Abstract
Genome sequencing can identify individuals in the general population who harbor rare coding variants in genes for Mendelian disorders1–7 – and who consequently may have increased disease risk. However, previous studies of rare variants in phenotypically extreme individuals have ascertainment bias and may demonstrate inflated effect size estimates8–12. We sequenced seven genes for maturity-onset diabetes of the young (MODY)13 in well-phenotyped population samples14,15 (n=4,003). Rare variants were filtered according to prediction criteria used to identify disease-causing mutations: i) previously-reported in MODY, and ii) stringent de novo thresholds satisfied (rare, conserved, protein damaging). Approximately 1.5% and 0.5% of randomly selected Framingham and Jackson Heart Study individuals carried variants from these two classes, respectively. However, the vast majority of carriers remained euglycemic through middle age. Accurate estimates of variant effect sizes from population-based sequencing are needed to avoid falsely predicting a significant fraction of individuals as at risk for MODY or other Mendelian diseases.
For personal genome sequencing to help identify at-risk individuals for preventative care1–7, the ascertainment bias typically employed in human genetic research presents a double-edged sword: studying individuals with extreme phenotype has efficiently identified disease variants but has also introduced an upwards bias in reported effect sizes10–12. Examples include initial inflation of reported BRCA1/BRCA2 (breast cancer; OMIM#114480)8,16 and HFE (hereditary hemochromatosis; OMIM#235200)9 mutation penetrance, remedied once adequately-sized population-based control groups were studied. Absent similar studies for other diseases, widespread personal genomic testing might exaggerate risk estimates, instigating needless intervention in low-risk individuals1,2,6,17.
Maturity-onset diabetes of the young (MODY, OMIM#606391)18,19 is a good candidate for personal genomic screening: (a) MODY is caused by dominant Mendelian mutations, such that heterozygous carriers develop disease20; (b) clinical presentation occurs early in life (<25 years) with non-ketotic hyperglycemia18,19; (c) MODY frequency is 0.1%-0.2% in European populations21,22, with the majority un- or misdiagnosed23; (d) MODY diagnosis can significantly impact on diabetes prognosis and treatment24,25 of the individual or affected family members26; (e) mutations in the MODY genes also influence late onset phenotypes, as common variants near many of these genes are associated with type 2 diabetes (T2D) risk in the general population27; and (f) this risk can be reduced via lifestyle intervention28,29. Thus, individuals identified through personal genomics to carry variants in MODY genes might be anticipated to exhibit elevated glycemic parameters by young adulthood, or at minimum by middle age, and could benefit from early intervention.
We aimed to characterize the spectrum of low frequency variation in MODY genes in the general population. Specifically: i) how many individuals carry rare variants in MODY genes of the sort that might be bioinformatically flagged in a personal genome sequencing context, and ii) what percentage of these carriers demonstrate an abnormal glycemic phenotype by middle age. In addition to randomly-ascertained individuals from population cohorts, we validated our methods using individuals selected for MODY or T2D diagnosis.
We focused on seven genes: four most-frequently (HNF1A30, GCK31,32, HNF4A33, HNF1B34), and three less-frequently (PDX135, INS36, NEUROD137), mutated in European MODY patients. Six other MODY genes were not studied: two because loss-of-function causes hypoglycemia rather than diabetes (ABCC8, KCNJ11)38, and four because sequence data were unavailable (PAX439, BLK40, KLF1141, CEL42; see Methods).
Primary analysis focused on 4,003 individuals drawn from three population-based cohorts (Table 1). First, we randomly ascertained 1,541 individuals of European ancestry from the Framingham Heart Study Offspring Cohort14 (FHS cohort) and 1,691 of African-American ancestry from the Jackson Heart Study (JHS cohort)15; these individuals are referred to as ‘unselected’ (Supplementary Figs. 1–3). Second, from Finnish and Swedish cohorts (>27,500 individuals), we ascertained 771 individuals from the extremes of T2D genetic risk (T2D cohort; 362 young lean T2D cases; 409 elderly obese euglycemic controls11); these individuals are referred to as ‘extreme’ (Supplementary Figs. 1,3–5; Methods). Target capture and DNA sequencing43 of the seven analyzed genes (Methods) were used to identify sequence variants: >93% of bases were covered with at least 20 reads in all genes except INS (79.1%) and PDX1 (37.7%; Supplementary Table 1). Genotyping of select variants, including those observed in single individuals, suggested very few (<0.05%) genotype calls were incorrect (Methods, Supplementary Data 1).
Table 1.
Description of studied individuals.
| Unselected cohorts | Extreme T2D cohort |
MODY patients |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| FHS cohort | JHS cohort | ||||||||
| No DM | DM/No Med | DM/Med | No DM | DM/No Med | DM/Med | No DM | DM | ||
| Number | 1435 | 54 | 52 | 1345 | 54 | 292 | 409 | 362 | 250 |
| Male (%) | 48.0 | 61.1 | 65.4 | 38.7 | 33.3 | 31.2 | 50.6 | 45.6 | 46.5 |
| Age (yr) | 55.1 ± 9.5 | 59.0 ± 7.8 | 60.4 ± 8.1 | 55.5 ± 11.5 | 57.7 ± 10.0 | 60.5 ± 10.1 | 67.4 ± 7.3 | 53.0 ± 9.2 | 22.3 ± 12.4 |
| BMI (kg/m2) | 25.1 ± 5.1 | 27.7 ± 5.5 | 28.6 ± 5.4 | 31.3 ± 7.1 | 33.3 ± 5.6 | 34.2 ± 7.2 | 33.6 ± 3.6 | 23.6 ± 2.0 | 23.5 ± 4.9 |
| FPG (mg/dL) | 95.5 ± 9.6 | 171.9 ± 55.3 | 183.3 ± 71.6 | 91.5 ± 9.3 | 183.8 ± 66.7 | 141.9 ± 56.7 | 92.2 ± 6.0 | 167.9 ± 51.1 | 134.2 ± 39.2 |
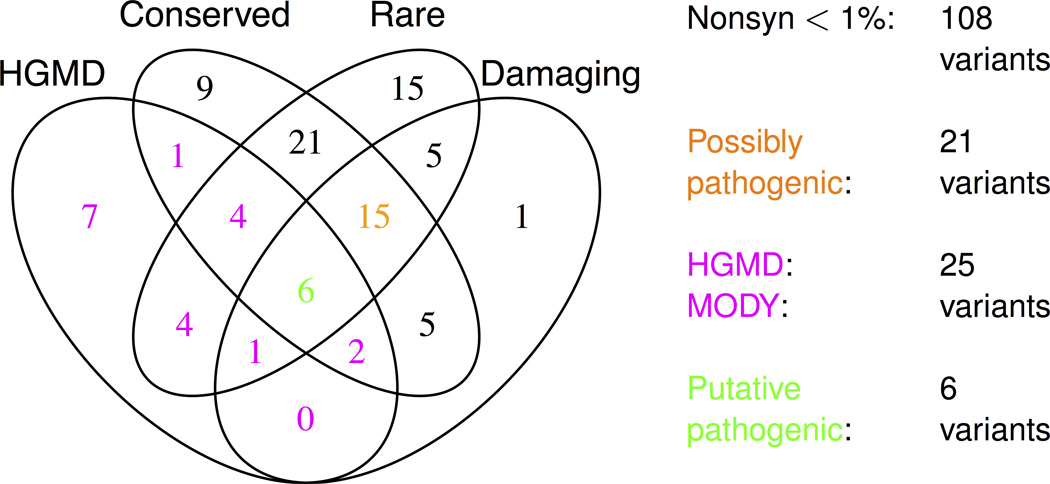
In total, 121 non-synonymous variants were identified across the seven genes (Supplementary Fig. 6; Supplementary Tables 2–3). Variants with minor allele frequency (MAF) >1% were omitted from analyses as these have been well-studied in larger epidemiological cohorts44,45. Following the model increasingly used in sequence interpretation7,17, we categorized the remaining 108 variants into four non-exclusive classes (Fig. 1, Table 2; Methods):
low frequency non-synonymous: resulting in altered protein sequence
possibly pathogenic: (a) evolutionarily conserved site; (b) private to one study individual, not observed in 1000 genomes project46; and (c) computationally predicted as protein-damaging by mutation analysis tools SIFT47 and PolyPhen-248
HGMD MODY: previously reported as causal for MODY in the Human Gene Mutation Database Professional v2012.149
putative pathogenic: meeting criteria for possibly pathogenic and HGMD MODY.
