read count plot for AR ChIP-seq signal in control or ‘FOXA1 high’ cells at differentially bound regions. D. Heatmap of raw AR reads for ChIP-seq in control or ‘FOXA1 high’ cells at the AR binding sites (ARBS) shared or unique to ‘FOXA1 high’ cells. Black triangle denotes the summit of peak and a 10kb window surrounding the summit is shown. E. Genome browser snapshot showing AR ChIP-seq signal in control and ‘FOXA1 high’ cells in 1MB region of chromosome 11.
In general, there was a significant increase in AR peak strength across all sites in ‘FOXA1 high’ cells compared to control (Figure 1B). Shared regions identified in both control and FOXA1 over-expressing cells have a significantly higher mean read count at the AR peak summit in ‘FOXA1 high’ cells (8.34 reads) compared to control (6.03 reads; Figure 1C). Additionally, there are a large number of sites (28,100) where an ARBS was only detected in ‘FOXA1 high’ cells, termed ‘gained’ sites (Figure 1C). Interestingly, the new AR binding sites that occur in the presence of elevated FOXA1 are regions that have some weak binding in control cells, implying an amplification of AR-DNA binding at suboptimal binding domains (mean read count 2.76 in ‘FOXA1 high’ versus 1.56 in Control cells; Figure 1C). Overall our data suggests a general amplification of AR binding in the presence of higher FOXA1 levels as illustrated by a heatmap of the shared and gained ARBS (Figure 1D) and an example genomic region (Figure 1E). This phenomenon was confirmed in another AR driven cancer cell line model, the ERα-AR+ MDA-MB-453 cell line, which is used as a model of molecular apocrine breast cancer. AR ChIP-seq in ‘FOXA1 high’ MDA-MB-453 cells also showed greater AR binding intensity than in control cells (Supplementary Figure 2A&B and Supplementary Table 1).
Two recently published studies reported reprogramming of AR to de novo regions in the genome following loss of FOXA1 in LNCaP prostate cancer cells (10, 21). We performed hierarchical cluster analysis to compare the sites of AR occupancy in our GFP control and ‘FOXA1 high’ LNCaP cells with those in the siControl and siFOXA1 treated LNCaP cells from the published studies. By directly comparing the changes that occur in AR binding in conditions where FOXA1 is both absent and over-expressed, we see that the ARBS determined in the siFOXA1 treatment group cluster separately from those in ‘FOXA1 high’ cells. This indicates that the novel ARBS seen in ‘FOXA1 high’ cells are not the same locations as the de novo AR binding seen after loss of FOXA1 (Supplementary Figure 1B & C).
Increase in AR Occupancy after FOXA1 over-expression is likely due to increased chromatin accessibility
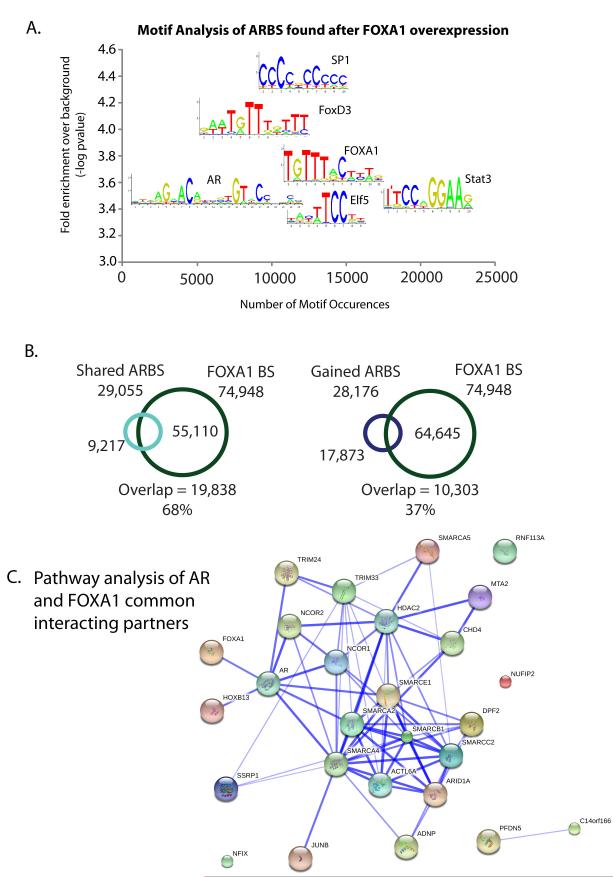
In order to infer the mechanism behind the increased AR binding seen after FOXA1 over-expression, we assessed whether there was a global increase in AR protein levels, but we did not detect an appreciable change in AR levels following FOXA1 over-expression (Supplementary Figure 1A). Therefore to identify alternate factors that may be redirecting AR to the chromatin, we conducted de novo motif analysis on the unique ARBS associated with FOXA1 over-expression. We found significant enrichment for motifs representing STAT3, SP1 and the ETS (ELF5) transcription factors (Figure 2A). We also detected a strong enrichment for forkhead motifs suggesting a large majority of the new ARBS may be directed by FOXA1 itself. Comparison of our data with previously published LNCaP FOXA1 ChIP-seq data (27) revealed that FOXA1 is present at a large number (37%) of the gained ARBS sites (Figure 2B). Therefore over-expression of FOXA1 may directly mediate AR binding to ‘FOXA1 high’ gained regions. This is supported by the visible but relatively weak FOXA1 binding at these gained AR binding regions, which one could postulate is increased upon FOXA1 over-expression (Supplementary Figure 3).
Figure 2. AR may increase binding via its interactions with a number of chromatin remodelling proteins.
A. Motif analysis of ARBS unique to ‘FOXA1 high’ cells reveals Forkhead, ARE, Stat and Ets motifs B. Overlap of ARBS shared or unique to ‘FOXA1 high’ cells with published FOXA1 ChIP-seq data (27), regions must overlap by at least 1 bp. C. Protein functional network of common AR and FOXA1 interacting proteins identified by proteomic profiling of the transcription factor complexes. Blue line denotes published evidence of interaction between two proteins and the width of the line is indicative of the confidence of the interaction.
