Abstract
The ability to reversibly control protein structure and function with light would offer high spatiotemporal resolution to investigate biological processes. To enable photoresponsiveness to general proteins, we genetically incorporated into proteins a set of photoswitchable click amino acids (PSCaas), which contain both a reversible photoswitch and an additional click functional group for further modifications. Orthogonal tRNA-synthetases were evolved to genetically encode PSCaas bearing azobenzene and alkene, keto or benzylchloride in E. coli and in mammalian cells. The benzylchloride PSCaa, after incorporated into calmodulin, spontaneously generated a covalent protein bridge by reacting with a nearby cysteine via proximity-enabled bioreactivity. The resultant azobenzene bridge isomerized in response to light changing calmodulin conformation. These genetically encodable PSCaas will prove valuable to engineer photoswitchable bridges into proteins for reversible optogenetic regulation.
Keywords: photoswitchable click amino acids, expanded genetic code, azo compounds, optogenetics, isomerization
Photoswitchable biomolecules, most commonly peptides and proteins, are finding increasing utility in the in vivo investigation of biological processes, where they confer a minimally invasive means for precise spatiotemporal control.[1] Of particular interest are photoisomerizable functionalities, which can induce reversible changes in protein conformation and bioactivity.[2] Azobenzene photoswitches are very effective for this application, because photoinduced trans↔cis isomerization around the central N=N bond occurs with high yield and can drive remarkable structural changes in vitro and even in vivo.[3-4] Until now, optical control of protein conformation required the use of a bifunctional, thiol-reactive azobenzene reagent, which can crosslink two suitably situated cysteines within a protein generating a photoswitchable bridge of the side chains.[5-6] However, the utility of this approach in vivo has been limited by the high reactivity of the crosslinking reagents and the lack of specificity in targeting the modification, factors that lead to the abundant formation of undesired products. To overcome these limitations, we recently developed a photoswitchable click amino acid (PSCaa) 1 (Figure 1A) that allows the site-specific introduction of a photoswitchable bridge. Through thiol-click chemistry, the bridge can be formed under mild conditions, and selectively between the alkene of 1 and the thiol group of a nearby cysteine, even in the presence of other thiols.[7] Previously, we chemically synthesized peptides that incorporate a PSCaa-mediated photocontrollable bridge, and demonstrated the helical conformation of the peptides could be optically regulated, leading to differing bioactivities.[7] The site-specific incorporation of PSCaas into recombinant proteins in situ will enable the study of protein function and biological processes via light modulation in live cells.[8] Although phenylazophenylalanine (AzoPhe) has been incorporated into proteins in E. coli,[9] the tRNA-synthetase pair evolved for AzoPhe cannot be used in mammalian cells. In addition, the AzoPhe side chain lacks a reactive functional group for further derivatization and thus cannot mediate bridge formation. Therefore, light-induced isomerization of AzoPhe may induce only localized structural perturbations and not large-scale changes in protein conformation. Currently there have been no reports of the use of azobenzene units to photocontrol protein structural states in living cells.
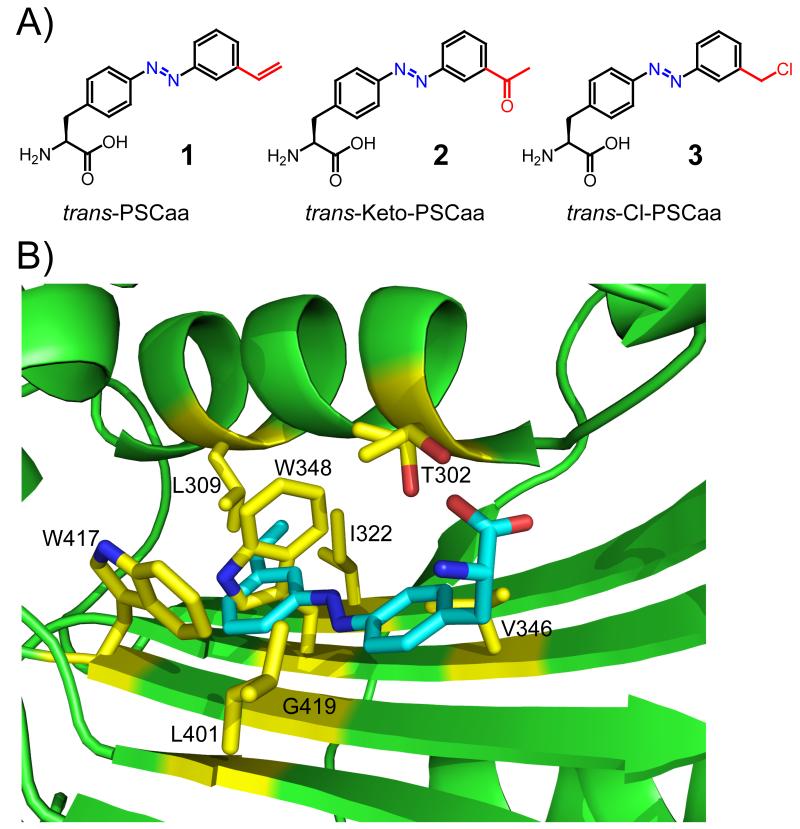
Figure 1.
