Abstract
The accumulation of RCY1 protein, which is encoded by RESISTANCE TO CMV(Y) (RCY1), a CC-NB-LRR class R-gene, is tightly correlated with the strength of the resistance to a yellow strain of Cucumber mosaic virus [CMV(Y)] in Arabidopsis thaliana. In order to enhance resistance to CMV by overexpression of RCY1, A. thaliana was transformed with intron-less RCY1 cDNA construct under the control of strong CaMV35S promoter. Remarkably, a relative amount of RCY1 protein accumulation in the transformants was much lower than that in plants expressing genomic RCY1 under the control of its native promoter. To identify a regulatory element of RCY1 that could cause such differential levels of RCY1 accumulation, a series of RCY1 cDNA and genomic RCY1 constructs were transiently expressed in Nicotiana benthamiana leaves by the Agrobacterium-mediated infiltration method. Comparative analysis of the level of RCY1 accumulation in the leaf tissues transiently expressing each construct indicated that the intron located in the RCY1-coding region of genomic RCY1, but not the native RCY1 genomic promoter or the 5′-and 3′-untranslated regions of RCY1, was indispensable for high level RCY1 accumulation. The increased levels of RCY1 accelerated plant disease defense reactions. Interestingly, such intron-mediated enhancement of RCY1 accumulation depended neither on the abundance of the RCY1 transcript nor on the RCY1 specific-intron sequence. Taken together, intron-mediated RCY1 expression seems to play a key role in the expression of complete resistance to CMV(Y) by maintaining RCY1 accumulation at high levels.
Introduction
Over the past 20 years, more than 80 resistance (R) genes that confer resistance to plant pathogens via a defense reaction have been cloned [1]–[3]. Common features of these R proteins include the conserved nucleotide-binding (NB) and leucine-rich-repeat (LRR) domains that they contain [4], [5]. The NB-LRR domains-containing R proteins are further separated into two subclasses depending on whether they have a coiled-coil (CC) domain or a toll-interleukin-1 receptor (TIR)-like region at their amino terminus [6]. To date, 149 NB-LRR-encoding genes (including 55 CC-NB-LRR-encoding and 94 TIR-NB-LRR-encoding genes) and another 58 related genes that do not encode an LRR domain have been identified in the genome sequence of Arabidopsis thaliana ecotype Columbia (Col-0) [6]. Nearly half of the NB-LRR-encoding genes are scattered throughout the genome as simple loci, whereas the remaining NB-LRR-encoding genes are located in gene family clusters that might have become duplicated and expanded due to unequal crossing-over between mispaired tandem copies during meiosis [6]–[8].
Although an increased number of NB-LRR-encoding genes and related genes in Arabidopsis genome were classified based on protein motifs, intron positions and sequence conservation, our understanding of the regulation of their gene expression is still limited. Global expression analysis of NB-LRR-encoding and related genes in Arabidopsis suggests that most of their transcripts are present at low levels [9]. The expression of a limited number of these genes can be induced by the bacterial flagellin peptide, flg22 or by salicylic acid (SA) treatment [9]. However, significant induction of known R gene expression during the plant defense response seems to occur only in a minority of the cases examined so far [9]–[15]. The transcript levels of other R genes do not change in response to pathogen attack [16]–[19]. Furthermore, global expression analysis of NB-LRR-encoding and related genes in Arabidopsis suggests that most NB-LRR-encoding genes tend to be more responsive to pathogens than randomly selected genes, but less responsive to pathogens than common defense-related marker genes [9], [15]. Therefore, in general, R genes may not generally need to be induced at the transcriptional level to regulate resistance to pathogens.
However, for some NB-LRR-encoding genes in Arabidopsis, there is evidence of significant correlation between the level of NB-LRR protein accumulation before pathogen infection and enhanced resistance to pathogens [20], [21]. Along similar lines, overexpression of R genes often triggers autoactivation of the defense reaction and spontaneous cell death independent of pathogen infection [22]–[24]. Thus, appropriate control of R gene expression seems to be important for a fully functional defense system.
Further, alternative splicing has been reported for a certain number of known NB-LRR-encoding R genes: e.g., N, L6, RPP5, RPS4, M, RAC1, Bs4, and Mla6 [17], [25]–[32]. Alternative splicing is known to be a requirement for the function of both of the plant R protein N and RPS4, govern the resistance to Tobacco mosaic virus (TMV) and to Pseudomonas syringae pv tomato carrying AvrRPS4, respectively [29], [33]. For a certain number of NB-LRR-encoding genes, alternative splicing may be indispensable for complete resistance to pathogens, although the mechanism and role of alternative splicing in NB-LRR-encoding gene-mediated disease resistance remains to be understood [34]. Overall, the regulatory system for controlling R gene expression requires much further analysis.
We have previously demonstrated that RESISTANCE TO CMV(Y) (RCY1), which encodes a CC-NB-LRR class resistance protein in A. thaliana, confers the resistance to a yellow strain of Cucumber mosaic virus [CMV(Y)] in an RCY1 protein accumulation-dependent manner [21], [24], [35]–[36]. RCY1-mediated resistance to CMV(Y) is accompanied by the development of necrotic local lesions at the primary infection sites, elevated expression of defense-related genes, such as Pathogenesis-Related 1a (PR-1a), and accumulation of SA [37], [38]. Interestingly, increased expression of RCY1 converts the hypersensitive resistance response (HR) to an extreme resistance response (ER) to CMV(Y) [21], suggesting that the level of RCY1 protein regulates the strength of resistance to CMV(Y) in Arabidopsis. Therefore, analysis of the mechanisms of regulation of RCY1 gene expression should provide new approaches to modulate NB-LRR-class R-protein-mediated defense reaction against pathogen attack. In this study, we identified a genomic element of RCY1 that enhances RCY1-conferred resistance to CMV(Y) by elevating level of RCY1 accumulation in A. thaliana and N. benthamiana.
Results
Detection of RCY1 transcript in CMV(Y)-inoculated leaves of Arabidopsis thaliana
RCY1 transcript was detected in the resistant ecotype C24 carrying RCY1, but not in the control susceptible ecotype Columbia-0 (Col-0) without RCY1, by northern hybridization with a DIG-labeled RCY1 DNA probe (Figure 1). The amount of RCY1 transcript did not significantly increase during the progress of the HR in CMV(Y)-inoculated C24 leaves (Figure 1). RCY1 transgene transcripts could also be detected in Col-0 transformed with genomic RCY1 tagged with hemagglutinin (HA)-epitope sequence at its 3′-end (Col::pRCY1-HA#12) [21]. However, the level of RCY1 transgene expression did not change in CMV(Y)-inoculated leaves of Col::pRCY1-HA#12 as much as in CMV(Y)-inoculated C24 (Figure 1), in contrast, the expression of PR-1a a typical marker gene for the HR induction, was clearly up-regulated in CMV(Y)-inoculated leaves of both C24 and Col::pRCY1-HA#12 (Figure 1), suggesting that the expression of neither endogenous nor transformed RCY1 is inducible in response to CMV(Y) infection. Therefore, the induction of RCY1 expression alone seems to be insufficient to activate the plant defense system against CMV(Y) in A. thaliana.
Figure 1. Detection of RCY1 and Pathogenesis-Related 1a transcripts in Arabidopsis thaliana ecotypes and RCY1-transformants.
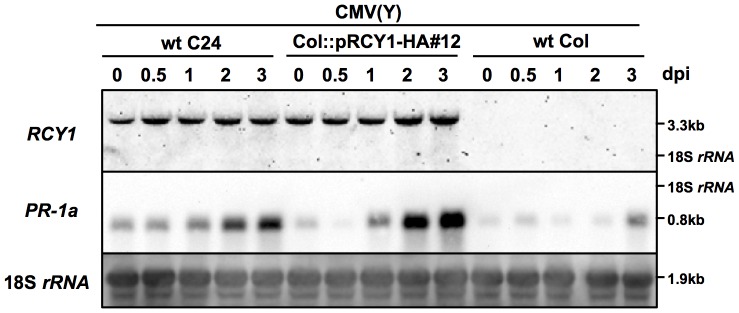
RESISTANCE TO CMV(Y) (RCY1) and Pathogenesis-Related 1a (PR-1a) transcripts in CMV(Y)-inoculated leaves of wild-type A. thaliana ecotypes C24 (wt C24) and Col-0 (wt Col) and HA epitope-tagged RCY1-transformed Col-0 (Col::pRCY1-HA#12) were detected by northern hybridization at 0, 0.5, 1, 2, and 3 days post-inoculation (dpi). As an internal control for RNA sample loading, 18S rRNA is shown. The size of each band and the position of 18S rRNA were shown at right side of the panel. For all experiments, three independent plants per line were used and a photograph of a representative plant is shown.