Figure 1. Description of low frequency non-synonymous variants.
Shown is a summary of all low frequency (MAF<1%) non-synonymous variants identified in the three cohorts (Supplementary Tables 1–3 show lists of all variants, not simply those of low frequency). MAF is calculated as the maximum frequency across the three cohorts. Shown also is the number of variants fitting each annotation (HGMD, conserved, rare, and damaging) and each variant class: low frequency non-synonymous (black, magenta, orange, or green), possibly pathogenic (orange or green), HGMD MODY (magenta or green), and putative pathogenic (green). Twelve low frequency variants fit no annotations. Low frequency non-synonymous is abbreviated to ‘Nonsyn <1%’.
Table 2.
Variant counts in each of the seven sequenced genes.
| Gene | Total | HGMD | Conserved | Rare | Damaging |
|---|---|---|---|---|---|
| GCK | 13 | 3 | 9 | 9 | 4 |
| HNF1A | 27 | 12 | 17 | 19 | 11 |
| HNF1B | 17 | 4 | 12 | 9 | 6 |
| HNF4A | 23 | 6 | 12 | 20 | 6 |
| INS | 6 | 0 | 0 | 2 | 0 |
| NEUROD1 | 11 | 0 | 8 | 9 | 3 |
| PDX1 | 11 | 0 | 5 | 3 | 5 |
| Total | 108 | 25 | 63 | 71 | 35 |
Despite the recognized limitations of bioinformatics criteria alone for disease risk prediction7,50,51, variants similar to those in the HGMD MODY and putative pathogenic classes are likely to be reported from personal genome sequencing52–55. Possibly pathogenic variants are also relevant in personal genomics as they fit criteria used to ascribe pathogenicity to variants identified via next-generation sequencing in Mendelian disease studies56,57.
For validation, we applied these bioinformatic criteria to DNA sequence data obtained from 250 Norwegian patients fitting MODY diagnostic criteria (Table 1, Supplementary Fig. 7). In total, 48% of these patients carried variants meeting the low frequency non-synonymous criteria (Supplementary Tables 4–5), 32% carried variants meeting the possibly pathogenic or HGMD MODY criteria, and 19% carried variants in the putative pathogenic class. As expected, a substantial fraction of patients fitting classic MODY diagnostic criteria carry variants that meet these annotation classes.
We used the same bioinformatics approach to analyze variants found in unselected individuals. For FHS and JHS: 4.4% and 5.7% of individuals carried a variant in the low frequency non-synonymous class (Supplementary Table 5), 1.5% carried variants in the HGMD MODY class, and 0.5% carried variants in the possibly pathogenic class. While these frequencies are a tenth the rate observed in MODY patients, they are at least an order of magnitude greater than estimates of MODY prevalence in the general population22,58.
We asked whether these variant carriers possessed clinical features associated with MODY (Methods): diabetes diagnosis ≤25 years (proband or family members), lean (BMI<25), and family history of diabetes (≥two generations; typically early-onset)13,20,59. One variant carrier in the JHS cohort, and none in the FHS cohort, fit these criteria (although several non-carriers did; Methods). These results suggest that only a small minority of individuals carrying such mutations demonstrate clinical characteristics consistent with MODY.
Despite lacking signs of MODY, these variant carriers might nonetheless be at elevated risk for T2D or hyperglycemia. We calculated among variant carriers the prevalence of: i) diabetes (‘receiving medication for diabetes’ or fasting plasma glucose (FPG) levels>126mg/dL), or ii) impaired fasting glucose60 (IFG; FPG 100–126mg/dL).
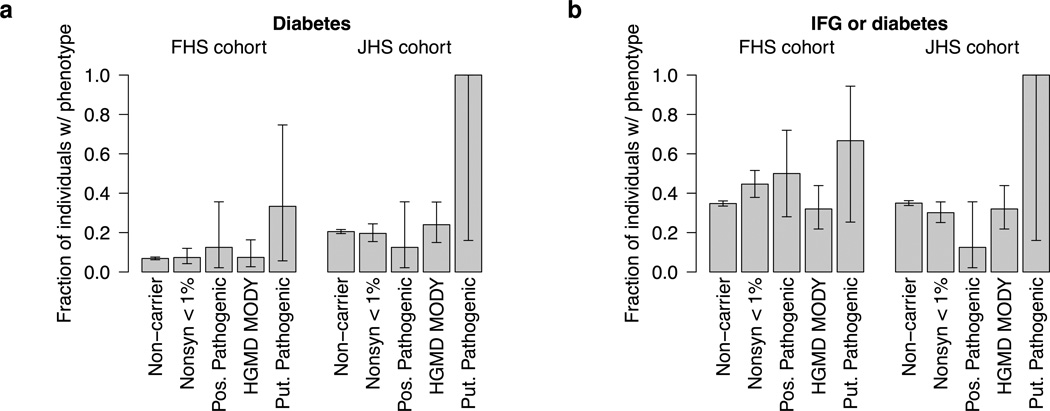
For carriers in the unselected cohorts, point estimates of risk for diabetes were not elevated relative to non-carriers (Fig. 2a, Supplementary Table 6). Carriers of low frequency non-synonymous variants showed no trend toward increased risk (FHS OR=1.1, JHS OR=0.9, p>.1), with HGMD MODY or putative pathogenic variants having similar effect size estimates. The majority of variant carriers in these two classes (>90% of the FHS and near 80% of the JHS) did not develop diabetes, and only two of four carriers (50%) of variants in the most stringent putative pathogenic class developed diabetes (p=0.049).
Figure 2. Phenotypic impact of variants in unselected cohorts.
Shown is the fraction of variant carriers in the two unselected cohorts with (a) diabetes and (b) IFG or diabetes. Separate fractions are given for each of the four defined variant classes, with the fraction of non-carriers with each phenotype shown as reference. Error bars reflect 68% confidence intervals in the estimated fractions and are computed via the Clopper-Pearson method. The number of analyzed samples is given in Supplementary Table 6. Fewer individuals had FPG measurements than diabetes measurements; thus the number of analyzed individuals for these two phenotypes differs. ‘Nonsyn <1%’ is low frequency non-synonymous, ‘Pos. Pathogenic’ is possibly pathogenic, and ‘Put. Pathogenic’ is putative pathogenic.