A) The trans forms of the photoswitchable click amino acids. PSCaa 1: 2-amino-3-(4-((3-vinylphenyl)diazenyl)phenyl)propanoic acid; Keto-PSCaa 2: 2-amino-3-(4-((3-(2-acetyl)phenyl)diazenyl)phenyl)propanoic acid; Cl-PSCaa 3: 2-amino-3-(4-((3-(chloromethyl)phenyl)diazenyl)phenyl)propanoic acid. B) X-ray crystal structure of the active site cavity of the MmOmePylRS (PDB 3QTC), in which the structure of PSCaa 1 (cyan) was superimposed. Residues mutated in the synthetase library are shown in yellow stick.
Here, we describe the generation of orthogonal tRNA-synthetase pairs that incorporate azobenzene-based photoswitchable click amino acids site-specifically into proteins in both E. coli and mammalian cells. Using a semi-rational approach for the construction of a mutant library of the synthetase (Methanosarcina mazei PylRS, MmPylRS), we were able to simultaneously mutate a greater number of residues in comparison to a more conventional approach that employs full randomization at each site. The increased diversity of the MmPylRS library enabled us to evolve mutant synthetases specific for AzoPhe derivatives bearing an additional alkene (PSCaa, 1), keto (Keto-PSCaa, 2), or benzylchloride (Cl-PSCaa 3) functionality (Figure 1A). In addition, we generated a protein bridge utilizing the proximity-enabled bioreactivity[10] of Cl-PSCaa 3 toward cysteine and showed photoswitching of the resultant azobenzene-containing bridge to alter protein conformation. This work demonstrates the first genetic incorporation into proteins of an unnatural amino acid (Uaa) containing both the azobenzene photoswitch and an additional click functionality, and forms the foundation for the generation in live cells of a designed recombinant protein carrying a photocontrollable bridge.
To evolve a mutant synthetase specific for 1, we initially generated 5 different synthetase libraries by fully randomizing 5 or 6 residues in the active site of MmPylRS using conventional NNK codon-randomization.[11] Although these libraries yielded mutants specific for other Uaas,[12] no mutant could be identified for 1, presumably because more substantial changes in the synthetase active site are required for accommodation of the greater length and rigidity of 1. Simultaneous mutation of a larger number of synthetase residues in E. coli necessitated that we use a semi-rational strategy, in which certain sites are restricted to a selected subset of the twenty amino acids. Using the structure of the MmOmeRS/O-methyl-L-tyrosine (Ome) complex as a guide,[12] we superimposed 1 on Ome and decided to mutate residues 302, 309, 322, 346, 348, 401, 417, and 419 (Figure 1B). Positions 309, 322 and 417 were fully randomized with the NNK codon. Based on its known importance in the recognition of Phe analogues,[12,15] residue 302 was allowed Thr, Ser, or Ala. Positions neighboring the azobenzene moiety were restricted to small, aliphatic residues: Val/Leu/Ala for 346 and 401, and Gly/Val/Ala for 348 and 419. The theoretical gene library size was 4 × 107 mutants, and an E. coli library of 6 × 108 colony forming units was made.
One round of positive selection was performed in DH10β carrying the selection plasmid pREP, which encodes both green fluorescent protein (GFP) and chloramphenicol acetyltransferase reporter cassettes containing the TAG stop codon.[13] Cells were grown on minimal medium containing 1 mM PSCaa 1 and 40 μg mL−1 chloramphenicol. A total of 60 green fluorescent colonies were obtained and subsequently screened by streaking on minimal-medium plates supplemented with chloramphenicol (60 μg mL−1) and in the presence or absence of 1 mM PSCaa. Thirteen clones showed PSCaa-dependent survival and green fluorescence (Figure S1A). To determine the efficiency of PSCaa incorporation by these clones, a mutant GFP gene containing a stop codon TAG at position 182 (GFP_182TAG) was expressed in E. coli in 2×YT medium using the corresponding -PSCaaRS pairs. PSCaa-dependent GFP expression was observed for 12 clones (Figure S1B).