Expression of genomic RCY1 and CaMV35S promoter-driven RCY1 cDNA transgenes in A. thaliana
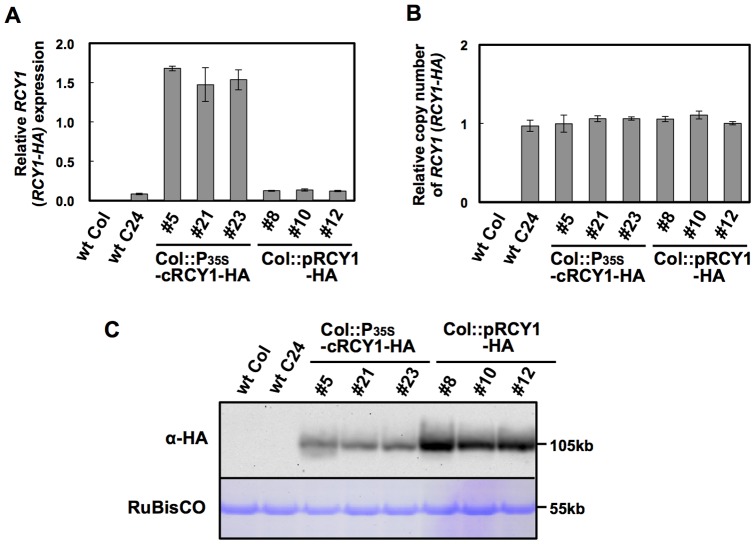
We previously found endogenous levels of RCY1 protein accumulation to be correlated with the degree of the resistance to CMV(Y) in A. thaliana [21]. Thus, to elevate the basal level of RCY1 transcript and thereby enhance the resistance to CMV(Y) in A. thaliana, we expressed the P35S-cRCY1-HA construct under the control of 35S promoter of Cauliflower mosaic virus (CaMV) in three independent transgenic lines (Col::P35S-cRCY1-HA#5, #21 and #23). The P35S-cRCY1-HA construct consisted of an intron-less RCY1 cDNA tagged with an HA-epitope sequence at its 3′-end, and included a 66-bp sequence 5′ upstream of the start codon of RCY1 (66-bp-5′-UTR) and a 71-bp sequence 3′-downstream from the stop codon of RCY1 (71-bp-3′-UTR) (Figure 2). As a control, three independent lines of Col::pRCY1-HA, #8, #10 and #12, each carrying the intron-containing genomic RCY1 transgene tagged with an HA-epitope sequence at its 3′-end [21] were grown under the same growth conditions. We assumed that RCY1 would be expressed at much higher levels in these Col::P35S-cRCY1-HA lines than in the Col::pRCY1-HA lines, as the activity of the CaMV35S promoter is generally stronger than that of the native genomic RCY1 promoter. As expected, the level of RCY1 transcript in the three independent Col::P35S-cRCY1-HA lines was much higher than that in three independent Col::pRCY1-HA lines (Figure 3A). Normalization of PCR-based quantification of genomic RCY1 or RCY1 cDNA transgene with ubiquitin 5 gene (UBQ5) suggested that each transgenic line carried the same or similar transgene copy numbers (Figure 3B). Therefore, the increased RCY1 transcript levels in Col::P35S-cRCY1-HA were due not to differential transgene copy numbers in each transgenic lines, but to the strength of the CaMV35S promoter. Next, when the HA-epitope-tagged RCY1 protein was immunologically detected in both transgenic lines using an anti-HA monoclonal antibody, remarkably, the abundance of RCY1 protein in three Col::pRCY1-HA lines was much higher than that in three Col::P35S-cRCY1-HA lines (Figure 3C). This inverse relationship between RCY1 transcript level and RCY1 protein accumulation between genomic RCY1-transformed Col::pRCY1-HA lines and RCY1 cDNA-transformed Col::P35S-cRCY1-HA lines could be due to distinctions between the regulatory elements such as intron sequence controlling genomic RCY1 expression versus RCY1 cDNA expression.
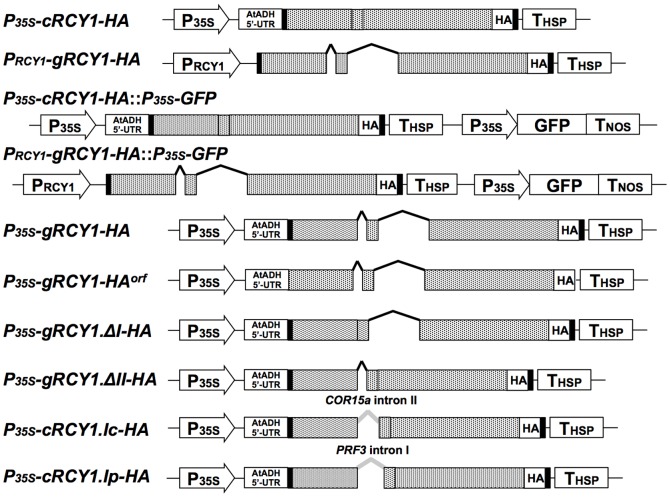
Figure 2. Schematic structure of vector constructs encoding RCY1.
P35S-cRCY1-HA contains the RCY1 cDNA with no introns but including an HA-epitope tag (HA) at its 3′-end, the 66-bp 5′ sequence upstream of the RCY1 start codon of (66-bp 5′-UTR), and the 71-bp 3′-downstream sequence from the RCY1 stop codon (71-bp 3′-UTR). All elements were cloned between the CaMV 35S promoter (P35S) and the 5′-UTR sequence of the Arabidopsis Alcohol Dehydrogenase gene (ADH5′-UTR) and terminator sequence of the Heat Shock Protein gene (THSP) in the pRI201-AN binary vector. PRCY1-gRCY1-HA, contains the 1.5 kb genomic RCY1 promoter (PRCY1), the genomic RCY1-coding regions including two introns and tagged with HA at its 3′-end, and the RCY1 66-bp 5′-UTR region and the 71-bp 3′-UTR cloned into the pRI201-AN binary vector. P35S-gRCY1-HA::P35S-GFP and PRCY1-gRCY1-HA::P35S-GFP include P35S, the GFP-coding region, and the nos terminator (TNOS) cloned into the vector pRI201-AN. P35S-gRCY1-HA, includes the genomic RCY1-coding region with two introns tagged with HA at its 3′-end, and the RCY1 66-bp 5′-UTR and 71-bp 3′-UTR, cloned downstream of P35S between ADH5′-UTR and THSP in pRI201-AN. P35S-gRCY1-HAorf was constructed by deletion of 66-bp-5′-UTR and 71-bp-3′-UTR from P35S-gRCY1-HA. P35S-gRCY1.ΔI-HA and P35S-gRCY1.ΔII-HA were constructed by deletion of the first or second intron from P35S-gRCY1-HA, respectively. P35S-cRCY1.Ic-HA was constructed by insertion of the COLD-REGULATED 15A (COR15a) intron II into the first splice junction site in P35S-cRCY1-HA. P35S-cRCY1.Ip-HA was created by insertion of the PROFILIN 3 (PRF3) intron I into the first splice junction site in P35S-cRCY1-HA. In all vector constructs, the 66-bp 5′-UTR and 71-bp 3′-UTR of RCY1 are shown as black boxes. The RCY1-coding region is indicated by gray boxes in which the two splice junction sites are indicated by vertical lines in the boxes. The two RCY1 introns are shown as black dashed lines and the COR15a or PRF3 introns are shown as gray dashed lines.
Figure 3. Detection of RCY1 protein, the RCY1 transcript, and the RCY1 transgene in three independent Col-0 lines transformed with HA-tagged genomic RCY1 or HA-tagged RCY1 cDNA without introns.
Relative amounts of RCY1 transcripts in wild-type A. thaliana ecotypes Col-0 (wt Col) and C24 (wt C24), and three independent lines transformed with HA-tagged genomic RCY1 (Col::pRCY1-HA #8, #10, and #12) or HA-tagged RCY1 cDNA without introns (Col::cRCY1-HA #5, #21 and #23), were measured by quantitative RT-PCR (A). Relative amounts of RCY1-coding transgene DNA in wild-type ecotypes (wt Col and wt C24) and three independent lines of Col::gRCY1-HA and Col::cRCY1-HA were measured by quantitative PCR using each genomic DNA as a template (B). HA-epitope-tagged RCY1 protein (α-HA) in wild-type ecotypes (wt Col and wt C24); Col::gRCY1-HA #8, #10 and #12 lines; and Col::cRCY1-HA #5, #21 and #23 lines was immunologically detected using monoclonal anti-HA epitope antibody. As an internal control for protein sample quantities, the large subunit of RuBisCO was visualized by staining by Coomassie Brilliant Blue R-250 (CBB) (C). For all experiments, three independent plants per vector were analyzed. The averages of relative RCY1 transcript amounts ±SE are shown in A and B. In C, a representative photograph is shown. The size of each band was shown at right side of the panel.
Transient expression of genomic RCY1 and CaMV35S promoter-driven RCY1 cDNA transgenes in Nicotiana benthamiana by Agrobacterium-mediated infiltration method
To further confirm increased RCY1 accumulation upon expression of genomic RCY1, genomic RCY1 tagged with an HA-epitope sequence at its 3′-end was cloned into the HindIII-SalI sites of pRI201-AN and designated PRCY1-gRCY1-HA (Figure 2). In PRCY1-gRCY1-HA, the genomic RCY1 contains its coding region with two introns, ∼1.5 kbp promoter region and 71-bp 3′-UTR. As an internal control, a DNA fragment containing the Green Fluorescence Protein (GFP)-coding region fused between the CaMV35S promoter (P35S) and the nopaline synthase (NOS) terminator (TNOS), was inserted downstream of PRCY1-gRCY1-HA and P35S-cRCY1-HA, respectively. When these two vector constructs, P35S-cRCY1-HA::P35S-GFP and PRCY1-gRCY1-HA::P35S-GFP (Figure 2), were transiently expressed in N. benthamiana leaves by Agrobacterium-mediated infiltration (agro-infiltration) method, the level of RCY1 accumulated in leaf tissues expressing PRCY1-gRCY1-HA::P35S-GFP was reproducibly much higher than those expressing P35S-cRCY1-HA::P35S-GFP at 48 and 72 hr after agro-infiltration. In contrast, there was no significant difference in the level of GFP accumulated in leaf tissues expressing each type of constructs (Figure 4). This results indicates that greater RCY1 accumulation in N. benthamiana leaves transiently expressing PRCY1-gRCY1-HA::P35S-GFP is not caused by differential efficiency of T-DNA transfer into the cells of the agro-infiltrated tissues, but by differences in the regulatory elements controlling genomic RCY1 versus CaMV35S promoter-driven RCY1 cDNA expression in N. benthamiana, just as in transgenic Arabidopsis.