Furthermore, we did not observe a trend towards IFG in variant carriers (Fig. 2b, Supplementary Table 6), despite many individuals being middle aged (Supplementary Fig. 8) and subjected to long-term follow-up14. Only 32% of HGMD MODY variant carriers, and 31% of proposed pathogenic variant carriers, had IFG or diabetes (compared to ~35% of non-carriers). These results are consistent with previous reports showing lack of association between PDX1 variation and diabetes61, although they apply more broadly across all seven analyzed genes.
We performed several additional analyses to investigate the observed low penetrance of variants in the unselected cohorts. To further validate our bioinformatics criteria, we computed carrier frequencies in the extreme T2D cohort (Supplementary Tables 5,7). Relative to old obese controls, young lean T2D cases carried a three-fold excess of low frequency non-synonymous variants (4.7% vs. 1.5%, OR=3.2, p=0.011) and an apparent excess of possibly pathogenic (four observations exclusive to cases, p=0.04) or HGMD MODY variants (four case observations, one control observation). Absolute variant frequencies in the extreme cohort are not directly comparable to those in the unselected cohorts due to differences in ethnic composition. The relative frequencies between cases and controls, however, validate the ability of bioinformatics criteria to identify an enrichment of rare mutations not only in individuals with MODY but also in those with T2D – provided that these individuals are pre-selected for phenotype rather than drawn at random from the population.
We next investigated whether analyzing multiple genes at once might influence our results, as MODY clinical presentation varies with gene affected and observed mutation13,20. While 64 individuals carried possibly pathogenic or HGMD MODY variants in the four most commonly-mutated MODY genes, only four carried variants in INS, PDX1, or NEUROD1. Thus, inclusion of these three additional genes had a minimal impact on our results (Supplementary Tables 8–9).
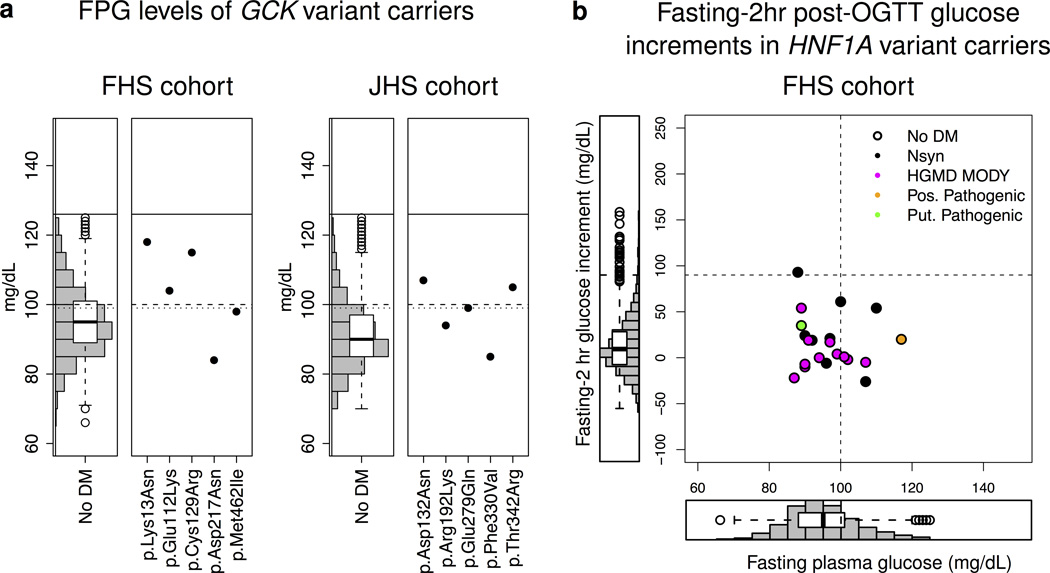
Next, we analyzed variants specific to GCK, where mutations cause only mild and typically stable elevations in FPG (99–144 versus 72–108mg/dL in normoglycemia)20,62. While only two carriers of low frequency non-synonymous GCK mutations met diabetes criteria, 67% (8/12) had FPG levels ≥99mg/dL (compared to 35% of non-carriers, combined p=0.054; Fig. 3a; Supplementary Table 10). Thus, individuals who carry GCK variants may display mild hyperglycemia. This result would need to be precisely communicated in the clinic as GCK-MODY patients are typically treated by diet modification alone62 and do not show accelerated decline in beta-cell function63.
Figure 3. Phenotypes of GCK and HNF1A variant carriers.
(a) Shown are FPG values for GCK-variant carriers in the FHS cohort (left) and JHS cohort (right). Three dashed lines correspond to defined FPG thresholds: top (solid) line represents diabetes (126 mg/dL), middle (dashed) line IFG (100 mg/dL), bottom (dotted) line GCK-MODY (99mg/dL). A histogram and box plot representing FPG levels in the non-diabetic population (computed separately for each cohort) is shown for comparison. Two GCK variant carriers were on medication for diabetes and are thus excluded from the plot. Tabular form of these results (including the two carriers with diabetes) is shown in Supplementary Table 10. (b) The scatter plot shows fasting-2hr post-OGTT plasma glucose increment (y-axis) and FPG (x-axis) for each HNF1A-variant carrier in the FHS cohort (OGTT information was unavailable for the JHS cohort). Histograms showing FPG and fasting-2hr post-OGTT plasma glucose increments for individuals in the FHS cohort without diabetes are shown on the left and below the scatter plot respectively. FPG values for individuals receiving treatment for diabetes were omitted from the plot. The vertical and horizontal dashed lines represent the IFG threshold (100mg/dL) and a plasma glucose increment consistent with HNF1A-MODY/beta-cell dysfunction (≥90mg/dL) respectively. Points are colored corresponding to the annotation class of the variant; for variants that belong to multiple classes, colors are chosen according to the following precedence: putative pathogenic, HGMD MODY, possibly pathogenic, low frequency non-synonymous.
Finally, we expanded the set of clinical read-outs used in our analysis of specific genes. For HNF1A variant carriers, we asked whether glucose tolerance is affected even if FPG remains within euglycemic range – a possibility suggested as i) HNF1A-MODY patients can demonstrate elevated fasting-2hr blood glucose increments (≥90mg/dL) following an oral glucose tolerance test (OGTT)63,64, and ii) the common HNF1A c.293C>T (p.Ala98Val) variant is associated with altered beta-cell function65. Only one (of 17) HNF1A-variant carriers had an elevated fasting-2hr glucose increment (Fig. 3b; Supplementary Fig. 9). Likewise, HNF1B-MODY can be accompanied by renal dysfunction34; none of the HNF1B variant carriers had abnormal creatinine values66 (Supplementary Fig. 10).
In summary, a substantial proportion of individuals in the general population carry low frequency non-synonymous variants in one of seven MODY genes. Two classes of bioinformatics criteria (either previously reported as causing MODY or rare, conserved, and computationally predicted damaging) are each sufficient to identify a substantial enrichment of variants in individuals diagnosed with MODY or selected for an extreme diabetic phenotype. However, for each class, the majority of variant carriers observed in the general population remain euglycemic through middle age. These results highlight the limitations of disease variant databases, as well as objective and stringent bioinformatics criteria, in ascribing pathogenicity to rare variants.
Our study has multiple limitations. As the individuals are drawn from different genetic and environmental backgrounds, frequencies and effect sizes are not directly comparable across cohorts. This reflects one of the potential challenges of personal genomics: that the personal genome analyzed may not match the datasets from which effect size estimates are drawn. Also, because rare coding variants have low counts even in a study of thousands of participants, power is limited for statistical assessment of heterogeneity across genes or variants; it is possible that a subset of the variants identified in fact have very large effects. Expert interpretation of each gene and variant, information about other family members, and functional characterization of variants may identify a subset with large and robust effects – but these are not yet practical, let alone standardized7, in the automated analysis of personal genome sequences.