DNA sequencing revealed that these clones represent seven distinct synthetase mutants (Table S1). All seven mutants carried same amino acid substitution at positions 348, 401, 417 and 419, whereas some variability was seen at positions 302, 309, 322 and 346. Based on GFP expression in the presence or absence of PSCaa (Figure S1B), we chose the clone that showed the highest selectivity for incorporating PSCaa over natural amino acids. The mutant synthetase encoded by this clone, named MmPSCaaRS, carried the amino acid identities Thr302, Ser309, Ile322, Val346, Gly348, Tyr384, Val401, Trp417, and Gly419, and was characterized further.
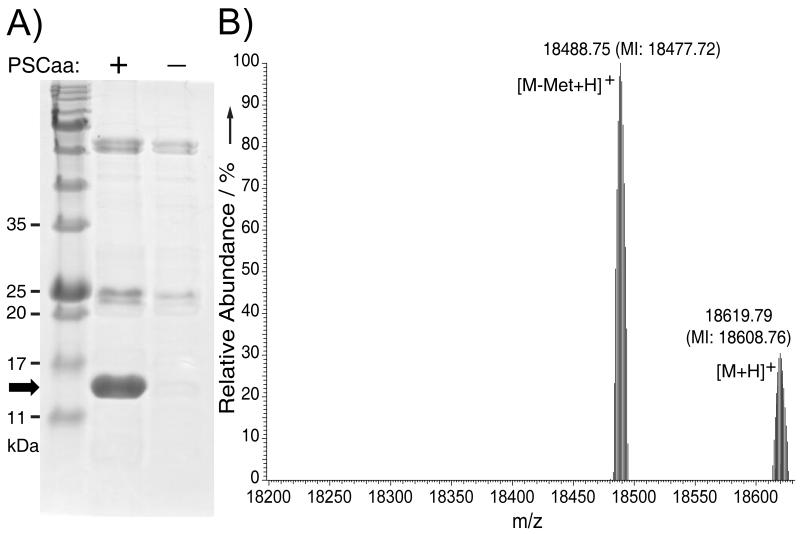
To evaluate the efficiency and fidelity of the evolved MmPSCaaRS to incorporate PSCaa 1, we expressed in BL21 cells a gene for sperm whale myoglobin (Myo_4TAGHis6, encoding an amber TAG codon at position 4 and a C-terminal Hisx6 tag) together with the -MmPSCaaRS. From SDS-PAGE analysis of the expressed products, full-length myoglobin was obtained in good yield (ca. 1.8 mg mL−1) in the presence of 1 mM PSCaa, but was undetectable in the absence of PSCaa (Figure 2A). The myoglobin purified from the former expression was analyzed by electrospray ionization Fourier transform ion trap mass spectrometry (ESI-FTMS). An observed peak with a monoisotopic mass of 18608.76 Da (Figure 2B) corresponds to intact myoglobin having a single PSCaa residue at position 4 (expected [M+H]+ = 18608.81 Da). A second observed peak corresponds to the PSCaa-containing myoglobin lacking the initiator Met (expected [M-Met+H]+ = 18477.77 Da, measured 18477.72 Da). No peaks were observed corresponding to myoglobin proteins containing any of the natural amino acids at the amber codon position. Furthermore, no hydrazo myoglobin resulting from the reduction of the azo bond was detected, indicating that the PSCaa incorporated into myoglobin remains stable under the reducing conditions of the E. coli cytosol.
Figure 2.
A) SDS-PAGE stained with Coomassie shows that -MmPSCaaRS specifically incorporated PSCaa 1 into myoglobin in E. coli by suppression of the amber codon. Samples were normalized for constant cell numbers for each lane. The arrow indicates the position of full-length myoglobin protein. B) High resolution ESI-FTMS analysis of intact myoglobin expressed in the presence of -MmPSCaaRS supplemented with 1 mM PSCaa 1 in nutrient rich growth media. Only the Uaa was found to be incorporated at the TAG encoded position. Average and monoisotopic (MI) masses are labeled.