Figure 4. Detection of HA-epitope-tagged RCY1 in N. benthamiana leaves transiently expressing genomic RCY1-HA under control of the native RCY1 promoter or the RCY1 cDNA without introns under control of the CaMV 35S promoter.
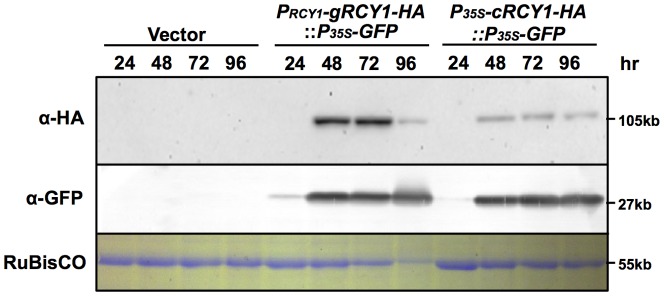
HA-epitope-tagged RCY1 protein (α-HA) in N. benthamiana leaf tissues transiently expressing PRCY1-gRCY1-HA::P35S-GFP or P35S-gRCY1-HA::P35S-GFP was immunologically detected using monoclonal antibody against the HA epitope at 24, 48, 72, and 96 h after agro-infiltration. GFP accumulation (α-GFP) was also immunologically detected at the same time points using polyclonal antibody against GFP as an internal standard. As an internal control for protein sample quantities, the large subunit of RuBisCO was visualized by staining with CBB. The size of each band was shown at right side of the panel. For all experiments, three independent plants per vector were analyzed and representative data are shown.
Comparison of RCY1 protein and RCY1 transcript accumulation in N. benthamiana transiently expressing a series of native genomic RCY1 and CaMV35S promoter-driven RCY1 cDNA constructs
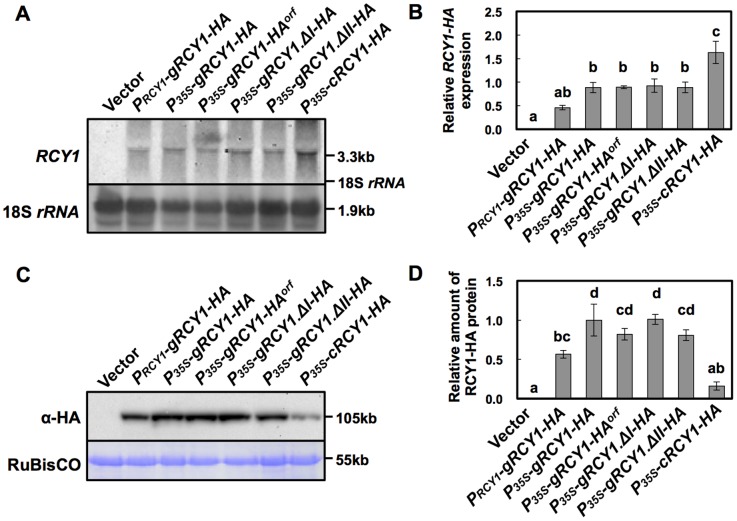
To compare the activities of the native RCY1 genomic and CaMV35S promoters for expressing the RCY1-coding region, PRCY1-gRCY1-HA and the construct P35S-gRCY1-HA, consisting of an HA-tagged genomic RCY1-coding region under the control of CaMV35S promoter (Figure 2) and including the 66-bp 5′-UTR and 71-bp 3′-UTR were each transiently expressed in N. benthamiana. As shown in Figure 5, the level of both RCY1 transcript and RCY1 protein accumulation in leaf tissues expressing P35S-gRCY1-HA were significantly higher than those of PRCY1-gRCY1-HA, suggesting that the activity of the CaMV35S promoter is much stronger than that of the genomic RCY1 promoter, just as we predicted. Therefore, increased RCY1 accumulation in leaf tissues of A. thaliana transformed with the genomic RCY1 construct (Figure 3) or in N. benthamiana transiently expressing genomic RCY1 (Figure 4) is not simply caused by the higher activity of the genomic RCY1 promoter than the CaMV35S promoter.
Figure 5. Detection of HA-epitope-tagged RCY1 protein and RCY1 transcript in N. benthamiana leaves transiently expressing a series of RCY1-HA constructs under control of the RCY1 or CaMV 35S promoters.
RCY1 transcripts in N. benthamiana leaves agro-infiltrated with PRCY1-gRCY1-HA, P35S-gRCY1-HA, P35S-gRCY1-HAorf, P35S-gRCY1.ΔI-HA, P35S-gRCY1.ΔII-HA, or P35S-cRCY1-HA were detected by northern hybridization. pRI201-AN (Vector) was used as an empty-vector control for agro-infiltration. As an internal control for RNA sample quantities, 18S rRNA is shown (A). Relative amounts of RCY1 transcripts in each line were measured by quantitative RT-PCR. EFα gene expression was used as a standard for normalization of RCY1 expression (B). HA-epitope-tagged RCY1 protein (α-HA) in N. benthamiana leaves transiently expressing PRCY1-gRCY1-HA, P35S-gRCY1-HA, P35S-gRCY1-HAorf, P35S-gRCY1.ΔI-HA, P35S-gRCY1.ΔII-HA, or P35S-cRCY1-HA was immunologically detected using anti-HA monoclonal antibody. As an internal control for protein sample quantities, the large subunit of RuBisCO was visualized by staining with CBB (C). RCY1-HA protein amounts in each line were quantified by band intensity using Quantity One software. For all experiments, four independent plants transiently expressing each vector construct were analyzed (D). The averages of relative RCY1 transcript amounts ±SE are shown in B and D. In A and C, representative photographs are shown. The size of each band and the position of 18S rRNA were shown at right side of the panels. Data were subjected to analysis of variance and treatment means were compared by Tukey's test. Different letters indicate a statistically significant difference in the relative amount of RCY1 transcript (n = 4, P<0.05).
Next, to analyze the roles of the 66-bp 5′-UTR and the 71-bp 3′-UTR of RCY1 in P35S-gRCY1-HA in increased RCY1 accumulation, P35S-gRCY1-HAorf was constructed by deletion of the 5′- and 3′-UTRs from P35S-gRCY1-HA (Figure 2). When P35S-gRCY1-HA and P35S-gRCY1-HAorf were each agro-infiltrated into different regions of a single N. benthamiana leaf, RCY1 transcript and RCY1 protein accumulated at similar levels between leaf tissues transiently expressing P35S-gRCY1-HA or P35S-gRCY1-HAorf (Figure 5). This result suggests that the 5′- and 3′-UTR regions of RCY1 might not be associated with difference in RCY1 accumulation in N. benthamiana leaf tissues expressing genomic RCY1.
However, the accumulation of RCY1 in leaf tissues transiently expressing P35S-gRCY1-HA was much higher than that in leaf tissue expressing P35S-cRCY1-HA (Figure 5C and D). But the level of RCY1 transcript in leaf tissues expressing P35S-gRCY1-HA was significantly lower than that in leaf tissues transiently expressing P35S-cRCY1-HA (Figure 5A and B). The vector P35S-gRCY1-HA contains the genomic intron-containing RCY1 with all of its introns, whereas the vector P35S-cRCY1-HA contains the RCY1 cDNA, which has no introns, both under the control of CaMV35S promoter (Figure 2). Together, these facts suggest that the existence of introns in RCY1-coding region could be indispensable for elevated level of RCY1 accumulation accompanied by decreased RCY1 transcript level.
To further analyze the role of two introns in the RCY1-coding region for this intron-mediated enhancement (IME) of RCY1 accumulation, constructs with RCY1 including either first or second intron in its coding region, P35S-gRCY1.ΔI-HA and P35S-gRCY1.ΔII-HA (Figure 2), were each transiently expressed in N. benthamiana leaves under control of the CaMV35S promoter. The level of RCY1 accumulation in each leaf tissue agro-infiltrated with P35S-gRCY1.ΔI-HA or P35S-gRCY1.ΔII-HA was similar to that in tissues expressing P35S-gRCY1-HA but was significantly higher than that in tissues expressing P35S-cRCY1-HA (Figure 5C and D). However, the transcript abundance of RCY1 in leaf tissue agro-infiltrated with P35S-gRCY1.ΔI-HA or P35S-gRCY1.ΔII-HA was the same as that of tissues expressing P35S-gRCY1-HA, but much lower than that of P35S-cRCY1-HA transcript (Figure 5 A and B). Thus, both introns of genomic RCY1 seem to be equally important for IME of RCY1 accumulation.