In an effort to identify additional regulatory mechanisms that impinge on the AR-FOXA1 complex, we performed RIME (Rapid IMmunoprecipitation of Endogenous proteins) proteomic analysis (28) of each transcription factor. Three independent replicates of AR and two independent replicates of FOXA1 RIME were conducted. Proteins must have been detected in at least two replicates, but in none of the matched IgG control immunoprecipitations to be considered an interacting protein. This led to the identification of 139 AR and 236 FOXA1 interacting proteins (Supplementary Table 2). In total, 56 proteins interact with both AR and FOXA1 in our data. Pathway analysis of these common interacting partners revealed a highly significant enrichment for proteins involved in chromatin remodelling, in particular for the locus control region (LCR)-associated remodelling complex, LARC (Figure 2C, Supplementary Table 2). LARC is a complex that controls accessibility of regulatory DNA sequences situated many kilobases away from their cognate promoters (29) that AR and FOXA1 co-occupy. A number of these proteins have been validated in other studies to interact with AR (30-33). FOXA1’s intrinsic pioneer factor function coupled with its interaction with a large number of histone modifying enzymes suggests that increased binding of AR under FOXA1 high conditions may occur because of enhanced chromatin accessibility at these regions.
FOXA1 over-expression increases proliferation in AR-driven cancers and correlates with poor outcome in prostate cancer
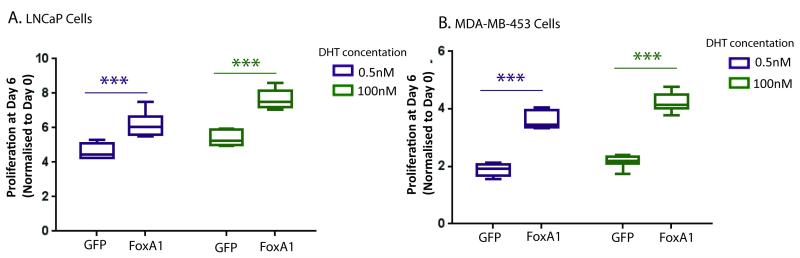
The effect of high levels of FOXA1 on prostate cancer proliferation was assessed in AR+ LNCaP prostate cancer cells and our second AR-driven cancer model, the MDA-MB-453 molecular apocrine breast cancer cell line. Western blot analysis confirmed FOXA1 over-expression in MDA-MB-453 cells with no alteration in AR protein levels (Supplementary Figure 1A). Proliferation of both cell lines was assessed following treatment with either high (100nM DHT) or low (0.5nM DHT) androgen concentrations, the latter to mimic androgen depleted conditions observed following ADT. ‘FOXA1 high’ cells exhibited significantly higher levels of proliferation than GFP control transfected cells at both DHT concentrations (Figure 3A). Interestingly, a similar effect of FOXA1 over-expression was observed in the MDA-MB-453 breast cancer cells following treatment with high and low doses of androgen (Figure 3B), suggesting that higher expression of FOXA1 enhances the growth promoting effects of AR in a ubiquitous, non-tissue specific manner in AR-driven cancers.
Figure 3. FOXA1 over-expression increases DHT stimulated growth in AR-driven cancers.
CellTiter-Blue cell viability assay assessing the cell density of A. LNCaP prostate cancer and B. MDA-MB-453 breast cancer cell lines which were hormone deprived, transfected with GFP control or FOXA1 plasmids and then stimulated with low 0.5nM DHT or high 100nM DHT levels of androgen. The data shown is an average of 8 replicates from one of three independent experiments, *** denotes p value <0.001. Error bars indicate standard deviations.
To address some of the conflicting data regarding the prognostic value of FOXA1 expression in prostate cancer, we undertook an immunohistochemical analysis of FOXA1 in a tissue microarray (TMA) of 102 prostate cancer cases. Our study with a median follow up time of 86 months after radical prostatectomy (range 3 to 151 months) has longer follow up than other published studies. Examples of FOXA1 immunostaining in prostate cancer are shown in Figure 4 A-F. We found FOXA1 expression to be significantly higher in tumors compared to matched benign and normal prostate tissues (median H-score cancer = 69.44 versus benign = 0, p value <0.0001, Supplementary Figure 4).
Figure 4. FOXA1 expression correlates with poor outcome in prostate cancer.
FOXA1 immunohistochemistry was performed on a prostate cancer TMA. For all sections, nuclei are shown in blue and FOXA1 staining in brown. Examples of staining criteria used are shown. A. Benign, B&C. Gleason Grade 3, D&E. Grade 4, F. Grade 5 Magnification is 20X. G. Kaplan Meier survival curve indicating time to biochemical recurrence for patients with no/low FOXA1 staining (H score ≤4) or high FOXA1 staining (H-score 5-300). Survival information was available for 84 patients of whom 30 (36%) experienced biochemical recurrence.
In evaluating the association between FOXA1 H-score and biochemical recurrence, we divided patients into two groups based on the first quartile given that this cut-point showed the most significant difference. There was a significantly higher risk of biochemical recurrence for patients with high FOXA1 staining (H-score >4.0, second through fourth quartile) compared to those with no or low FOXA1 staining, H-score ≤4.0 (HR 5.0, 95% CI 1.2-21.1, p=0.028, Figure 4G). In particular, we noted a marked difference in the ten year biochemical recurrence free survival rate; 94.1% (95% CI 83.6%-100%) for patients with low FOXA1 staining compared to 53.6% (95% CI 42.2%-68.0%) for those with high FOXA1 staining. As detailed in Table 1, high FOXA1 staining also significantly correlated with higher pathological stage (p <0.0001) and positive surgical margins (p = 0.021), which are markers of poor disease outcome (Supplementary Table 3). There was no statistically significant evidence of an association with Gleason Score (p = 0.25). Additionally, we assessed whether FOXA1 predicted outcome independently of other known prostate cancer markers. We observed a greater than three-fold increased risk of biochemical recurrence in patients with high FOXA1 staining compared to those with low staining when adjusting individually for pathological stage (HR: 3.60, p=0.084, n=79) Gleason Score (HR: 3.02, p=0.13, n=74) and PSA (HR: 4.58, p=0.039, n=76), although this was not always significant in these smaller samples with missing clinicopathologic data.
Table 1.