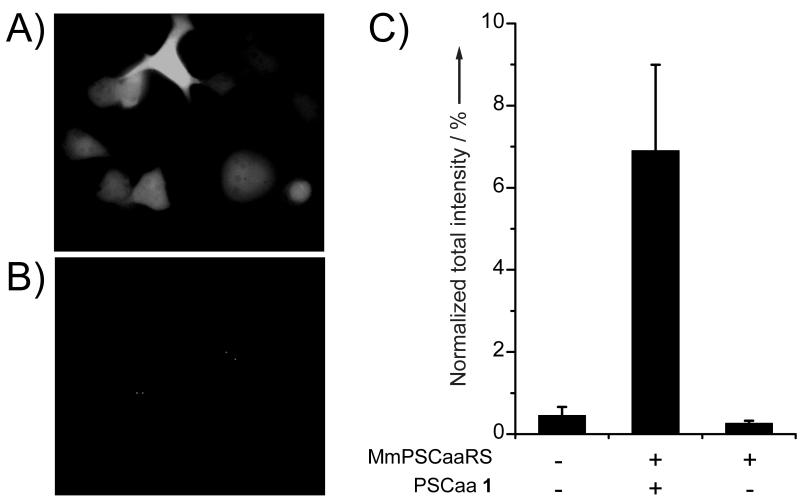
To investigate whether PSCaa 1 could be incorporated into proteins in mammalian cells, we transfected genes for the -MmPSCaaRS pair into a HeLa-GFP reporter cell line, in which the GFP_182TAG gene is stably integrated into the genome.[12,14] Green fluorescence of GFP appeared only when cells were grown in the presence of PSCaa 1 (Figure 3A and 3B), indicating the selective incorporation of 1 into GFP. The relatively low concentration (0.1 mM) of 1 in the growth medium that was sufficient to support GFP expression suggests efficient cellular uptake of this hydrophobic Uaa into mammalian cells. GFP expression was quantified by measurement of cellular fluorescence using flow cytometry (Figure 3C). Total fluorescence intensity of each sample was normalized to that of the HeLa-GFP reporter cells transfected with the M. maizei -PylRS (WT) pair and supplemented with Nε-tert-butyloxycarbonyl-L-lysine (Boc-Lys), which is efficiently incorporated by the WT MmPylRS. The incorporation efficiency for PSCaa 1 by the -MmPSCaaRS pair was ~7% of WT, with a background suppression of only 0.2% detected in the absence of PSCaa.
Figure 3.
Incorporation of PSCaa 1 into GFP in mammalian cells by the -MmPSCaaRS pair. A) Fluorescent imaging of the HeLa-GFP(182TAG) reporter cell line transfected with the -MmPSCaaRS and grown in the presence of 0.1 mM PSCaa 1. Green fluorescence was detected indicating in vivo incorporation of PSCaa into GFP. B) No fluorescence was detected in cells cultured in medium lacking PSCaa. C) Incorporation efficiency of PSCaa 1 measured with flow cytometry. Total fluorescence intensity of cells was measured and normalized to cells transfected with the -PylRS (WT) pair and supplemented with Boc-Lys. The same number of cells was analyzed for each sample. Error bars represent standard error of mean (s.e.m.). The value (mean ± s.e.m.) is 6.9 ± 2.5% (n = 3) for PSCaa 1.
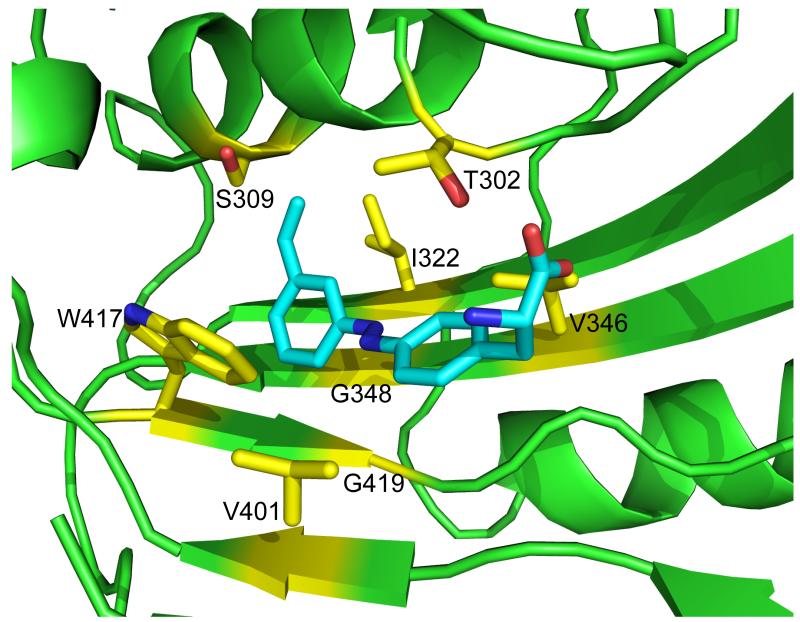
To understand how the PSCaa is recognized by the evolved synthetases, we structurally modeled MmPSCaaRS and the binding of PSCaa using the MmOmeRS/Ome complex structure as the template (Figure 4). In agreement with previously characterized PylRS mutants evolved to recognize other Phe analogues,[12,15] Thr302 in MmPSCaaRS (or the equivalent Ser302 in other PSCaa-incorporating mutants) hydrogen bonds to the α-carboxylate group of the PSCaa. The azobenzene moiety of the PSCaa occupies a hydrophobic pocket formed by G348, V401, W417 and G419, which are conserved in all of the identified mutants (Table S1). The alkene group of PSCaa is in contact with residues 309 and 322, for which diverse substitutions were identified in the PSCaa-incorporating mutants.