As shown in Figure 5, the level of full-length RCY1 mRNA was inversely related to the accumulation of RCY1 in the leaf tissues transiently expressing either the intron-containing genomic RCY1 or those expressing the intron-less RCY1 cDNA. For some other R genes, alternative transcripts are required to induce complete resistance against pathogens [34]. However, analysis of RCY1 transcript by northern hybridization using DIG-labeled cDNA probes corresponding to the sequence coding the LRR domain of RCY1 suggests that no alternative RCY1 transcript but the full-length RCY1 mRNA was observed in N. benthamiana leaf tissues transiently expressing either P35S-gRCY1-HA or P35S-cRCY1-HA (Figure S1).
Acceleration of the defense reaction in N. benthamiana transiently expressing genomic RCY1
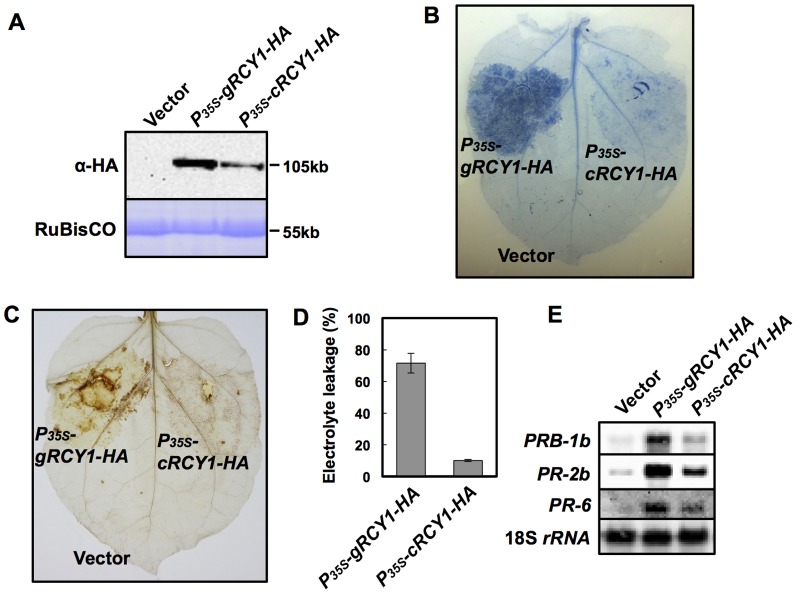
In N. benthamiana leaf tissues transiently accumulating RCY1, HR-like cell death is spontaneously induced [24]. Pathogen-independent autoactivation of defense reaction triggered by overexpression of some other R-gene has been reported [22]–[24]. HR-like cell death developed in N. benthamiana leaf tissues accumulating RCY1 can be considered as autoactivation of defense reaction [24]. When different regions of one fully expanded N. benthamiana leaf were infiltrated with P35S-gRCY1-HA or P35S-cRCY1-HA, leaf tissues expressing P35S-gRCY1-HA, which accumulated increased levels of RCY1-HA exhibited a greater magnitude of HR-like cell death than did leaf tissues transiently expressing P35S-cRCY1-HA, which accumulated relatively lower amounts of RCY1-HA (Figure 6A and B). The levels of electrolyte leakage and H2O2 production also increased in leaf tissues transiently expressing P35S-gRCY1-HA in comparison with P35S-cRCY1-HA (Figure 6C and D). Furthermore, the expression levels of several defense-related genes including Pathogenesis-Related 1b (PRB-1b), PR-2b and PR-6 were much more induced in leaf tissues expressing P35S-gRCY1-HA than in those expressing P35S-cRCY1-HA (Figure 6E). Thus, intron-mediated enhancement (IME) of RCY1 accumulation seems to result in the acceleration of the defense reaction.
Figure 6. Activation of defense reaction in N. benthamiana leaves transiently expressing P35S-gRCY1-HA and P35S-cRCY1-HA under control of the CaMV 35S promoter.
HA-epitope-tagged RCY1 protein (α-HA) (A) in N. benthamiana leaves transiently expressing P35S-gRCY1-HA, P35S-cRCY1-HA, or pRI201-AN (Vector) as an empty-vector control was immunologically detected using anti-HA monoclonal antibody. As an internal control for protein sample quantities, the large subunit of RuBisCO was visualized by staining with CBB. The size of each band was shown at right side of the panel. In N. benthamiana leaves transiently expressing P35S-gRCY1-HA or P35S-cRCY1-HA, hypersensitive response (HR) cell death was visualized by trypan blue staining (B), and H2O2 production was detected by DAB staining (C). To evaluate HR-cell death quantitatively, electrolyte leakage (D) in N. benthamiana leaves transiently expressing P35S-gRCY1-HA, P35S-cRCY1-HA, or empty-vector control was measured. Expression of the defense-related genes PRB-1b, PR-2b, and PR-6 in N. benthamiana leaf tissue transiently expressing P35S-gRCY1-HA, P35S-cRCY1-HA, or the empty-vector control was analyzed by northern hybridization (E). As an internal control for RNA sample quantities, 18S rRNA was shown.
Ability of each intron to increase RCY1 accumulation is not sequence specific
We constructed RCY1intI+II-P35S-GUS by inserting the DNA fragment containing the first and second introns of RCY1 upstream of the CaMV35S promoter fused to theβ-glucronidase-coding region (P35S-GUS) (Figure S2A). This construct was used to analyze the possible function of the introns in genomic RCY1 for IME of RCY1 accumulation. When P35S-GUS and RCY1intI+II-P35S-GUS were transiently expressed in different tissues within a single N. benthamiana leaf, the presence of neither RCY1 introns upstream of the promoter affected the accumulation of RCY1 (Figure S2B). Therefore, these two RCY1 introns may not function in IME of RCY1 accumulation through activation of promoter-mediated transcriptional process.
In general, the introns seem to be required to drive the correct expression pattern of the endogenous genes [39]. Some introns are known to be required to regulate gene expression at high level, while others have no effect or negative effects on gene expression [40]. To determine whether the ability of intron to increase RCY1 accumulation is specific to these RCY1 introns, the native RCY1 introns were replaced by other introns with no sequence homology to RCY1 introns. The insertion of the intron of either COLD-REGULATED 15A (COR15a) or PROFILIN 3 (PRF3) into the position near the transcriptional starting site of GUS reporter construct does not enhance GUS expression in A. thaliana [40], [41]. Thus, we first confirmed that the introns of COR15a and PRF3 had no effect on the expression of those genes. Indeed, as shown with Figure S3 and S4, we compared COR15a transcript levels and COR15a accumulation between leaf tissue transiently expressing HA epitope-tagged COR15a with or without introns (P35S-gCOR15-HA or P35S-cCOR15-HA) under the control of CaMV35S promoter in N. benthamiana leaves. Our results suggested that neither COR15a transcript levels nor COR15a accumulation differed depending on the presence or absence of these introns (Figure S4A, C and E). Furthermore, when HA-tagged PRF3 constructs (P35S-gPRF3-HA and P35S-cPRF3-HA) with or without introns were transiently expressed in N. benthamiana leaf tissues, the accumulation of PRF3 transcripts and PRF3 protein were also unaffected (Figure S4B, D and E). These results suggest that the COR15a or PRF3 introns do not function in IME of expression in either A. thaliana or N. benthamiana.
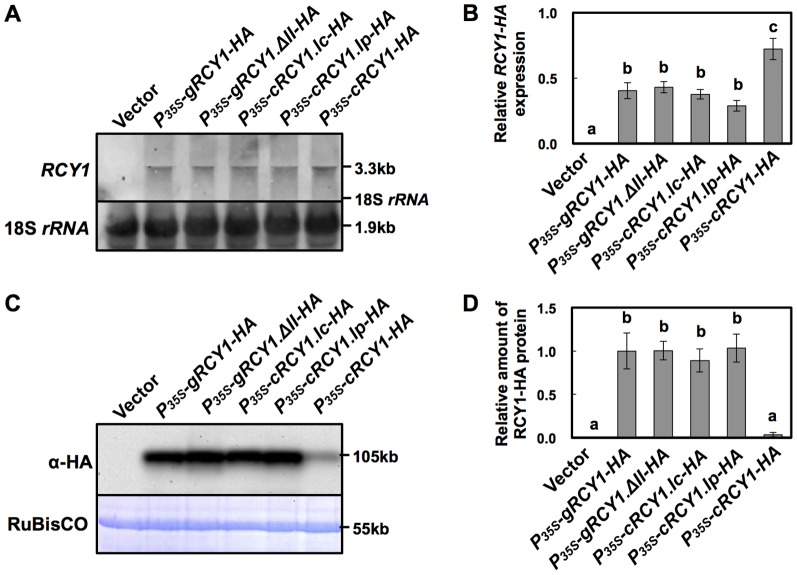
Subsequently, the COR15a second intron or the PRF3 first intron was integrated into the first exon junction position of P35S-cRCY1-HA, and the resulting constructs were designated P35S-cRCY1.Ic-HA and P35S-cRCY1.Ip-HA, respectively (Figure 2). The accumulation of RCY1-HA in leaf tissue transiently expressing P35S-cRCY1.Ic-HA or P35S-cRCY1.Ip-HA was very similar to that in leaf tissue expressing P35S-gRCY1-HA containing introns I and II or P35S-gRCY1.ΔII-HA containing intron I, but was much higher than that in leaf tissues expressing P35S-cRCY1-HA (Figure 7C and D). In contrast, RCY1 transcript levels were also similar among leaf tissues transiently expressing intron-containing RCY1-HA constructs (P35S-gRCY1-HA, P35S-gRCY1.ΔII-HA, P35S-cRCY1.Ic-HA and P35S-cRCY1.Ip-HA), but were much lower than in leaf expressing intron-less RCY1-HA (P35S-cRCY1-HA) (Figure 7A and B). These results indicate that IME of RCY1 accumulation does not dependent on the specific sequence of RCY1 introns, which are exchangeable for other introns from other genes having no IME activity.