Association of FOXA1 Staining with Clinical Parameters
| FOXA1 Low (n=26) |
FOXA1 High (n=76) |
p value | |
|---|---|---|---|
|
Age at surgery
(years) |
62 (58-70) | 63 (45-69) | 0.523 |
| Gleason Grade | 0.247 | ||
| G6 | 14 (53.8%) | 31 (40.8%) | |
| G7 | 11 (42.3%) | 39 (51.3%) | |
| G8 | 1 (3.9%) | 6 (7.9%) | |
| Pathological Stage | <0.0001 | ||
| T2 | 14 (73.7%) | 30 (49.2%) | |
| T3 & T4 | 5 (26.3%) | 31 (50.8%) | |
|
PSA at diagnosis
(ng/mL) |
7.2 (3.9-16.2) | 6.9 (2.1- 17.2) | 0.273 |
| Surgical Margins | 0.021 | ||
| Clear | 13 (68.4%) | 24 (37.5%) | |
| Positive | 6 (31.6%) | 40 (62.5%) |
Patients were divided into quartiles based on the FOXA1 H-score; first quartile = FOXA1 low, second, third and fourth quartiles = FOXA1 high. Median and range is given for age and PSA values. Information was unavailable for some patients regarding pathological stage (n=80), PSA (n= 77) and surgical margins (n=83).
High FOXA1 induce a gene expression program similar to CPRC tissue signature
To elucidate the downstream effects of elevated FOXA1 expression, we conducted microarray analysis on Control versus ‘FOXA1 high’ asynchronous LNCaP cells. Using six biological replicates, we found 124 differentially regulated genes (DEG) with an FDR <0.05; this included 51 up-regulated and 73 down-regulated genes in ‘FOXA1 high’ cells (Figure 5A). The majority of these genes have an ARBS within 25kb of their start site in ‘FOXA1 high’ cells (93/124 DEG, 75%) implying that AR may be directly regulating expression of these differential genes. However, these genes are not typically regulated in vitro by DHT in the context of normal endogenous levels of FOXA1, with only 5.6% of the differentially expressed genes in ‘FOXA1 high’ cells considered to be androgen-regulated in an independent dataset of DHT-treated LNCaP cells (34).
Figure 5. FOXA1 over-expression DEG correlate with CRPC gene expression program.
A. Microanalysis of gene expression changes in LNCaP cells transfected with FOXA1 over-expression plasmid for 48h. Only genes with an FDR <0.05 were considered which resulted in 124 FOXA1-high differentially expressed genes. B. The 124 FOXA1-high genes were analyzed for enriched biological pathways. C. Gene Set Enrichment analysis for genes determined in CRPC signature (24) using expression data from FOXA1 high LNCaPs. NES = Normalized Enrichment Score.
Pathway analysis of the ‘FOXA1 high’ differentially expressed genes (Figure 5B) revealed a number of networks focused on signalling pathways known to be perturbed in prostate cancer (PTEN null, polycomb targets and TNC targets which signal through WNT) as well as differentiation pathways (Trogatizalone targets, poorly differentiated carcinoma and mature luminal cell program). Due to the high expression of FOXA1 in CRPC and metastases (15, 22), we used Gene Set Enrichment Analysis to correlate the CRPC gene program identified by Sharma et al (24) from primary CRPC tissue with the data from our ‘FOXA1 high’ microarray. There was a significant enrichment of the CRPC gene set in our data set (p value 0.018, normalized enrichment score 1.14, Figure 5C) implying that FOXA1 may be in part responsible for the expression of a number of the genes expressed in CRPC. In support of this concept, others have shown that silencing of FOXA1 in LNCaP-abl cells (one model of CRPC) has both AR-dependent and –independent effects on cell proliferation (25), and that loss of FOXA1 inhibits ligand-dependent AR chromatin binding in C4-2B cells (another LNCaP-derived model of CRPC (23). Importantly we found that silencing of FOXA1 in C4-2B cells markedly inhibited their proliferative capacity (Supplementary Figure 5). Our data suggests that high levels of FOXA1 can drive a gene expression program that is similar to that seen in patients who develop castrate resistant disease, implying a central role for FOXA1 in the development of CRPC.
Discussion
FOXA1 is a well-established member of the AR transcription complex, where it is required to direct AR binding to tissue specific sites by opening up chromatin (11, 14, 17, 18). In addition, FOXA1 is essential for optimal proliferative capacity of AR-positive LNCaP-derived prostate cancer cell lines (20, 25). Herein, we model for the first time the genomic consequences of elevated levels of FOXA1 in LNCaP prostate cancer cells and show that abnormally high FOXA1 levels induce a genome-wide increase in AR chromatin binding at 29,055 “expected” genomic locations, without a concomitant increase in AR protein levels. In addition to enhancement of AR binding to traditional AR regulatory elements with overexpression of FOXA1, there was a significant increase in AR binding at 28,100 ARBS that exhibited weak binding under endogenous levels of FOXA1 expression. These findings suggest that elevated FOXA1 levels, which are characteristic of CRPC, facilitate AR binding to secondary sites where AR typically binds with low affinity and is not transcriptionally active. Importantly, the gained AR sites observed under elevated FOXA1 conditions contain both canonical forkhead and ARE motifs. When FOXA1 levels are increased, FOXA1 binds to these regions, making them more accessible in a greater proportion of the cells, resulting in an overall increase in AR binding. It is important to note that in the opposite condition, where FOXA1 is lost, AR is also reprogrammed (10, 21), but the novel AR binding sites that occur in the absence of FOXA1 are not the same as those that arise with over-expression of FOXA1. The simplest hypothesis for this difference is that in the absence of FOXA1, alternative transcription factors mediate AR binding to chromatin. In the presence of FOXA1, relative levels of this pioneer factor appear to be the rate-limiting determinant of AR chromatin binding in prostate cancer cells. Under conditions where FOXA1 is abnormally elevated, AR gains the capacity for enhanced chromatin binding at non-classical sites, as observed in CRPC. Intriguingly, these alterations in AR and FOXA1 binding elicit a gene expression program that is reminiscent of that seen in CRPC primary tissues, which also exhibit high FOXA1 expression (15, 22). Resistance to ADT and progression to CRPC involves a diverse range of adaptive mechanisms to enhance AR signaling (4). Herein we provide evidence that one such mechanism could be via up-regulation of FOXA1, resulting in an expanded AR chromatin binding landscape. Tumors with high FOXA1 levels may have greater intrinsic AR activity and therefore are better adapted to survive at low levels of androgen. The increased proliferation that ‘FOXA1 high’ cells exhibited with low androgen concentrations in our study supports this theory. Further in vivo investigation of the effect of increased FOXA1 levels on tumor growth is required. To date, two reports have been published investigating the effects of high FOXA1 levels on tumor growth, but the conclusions were inconclusive. The first of these studies concluded that elevated FOXA1 inhibit prostate cancer metastasis in an orthotopic mouse xenograft model (19). However, in that study, the AR negative PC3-M cell line, which is not representative of the vast majority of prostate tumors, including CRPC, was used as the experimental model, Another xenograft study that examined the consequence of FOXA1 over-expression in AR+ LNCaP cells found a significant increase in tumor size compared to parental control cell xenografts (26), consistent with the results of the current study.