MODY is a useful model for studying the application of personal genome sequencing to disease risk prediction13,26: a number of causal genes are well established and a dominant pattern of inheritance predicts that heterozygous mutation carriers will have a phenotype. However, even for this one disease, extrapolation of effect size estimates from extreme individuals to unselected individuals (in the FHS and JHS cohorts) might falsely predict a significant fraction as at higher risk for diabetes: 3 in 200 individuals carry a variant previously reported to cause MODY and yet exhibit no trend toward even late-onset T2D or impaired fasting glucose. Even objective bioinformatics criteria might incorrectly classify a significant, although three-fold lower, fraction of individuals as at-risk. The view that rare variants have deterministic effects, whereas common variants have modest effects, reflects in part the ascertainment bias of study designs used in Mendelian genetic research as well as the true penetrance of rare mutations.
Methods
Sample selection
To obtain individuals for the unselected cohorts, we drew from the Framingham Heart Study (FHS) Offspring cohort14 and from the Jackson Heart Study (JHS)15 cohort. As previously described, the FHS is a three generation prospective, community-based, family study which began in 1948 and was designed to identify factors that contributing to cardiovascular disease74; the Offspring cohort consists of 5,124 of the adult children and spouses (enrolled in 1971) of the original participants14. The JHS is a large, community-based, observational study whose participants were recruited from urban and rural areas of the Jackson Mississippi metropolitan statistical area (MSA)15. These studies were performed using protocols approved by FHS, JHS, and institutional ethics committees, and with informed consent from all participants.
To select individuals for the T2D cohort, we drew from 27,500 individuals in three prospective cohorts: the Malmö Preventive Project75 and Scania Diabetes Registry76 (from Sweden), and the Botnia Study77 (from Finland). Individuals were ranked according to a liability model that measured risk for T2D as previously described11. Briefly, liability scores were computed as the difference between diabetes status and the predicted risk based on age, BMI, and gender; extreme cases were selected to have the highest liability scores (with diabetes but with low predicted risk for diabetes), and extreme controls were selected to have the lowest liability scores (without diabetes but with high predicted risk for diabetes). Individuals with age of diabetes diagnosis below 35 were excluded in an attempt to avoid consideration of patients with type 1 diabetes or MODY. The participants gave their written informed consent and the study protocol was approved by the Ethics Committees of Helsinki University Hospital, Finland, and Lund University.
Clinical and phenotypic parameters in unselected cohorts
Phenotype information for the unselected cohorts was contained in the NHLBI Framingham Cohort, dbGAP dataset: phs000007.v16.p6 (FHS) and the NHLBI Jackson Heart Study Candidate Gene Association Resource dbGAP dataset: phs000286.v2.p1 (JHS). We used phenotypic data from exam 5 for individuals from the FHS and phenotypic data from exam 1 for individuals from the JHS.
Individuals were classified as having diabetes if they were i) documented as such in the FHS or JHS exams, or ii) had FPG levels ≥126mg/dL. Individuals with FPG values 100-126mg/dL were said to display IFG. The euglycemic FPG range was consequently defined as 72–99mg/dL60. As GCK-MODY patients exhibit only mildly-elevated FPG versus other MODY subtypes, the FPG range fitting this specific phenotype was defined as 99–144mg/dL20,64. In the case of HNF1A variant carriers, aberrant fasting-2hr post-OGTT plasma glucose increments were defined as those ≥90mg/dL (this indicative of beta-cell dysfunction)64,78. HNF1B variant carriers were assessed for signs of renal dysfunction by evaluating serum creatinine levels, the normal range defined as 0.7–1.3mg/dL66.
Sequencing, quality control, and variant annotation
Although individuals in the T2D, FHS, and JHS cohorts spanned multiple cohorts, all analysis (target capture, sequencing, variant calling, quality control, annotation, and association analysis) was performed in an identical fashion using the same statistical pipeline. MODY patients were sequenced separately, but annotation and association was performed identically to the other three cohorts.
To sequence individuals in the T2D, FHS, and JHS cohorts, we designed two custom hybrid capture arrays, each using the same previously described technology43,79, to sequence two sets of genes as part of two larger studies. Individuals from the FHS and JHS cohorts were sequenced for 181 genes previously associated with cardiovascular disease risk factors, including 37 genes associated with diabetes. Individuals from the T2D cohort were sequenced for 257 genes previously associated with diabetes or heart attack (either identified through genome-wide associations or reported to cause Mendelian disorders). These arrays had in common nine genes reported to harbor variants that cause MODY: the seven genes analyzed for this study as well as ABCC8 and KCNJ11.
DNA libraries were barcoded using the Illumina index read strategy and sequenced with an Illumina HiSeq2000. Reads were mapped to the human genome hg19 with the BWA algorithm80,81 and processed with the Genome Analysis Toolkit (GATK) to recalibrate base quality-scores and perform local realignment around known indels82. Target coverage or each sample was also computed with the GATK. Single nucleotide polymorphisms (SNPs) and small insertions and deletions (indels) were called with the Unified Genotyper module of the GATK and filtered to remove SNPs with annotations indicative of technical artifacts (such as strand-bias, low variant call quality, or homopolymer runs)82. SNPs with differential call rates (p<1e-3 as computed by the PLINK software package83) were excluded from association analysis. Variant calls from sequence data were deposited in dbGAP (FHS: dataset phs000307.v4.p7; JHS: dataset phs000498.v2.p1).
Samples were also genotyped on one of three genome-wide SNP arrays: the Affymetrix 500k GeneChip Mapping Set (FHS cohort), the Affymetrix Genome-Wide Human SNP Array 6.0 (JHS cohort), and the Metabochip (T2D cohort). We computed concordance of sequence genotypes with these SNP array genotypes using the PLINK software package83. Principal component analysis was performed on a set of SNPs common to all three platforms using PLINK and EIGENSTRAT84. These analyses verified that all individuals were unrelated (<25% of their genomes identical by descent) and confirmed the distinct genetic background of each cohort (Supplementary Fig. 3). Quality control showed high (>96%) concordance between sequence and SNP array genotypes for the same individuals.
To ensure that all analyzed variants had genotypes strongly supported by sequence data, we used three strategies. First, we ignored all genotypes supported by fewer than 10 reads (e.g. set as “missing”). Second, we examined the raw read data of every called variant and excluded any from analysis that had visual signatures of sequencing artifacts, such as reads of poor mapping quality, evidence for variation supported by only reads on one strand of the genome, or additional called variants nearby; screenshots of all variants are available as Supplementary Data 1. Third, to evaluate the extent to which erroneous sequence genotypes might impact our analysis, we genotyped 143 SNPs called in the T2D cohort (the first cohort sequenced and with the lowest depth of coverage) using a Sequenom iPLEX Assay. This included nine SNPs observed in only a single individual (singletons) and 27 SNPs observed in two individuals (doubletons). Of the 1573 individuals identified as carrying one of these 143 SNPs based on sequence genotypes, only nine had different Sequenom genotypes (>99.4% non-reference concordance). All (100% of) the 63 identified carriers of singleton or doubleton variants were confirmed based on Sequenom genotypes. These analyses collectively suggested that the analyzed genotypes were of high quality and that any errors had at most a minimal impact on our results.