Figure 7. Detection of HA-epitope-tagged RCY1 protein and RCY1 transcript in N. benthamiana leaves transiently expressing RCY1-HA constructs in which the RCY1 introns were replaced with COR15a or PRF3 introns.
RCY1 transcripts in N. benthamiana leaves agro-infiltrated with P35S-gRCY1-HA, P35S-gRCY1.ΔII-HA, P35S-cRCY1.Ic-HA, P35S-cRCY1.Ip-HA, or P35S-cRCY1-HA were detected by northern hybridization. pRI201-AN (Vector) was used as an empty-vector control for agro-infiltration. As an internal control for RNA sample quantities, 18S rRNA is shown (A). Relative amounts of RCY1 transcripts in each line were measured by quantitative RT-PCR (B). HA-epitope-tagged RCY1 protein (α-HA) in N. benthamiana leaves transiently expressing P35S-gRCY1-HA, P35S-gRCY1.ΔII-HA, P35S-cRCY1.Ic-HA, P35S-cRCY1.Ip-HA, or P35S-cRCY1-HA was immunologically detected using anti-HA monoclonal antibody. As an internal control for protein sample quantities, the large subunit of RuBisCO was visualized by staining with CBB (C). RCY1-HA protein amounts in each line were quantified by band intensity using Quantity One software (D). For all experiments, four independent plants transiently expressing each vector construct were analyzed. The averages of relative amounts of RCY1 transcript and protein ±SE are shown in B and D, respectively. In A and C, representative photographs are shown. The size of each band and the position of 18S rRNA were shown at right side of the panels. Data were subjected to analysis of variance and treatment means were compared by Tukey's test. Different letters indicate a statistically significant difference in the relative amount of RCY1 transcript (n = 4, P<0.05).
Discussion
Gene expression is tightly regulated through a combination of transcriptional and post-transcriptional control mechanisms. Aside from epigenetic factors, many elements of a gene's sequence structure, including the promoter, enhancers, the introns, and the 5′ and 3′-UTRs contribute to the expression level of a given gene at any point in time [39]. In this study, the analysis of RCY1 accumulation in N. benthamiana leaves transiently expressing a series of RCY1 constructs reveals that the presence of the intron in the RCY1 coding region seems to be indispensable to achieve an appropriate level of RCY1 expression to provide complete resistance to CMV(Y), while the effect of the intron on gene expression is often eclipsed by the function of other elements. To our knowledge, this is the first report indicating that an intron in the coding region of an R gene is directly associated with the level of R-protein accumulation. The enhancement of the RCY1-mediated defense reaction via elevated RCY1 protein accumulation appears to indicate that maintenance of appropriate R-protein levels in host plants is important for suppression of virus multiplication, restriction of the spread of virus around the primary virus infection site [21], [24], and induction of the defense reaction (Figure 6). Therefore, the intron-mediated enhancement of RCY1 expression seems to play a key role in complete resistance to CMV(Y) in Arabidopsis.
Introns do seem to be required to drive correct expression patterns in diverse organisms including plants. The positive effect of introns on gene expression has been named intron-mediated enhancement (IME) [40], [42]. IME of gene expression in plants is generally associated with up-regulation of mRNA levels, which often increases the accumulation of the corresponding gene product. Actually, many plant gene introns have been shown to induce gene expression at the transcriptional level [40], [43]–[47]. However, it is still unclear exactly how the presence of an intron in a gene affects the level of gene expression. Splicing of introns may induce the modification of transcripts, such as capping or polyadenylation to increase transcript stability. IME signals, predicted by the IMEter algorithm for estimating the efficiency of IME in A. thaliana, have recently been reported to be most abundant in introns located in the 5′-UTRs and coding regions near the transcription start site [40], [48]. But remarkably, for IME of RCY1, RCY1 protein accumulation was not preceded by increases in levels of RCY1 transcript, but by relatively decreased transcript levels in A. thaliana and N. benthamiana (Figures 3 and 5). GUS expression was not enhanced by insertion of either of two RCY1 introns upstream of the CaMV 35S promoter (Figure S2). However, IME of RCY1 accumulation was observed even when the introns of RCY1 were replaced with COR15a or PRF3 introns that lack IME signals (Figure 7) [40]. These facts suggest that IME of RCY1 accumulation does not operate by either a typical transcriptional enhancer in the introns or by an increase in the steady state quantity of mature mRNA. The mechanism here may differ from IME observed to date, which is typified by an increase in mRNA accumulation controlled by IME signal-containing introns located near the 5′-end of a gene.
Alternative transcripts have been detected in a certain number of known NB-LRR-encoding R genes [17], [25]–[32]. However, the direct evidence of the requirement of alternative splicing for R-gene-mediated complete resistance is just limited in some well-characterized R-genes: e.g. N and RPP4 [29], [33], thus I think that the role of alternative splicing in R-gene-mediated disease resistance is still unclear. Intron retention is a major event in alternative splicing. However, since any alternative RCY1 transcript was not detected by northern blot analysis (Figure S1), we speculate less possibility that alternative splicing occurs in RCY1 gene expression. Although it seems to be not easy to distinguish alternative transcript from incompletely spliced heteronuclear RNA by RT-PCR, it is necessary to further determine whether splice variants reside with ribosomes in cytoplasm and are required for RCY1-conferred resistance to CMV(Y).
Following intron splicing, the exon junction complex (EJC), a protein complex that localizes near the junction of two exons (20–25 nucleotides upstream), is deposited on the mRNA, and functions from intracellular transport of mRNA through translation of mRNA [49]. In animals, the EJC, which contains several proteins including core components Y14, Mago, eIF4AIII, and MLN51, is required for increased translation of spliced mRNA via interaction between Y14, Mago, and PYM (Partner of Y14 and mago), which then associate with the 40S ribosomal subunit in the cytoplasm [49], [50]. In plants, there is insufficient evidence to suggest that EJC plays an important role in the translation of mRNA. However, homologues of Y14, Mago, and PYM have been identified in A. thaliana and some other plant species [51]–[53]. Therefore, a translational control system mediated by EJC with its interactor PYM might function in IME of RCY1 accumulation in A. thaliana and N. benthamiana. In next step, it is necessary to confirm the physical interaction of EJC with the region near the junction of the exon of RCY1 mRNA or identify other host factors interacting with the exon junction sequence.
In A. thaliana, 118 out of 149 NB-LRR-encoding genes and 7 out of 58 related genes contain variable numbers of introns, whereas the remaining 31 NB-LRR-encoding genes and 51 related genes contain no introns [6]. Therefore, the role of introns is under focus for understanding the regulatory mechanisms of NB-LRR-class R-gene expression. Molecular dissection of the machinery for regulating intron-mediated RCY1 expression will provide a new platform for the design of gene structures that are more effective than simple overexpression of R genes for regulating R-gene expression and thereby conferring complete resistance to pathogens.
Materials and Methods
Plant and Virus
A yellow strain of Cucumber mosaic virus [CMV(Y)] [54] was propagated on N. tabacum ‘Xanthi nc’, and the virus was purified as previously described [55]. Nicotiana benthamiana was grown on conventional soil in a growth chamber (KG-50HLA; Koito Manufacturing Co. Ltd., Tokyo) at day and night temperatures of 25°C and 22°C, respectively, under a 14 h photoperiod at 10,000 lux. A. thaliana ecotype Columbia (Col-0): three independent lines of Col::pRCY1-HA [21] #8, #10 and #12; and three independent lines of Col::P35S-cRCY1-HA: #5, #21 and #23 which were generated in this study, were grown on soilless mix (PRO-MIX B; Premier Horticulture, Ltd., Quakertown, PA, USA) at 25°C under continuous illumination (8,000 lux).
Vector construction
Ten vector constructs encoding HA-tagged RCY1 are shown in Figure 2. For all cloning manipulations, the In-Fusion HD Cloning System (Clontech-TAKARA, Kyoto, Japan) was used according to the manufacturer's instruction manual. To generate PRCY1-gRCY1-HA, a genomic DNA fragment of RCY1 including the promoter region from approximately 1.5 kb upstream of the start codon and the 71-bp 3′ sequence downstream of the RCY1 stop codon (71-bp 3′-UTR) was amplified by PCR. For this reaction, the primers RI201AN.HindIII15.gRCY1 and RI201AN.SalI15.RCY1utr3 (Table S1) were used with pBS+SK/RCY1-HA [21] as a template, in which a HA-epitope tag sequence was inserted at the 3′-end of the RCY1-coding region. After purification of the PCR product according to the standard protocol [56], the purified DNA fragment was cloned upstream of the Heat Shock Protein gene (THSP) terminator in pRI201-AN (Takara-BIO, Mie, Japan) that had been linearized by HindIII and SalI digestion. pRI201::RCY1-HA which we constructed previously [24] was renamed P35S-cRCY1-HA in this study.