FOXA1 has been implicated as a marker of poor outcome in a number of tumor types, including lung (35), thyroid (36) esophageal (37) and malignant glioma (38). Our data supports the growing bank of evidence suggesting that FOXA1 is a marker of poor outcome in prostate cancer patients, where high expression levels correlate with a shorter time to biochemical relapse (15, 20, 22, 39). Furthermore, high FOXA1 expression robustly correlates with higher pathological stage, higher Gleason Score and AR expression (15, 20, 21), implying a central role for FOXA1 in the progression of prostate cancer. In contrast, high FOXA1 mRNA levels correlate with better patient outcome measures, such as a longer time to biochemical recurrence (19, 40). A study of the concordance between changes seen by mRNA microarray analysis and of high-throughput proteomic profiling of primary prostate cancer tissue showed only a 48% - 61% agreement (41). Given the role of FOXA1 as a transcription factor that modulates chromatin accessibility and binding of other proteins, it is prudent to consider the correlations that occur with protein levels rather than mRNA levels.
The exact role that FOXA1 plays in CRPC is still unclear, but interestingly it was recently observed that FOXA1 is mutated in 3-5% of prostate cancers, suggesting that altered FOXA1 function may be a feature of CRPC (26, 42). Some in vitro data suggest that FOXA1 may not play an important part of the AR transcriptional complex in CRPC. Unique castration-resistant ARBS detected in the C4-2B cell line (23) do not significantly overlap with FOXA1 binding sites or contain the Forkhead motif. However, our results show that ablating FOXA1 in the C4-2B cell line robustly diminishes cell growth, indicating a persistent role for FOXA1. Additionally, our gene expression data for FOXA1 over-expressing LNCaP cells correlated well with a published CRPC gene signature (24), indicating that FOXA1 may be an important player in the switch to a more aggressive gene expression profile like that seen in CRPC. Thus, it will be critical to map AR and FOXA1 binding in matched primary and CRPC tissues in order to definitively assess the role of FOXA1 in prostate cancer progression.
In summary, FOXA1 is a key component of the AR transcription factor complex. High levels of FOXA1 result in increased AR binding, a transcription profile akin to CRPC and proliferation in the presence of low levels of androgenic hormones. These findings suggest a pivotal role for FOXA1 in the progression of prostate cancer.
Materials and methods
Cell Culture and Transfection
LNCaP and C4-2B prostate cancer and MDA-MB-453 breast cancer cell lines were grown in RPMI or DMEM media, respectively, supplemented with 10% FBS and standard antibiotics. For hormone deprivation conditions, phenol-red free RPMI or DMEM medium was supplemented with 5% charcoal dextran treated FBS. Cells were transfected with 3ug FOXA1 expression plasmid per 10cm dish using Lipofectamine 2000 (Invitrogen), according to manufacturer’s instructions. Full length FOXA1 cDNA was ligated into pcDNA3.1 (43). For FOXA1 knockdown experiments, C4-2B cells were transfected with 50nM Allstars Negative Control siRNA (Qiagen) or a custom siRNA (Thermo Scientific) targeted to FOXA1 (GAGAGAAAAAAUCAACAGC) using Lipofectamine RNAiMAX (Invitrogen).
Cell growth assay
LNCaP and MDA-MB-453 cells were hormone deprived for two days then transfected with GFP or FOXA1 expression plasmid. The following day cells were trypsinised and re-plated at 4×103 (MDA-MB-453) or 5×103 (LNCaP) cells/well of 96 well plate in steroid free media and the next day cells were treated with 1nM DHT, 100nM DHT or vehicle control for 6 days. The number of live cells was quantified at day 0 and day 6 using CellTiter-Blue assay (Promega) with 8 replicates per condition. Three independent experiments were performed. C4-2B cells were plated at equal confluence and transfected with siControl or siFoxA1. The number of live cells was quantified using a hemocytometer after Trypan Blue staining at days 0, 4 and 7.
Western Blots
Whole cell lysate was extracted for western blots and protein quantified using Bradford assay. Antibodies used were anti-AR (sc-816) purchased from Santa Cruz Biotechnologies and anti-FOXA1 (ab5089) and anti-β-actin (ab6276) from Abcam. Secondary antibodies were used at a concentration of 1:2000.
Chromatin Immunoprecipitation coupled to high-throughput sequencing (ChIP-seq)
AR Chromatin immunoprecipitation (ChIP) experiments were conducted as described previously (44) using rabbit polyclonal anti-AR antibody (Santa Cruz Biotechnologies, sc-816). Briefly, 48 hours after transfection with GFP or FOXA1 expression plasmid cells were crosslinked with 1% formaldehyde and harvested. Cells were lysed, nucleus extracted and DNA sonicated using Diaganode Biorupter before immunopreciptation with AR antibody conjugated beads. Beads were thoroughly washed and reversed crosslinked at 65°C before amplification using TruSeq kit (Illumina). Single end 36-bp ChIP-seq data were generated by the Illumina analysis pipeline version 1.6.1, and reads were aligned to the Human Reference Genome (assembly hg19, GRCh37, Feb 2009) using bwa 0.5.9. Reads with MapQ scores less than 16 or falling within Duke’s Excluded Regions (45) were filtered from further analysis. Peaks were called using MACS, version 1.4.1. Heatmaps were created using python script as described in (46). For all ChIP-seq experiments, two biological replicates were performed and only reproducible peaks (i.e. those that occur in both replicates) were considered in downstream analysis if it occurred in both replicates. Motif analysis was performed using MEME-ChIP suite (47). Hierarchical clustering analysis of peaksets was performed using the DiffBind package (version 1.4.2) in Bioconductor version 2.11 (48).
Rapid IMmunoprecipitation of Endogenous proteins (RIME)
LNCaP cells were crosslinked with 1% EM-grade formaldehyde for 7 min before they were harvested. RIME experiments were conducted as described in (28). Antibodies used for immunoprecipitation were anti-AR (sc-816), anti-rabbit IgG (sc-2027) and anti-goat IgG (sc-2028) from Santa Cruz Biotechnologies and anti-FOXA1 (ab5089) from Abcam. Mass Spectrometry was performed using LTQ Orbitrap Velos (Thermo Scientific).