Figure 4.
Molecular docking of PSCaa into a model of MmPSCaaRS, which was built from 3QTC bearing the identified substitutions. Most notably, position 348 and 419 at the bottom of the substrate binding pocket were restricted to Gly to allow the long planar azobenzene moiety to extend into the active site. Mutation of 309 into A/S/G shortens the side chain to possibly accommodate the extended alkene group. T or S at position 302 hydrogen bonds to the α-carboxyl group of PSCaa.
The tolerance of multiple substituting residues at the 309 and 322 sites suggests that the alkene group of PSCaa can be accommodated by several distinct combinations of amino acid residues and its binding interactions contribute secondarily to the recognition of the PSCaa. We therefore reasoned that azobenzene Uaas containing alternative functional groups in place of the alkene might also be accommodated by one or more of the identified PSCaa-incorporating synthetase mutants. To test this hypothesis, we assessed the set of previously identified PSCaa-specific synthetase mutants for their ability to incorporate the meta substituted keto-azobenzene-amino acid Keto-PSCaa 2 and the benzylchloride substituted azobenzene amino acid Cl-PSCaa 3 (Figure 1A).
The keto group in 2 is bio-orthogonal and valuable for selective protein modification via reaction with hydrazide or hydroxylamine derivatives.[16] We found that three clones were able to incorporate 2 into GFP by Western analysis (Supporting Information, Figure S2). One synthetase mutant named MmKetoPSCaaRS (Thr302, Ala309, Thr322, Ala346, Gly348, Val401, Trp417, Gly419) was used for determining translation efficiency by co-expression analyses with the Myo_4TAGHis6 gene. Full-length myoglobin was produced only in the presence of 1 mM 2 with good yield (ca. 1.5 mg mL−1), and ESI-FTMS confirmed the site-specific incorporation of 2 into position 4 of myoglobin with high fidelity (Supporting Information, Figure S3, Figure S4).
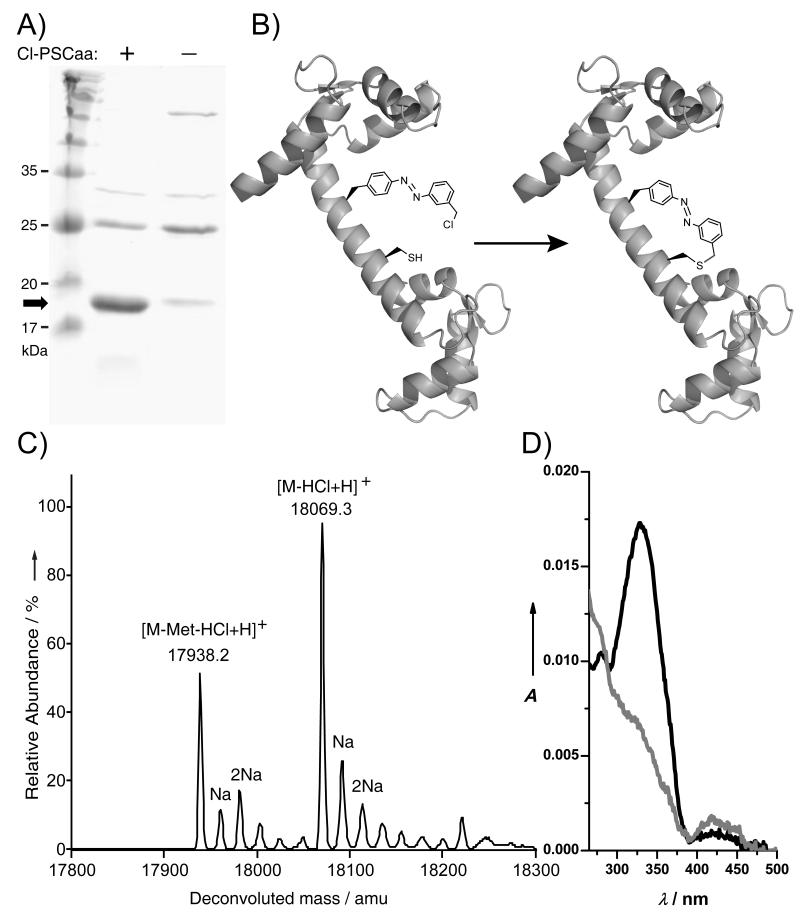
The benzylchloride group of Cl-PSCaa 3 may react with the thiol group of a nearby cysteine through proximity-enabled bioreactivity [10] to build a covalent protein bridge in situ. To test the reactivity, we incubated Cl-PSCaa 3 and Boc-protected cysteine at pH 7.5. As monitored by HPLC-MS, the crosslinking between 3 and cysteine was complete after ~2 h (Figure S5). We next found that Cl-PSCaa 3 could be genetically incorporated into proteins by the MmPSCaaRS. To investigate whether Cl-PSCaa 3 could be used to build a protein bridge through reacting with cysteine, we incorporated Cl-PSCaa 3 into the central helix of calmodulin (CaM) at residue 76 and placed cysteine at the i+7 position (residue 83) (Figure 5B). Using the -MmPSCaaRS pair, Cl-PSCaa 3 was incorporated into CaM in E. coli yielding 1.2 mg/mL of CaM after Ni-NTA purification (Figure 5A). Mass spectrometric analysis of this purified intact protein indicated that only Cl-PSCaa 3 was incorporated at position 76 and that an intramolecular bond was formed between Cl-PSCaa 3 and the cysteine at i+7 position (Figure 5C). A peak at 18069.3 Da was detected, corresponding to the CaM containing the covalent azobenzene bridge. A second peak measured at 17938.2 Da corresponds to CaM lacking the initiating Met and containing the covalent bridge. No peak corresponding to non-crosslinked 3 and cysteine was detected, indicating complete bridge formation.