To generate modified P35S-cRCY1-HA and PRCY1-gRCY1-HA vectors carrying the CaMV35S promoter (P35S)-GFP coding region (GFP)-TNOS as an internal control, a P35S-GFP-TNOS fragment was amplified by PCR using the primers pUC.-40+NotI.15R and pUC.RV+NotI.15F (Table S1) with the 35Spro:GFP construct [57] as a template, and then purified according to the standard protocol [56]. The purified P35S-GFP-TNOS fragment was cloned into the NotI site of P35S-cRCY1-HA and PRCY1-gRCY1-HA. The resulting vector constructs were designated P35S-cRCY1-HA::P35S-GFP and PRCY1-gRCY1-HA::P35S-GFP, respectively.
To construct P35S-gRCY1-HA, a DNA fragment containing the RCY1-coding genomic region tagged with an HA-epitope sequence at its 3′-end, and including both the 66-bp 5′ sequence upstream of the RCY1 start codon (66-bp 5′-UTR), and the 71-bp 3′-UTR, was amplified by PCR with the primers RI201AN.NdeI15.RCY1utr5 and RI201AN.SalI15.RCY1utr3 (Table S1) using pGA482/RCY1-HA [21] as a template. After purification by the standard protocol [56], the PCR product was inserted between the CaMV 35S promoter (P35S) fused to the 5′-UTR sequence of Arabidopsis Alcohol Dehydrogenase gene (ADH5′-UTR) and the THSP in pRI201-AN that had been linearized with NdeI and SalI. To create the P35S-gRCY1-HAorf construct containing the HA-tagged RCY1 coding region under the control of CaMV 35S promoter but without it 66-bp 5′-UTR and 71-bp 3′-UTR, the PCR product amplified using the primers RI201AN.NdeI15bp and RI201AN.SalI15bp (Table S1) with pGA482/RCY1-HA [21] as a template was cloned downstream of P35S between the ADH5′-UTR and the THSP terminator of NdeI- and SalI-digested pRI201-AN.
P35S-gRCY1.ΔI-HA and P35S-gRCY1.ΔII-HA were derived from P35S-gRCY1-HA, but lack either the first or second intron of the RCY1-coding region, respectively. The inserts for each of these constructs were generated by fusing two other PCR fragments in PCR. For P35S-gRCY1.ΔI-HA, a PCR fragment containing the 5′ half of the cDNA-derived RCY1-coding sequence with no introns was amplified with the primers RI201AN.NdeI15.RCY1utr5 and RCY1.ExonII-R (Table S1) using P35S-cRCY1-HA as a template. The second PCR fragment containing the 3′ half of the RCY1-coding sequence including the second intron was amplified by PCR with the primers RCY1.ExonII-F (Table S1) and RI201AN.SalI15.RCY1utr3 using P35S-gRCY1-HA as a template. The final PCR fragment was amplified with the primers RI201AN.NdeI15.RCY1utr5 and RI201AN.SalI15.RCY1utr3 using a mixture of the first and second PCR fragments above as templates, was purified, then was cloned downstream of P35S between the ADH5′-UTR and THSP terminator in pRI201-AN linearized by NdeI and SalI-digestion. To generate P35S-gRCY1.ΔII-HA lacking the second RCY1 intron, two PCR fragments were amplified with two different sets of primers: RI201AN.NdeI15.RCY1utr5 and RCY1.ExonII-R using P35S-gRCY1-HA as a template; and RCY1.ExonII-F and RI201AN.SalI15.RCY1utr3 using P35S-cRCY1-HA as a template, respectively. A mixture of these two PCR products was used as a template for final PCR with the primers RI201AN.NdeI15.RCY1utr5 and RI201AN.SalI15.RCY1utr3. The resulting PCR product was purified and cloned downstream of P35S between the ADH5′-UTR and the THSP terminator in pRI201-AN.
Introns of Arabidopsis COLD-REGULATED 15A (COR15a, At2g42540) [58] and PROFILIN 3 (PRF3, At5g56600) [41] do not have the activity of intron-mediated enhancement for their expression [41], [40]. The HA-epitope-tagged sequence of COR15a including its intron was amplified by PCR with the primers RI201AN.NdeI.COR15a-F and RI201AN.SalI.V.COR15a-R (Table S1) for COR15a using genomic DNA extracted from A. thaliana ecotype Col-0 as template. After purification, this fragment was cloned into the NdeI and SalI sites of pRI201-AN, and the resulting construct was named P35S-gCOR15a-HA (Figure S3). The HA-epitope-tagged sequence of PRF3 including its introns was amplified by PCR with the primers RI201AN.NdeI.PRF3-F and HA.BstZ17I.PRF3-R (Table S1) using A. thaliana ecotype Col-0 genomic DNA as template. The purified PCR product was then cloned into the NdeI and BstZ17I sites of P35S-gCOR15a-HA (Figure S3), and the resulting construct was designated P35S-gPRF3-HA. The cDNAs of COR15a and PRF3 without introns were amplified by PCR with the same primers as immediately above using first-strand cDNA as a template, which had been reverse-transcribed from total RNA extracted from cold-assimilated Col-0. These purified PCR products were then cloned into either the NdeI and SalI sites of pRI201-AN or the NdeI and BstZ17I sites of P35S-gCOR15a-HA, respectively. The resulting constructs were designated as P35S-cCOR15a-HA and P35S-cPRF3-HA, respectively (Figure S3).
An RCY1-coding DNA fragment containing the COR15a intron at its first splice junction site, was constructed by fusing three purified DNA fragments individually amplified by PCR separate reactions with the following three sets of primers: RI201AN.NdeI15.RCY1utr5 and nE1rcy.cIcor-R (Table S1) for the first exon of RCY1; nE1rcy.cIcor-F and nIcor.cE2rcy-R (Table S1) for the COR15a second intron; and nIcor.cE2rcy-F (Table S1) and RI201AN.SalI15.RCY1utr3 for second exon of RCY1. The reactions to produce the above three fragments were then followed by second PCR with a new set of primers: RI201AN.NdeI15.RCY1utr5 and RI201AN.SalI15.RCY1utr3, using the purified PCR products above as a template.
The RCY1-coding DNA fragment with the PRF3 intron added at its first splicing junction site was also used as template for amplification in three individual PCR with the following three sets of primers: RI201AN.NdeI15.RCY1utr5 and nE1rcy.cIprf3-R (Table S1) for the first exon of RCY1; nE1rcy.cIprf3-F and nIprf3.cE2rcy-R (Table S1) for the PRF3a first intron; and nIprf3.cR2rcy-F (Table S1) and RI201AN.SalI15.RCY1utr3 for the second exon of RCY1 (Table S1). The reactions to produce the above three fragments were then followed by a second PCR with the primers RI201AN.NdeI15.RCY1utr5 and RI201AN.SalI15.RCY1utr3 using the purified PCR products immediately above as a template. The two resulting purified DNA fragments were then cloned into the NdeI and SalI sites of pRI201-AN, respectively, and the resulting constructs were designated as P35S-cRCY1.Ic-HA and P35S-cRCY1.Ip-HA (Figure 2).
Genomic DNA to be used as PCR template was extracted from the leaves of A. thaliana ecotype Col-0 by the CTAB method [59]. Total RNA to be used as template for reverse-transcriptional PCR was extracted from Col-0 leaves using an RNeasy Plant Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions. First-strand RNA was reverse transcribed from total RNA using Thermoscript RT-PCR System (Invitrogen, Carlsbad, California, USA) containing oligo(dT)20 primer according to the manufacturer's instruction manual.
All desired constructs were confirmed by restriction patterns and the Sanger sequencing method using a CEQ 8000 Automated DNA Sequencer (Beckman Coulter, Brea, CA, USA). Each plasmid (100 ng) was introduced into Agrobacterium tumefaciens LBA4404 (Takara-BIO) by electroporation according to the standard protocol [56].
Transformation of A. thaliana
A. thaliana ecotype Col-0 plants were transformed with Agrobacterium tumefaciens LBA4404 containing P35S-cRCY1-HA by vacuum infiltration [60]. Transgenic plants were screened on 0.5× MS medium [61] with 0.8% agar and 50 µg of kanamycin per ml. Transformation of P35S-cRCY1-HA into Col-0 plants was confirmed in second and third generation (T2 and T3) plants by rat anti-HA monoclonal antibodies (clone 3F10, dilution 1∶10,000; Roche, Indianapolis, IN, USA), according to the method described previously [21]. These transgenic plants were designated as line Col::P35S-cRCY1-HA. The RCY1 transgene was detected by quantitative PCR using the 7300 Real-Time PCR System (Applied Biosystems, Foster City, CA, USA) with the primers RCY1-HA.F and RCY1-HA.R (Table S2) and genomic DNA extracted from each transformant as a template.