Raw MS data files were processed using Proteome Discoverer v.1.3 (Thermo Scientific). Processed files were searched against the SwissProt human database using the Mascot search engine version 2.3.0, allowing up to one tryptic miscleavage and a tolerance on mass measurement of 10 ppm in MS mode and 0.6 Da for MS/MS ions. Identified proteins have at least 2 unique peptides and not occurred in any of the matched IgG samples. Pathway analysis of the common AR and FOXA1 partners was performed using GSEA molecular signature database tool version 3.1 (49) and STRING (version 9.05) produced the protein interaction network (50).
Microarray Analysis
Cells were transfected with GFP or FOXA1 expression plasmid for 48h. RNA was collected from six biological replicates. The Illumina BeadChIP (HumanWG-12 version 4) bead-level data were preprocessed, log2-transformed, and quantile-normalised using the bead array package (51, 52) in Bioconductor (53). Differential expression analysis was performed using limma eBayes (Smyth 2004 with a Benjamini and Hochberg multiple test correction procedure (54) to identify statistically significant differentially expressed genes (FDR 0.05). GSEA molecular signature database tool version 3.1 (49) was used for pathway analysis of differentially expressed genes.
Patient Cohort
Full ethical approval was obtained for all human sample collections from Addenbrooke’s Hospital Research Ethics Committee (MREC 01/4/061). The tissue microarray (TMA) consisted of selected cores at least two distinct regions of tumor from each of the 102 men undergoing open radical retropubic prostatectomy. Matched cores from normal/benign regions (>3) were also taken as well as prostatic intraepithelial neoplasia (PIN) where available (55). We define biochemical recurrence (BCR) as a single PSA value of ≥0.2ng/ml, with persistent elevation on subsequent PSA measurement. We included a “triggered treatment” group where the decision to initiate treatment with modalities such as radiotherapy occurred before the PSA value had reached this threshold. Time to relapse was calculated as the time from radical prostatectomy to first PSA of 0.2ng/ml or treatment, whichever occurred first.
Immunohistochemistry
Paraffin-embedded TMA blocks were freshly cut before immunohistochemistry which was performed on a Bond automated system (Leica). Primary goat anti-FOXA1 antibody (Abcam, ab5089), dilution 1:800 in Sanger diluent, was used with the F DABe protocol and Bond epitope retrieval solution 1 for 20min. Evaluation of the stained TMAs was undertaken by two independent observers, one of which was a specialized uropathologist (A.W.). Analysis was conducted using a multi-headed microscope and neither had any knowledge or information pertaining to the patient’s clinical status. Staining intensity for FOXA1 was evaluated on a four-tiered scale: 0 (negative), 1 (weak), 2 (moderate) and 3 (strong) as well as percentage of nuclei stained. The resultant H-score incorporates both pieces of data (H-score = intensity X % positive stained cells).
Statistical analysis
In order to compare the difference in H-score between tumor and matched normal/benign, the Wilcoxon matched-pairs signed rank test was performed. To assess the effect of FOXA1 expression on prostate cancer patient outcome, patients were divided into groups based on quartiles of their FOXA1 H-score, quartiles 2-4 were combined for analysis. The Kaplan-Meier method was used to estimate the proportion of patients free of biochemical recurrence after radical prostatectomy, censoring at the last date of follow up. A univariable Cox proportional hazards regression model was used to evaluate the association between FOXA1 staining and time to relapse. Clinical and pathological information was compared to FOXA1 staining using either a Mann-Whitney test for continuous data or Fisher’s exact test for categorical data.
Wilcoxon matched-pair signed rank test was used to assess of the significance in the average read count for AR ChIP-seq samples in control and ‘FOXA1 high’ cells. All other analyses unless stated were performed using unpaired Student’s T-Test or Fisher’s Exact Test. Only values with a p-value <0.05 were considered significant. Statistical analyses were performed using R Statistical software (version 2.14.0) or Graphpad Prism 6.
Data deposition
ChIP-seq sequencing data are available in the ArrayExpress database (www.ebi.ac.uk/arrayexpress) under the accession number E-MTAB-1749. Microarray data has been deposited in the GEO database (accession number is pending).
Supplementary Material
Supplementary Figure 1: A. Western blot showing the levels of FOXA1 and AR in transfected LNCaP and MDA-MB-453 cells, β-actin was used as a loading control. B. Correlation heatmap showing the similarity between ARBS peaksets in control or ‘FOXA1 high’ cells with published data for ARBS in FOXA1 knockdown and control knockdown conditions (10, 21). C. Comparison of Control ARBS or ARBS unique to ‘FOXA1 high’ cells with AR reprogrammed binding events which occur after the loss of FOXA1. Percentage of the smallest peakset is shown. Darker orange colors indicate a greater level of concordance.
Supplementary Figure 2: AR binding is also increased in MDA-MB-453 breast cancer cell line when FOXA1 expression is increased. A&B. Example tracks from AR ChIP-seq in Control and ‘FOXA1 high’ MDA-MB-453 and LNCaP cell lines.
Supplementary Figure 3: FOXA1 ChIP-seq average binding intensity plot at ARBS shared with control or gained in ‘FOXA1 high’ cells.
Supplementary Figure 4: FoxA1 H-scores for cancer versus matched benign/normal prostate tissue. Median is indicated by a line and *** denotes a p value <0.0001.
Supplementary Figure 5: The C4-2B cell line, a LNCaP-derived CRPC model, was transfected with siControl or siRNA specifically targeted to FOXA1. A. Western blot showing the levels of FOXA1 and AR 4 days after siRNA transfection; β-actin was used as a loading control. B. Cell counting using Trypan blue exclusion to assess viability was performed at days 0, 4 and 7. Data from two biological replicates are shown; *** denotes a p value <0.001.
Acknowledgements
We would like to thank the members of the genomic, proteomic and histopathology core facilities at Cancer Research UK. We would like to acknowledge the support of The University of Cambridge, Cancer Research UK and Hutchison Whampoa Limited. We are grateful to study volunteers for their participation and staff at the Welcome Trust Clinical Research Facility, Addenbrooke’s Clinical Research Centre, Cambridge. W.D.T. is supported by grants from the National Health and Medical Research Council of Australia (ID 627185), Cancer Australia (ID 627229) and the Prostate Cancer Foundation of Australia. T.E.H holds a Postdoctoral Fellowship Award from the US Department of Defense Breast Cancer Research Program (BCRP; #W81XWH-11-1-0592). J.S.C. is supported by an ERC starting grant and an EMBO Young Investigator Award.