Figure 5.
Incorporation of Cl-PSCaa 3 into CaM to form a photoisomerizable covalent protein bridge. A) SDS-PAGE stained with Coomassie shows that -MmPSCaaRS specifically incorporated the Cl-PSCaa 3 into CaM in E. coli by suppression of the amber codon introduced at position 76. Samples were normalized for constant cell numbers for each lane. The arrow indicates the position of full-length protein. B) Scheme showing intramolecular bridge formation in the central helix of CaM via Cl-PSCaa 3 reacting with cysteine through proximity-enabled bioreactivity. C) ESI-MS analysis of CaM expressed in the presence of -MmPSCaaRS supplemented with 1 mM Cl-PSCaa 3. The crosslinking reaction of the Cl-PSCaa 3 with cysteine results in a loss of HCl. Crosslinked products containing the covalent azobenzene bridge: [M-HCl+H], expected 18069.8 Da, measured 18069.3 Da; [M-Met-HCl+H], expected 17938.6 Da, measured 17938.2 Da. Na+ adducts of these two species were also detected and labeled. Non-crosslinked products: [M+H], expected 18106.2 Da, not detected; [M-Met+H], expected 17975.0 Da, not detected. D) UV/Vis spectra of CaM containing the azobenzene bridge before (black line) and after (grey line) light illumination at 365 nm.
To determine whether the azobenzene bridge built in CaM could be photoisomerized, we measured UV/visible spectra of the purified CaM protein. Before light illumination, the protein showed a strong absorption at 330 nm and a weak peak at 424 nm, representing π-π* and n-π* transitions, respectively, which is characteristic of the trans-azobenzene (Figure 5D). After illumination at 365 nm, the absorbance at 330 nm decreased along with a concomitant slight increase in absorbance at 424 nm, indicating a clear trans-to-cis photoisomerization of the azobenzene bridge. To study the effect of bridge photoisomerization on protein conformation, circular dichroism spectra of the bridged CaM were recorded (Figure S6). Consistent with that the cis form of PSCaa is less compatible with an i+7 helical spacing than the trans form,[7b] trans-cis photoisomerization of the azobenzene bridge resulted in a decrease in helix content of CaM in the cis state.
In summary, azobenzene-based photoswitchable click amino acids were successfully incorporated into recombinant proteins in E. coli and mammalian cells through the use of an expanded genetic code. These unnatural amino acids contain both a reversible photoswitch and an additional click functional group for further modifications. A covalent protein bridge was spontaneously generated between Cl-PSCaa and an appropriately positioned cysteine in CaM, which photoisomerized upon light activation and altered the protein conformation. Furthermore, the demonstrated promiscuity of the evolved synthetases may allow various alternative azobenzene-based amino acids to be incorporated into proteins. We expect that the genetic incorporation of these photoswitchable click amino acids into proteins will serve as a basis for the design of proteins carrying a photoswitchable bridge, through which protein conformation and bioactivity can be photo-modulated in live cells to provide a novel avenue for the minimally invasive investigation of biological processes in vivo.
Supplementary Material
Footnotes
C.H. gratefully acknowledges a fellowship from the Deutsche Forschungsgemeinschaft DFG (HO 4778/1-1) and partial funding from the Pioneer Fund. L.W. acknowledges support from California Institute for Regenerative Medicine (RN1-00577-1) and US National Institutes of Health (1DP2OD004744-01 and P30CA014195).
Supporting information for this article is available on the WWW under http://www.angewandte.org.