Transient expression of RCY1 constructs in N. benthamiana
Agrobacterium-mediated transient expression of the following ten vector constructs: P35S-cRCY1-HA, PRCY1-gRCY1-HA, P35S-cRCY1-HA::P35S-GFP, PRCY1-gRCY1-HA::P35S-GFP, P35S-gRCY1-HA, P35S-gRCY1-HAorf, P35S-gRCY1.ΔI-HA, P35S-gRCY1.ΔII-HA, P35S-cRCY1.Ic-HA, and P35S-cRCY1.Ip-HA (Figure 2) in N. benthamiana leaves was performed by the method described previously [24]. To compare levels of RCY1-HA accumulation among leaves expressing a set of vector constructs, independent regions of single fully expanded of N. benthamiana leaves were infiltrated with each Agrobacterium suspension containing each vector construct. RCY1-HA protein was detected by immunoblotting with rat anti-HA monoclonal antibody (clone 3F10, dilution 1∶10,000; Roche, Indianapolis, IN, USA) according to the method described previously [21]. The intensity of bands visualized with the ECL Prime chemiluminescent substrate (GE Healthcare, Piscataway, NJ, USA) was measured by Quantity One software using a VersaDoc MP 4000 system (Bio-Rad, Hercules, CA, USA). Four independent protein samples per experiment were used to quantitatively analyze HA-tagged protein levels, and the average band intensities ±SE are shown. GFP accumulation was immunologically detected by the method described previously [62] using antiserum against GFP (Medical and Biological Laboratories: MBL, Nagoya, Japan). As an internal control, the large subunit of RuBisCO was detected by staining with Coomassie Brilliant Blue R-250 (CBB) according to the standard protocol.
Northern blot and quantitative RT-PCR
Total RNA was extracted from the leaves of A. thaliana and N. benthamiana, using an RNeasy Plant Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer's instruction. Transcripts of the RCY1 transgene and the PR-1a gene of A. thaliana were detected by northern hybridization according to the procedure described previously [37]. DIG-labeled probe for Arabidopsis PR-1a was prepared according to the procedure described previously [63]. DIG-labeled probes specific to the RCY1 transgene in A. thaliana were amplified by PCR with the primers RCY1utr3-F and RCY1utr3-R (Table S3). To detect the transcripts in N. benthamiana, DIG-labeled probes for the RCY1 transgene, and PRB-1b, PR-2b, and PR-6 from N. benthamiana were amplified by PCR with the following sets of primers: RCY1.ExonI-F and rpp8-R3 for the RCY1 transgene; PRB-1b-F and PRB-1b-R for PRB-1b; PR-2b-F and PR-2b-R for PR-2b; and PR-6-F and PR-6-R for PR-6 (Table S3). Transcripts of COR15a and PRF3 were detected with DIG-labeled probes amplified by PCR with the primers COR15a-F and COR15a-R for COR15a (Table S3); or PRF3-F and PRF3-R for PRF3 (Table S3), respectively. All DNA probes were labeled with digoxigenin (DIG)-11-dUTP by PCR using a PCR DIG Synthesis Kit (Roche, Penzberg, Germany) according to the manufacturer's instructions. DIG-labeled probes were detected using an alkaline phosphatase-conjugated anti-DIG antibody and were visualized with the CDP-Star Reagent (New England Biolabs, Beverly, MA, USA) according to the instruction manuals.
For quantitative measurement of RCY1 transgene transcripts, and the controls UBQ5 of A. thaliana, and EF-1a in N. benthamiana, 1 µg total RNA was reverse transcribed into cDNA using the PrimeScript RT Reagent Kit with gDNA Eraser (Takara-Bio) containing random hexamer primers according to the manufacturer's instruction manual. Quantitative RT-PCR amplification for detecting RCY1 transcripts was performed in triplicate 20 µl reactions containing template cDNA (2 µl), 0.4 µM RCY1-specific primers RCY1-HA.F and RCY1-HA.R (Table S2), 1xROX Reference Dye, and 1xSYBR Premix Ex Taq II (Tli RNase H plus) (Takara-Bio) using the 7300 Real-Time PCR System (Applied Biosystems). PCR conditions and data analysis were performed according to the procedure described previously [64]. As a standard control, the level of the A. thaliana UBQ5 transcript or the N. benthamiana EF1a transcript was quantified by RT-PCR using SYBR with the primers RTUBQ5-F1 and RTUBQ5-R1 for UBQ5 (Table S2) or NbEF1a-F and NbEF1a-R for EF-1a (Table S2). RCY1 mRNA quantities were normalized relative to the values of constitutively expressed UBQ5 or EF-1a mRNAs. In each experiment, three independent A. thaliana and four independent N. benthamiana plants were used for quantitative measurement of each transcript, and the level of gene expression is shown as the average ± SE of the value of RCY1 mRNA relative to UBQ5 or EF-1a mRNA. Data were subjected to analysis of variance and treatment means were compared by Tukey's test.
Detection of cell death and H2O2 production
The development of HR cell death at 72 h after agro-infiltration with RCY1 constructs was visualized by trypan blue staining according to a standard protocol [65]. To evaluate HR cell death quantitatively, electrolyte leakage was measured a method modified from that described by Kim et al. (2003) [66]. At 80 h after agro-infiltration with RCY1 constructs, ten leaf discs 0.6 mm in diameter were floated on 11 ml of 0.4 M sorbitol and incubated at 25°C. After 20 h of incubation, the incubation solution was removed from the leaf discs, and the conductivity of the incubation solution was measured as sample conductivity with a conductivity meter (ES-51, HORIBA, Ltd., Kyoto, Japan). Ten leaf discs were then combined with the incubated solution and boiled for 10 min. The conductivity of the boiled solution was measured as total conductivity. Relative electrolyte leakage was expressed as the percentage of sample conductivity to total conductivity. Four independent plants were used for measurement of electrolyte leakage and the averages of relative amounts ±SE are shown.
H2O2 evolved due to HR cell death was detected by 3,3′-dianimobenzidine (DAB) staining. At 72 h after agro-infiltration with RCY1 constructs, the infiltrated leaves were incubated with 1 mg/ml DAB solution for 12 h at 25°C in dark condition, and then washed in 95% ethanol at 94°C for 10 min.
Analysis of transcriptional enhancer activity from introns of RCY1
A DNA fragment spanning the first intron to the second intron of RCY1 genomic DNA was cloned upstream of the CaMV P35S in pSMAHdN632L-M2GUS [67], which was then renamed P35S-GUS in this study. The intron fragment was amplified by PCR with the primers: SMAH.SbfIXhoI.RCYintron-F (5′-GTTAAGGAATTGCCCTGCA- GGCTCGAGTTCCACGGAAAAGAGGTG-3′, in which the 15-bp vector sequence for In-Fusion Cloning is underlined) and SMAH.SbfI.RCY1intron-R (5′-GGCTAATCT- GGGGACCTGCACATCAAGCCTTACTTCTGC-3′), using P35S-gRCY1-HA as a template. After purification of the PCR product according to the standard protocol [56], the fragment was cloned into pSMAHdN632L-M2GUS linearized by digestion with SbfI, and the resulting vector was named RCY1intI+II-P35S-GUS.
GUS reporter accumulation was measured by ELISA. Leaf tissues transiently expressing each gene were homogenized in 100-fold volumes of 0.05 M Na2CO3. The protein concentration in the homogenate was determined by the Bradford method [68]. The homogenate was diluted 10-fold with 0.05 M Na2CO3 and subjected to ELISA according to the standard protocol [56] using a polyclonal anti-GUS antibody (Molecular Probes, Eugene, Oregon, USA). GUS accumulation is shown as absorbance at 405 nm per 0.22 mg/ml of total protein. Four independent leaf samples transiently expressing vector constructs were used for detection of GUS accumulation. To present quantitative measurement of GUS accumulation, the average absorbance ±SE values are shown.
Supporting Information
Detection of RCY1 transcripts in N. benthamiana leaves transiently expressing P35S-cRCY1-HA and P35S-gRCY1-HA . RCY1 transcripts in N. benthamiana leaves agro-infiltrated with the intron-containing genomic RCY1 coding region (P35S-gRCY1-HA), RCY1 cDNA without introns (P35S-cRCY1-HA), or pRI201-AN (Vector) as an empty vector control were detected by northern hybridization. Full-length RCY1 transcripts are indicated as bands marked by the arrow. RNA was extracted from three independent plants (1, 2, and 3) per vector-infiltrated plants.
(TIFF)
Assay for the enhancement of promoter activity by the RCY1 intron sequence. Schematic of the β-glucronidase (gusA)-coding vector constructs: P35S-GUS with gusA under control of the CaMV 35S promoter (P35S) and nopaline synthase terminator (TNOS); RCY1intI+II-P35S-GUS in which RCY1 intron sequences shown by the black fold lines, were inserted into SbfI site upstream of P35S of P35S-GUS (A). The position of insertion of the intron–containing fragment from P35S-gRCY1-HA into RCY1intI+II-P35S-GUS is indicated by dotted lines. Relative GUS protein quantities in N. benthamiana leaves transiently expressing P35S-GUS or RCY1intI+II-P35S-GUS were measured by ELISA at 0, 24, 48, and 72 h after agro-infiltration (B). Four independent plants transiently expressing each vector construct were analyzed. The averages of relative GUS protein amounts ±SE are shown in B.
(TIFF)
Schematic structure of the COR15a and PRF3 vector constructs under control of the CaMV 35S promoter. COR15a or PRF3 cDNA without introns but with HA-epitope tags (HA) at their 3′-ends were cloned between the CaMV 35S promoter with the 5′-UTR sequence of the Arabidopsis Alcohol Dehydrogenase gene (ADH5′-UTR) and Heat Shock Protein gene terminator (THSP) in the pRI201-AN binary vector. The resulting constructs were named P35S-cCOR15a-HA and P35S-cPRF3-HA, respectively. The COR15a- or PRF3-coding regions are indicated by gray boxes and the two splice junction sites are indicated by vertical lines in the boxes. Genomic COR15a or PRF3 tagged with HA at its 3′-end under control of the CaMV 35S promoter (P35S-gCOR15a-HA and P35S-gPRF3-HA) contains intron sequences indicated by the black dashed lines.