We annotated variants previously reported to cause MODY using a list of mutations given in the Human Gene Mutation Database Professional v2012.149. Evolutionarily conserved variants were defined as those at sites conserved across 46 vertebrates, based on PhyloP scores (LOD >3) downloaded from the UCSC genome browser database85. Rare variants were defined as those observed in only a single study individual across the T2D, FHS, and JHS cohorts, and furthermore were not identified as part of the 1000 genomes project46. Predicted protein-damaging variants were defined as those either (a) deleterious according to SIFT47 and possibly or probably damaging according to Polyphen-248, or (b) nonsense, frame-shift, or essential splice site mutations according to the Variant Effect Predictor67. Protein and nucleotide changes, and SIFT and Polyphen-2 scores, were produced for each variant with the Variant Effect Predictor, using the biologically-relevant transcripts for each gene (as listed in the HGMD and shown in the manuscript ‘Accession Numbers’ section).
For analyses that required variants below a given frequency, variants that exceeded the threshold in any of the cohorts were removed from analysis.
Analysis of MODY patients
As a technical validation of the annotation protocol used for the population-based cohorts, variants in 250 subjects with MODY were analyzed using the same methodology. As previously described21,36,86–93, subjects with MODY were recruited from the Norwegian MODY Registry, a national, population-based registry established in 1997. Patients are referred to the registry if they fit the following criteria: i) a first-degree relative with diabetes; ii) onset of diabetes before age 25 years in at least one family member; iii) a low dose insulin requirement; iv) unusual type 1 diabetes (insulin requirement below 0.5 U/kg/day, no antibodies, atypical history). The four most common MODY genes are routinely sequenced in the first instance: HNF1A, HNF4A, GCK and INS. If fasting glucose is >100 mg/dL and HbA1c is less than 7.5 %, GCK is investigated first followed by the other genes from the list above. If renal cysts or renal failure presents before diabetes, HNF1B is investigated. NEUROD1 and PDX1 were investigated as part of a screening program and are not routinely tested. Informed consent was obtained from all participants and the study was approved by the Regional Committee for Research Ethics and the Norwegian Data Inspectorate.
To screen patients, genomic DNA was first extracted from peripheral leukocytes using standard procedures. The coding exons and intron/exon boundaries of HNF1A, HNF4A, GCK, HNF1B, PDX1, NEUROD1 and/or INS were then PCR-amplified and sequenced using an Applied Biosystems 3730 capillary sequencer (Applied Biosystems, Foster City, CA). We imported the samples into the SeqScape Software (Applied Biosystems) and analysed for sequence variations by comparing with published reference sequences (NM_000545.5, NM_175914.3, NM_000162.3, NM_000458.2, NM_000209.3, NM_002500.3 and NM_000207.2, respectively). Variants were annotated and classified using the same procedure as for the three population-based cohorts.
Assessment of MODY-relevant clinical criteria in unselected cohorts
We examined variant carriers in the FHS and JHS cohorts to see if any fit the following phenotypic criteria associated with MODY: diabetes diagnosis ≤25 years (for proband or family members) lean (BMI<25), and family history of diabetes (at least two generations; typically early-onset). We did not have access to age of diabetes diagnosis for all family members and subsequently could only examine diagnosis age of variant carriers.
In the FHS cohort, no variant carriers simultaneously fit criteria for early diabetes diagnosis and low BMI (4/116 variant carriers had diabetes with BMI below 25, but each had age of onset above 55, and only one variant carrier had BMI below 30 and age of onset below 40). The frequency of carriers fitting these criteria was thus comparable to that for non-carriers: one of the 1349 non-carriers had BMI below 25 and age of onset below 30, while eight had BMI below 30 and age of onset below 40.
In the JHS cohort, 1/235 variant carriers had a family history of diabetes, age of onset 25, and BMI of 29.8; the rest had BMI above 30 or age of onset above 42. Similar to the FHS cohort, the frequency of carriers fitting these criteria was similar to that for non-carriers: two of the 1405 non-carriers had a family history of diabetes, age of onset below 25, and BMI below 25, with four more having a family history, age of onset below 40, and BMI below 30.
Statistical analysis
We tested for association between phenotype (whether extreme T2D status, diabetes, IFG, or a gene-specific phenotype) and variant carrier status using the Cochran-Mantel-Haenszel test as implemented in the PLINK software package83. Randomized permutation of phenotypes was used to obtain one-sided p-values. For the T2D cohort, stratified permutation of phenotypes (separately for individuals of Finnish and Swedish descent) was used to control for ethnic differences across individuals; due to the small number of variant counts in this cohort, further investigation of population stratification was statistically challenging. For the unselected cohorts, tests were first run separately for individuals from each of the FHS and JHS cohorts and then jointly for individuals from both cohorts (with stratified permutation of phenotypes within cohort) to obtain combined p-values in all figures or tables (including Figure 2). For all figures, confidence intervals in were obtained via the Clopper-Pearson method as implemented in the R software package.
Supplementary Material
Acknowledgements
We gratefully acknowledge the contribution of Framingham and Jackson Heart Study cohort participants, as well as participants from the Malmö Preventive Project, the Scania Diabetes Registry, and the Botnia Study. This work was supported by grants from the National Human Genome Research Institute of NIH (Medical Sequencing Program grant U54 HG003067 to the Broad Institute PI, Lander) and the Howard Hughes Medical Institute, as well as funding from Pfizer Inc. J. Flannick was supported in part by NIH Training Grant 5-T32-GM007748-33. NLB was supported by a Fulbright Diabetes UK Fellowship (BDA 11/0004348). DA was supported by funding from the Doris Duke Charitable Foundation (2006087). JM acknowledges support from NIDDK K24 DK080140. JM and JCF acknowledge support from R01 DK078616. AGB and VA were supported by NIH Medical Scientist Training Program fellowship T32GM007753. JGS and CS were supported by NIH RO1 2R01HL080494, NHLBI, and the LeDucq Foundation. The Jackson Heart Study is supported by Contracts N01-HC-95170, N01-HC-95171, and N01-HC-95172 from the National Heart, Lung, and Blood Institute, the National Institute for Minority Health and Health Disparities, and additional support from the National Institute of Biomedical Imaging and Bioengineering. The Framingham Heart Study was supported by Contracts N01-HC-25195 and 6R01-NS 17950 from the National Heart, Lung and Blood Institute, and genotyping services from Affymetrix, Inc. (Contract No. N02-HL-6-4278 for the SNP Health Association Resource, SHARe, project). The Malmö Preventive Project and the Scania Diabetes Registry were supported by a Swedish Research Council Grant (Linné) to Lund University Diabetes Centre. The Botnia study was supported by funding from Sigrid Juselius Foundation and Folkhälsan Research Foundation, as well as an ERC advanced research grant to LG (GA 269045). The MODY study was supported by grants from the KG Jebsen Foundation, the Norwegian Research Council, the University of Bergen, Helse Vest, Innovest and the European Research Council (AdG).