References
- [1].Brieke C, Rohrbach F, Gottschalk A, Mayer G, Heckel A. Angew. Chem. 2012;124:8572–8604. doi: 10.1002/anie.201202134. [DOI] [PubMed] [Google Scholar]; Angew. Chem. Int. Ed. 2012;51:8446–8476. doi: 10.1002/anie.201202134. [DOI] [PubMed] [Google Scholar]
- [2].Szymański W, Beierle JM, Kistemaker HAV, Velema WA, Feringa BL. Chem. Rev. 2013 doi: 10.1021/cr300179f. DOI: 10.1021/cr300179f. [DOI] [PubMed] [Google Scholar]
- [3].for review see Beharry AA, Woolley GA. Chem. Soc. Rev. 2011;40:4422–4437. doi: 10.1039/c1cs15023e.
- [4].for some examples see: Polosukhina A, Litt J, Tochitsky I, Nemargut J, Sychev Y, De Kouchkovsky I, Huang T, Borges K, Trauner D, Van Gelder RN, Kramer RH. Neuron. 2012;75:271–282. doi: 10.1016/j.neuron.2012.05.022. Beharry AA, Wong L, Tropepe V, Woolley GA. Angew. Chem. 2011;123:1361–1363. doi: 10.1002/anie.201006506. Angew. Chem. Int. Ed. 2011;50:1325–1327. doi: 10.1002/anie.201006506. Zhang F, Timm KA, Arndt KM, Woolley GA. Angew. Chem. 2010;122:4035–5038. doi: 10.1002/anie.201000909. Angew. Chem. Int. Ed. 2010;49:3943–3946. doi: 10.1002/anie.201000909. Hoppmann C, Schmieder P, Domaing P, Vogelreiter G, Eichhorst J, Wiesner B, Morano I, Rück-Braun K, Beyermann M. Angew. Chem. 2011;50:7699–7702. doi: 10.1002/anie.201101398. Wyart C, Del Bene F, Warp E, Scott EK, Trauner D, Baier H, Isacoff EY. Nature. 2009;461:407–410. doi: 10.1038/nature08323. Kramer RH, Chambers JJ, Trauner D. Nat. Chem. Biol. 2005;1:360–365. doi: 10.1038/nchembio750. Fortin DL, Banghart MR, Dunn TW, Borges K, Wagenaar DA, Gaudry Q, Karakossian MH, Otis TS, Kristan WB, Trauner D, Kramer RH. Nat. Methods. 2008;5:331–338. doi: 10.1038/nmeth.1187. Cattani-Scholz A, Renner C, Cabrele C, Behrendt R, Oesterhelt D, Moroder L. Angew. Chem. 2002;114:299–302. doi: 10.1002/1521-3773(20020118)41:2<289::aid-anie289>3.0.co;2-2. Angew. Chem. Int. Ed. 2002;41:289–292. Schutt M, Krupka SS, Milbradt AG, Deindl S, Sinner EK, Oesterhelt D, Renner C, Moroder L. Chem. Biol. 2003;10:487–490. doi: 10.1016/s1074-5521(03)00128-5. Zhang Y, Erdmann F, Fischer G. Nat. Chem. Biol. 2009;5:724–726. doi: 10.1038/nchembio.214. Volgraf M, Gorostiza P, Numano R, Kramer RH, Isacoff EY, Trauner D. Nat. Chem. Biol. 2006;2:47–52. doi: 10.1038/nchembio756. Hoppmann C, Seedorff S, Richter A, Fabian H, Schmieder P, Rück-Braun K, Beyermann M. Angew. Chem. 2009;121:6763–6766. doi: 10.1002/anie.200901933. Angew. Chem. Int. Ed. 2009;48:6636–6639. doi: 10.1002/anie.200901933. Stein M, Middendorp SJ, Carta V, Pejo E, Raines DE, Forman SA, Sigel E, Trauner D. Angew. Chem. 2012;125:10652–10656. doi: 10.1002/anie.201205475. Angew. Chem. Int. Ed. 2012;51:10500–10504. doi: 10.1002/anie.201205475. Hoppmann C, Barucker C, Lorenz D, Multhaup G, Beyermann M. ChemBioChem. 2012;13:2657–2660. doi: 10.1002/cbic.201200605.