Abbreviations
- ADT
androgen deprivation therapy
- AR
androgen receptor
- ARBS
androgen receptor binding site
- ChIP
chromatin immunoprecipitation
- CRPC
castrate resistant prostate cancer
- DEG
differentially regulated genes
- DHT
5α-dihydrotestosterone
- LARC
LCR-associated remodelling complex
- RIME
rapid immunoprecipitation of endogenous proteins
Footnotes
Conflict of Interest The authors declare no conflict of interest.
References
- 1.Siegel R, Naishadham D, Jernal A. Cancer Statistics, 2012. CA, A Cancer Journal for Clinicians. 2012;62:10–29. doi: 10.3322/caac.20138. [DOI] [PubMed] [Google Scholar]
- 2.Scher HI, Buchanan G, Gerald W, Butler LM, Tilley WD. Targeting the androgen receptor: improving outcomes for castration-resistant prostate cancer. Endocrine-Related Cancer. 2004 Sep 1;11(3):459–76. doi: 10.1677/erc.1.00525. 2004. [DOI] [PubMed] [Google Scholar]
- 3.de Bono JS, Logothetis CJ, Molina A, Fizazi K, North S, Chu L, et al. Abiraterone and Increased Survival in Metastatic Prostate Cancer. New England Journal of Medicine. 2011;364(21):1995–2005. doi: 10.1056/NEJMoa1014618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Scher HI, Fizazi K, Saad F, Taplin M-E, Sternberg CN, Miller K, et al. Increased Survival with Enzalutamide in Prostate Cancer after Chemotherapy. New England Journal of Medicine. 2012;367(13):1187–97. doi: 10.1056/NEJMoa1207506. [DOI] [PubMed] [Google Scholar]
- 5.Chen CD, Welsbie DS, Tran C, Baek SH, Chen R, Vessella R, et al. Molecular determinants of resistance to antiandrogen therapy. Nat Med. 2004;10(1):33–9. doi: 10.1038/nm972. [DOI] [PubMed] [Google Scholar]
- 6.Taplin M-E, Balk SP. Androgen receptor: A key molecule in the progression of prostate cancer to hormone independence. Journal of Cellular Biochemistry. 2004;91(3):483–190. doi: 10.1002/jcb.10653. [DOI] [PubMed] [Google Scholar]
- 7.Waltering KK, Urbanucci A, Visakorpi T. Androgen receptor (AR) aberrations in castration-resistant prostate cancer. Molecular and Cellular Endocrinology. 2012;360(1-2):38–43. doi: 10.1016/j.mce.2011.12.019. [DOI] [PubMed] [Google Scholar]
- 8.Li Y, Alsagabi M, Fan D, Bova GS, Tewfik AH, Dehm SM. Intragenic rearrangements and altered RNA splicing of androgen receptor in a cell-based model of prostate cancer progression. Cancer Res. 2011;71:2108–17. doi: 10.1158/0008-5472.CAN-10-1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chmelar R, Buchanan G, Need EF, Tilley W, Greenberg NM. Androgen receptor coregulators and their involvement in the development and progression of prostate cancer. International Journal of Cancer. 2007;120(4):719–33. doi: 10.1002/ijc.22365. [DOI] [PubMed] [Google Scholar]
- 10.Wang D, Garcia-Bassets I, Benner C, Li W, Su X, Zhou Y, et al. Reprogramming transcription by distinct classes of enhancers functionally defined by eRNA. Nature. 2011;474(7351):390–4. doi: 10.1038/nature10006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gao N, Zhang J, Rao MA, Case TC, Mirosevich J, Wang Y, et al. The Role of Hepatocyte Nuclear Factor-3a (Forkhead Box A1) and Androgen Receptor in Transcriptional Regulation of Prostatic Genes. Molecular Endocrinology. 2003 Aug 1;17(8):1484–507. doi: 10.1210/me.2003-0020. 2003. [DOI] [PubMed] [Google Scholar]
- 12.Carroll JS, Meyer CA, Song J, Li W, Geistlinger TR, Eeckhoute J, et al. Genome-wide analysis of estrogen receptor binding sites. Nat Genet. 2006;38(11):1289–97. doi: 10.1038/ng1901. [DOI] [PubMed] [Google Scholar]
- 13.Clarke CL, Graham JD. Non-Overlapping Progesterone Receptor Cistromes Contribute to Cell-Specific Transcriptional Outcomes. PLoS ONE. 2012;7(4):e35859. doi: 10.1371/journal.pone.0035859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lupien M, Eeckhoute J, Meyer CA, Wang Q, Zhang Y, Li W, et al. FoxA1 Translates Epigenetic Signatures into Enhancer-Driven Lineage-Specific Transcription. Cell. 2008;132(6):958–70. doi: 10.1016/j.cell.2008.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gerhardt J, Montani M, Wild P, Beer M, Huber F, Hermanns T, et al. FOXA1 Promotes Tumor Progression in Prostate Cancer and Represents a Novel Hallmark of Castrate-Resistant Prostate Cancer. The American Journal of Pathology. 2012;180(2):848–61. doi: 10.1016/j.ajpath.2011.10.021. [DOI] [PubMed] [Google Scholar]
- 16.Gao N, Ishii K, Mirosevich J, Kuwajima S, Oppenheimer SR, Roberts RL, et al. Forkhead box A1 regulates prostate ductal morphogenesis and promotes epithelial cell maturation. Development. 2005 Aug 1;132(15):3431–43. doi: 10.1242/dev.01917. 2005. [DOI] [PubMed] [Google Scholar]
- 17.Wang Q, Li W, Liu XS, Carroll JS, Janne OA, Keeton EK, et al. A Hierarchical Network of Transcription Factors Governs Androgen Receptor-Dependent Prostate Cancer Growth. Molecular Cell. 2007;27(3):380–92. doi: 10.1016/j.molcel.2007.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jia L, Berman BP, Jariwala U, Yan X, Cogan JP, Walters A, et al. Genomic Androgen Receptor-Occupied Regions with Different Functions, Defined by Histone Acetylation, Coregulators and Transcriptional Capacity. PLoS ONE. 2008;3(11):e3645. doi: 10.1371/journal.pone.0003645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jin HJ, Zhao JC, Ogden I, Bergan RC, Yu J. Androgen receptor-independent function of FoxA1 in prostate cancer metastasis. Cancer research. 2013 Jun 15;73(12):3725–36. doi: 10.1158/0008-5472.CAN-12-3468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Imamura Y, Sakamoto S, Endo T, Utsumi T, Fuse M, Suyama T, et al. FOXA1 Promotes Tumor Progression in Prostate Cancer via the Insulin-Like Growth Factor Binding Protein 3 Pathway. PLoS ONE. 2012;7(8):e42456. doi: 10.1371/journal.pone.0042456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sahu B, Laakso M, Ovaska K, Mirtti T, Lundin J, Rannikko A, et al. Dual role of FoxA1 in androgen receptor binding to chromatin, androgen signalling and prostate cancer. EMBO J. 2011;30(19):3962–76. doi: 10.1038/emboj.2011.328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jain RK, Mehta RJ, Nakshatri H, Idrees MT, Badve SS. High-level expression of forkhead-box protein A1 in metastatic prostate cancer. Histopathology. 2011;58(5):766–72. doi: 10.1111/j.1365-2559.2011.03796.x. [DOI] [PubMed] [Google Scholar]