Footnotes
URLs
Human Gene Mutation Database Professional v2012.149 (http://www.biobase-international.com/product/hgmd)
Variant Effect Predictor67 (http://www.ensembl.org/info/docs/variation/vep/index.html)
Sequence accession numbers
Author contributions
This manuscript describes an integrated analysis that draws together two studies that were initially independent: the Pfizer/MGH/Broad/Lund collaborative project “Towards Therapeutic Targets for Type 2 Diabetes and Myocardial Infarction in the Background of Type 2 Diabetes” (PMBL), supervised by DA, LG, TR, DRC, SK, JKT, and MJB, and the NHGRI funded project "Analyses of the Allelic Spectrum of Cardiovascular Disease Genes in the Framingham Heart Study and Jackson Heart Study Cohorts” (FHS/JHS), led by CS and SG. For the PMBL project, NPB was project manager and LG, TT, VL, and FB were responsible for clinical investigation and sample management. For the FHS/JHS project, NG was project manager; HT, EF, JW, and CJO were responsible for clinical investigation and sample management; and JW, CO, CNC, SK, JNH, JGS, DA, and SG, and CS supervised the project. All DNA sequencing and data processing for these two projects was performed at the Broad Institute. JM, HI, SJ, AM, and PRN were responsible for all clinical investigation, sample management, sequencing, and data processing for the MODY study. JF, NLB, and DA designed and conceived the joint analysis. JF, NLB, JCF, JM, and DA provided methodological expertise. NLB, AGB, JF, and VA defined and interpreted the clinical information included. JF performed all analysis of the three population-based cohorts and annotation and comparative analysis of the MODY patients. JF, NLB, and DA wrote the manuscript. All authors reviewed, edited, and approved the manuscript.
References
- 1.Collins FS. Shattuck lecture--medical and societal consequences of the Human Genome Project. The New England Journal of Medicine. 1999;341:28–37. doi: 10.1056/NEJM199907013410106. [DOI] [PubMed] [Google Scholar]
- 2.Collins FS. Genetics: an explosion of knowledge is transforming clinical practice. Geriatrics. 1999;54:41–47. quiz 48. [PubMed] [Google Scholar]
- 3.Roses AD. Pharmacogenetics and the practice of medicine. Nature. 2000;405:857–865. doi: 10.1038/35015728. [DOI] [PubMed] [Google Scholar]
- 4.Hall Y. Coming Soon: Your Personal DNA Map? National Geographic News. 2006 [Google Scholar]
- 5.Duncan DE. On a Mission to Sequence the Genomes of 100,000 People. New York Times. 2010 [Google Scholar]
- 6.Brunham LR, Hayden MR. Medicine. Whole-genome sequencing: the new standard of care? Science. 2012;336:1112–1113. doi: 10.1126/science.1220967. [DOI] [PubMed] [Google Scholar]
- 7.Ball MP, et al. A public resource facilitating clinical use of genomes. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:11920–11927. doi: 10.1073/pnas.1201904109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Begg CB. On the use of familial aggregation in population-based case probands for calculating penetrance. J Natl Cancer Inst. 2002;94:1221–1226. doi: 10.1093/jnci/94.16.1221. [DOI] [PubMed] [Google Scholar]
- 9.Beutler E, Felitti VJ, Koziol JA, Ho NJ, Gelbart T. Penetrance of 845G-->A (C282Y) HFE hereditary haemochromatosis mutation in the USA. Lancet. 2002;359:211–218. doi: 10.1016/S0140-6736(02)07447-0. [DOI] [PubMed] [Google Scholar]
- 10.Goring HH, Terwilliger JD, Blangero J. Large upward bias in estimation of locus-specific effects from genomewide scans. American Journal of Human Genetics. 2001;69:1357–1369. doi: 10.1086/324471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guey LT, et al. Power in the phenotypic extremes: a simulation study of power in discovery and replication of rare variants. Genet Epidemiol. 2011 doi: 10.1002/gepi.20572. [DOI] [PubMed] [Google Scholar]
- 12.Terwilliger JD, Weiss KM. Confounding, ascertainment bias, and the blind quest for a genetic 'fountain of youth'. Annals of Medicine. 2003;35:532–544. doi: 10.1080/07853890310015181. [DOI] [PubMed] [Google Scholar]
- 13.Molven A, Njolstad PR. Role of molecular genetics in transforming diagnosis of diabetes mellitus. Expert review of molecular diagnostics. 2011;11:313–320. doi: 10.1586/erm.10.123. [DOI] [PubMed] [Google Scholar]
- 14.Kannel WB, Feinleib M, McNamara PM, Garrison RJ, Castelli WP. An investigation of coronary heart disease in families. The Framingham offspring study. American Journal of Epidemiology. 1979;110:281–290. doi: 10.1093/oxfordjournals.aje.a112813. [DOI] [PubMed] [Google Scholar]
- 15.Sempos CT, Bild DE, Manolio TA. Overview of the Jackson Heart Study: a study of cardiovascular diseases in African American men and women. The American Journal of the Medical Sciences. 1999;317:142–146. doi: 10.1097/00000441-199903000-00002. [DOI] [PubMed] [Google Scholar]
- 16.Begg CB, et al. Variation of breast cancer risk among BRCA1/2 carriers. JAMA : the journal of the American Medical Association. 2008;299:194–201. doi: 10.1001/jama.2007.55-a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kohane IS, Hsing M, Kong SW. Taxonomizing, sizing, and overcoming the incidentalome. Genetics in Medicine. 2012;14:399–404. doi: 10.1038/gim.2011.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tattersall RB. Mild familial diabetes with dominant inheritance. The Quarterly Journal of Medicine. 1974;43:339–357. [PubMed] [Google Scholar]
- 19.Tattersall RB, Fajans SS. A difference between the inheritance of classical juvenile-onset and maturity-onset type diabetes of young people. Diabetes. 1975;24:44–53. doi: 10.2337/diab.24.1.44. [DOI] [PubMed] [Google Scholar]
- 20.Murphy R, Ellard S, Hattersley AT. Clinical implications of a molecular genetic classification of monogenic beta-cell diabetes. Nat Clin Pract Endocrinol Metab. 2008;4:200–213. doi: 10.1038/ncpendmet0778. [DOI] [PubMed] [Google Scholar]
- 21.Eide SA, et al. Prevalence of HNF1A (MODY3) mutations in a Norwegian population (the HUNT2 Study) Diabetic Medicine. 2008;25:775–781. doi: 10.1111/j.1464-5491.2008.02459.x. [DOI] [PubMed] [Google Scholar]
- 22.Ledermann HM. Is maturity onset diabetes at young age (MODY) more common in Europe than previously assumed? Lancet. 1995;345:648. doi: 10.1016/s0140-6736(95)90548-0. [DOI] [PubMed] [Google Scholar]
- 23.Shields BM, et al. Maturity-onset diabetes of the young (MODY): how many cases are we missing? Diabetologia. 2010;53:2504–2508. doi: 10.1007/s00125-010-1799-4. [DOI] [PubMed] [Google Scholar]
- 24.Shepherd M, et al. No deterioration in glycemic control in HNF-1alpha maturity-onset diabetes of the young following transfer from long-term insulin to sulphonylureas. Diabetes Care. 2003;26:3191–3192. doi: 10.2337/diacare.26.11.3191-a. [DOI] [PubMed] [Google Scholar]
- 25.Shepherd M, Hattersley AT. 'I don't feel like a diabetic any more': the impact of stopping insulin in patients with maturity onset diabetes of the young following genetic testing. Clinical medicine. 2004;4:144–147. doi: 10.7861/clinmedicine.4-2-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shepherd M, et al. Predictive genetic testing in maturity-onset diabetes of the young (MODY) Diabetic Medicine. 2001;18:417–421. doi: 10.1046/j.1464-5491.2001.00447.x. [DOI] [PubMed] [Google Scholar]