- [5].a) Zhang F, Zarrine-Afsar A, Al-Abdul-Wahid MS, Prosser RS, Davidson AR, Woolley GA. J. Am. Chem. Soc. 2009;131:2283–2289. doi: 10.1021/ja807938v. [DOI] [PubMed] [Google Scholar]; b) Schierling B, Noel A-J, Wende W, Hien LT, Volkov E, Kubareva E, Oretskaya T, Kokkinidis M, Rompp A, Spengler B, Pingoud A. Proc. Natl. Acad. Sci. U S A. 2009;107:1361–1366. doi: 10.1073/pnas.0909444107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].for thiol-reactive azobenzene photoswitches see: Kumita JR, Smart OS, Woolley GA. Proc. Natl. Acad. Sci. U S A. 2000;97:3803–3808. doi: 10.1073/pnas.97.8.3803. Woolley GA. Acc. Chem. Res. 2005;38:486–493. doi: 10.1021/ar040091v. Zhang F, Sadovski O, Woolley GA. ChemBioChem. 2008;9:2147–2154. doi: 10.1002/cbic.200800196. d); Kneissl S, Loveridge EJ, Williams C, Crump MP, Allemann RK. ChemBioChem. 2008;9:3046–3054. doi: 10.1002/cbic.200800502. Kusebauch U, Cadamuro SA, Musiol H-J, Lenz MO, Wachtveitl J, Moroder L, Renner C. Angew. Chem. 2006;118:7170–7173. doi: 10.1002/anie.200601432. Angew. Chem. Int. Ed. 2006;45:7015–7018. doi: 10.1002/anie.200601432. Kusebauch U, Cadamuro SA, Musiol H-J, Moroder L, Renner C. Chem.-Eur. J. 2007;13:2966–2973. doi: 10.1002/chem.200601162. Le Thi Hien B, Schierling A, Yu. Ryazanova T, Zatsepin S, Volkov EM, Kubareva EA, Velichko TI, Pingoud A, Oretskaya TS. Russ. J. Bioorg. Chem. 2009;35:549–555.
- [7].a) Hoppmann C, Schmieder P, Heinrich N, Beyermann M. ChemBioChem. 2011;12:2555–2559. doi: 10.1002/cbic.201100578. [DOI] [PubMed] [Google Scholar]; b) Hoppmann C, Kühne R, Beyermann M. Beilstein J. Org. Chem. 2012;8:884–889. doi: 10.3762/bjoc.8.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].a) Wang L, Schultz PG. Angew. Chem. 2005;117:34–68. [Google Scholar]; Angew. Chem. Int. Ed. 2005;44:34–66. [Google Scholar]; b) Wang Q, Parrish AR, Wang L. Chem. Biol. 2009;16:323–336. doi: 10.1016/j.chembiol.2009.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Liu CC, Schultz PG. Annu. Rev. Biochem. 2010;79:413–44. doi: 10.1146/annurev.biochem.052308.105824. [DOI] [PubMed] [Google Scholar]
- [9].Bose M, Groff D, Xie J, Brustad E, Schultz PG. J. Am. Chem. Soc. 2006;128:388–389. doi: 10.1021/ja055467u. [DOI] [PubMed] [Google Scholar]
- [10].a) Xiang Z, Ren H, Hu YS, Coin I, Wei J, Cang H, Wang L. Nat. Methods. 2013;10:885–88. doi: 10.1038/nmeth.2595. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Coin I, Katritch V, Sun T, Xiang Z, Siu FY, Beyermann M, Stevens RC, Wang L. Cell. 2013;155:1258–1269. doi: 10.1016/j.cell.2013.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Xiang Z, Lacey VK, Ren H, Xu J, Burban DJ, Jennings PA, Wang L. Angew. Chem. 2014:126. [Google Scholar]; Angew. Chem. Int. Ed. 2014;53 doi: 10.1002/anie.201308794. [Google Scholar]
- [11].Wang L, Brock A, Herberich B, Schultz PG. Science. 2001;292:498–500. doi: 10.1126/science.1060077. [DOI] [PubMed] [Google Scholar]
- [12].Takimoto JK, Dellas N, Noel JP, Wang L. ACS Chem. Biol. 2011;6:733–743. doi: 10.1021/cb200057a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Santoro SW, Wang L, Herberich B, King DS, Schultz PG. Nat. Biotechnol. 2002;20:1044–1048. doi: 10.1038/nbt742. [DOI] [PubMed] [Google Scholar]
- [14].Wang W, Takimoto JK, Louie GV, Baiga TJ, Noel JP, Lee KF, Slesinger PA, Wang L. Nat. Neurosci. 2007;10:1063–1072. doi: 10.1038/nn1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Lacey VK, Louie GV, Noel JP, Wang L. ChemBioChem. 2013;14:2100–2105. doi: 10.1002/cbic.201300400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].a) Wang L, Zhang Z, Brock A, Schultz PG. Proc. Natl. Acad. Sci. U S A. 2003;100:56–61. doi: 10.1073/pnas.0234824100. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Liu H, Wang L, Brock A, Wong C-H, Schultz PG. J. Am. Chem. Soc. 2003;125:1702–1703. doi: 10.1021/ja029433n. [DOI] [PubMed] [Google Scholar]; c) Mahal L, Yarema KJ, Bertozzi CR. Science. 1997;276:1125–1128. doi: 10.1126/science.276.5315.1125. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.