- 23.Decker KF, Zheng D, He Y, Bowman T, Edwards JR, Jia L. Persistent androgen receptor-mediated transcription in castration-resistant prostate cancer under androgen-deprived conditions. Nucleic Acids Research. 2012 Sep 27; doi: 10.1093/nar/gks888. 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sharma Naomi L, Massie Charlie E, Ramos-Montoya A, Zecchini V, Scott HE, Lamb Alastair D, et al. The Androgen Receptor Induces a Distinct Transcriptional Program in Castration-Resistant Prostate Cancer in Man. Cancer Cell. 2013;23(1):35–47. doi: 10.1016/j.ccr.2012.11.010. [DOI] [PubMed] [Google Scholar]
- 25.Zhang C, Wang L, Wu D, Chen H, Chen Z, Thomas-Ahner JM, et al. Definition of a FoxA1 Cistrome That Is Crucial for G1 to S-Phase Cell-Cycle Transit in Castration-Resistant Prostate Cancer. Cancer Research. 2011 Nov 1;71(21):6738–48. doi: 10.1158/0008-5472.CAN-11-1882. 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grasso CS, Wu Y-M, Robinson DR, Cao X, Dhanasekaran SM, Khan AP, et al. The mutational landscape of lethal castration-resistant prostate cancer. Nature. 2012;487(7406):239–43. doi: 10.1038/nature11125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Robinson JLL, MacArthur S, Ross-Innes CS, Tilley WD, Neal DE, Mills IG, et al. Androgen receptor driven transcription in molecular apocrine breast cancer is mediated by FoxA1. EMBO J. 2011;30(15):3019–27. doi: 10.1038/emboj.2011.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mohammed H, D Santos C, Serandour Aurelien A, Ali HR, Brown Gordon D, Atkins A, et al. Endogenous Purification Reveals GREB1 as a Key Estrogen Receptor Regulatory Factor. Cell Reports. 2013;3(2):342–9. doi: 10.1016/j.celrep.2013.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mahajan MC, Narlikar GJ, Boyapaty G, Kingston RE, Weissman SM. Heterogeneous nuclear ribonucleoprotein C1/C2, MeCP1, and SWI/SNF form a chromatin remodeling complex at the Î2-globin locus control region. Proceedings of the National Academy of Sciences of the United States of America. 2005 Oct 18;102(42):15012–7. doi: 10.1073/pnas.0507596102. 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kikuchi M, Okumura F, Tsukiyama T, Watanabe M, Miyajima N, Tanaka J, et al. TRIM24 mediates ligand-dependent activation of androgen receptor and is repressed by a bromodomain-containing protein, BRD7, in prostate cancer cells. Biochim Biophys Acta. 2009 Dec;1793(12):1828–36. doi: 10.1016/j.bbamcr.2009.11.001. [DOI] [PubMed] [Google Scholar]
- 31.Jung C, Kim RS, Zhang HJ, Lee SJ, Jeng MH. HOXB13 induces growth suppression of prostate cancer cells as a repressor of hormone-activated androgen receptor signaling. Cancer Res. 2004 Dec 15;64(24):9185–92. doi: 10.1158/0008-5472.CAN-04-1330. [DOI] [PubMed] [Google Scholar]
- 32.Liao G, Chen LY, Zhang A, Godavarthy A, Xia F, Ghosh JC, et al. Regulation of androgen receptor activity by the nuclear receptor corepressor SMRT. J Biol Chem. 2003 Feb 14;278(7):5052–61. doi: 10.1074/jbc.M206374200. [DOI] [PubMed] [Google Scholar]
- 33.Hodgson MC, Astapova I, Cheng S, Lee LJ, Verhoeven MC, Choi E, et al. The androgen receptor recruits nuclear receptor CoRepressor (N-CoR) in the presence of mifepristone via its N and C termini revealing a novel molecular mechanism for androgen receptor antagonists. J Biol Chem. 2005 Feb 25;280(8):6511–9. doi: 10.1074/jbc.M408972200. [DOI] [PubMed] [Google Scholar]
- 34.Wang Q, Li W, Zhang Y, Yuan X, Xu K, Yu J, et al. Androgen Receptor Regulates a Distinct Transcription Program in Androgen-Independent Prostate Cancer. Cell. 2009;138(2):245–56. doi: 10.1016/j.cell.2009.04.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Deutsch L, Wrage M, Koops S, Glatzel M, Uzunoglu FG, Kutup A, et al. Opposite roles of FOXA1 and NKX2-1 in lung cancer progression. Genes, Chromosomes and Cancer. 2012;51(6):618–29. doi: 10.1002/gcc.21950. [DOI] [PubMed] [Google Scholar]
- 36.Nucera C, Eeckhoute J, Finn S, Carroll JS, Ligon AH, Priolo C, et al. FOXA1 Is a Potential Oncogene in Anaplastic Thyroid Carcinoma. Clinical Cancer Research. 2009 Jun 1;15(11):3680–9. doi: 10.1158/1078-0432.CCR-08-3155. 2009. [DOI] [PubMed] [Google Scholar]
- 37.Lin L, Miller CT, Contreras JI, Prescott MS, Dagenais SL, Wu R, et al. The Hepatocyte Nuclear Factor 3a Gene, HNF3a (FOXA1), on Chromosome Band 14q13 Is Amplified and Overexpressed in Esophageal and Lung Adenocarcinomas. Cancer Research. 2002 Sep 15;62(18):5273–9. 2002. [PubMed] [Google Scholar]
- 38.Wang L, Qin H, Li L, Feng F, Ji P, Zhang J, et al. Forkhead-box A1 transcription factor is a novel adverse prognosis marker in human glioma. Journal of Clinical Neuroscience. 2013;20(5):654–8. doi: 10.1016/j.jocn.2012.03.055. [DOI] [PubMed] [Google Scholar]
- 39.Sahu B, Laakso M, Pihlajamaa P, Ovaska K, Sinielnikov I, Hautaniemi S, et al. FoxA1 Specifies Unique Androgen and Glucocorticoid Receptor Binding Events in Prostate Cancer Cells. Cancer Research. 2013 Mar 1;73(5):1570–80. doi: 10.1158/0008-5472.CAN-12-2350. 2013. [DOI] [PubMed] [Google Scholar]