- 27.McCarthy MI. Genomics, type 2 diabetes, and obesity. The New England Journal of Medicine. 2010;363:2339–2350. doi: 10.1056/NEJMra0906948. [DOI] [PubMed] [Google Scholar]
- 28.Knowler WC, et al. Reduction in the incidence of type 2 diabetes with lifestyle intervention or metformin. The New England journal of medicine. 2002;346:393–403. doi: 10.1056/NEJMoa012512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Knowler WC, et al. 10-year follow-up of diabetes incidence and weight loss in the Diabetes Prevention Program Outcomes Study. Lancet. 2009;374:1677–1686. doi: 10.1016/S0140-6736(09)61457-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yamagata K, et al. Mutations in the hepatocyte nuclear factor-1alpha gene in maturity-onset diabetes of the young (MODY3) Nature. 1996;384:455–458. doi: 10.1038/384455a0. [DOI] [PubMed] [Google Scholar]
- 31.Hattersley AT, et al. Linkage of type 2 diabetes to the glucokinase gene. Lancet. 1992;339:1307–1310. doi: 10.1016/0140-6736(92)91958-b. [DOI] [PubMed] [Google Scholar]
- 32.Froguel P, et al. Familial hyperglycemia due to mutations in glucokinase. Definition of a subtype of diabetes mellitus. The New England journal of medicine. 1993;328:697–702. doi: 10.1056/NEJM199303113281005. [DOI] [PubMed] [Google Scholar]
- 33.Yamagata K, et al. Mutations in the hepatocyte nuclear factor-4alpha gene in maturity-onset diabetes of the young (MODY1) Nature. 1996;384:458–460. doi: 10.1038/384458a0. [DOI] [PubMed] [Google Scholar]
- 34.Horikawa Y, et al. Mutation in hepatocyte nuclear factor-1 beta gene (TCF2) associated with MODY. Nature genetics. 1997;17:384–385. doi: 10.1038/ng1297-384. [DOI] [PubMed] [Google Scholar]
- 35.Stoffers DA, Ferrer J, Clarke WL, Habener JF. Early-onset type-II diabetes mellitus (MODY4) linked to IPF1. Nature genetics. 1997;17:138–139. doi: 10.1038/ng1097-138. [DOI] [PubMed] [Google Scholar]
- 36.Molven A, et al. Mutations in the insulin gene can cause MODY and autoantibody-negative type 1 diabetes. Diabetes. 2008;57:1131–1135. doi: 10.2337/db07-1467. [DOI] [PubMed] [Google Scholar]
- 37.Malecki MT, et al. Mutations in NEUROD1 are associated with the development of type 2 diabetes mellitus. Nature genetics. 1999;23:323–328. doi: 10.1038/15500. [DOI] [PubMed] [Google Scholar]
- 38.Flanagan SE, et al. Update of mutations in the genes encoding the pancreatic beta-cell K(ATP) channel subunits Kir6.2 (KCNJ11) and sulfonylurea receptor 1 (ABCC8) in diabetes mellitus and hyperinsulinism. Human mutation. 2009;30:170–180. doi: 10.1002/humu.20838. [DOI] [PubMed] [Google Scholar]
- 39.Plengvidhya N, et al. PAX4 mutations in Thais with maturity onset diabetes of the young. The Journal of clinical endocrinology and metabolism. 2007;92:2821–2826. doi: 10.1210/jc.2006-1927. [DOI] [PubMed] [Google Scholar]
- 40.Borowiec M, et al. Mutations at the BLK locus linked to maturity onset diabetes of the young and beta-cell dysfunction. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:14460–14465. doi: 10.1073/pnas.0906474106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Neve B, et al. Role of transcription factor KLF11 and its diabetes-associated gene variants in pancreatic beta cell function. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:4807–4812. doi: 10.1073/pnas.0409177102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Raeder H, et al. Mutations in the CEL VNTR cause a syndrome of diabetes and pancreatic exocrine dysfunction. Nature genetics. 2006;38:54–62. doi: 10.1038/ng1708. [DOI] [PubMed] [Google Scholar]
- 43.Gnirke A, et al. Solution hybrid selection with ultra-long oligonucleotides for massively parallel targeted sequencing. Nature biotechnology. 2009;27:182–189. doi: 10.1038/nbt.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dupuis J, et al. New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk. Nature Genetics. 2010;42:105–116. doi: 10.1038/ng.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Voight BF, et al. Twelve type 2 diabetes susceptibility loci identified through large-scale association analysis. Nature Genetics. 2010;42:579–589. doi: 10.1038/ng.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Abecasis GR, et al. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nature protocols. 2009;4:1073–1081. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- 48.Adzhubei IA, et al. A method and server for predicting damaging missense mutations. Nature methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stenson PD, et al. The Human Gene Mutation Database: 2008 update. Genome medicine. 2009;1:13. doi: 10.1186/gm13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xue Y, et al. Deleterious- and disease-allele prevalence in healthy individuals: insights from current predictions, mutation databases, and population-scale resequencing. American journal of human genetics. 2012;91:1022–1032. doi: 10.1016/j.ajhg.2012.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.MacArthur DG, et al. A systematic survey of loss-of-function variants in human protein-coding genes. Science. 2012;335:823–828. doi: 10.1126/science.1215040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Srinivasan BS, et al. A universal carrier test for the long tail of Mendelian disease. Reproductive biomedicine online. 2010;21:537–551. doi: 10.1016/j.rbmo.2010.05.012. [DOI] [PubMed] [Google Scholar]
- 53.Arthur C. Mapping the individual - cheaply. The Gaurdian. 2008 [Google Scholar]
- 54.Pinker S. My Genome, My Self. New York Times. 2009 [Google Scholar]
- 55.Rochman B. The DNA Dilemma: A Test That Could Change Your Life. TIME Magazine. 2012 [PubMed] [Google Scholar]
- 56.Lango Allen H, et al. GATA6 haploinsufficiency causes pancreatic agenesis in humans. Nature genetics. 2012;44:20–22. doi: 10.1038/ng.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Johansson S, et al. Exome sequencing and genetic testing for MODY. PloS one. 2012;7:e38050. doi: 10.1371/journal.pone.0038050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Eide SA, et al. Prevalence of HNF1A (MODY3) mutations in a Norwegian population (the HUNT2 Study) Diabetic medicine : a journal of the British Diabetic Association. 2008;25:775–781. doi: 10.1111/j.1464-5491.2008.02459.x. [DOI] [PubMed] [Google Scholar]
- 59.Ellard S, Bellanne-Chantelot C, Hattersley AT. Best practice guidelines for the molecular genetic diagnosis of maturity-onset diabetes of the young. Diabetologia. 2008;51:546–553. doi: 10.1007/s00125-008-0942-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.World Health Organisation, editor. Geneva, Switzerland: World Health Organisation Press; 2006. Definition and diagnosis of diabetes mellitus and intermediate hyperglycaemia: report of a WHO/IDF consultation. [Google Scholar]
- 61.Edghill EL, et al. Sequencing PDX1 (insulin promoter factor 1) in 1788 UK individuals found 5% had a low frequency coding variant, but these variants are not associated with Type 2 diabetes. Diabetic Medicine. 2011;28:681–684. doi: 10.1111/j.1464-5491.2011.03269.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gill-Carey O, Shields B, Colclough K, Ellard S, Hattersley A. Finding a glucokinase mutation alters patient treatment. Diabetic Medicine. 2007;24(Suppl. 1) [Google Scholar]
- 63.Pearson ER, et al. beta-cell genes and diabetes: quantitative and qualitative differences in the pathophysiology of hepatic nuclear factor-1alpha and glucokinase mutations. Diabetes. 2001;50(Suppl 1):S101–S107. doi: 10.2337/diabetes.50.2007.s101. [DOI] [PubMed] [Google Scholar]
- 64.Stride A, et al. The genetic abnormality in the beta cell determines the response to an oral glucose load. Diabetologia. 2002;45:427–435. doi: 10.1007/s00125-001-0770-9. [DOI] [PubMed] [Google Scholar]
- 65.Bergmann A, et al. The A98V single nucleotide polymorphism (SNP) in hepatic nuclear factor 1 alpha (HNF-1alpha) is associated with insulin sensitivity and beta-cell function. Experimental and Clinical Endocrinology & Diabetes. 2008;116(Suppl 1):S50–S55. doi: 10.1055/s-2008-1081492. [DOI] [PubMed] [Google Scholar]
- 66.Iwasaki N, et al. Liver and kidney function in Japanese patients with maturity-onset diabetes of the young. Diabetes Care. 1998;21:2144–2148. doi: 10.2337/diacare.21.12.2144. [DOI] [PubMed] [Google Scholar]
- 67.McLaren W, et al. Deriving the consequences of genomic variants with the Ensembl API and SNP Effect Predictor. Bioinformatics. 2010;26:2069–2070. doi: 10.1093/bioinformatics/btq330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bach I, et al. Cloning of human hepatic nuclear factor 1 (HNF1) and chromosomal localization of its gene in man and mouse. Genomics. 1990;8:155–164. doi: 10.1016/0888-7543(90)90238-p. [DOI] [PubMed] [Google Scholar]
- 69.Chartier FL, Bossu JP, Laudet V, Fruchart JC, Laine B. Cloning and sequencing of cDNAs encoding the human hepatocyte nuclear factor 4 indicate the presence of two isoforms in human liver. Gene. 1994;147:269–272. doi: 10.1016/0378-1119(94)90079-5. [DOI] [PubMed] [Google Scholar]
- 70.Abbott C, et al. Mapping of the gene TCF2 coding for the transcription factor LFB3 to human chromosome 17 by polymerase chain reaction. Genomics. 1990;8:165–167. doi: 10.1016/0888-7543(90)90239-q. [DOI] [PubMed] [Google Scholar]
- 71.Leonard J, et al. Characterization of somatostatin transactivating factor-1, a novel homeobox factor that stimulates somatostatin expression in pancreatic islet cells. Mol Endocrinol. 1993;7:1275–1283. doi: 10.1210/mend.7.10.7505393. [DOI] [PubMed] [Google Scholar]
- 72.Sanger F. Chemistry of insulin; determination of the structure of insulin opens the way to greater understanding of life processes. Science. 1959;129:1340–1344. doi: 10.1126/science.129.3359.1340. [DOI] [PubMed] [Google Scholar]
- 73.Tamimi R, et al. The NEUROD gene maps to human chromosome 2q32 and mouse chromosome 2. Genomics. 1996;34:418–421. doi: 10.1006/geno.1996.0306. [DOI] [PubMed] [Google Scholar]
- 74.Dawber TR, Meadors GF, Moore FE., Jr Epidemiological approaches to heart disease: the Framingham Study. American journal of public health and the nation's health. 1951;41:279–281. doi: 10.2105/ajph.41.3.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Berglund G, et al. Long-term outcome of the Malmo preventive project: mortality and cardiovascular morbidity. Journal of internal medicine. 2000;247:19–29. doi: 10.1046/j.1365-2796.2000.00568.x. [DOI] [PubMed] [Google Scholar]
- 76.Lindholm E, Agardh E, Tuomi T, Groop L, Agardh CD. Classifying diabetes according to the new WHO clinical stages. European journal of epidemiology. 2001;17:983–989. doi: 10.1023/a:1020036805655. [DOI] [PubMed] [Google Scholar]
- 77.Groop L, et al. Metabolic consequences of a family history of NIDDM (the Botnia study): evidence for sex-specific parental effects. Diabetes. 1996;45:1585–1593. doi: 10.2337/diab.45.11.1585. [DOI] [PubMed] [Google Scholar]
- 78.Byrne MM, et al. Altered insulin secretory responses to glucose in diabetic and nondiabetic subjects with mutations in the diabetes susceptibility gene MODY3 on chromosome 12. Diabetes. 1996;45:1503–1510. doi: 10.2337/diab.45.11.1503. [DOI] [PubMed] [Google Scholar]
- 79.Bick AG, et al. Burden of rare sarcomere gene variants in the Framingham and Jackson Heart Study cohorts. American journal of human genetics. 2012;91:513–519. doi: 10.1016/j.ajhg.2012.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.DePristo MA, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nature Genetics. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Purcell S, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. American journal of human genetics. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Price AL, et al. Principal components analysis corrects for stratification in genome-wide association studies. Nature genetics. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 85.Meyer LR, et al. The UCSC Genome Browser database: extensions and updates 2013. Nucleic acids research. 2013;41:D64–D69. doi: 10.1093/nar/gks1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lindner TH, et al. A novel syndrome of diabetes mellitus, renal dysfunction and genital malformation associated with a partial deletion of the pseudo-POU domain of hepatocyte nuclear factor-1beta. Human molecular genetics. 1999;8:2001–2008. doi: 10.1093/hmg/8.11.2001. [DOI] [PubMed] [Google Scholar]
- 87.Njolstad PR, et al. Permanent neonatal diabetes caused by glucokinase deficiency: inborn error of the glucose-insulin signaling pathway. Diabetes. 2003;52:2854–2860. doi: 10.2337/diabetes.52.11.2854. [DOI] [PubMed] [Google Scholar]
- 88.Bjorkhaug L, et al. Hepatocyte nuclear factor-1 alpha gene mutations and diabetes in Norway. The Journal of clinical endocrinology and metabolism. 2003;88:920–931. doi: 10.1210/jc.2002-020945. [DOI] [PubMed] [Google Scholar]
- 89.Sagen JV, et al. Diagnostic screening of NEUROD1 (MODY6) in subjects with MODY or gestational diabetes mellitus. Diabetic medicine : a journal of the British Diabetic Association. 2005;22:1012–1015. doi: 10.1111/j.1464-5491.2005.01565.x. [DOI] [PubMed] [Google Scholar]
- 90.Raeder H, et al. A hepatocyte nuclear factor-4 alpha gene (HNF4A) P2 promoter haplotype linked with late-onset diabetes: studies of HNF4A variants in the Norwegian MODY registry. Diabetes. 2006;55:1899–1903. doi: 10.2337/db05-1677. [DOI] [PubMed] [Google Scholar]
- 91.Sagen JV, et al. From clinicogenetic studies of maturity-onset diabetes of the young to unraveling complex mechanisms of glucokinase regulation. Diabetes. 2006;55:1713–1722. doi: 10.2337/db05-1513. [DOI] [PubMed] [Google Scholar]
- 92.Haldorsen IS, et al. Lack of pancreatic body and tail in HNF1B mutation carriers. Diabetic medicine : a journal of the British Diabetic Association. 2008;25:782–787. doi: 10.1111/j.1464-5491.2008.02460.x. [DOI] [PubMed] [Google Scholar]
- 93.Sagen JV, et al. Diagnostic screening of MODY2/GCK mutations in the Norwegian MODY Registry. Pediatric diabetes. 2008;9:442–449. doi: 10.1111/j.1399-5448.2008.00399.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.