- 40.Wang D, Garcia-Bassets I, Benner C, Li W, Su X, Zhou Y, et al. Reprogramming transcription by distinct classes of enhancers functionally defined by eRNA. Nature. 2011 Jun 16;474(7351):390–4. doi: 10.1038/nature10006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Varambally S, Yu J, Laxman B, Rhodes DR, Mehra R, Tomlins SA, et al. Integrative genomic and proteomic analysis of prostate cancer reveals signatures of metastatic progression. Cancer Cell. 2005;8(5):393–406. doi: 10.1016/j.ccr.2005.10.001. [DOI] [PubMed] [Google Scholar]
- 42.Barbieri CE, Baca SC, Lawrence MS, Demichelis F, Blattner M, Theurillat J-P, et al. Exome sequencing identifies recurrent SPOP, FOXA1 and MED12 mutations in prostate cancer. Nat Genet. 2012;44(6):685–9. doi: 10.1038/ng.2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hurtado A, Holmes KA, Ross-Innes CS, Schmidt D, Carroll JS. FOXA1 is a key determinant of estrogen receptor function and endocrine response. Nat Genet. 2011;43(1):27–33. doi: 10.1038/ng.730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schmidt D, Wilson MD, Spyrou C, Brown GD, Hadfield J, Odom DT. ChIP-seq: Using high-throughput sequencing to discover protein:DNA interactions. Methods. 2009;48(3):240–8. doi: 10.1016/j.ymeth.2009.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.ENCODE. Dunham I, Kundaje A, Aldred SF, Collins PJ, Davis CA, et al. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012 Sep 6;489(7414):57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ross-Innes CS, Stark R, Holmes KA, Schmidt D, Spyrou C, Russell R, et al. Cooperative interaction between retinoic acid receptor-α and estrogen receptor in breast cancer. Genes & Development. 2010 Jan 15;24(2):171–82. doi: 10.1101/gad.552910. 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Machanick P, Bailey TL. MEME-ChIP: motif analysis of large DNA datasets. Bioinformatics. 2011 Jun 15;27(12):1696–7. doi: 10.1093/bioinformatics/btr189. 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ross-Innes CS, Stark R, Teschendorff AE, Holmes KA, Ali HR, Dunning MJ, et al. Differential oestrogen receptor binding is associated with clinical outcome in breast cancer. Nature. 2012;481(7381):389–93. doi: 10.1038/nature10730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences of the United States of America. 2005 Oct 25;102(43):15545–50. doi: 10.1073/pnas.0506580102. 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jensen LJ, Kuhn M, Stark M, Chaffron S, Creevey C, Muller J, et al. STRING 8 - a global view on proteins and their functional interactions in 630 organisms. Nucleic Acids Research. 2009 Jan 1;37(suppl 1):D412–D6. doi: 10.1093/nar/gkn760. 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cairns JM, Dunning MJ, Ritchie ME, Russell R, Lynch AG. BASH: a tool for managing BeadArray spatial artefacts. Bioinformatics (Oxford, England) 2008 Dec 15;24(24):2921–2. doi: 10.1093/bioinformatics/btn557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dunning MJ, Smith ML, Ritchie ME, Tavare S. beadarray: R classes and methods for Illumina bead-based data. Bioinformatics. 2007 Aug 15;23(16):2183–4. doi: 10.1093/bioinformatics/btm311. 2007. [DOI] [PubMed] [Google Scholar]
- 53.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, Hornik K, Hothorn T, Huber W, Iacus S, Irizarry R, Leisch F, Li C, Maechler M, Rossini AJ, Sawitzki G, Smith C, Smyth G, Tierney L, Yang JYH, Zhang J. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80–R.16. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J R Stat Soc Series B Stat Methodol. 1995;57:289–300. [Google Scholar]
- 55.Whitaker HC, Kote-Jarai Z, Ross-Adams H, Warren AY, Burge J, George A, et al. The rs10993994 Risk Allele for Prostate Cancer Results in Clinically Relevant Changes in Microseminoprotein-Beta Expression in Tissue and Urine. PLoS ONE. 2010;5(10):e13363. doi: 10.1371/journal.pone.0013363. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1: A. Western blot showing the levels of FOXA1 and AR in transfected LNCaP and MDA-MB-453 cells, β-actin was used as a loading control. B. Correlation heatmap showing the similarity between ARBS peaksets in control or ‘FOXA1 high’ cells with published data for ARBS in FOXA1 knockdown and control knockdown conditions (10, 21). C. Comparison of Control ARBS or ARBS unique to ‘FOXA1 high’ cells with AR reprogrammed binding events which occur after the loss of FOXA1. Percentage of the smallest peakset is shown. Darker orange colors indicate a greater level of concordance.
Supplementary Figure 2: AR binding is also increased in MDA-MB-453 breast cancer cell line when FOXA1 expression is increased. A&B. Example tracks from AR ChIP-seq in Control and ‘FOXA1 high’ MDA-MB-453 and LNCaP cell lines.
Supplementary Figure 3: FOXA1 ChIP-seq average binding intensity plot at ARBS shared with control or gained in ‘FOXA1 high’ cells.
Supplementary Figure 4: FoxA1 H-scores for cancer versus matched benign/normal prostate tissue. Median is indicated by a line and *** denotes a p value <0.0001.
Supplementary Figure 5: The C4-2B cell line, a LNCaP-derived CRPC model, was transfected with siControl or siRNA specifically targeted to FOXA1. A. Western blot showing the levels of FOXA1 and AR 4 days after siRNA transfection; β-actin was used as a loading control. B. Cell counting using Trypan blue exclusion to assess viability was performed at days 0, 4 and 7. Data from two biological replicates are shown; *** denotes a p value <0.001.