(TIFF)
Comparison of HA-epitope-tagged COR15a and PRF3 transcript and protein levels among N. benthamiana leaf tissues transiently expressing intron-containing genomic COR15a or PRF3 or COR15a or PRF3 cDNAs without introns. COR15a (A) or PRF3 (B) transcripts in N. benthamiana leaf tissues transiently expressing either the intron-containing genomic COR15a (P35S-gCOR15a-HA) or PRF3 (P35S-gPRF3-HA), or the cDNAs for COR15a (P35S-cCOR15a-HA) or PRF3 (P35S-cPRF3-HA) without introns, were detected by northern hybridization. pRI201-AN (Vector) was used as an empty-vector control. As an internal control for RNA sample quantities, 18S rRNA is shown. The size of each band and the position of 18S rRNA were shown at right side of the panels. COR15a (C) or PRF3 (D) protein amounts in each line were quantified by band intensity using Quantity One software. Four independent plants transiently expressing each vector construct were analyzed. The averages of relative COR15a-HA and PRF3-HA protein amounts ±SE are shown. The COR15a-HA and PRF3-HA proteins in leaf tissues of each line were also detected by immunoblotting (E). As controls, pRI201-AN (Vector), P35S-gRCY1-HA, and P35S-cRCY1-HA were agro-infiltrated into N. benthamiana leaves. As an internal control for protein sample quantities, the large subunit of RuBisCO was visualized by staining with CBB. In this experiment, 1/50 volume of total protein sample of leaf accumulating COR15a-HA against that of RCY1-HA and PRF3-HA was applied on the gel, since the level of COR15a-HA accumulation was essentially much higher than others. The size of each band and the position of RuBisCO large subunit were shown at right side of the panel.
(TIFF)
(XLSX)
(XLSX)
(XLSX)
Acknowledgments
We thank H. Ichikawa at the National Institute of Agrobiological Sciences, Japan for kindly providing the pSMAHdN632L-M2GUS binary Ti plasmid.
Data Availability
The authors confirm that all data underlying the findings are fully available without restriction. All data are included within the manuscript and Supporting Information files.
Funding Statement
This research was supported by a grant-in-aid for Scientific Research (B) (22380028) and Challenging Exploratory Research (24658037) from the Ministry of Education, Culture, Sports, Science and Technology, Japan. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Martin GB, Bogdanove AJ, Sessa G (2003) Understanding the functions of plant disease resistance proteins. Annu Rev Plant Biol 54: 23–61. [DOI] [PubMed] [Google Scholar]
- 2. Liu J, Liu X, Dai L, Wang G (2007) Recent progress in elucidating the structure, function and evolution of disease resistance genes in plants. J Genet Genomics 34: 765–776. [DOI] [PubMed] [Google Scholar]
- 3. Gururani MA, Venkatesh J, Upadhyaya CP, Nookaraju A, Pandey SK, et al. (2012) Plant disease resistance genes: Current status and future directions. Physiol Mol Plant Pathol 78: 51–65. [Google Scholar]
- 4. Hammond-Kosack KE, Jones JDG (1997) Plant disease resistance genes. Annu Rev Plant Physiol Plant Mol Biol 48: 575–607. [DOI] [PubMed] [Google Scholar]
- 5. Dangl JL, Jones JDG (2001) Plant pathogens and integrated defence responses to infection. Nature 411: 826–833. [DOI] [PubMed] [Google Scholar]
- 6. Meyers BC, Kozik A, Griego A, Kuang H, Michelmore RW (2003) Genome-wide analysis of NBS-LRR-encoding genes in Arabidopsis. Plant Cell 15: 809–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Meyers BC, Dickerman AW, Michelmore RW, Sivaramakrishnan S, Sobral BW, et al. (1999) Plant disease resistance genes encode members of an ancient and diverse protein family within the nucleotide-binding superfamily. Plant J 20: 317–332. [DOI] [PubMed] [Google Scholar]
- 8. Richly E, Kurth J, Leister D (2002) Mode of amplification and reorganization of resistance genes during recent Arabidopsis thaliana evolution. Mol Biol Evol 9: 76–84. [DOI] [PubMed] [Google Scholar]
- 9. Tan X, Meyers BC, Kozik A, West MAL, Morgante M, et al. (2007) Global expression analysis of nucleotide binding site-leucine rich repeat-encoding and related genes in Arabidopsis. BMC Plant Biol 7: 56–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Yoshimura S, Yamanouchi U, Katayose Y, Toki S, Wang ZX, et al. (1998) Expression of Xa1, a bacterial blight resistance gene, is induced by bacterial inoculation. Proc Natl Acad Sci USA 95: 1663–1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Halterman DA, Wei F, Wise RP (2003) Powdery mildew-induced Mla mRNAs are alternatively spliced and contain multiple upstream open reading frames. Plant Physiol 131: 558–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Levy M, Edelbaum O, Sela I (2004) Tobacco mosaic virus regulates the expression of its own resistance gene N . Plant Physiol 135: 2392–2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gu K, Yang B, Tian D, Wu L, Wang D, et al. (2005) R gene expression induced by a type-III effector triggers disease resistance in rice. Nature 435: 1122–1125. [DOI] [PubMed] [Google Scholar]
- 14. Cao Y, Ding X, Cai M, Zhao J, Lin Y, et al. (2007) The expression pattern of a rice disease resistance gene xa3/xa26 is differentially regulated by the genetic backgrounds and developmental stages that influence its function. Genetics 177: 523–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mohr TJ, Mammarella ND, Hoff T, Woffenden BJ, Jelesko JG, et al. (2010) The Arabidopsis downy mildew resistance gene RPP8 is induced by pathogens and salicylic acid and is regulated by W box cis elements. Mol Plant-Microbe Interact 23: 1303–1315. [DOI] [PubMed] [Google Scholar]
- 16. Boyes DC, Nam J, Dangl JL (1998) The Arabidopsis thaliana RPM1 disease resistance gene product is a peripheral plasma membrane protein that is degraded coincident with the hypersensitive response. Proc Natl Acad Sci USA 95: 15849–15854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ayliffe MA, Frost DV, Finnegan EJ, Lawrence GJ, Anderson PA, et al. (1999) Analysis of alternative transcripts of the flax L6 rust resistance gene. Plant J 17: 287–292. [DOI] [PubMed] [Google Scholar]
- 18. Collins N, Drake J, Ayliffe M, Sun Q, Ellis J, et al. (1999) Molecular characterization of the maize Rp1-D rust resistance haplotype and its mutants. Plant Cell 11: 1365–1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mes JJ, van Doorn AA, Wijbrandi J, Simons G, Cornelissen BJ, et al. (2000) Expression of the Fusarium resistance gene I-2 colocalizes with the site of fungal containment. Plant J 23: 183–193. [DOI] [PubMed] [Google Scholar]
- 20. Cooley MB, Pathirana S, Wu HJ, Kachroo P, Klessig DF (2000) Members of the Arabidopsis HRT/RPP8 family of resistance genes confer resistance to both viral and oomycete pathogens. Plant Cell 12: 663–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sekine K-T, Kawakami S, Kubota M, Ichinose Y, Shah J, et al. (2008) High-level expression of a virus resistance gene, RCY1 confers extreme resistance to Cucumber mosaic virus in Arabidopsis thaliana . Mol Plant-Microbe Interact 21: 1398–1407. [DOI] [PubMed] [Google Scholar]
- 22. Bendahmane A, Farnham G, Moffett P, Baulcombe DC (2002) Constitutive gain-of-function mutants in a nucleotide binding site-leucine rich repeat protein encoded at the Rx locus of potato. Plant J 32: 195–204. [DOI] [PubMed] [Google Scholar]
- 23. Shirano Y, Kachroo P, Shah J, Klessig DF (2002) A gain-of-function mutation in an Arabidopsis Toll Interleukin1 receptor-nucleotide binding site-leucine-rich repeat type R gene triggers defense responses and results in enhanced disease resistance. Plant Cell 14: 3149–3162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Takahashi H, Shoji H, Ando S, Kanayama Y, Kusano T, et al. (2012a) RCY1-mediated resistance to Cucumber mosaic virus is regulated by LRR domain-mediated interaction with CMV(Y) following degradation of RCY1. Mol Plant-Microbe Interact 25: 1171–1185. [DOI] [PubMed] [Google Scholar]
- 25. Lawrence GJ, Finnegan EJ, Ayliffe MA, Ellis JG (1995) The L6 gene for flax rust resistance is related to the Arabidopsis bacterial resistance gene RPS2 and the tobacco viral resistance gene N . Plant Cell 7: 1195–1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Anderson PA, Lawrence GJ, Morrish BC, Ayliffe MA, Finnegan EJ, et al. (1997) Inactivation of the flax rust resistance gene M associated with loss of a repeated unit within the leucine-rich repeat coding region. Plant Cell 9: 641–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Parker JE, Coleman MJ, Szabò V, Frost LN, Schmidt R, et al. (1997) The Arabidopsis downy mildew resistance gene RPP5 shares similarity to the Toll and interleukin-1 receptors with N and L6 . Plant Cell 9: 879–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Gassmann W, Hinsch ME, Staskawicz BJ (1999) The Arabidopsis RPS4 bacterial-resistance gene is a member of the TIR-NBS-LRR family of disease-resistance genes. Plant J 20: 265–277. [DOI] [PubMed] [Google Scholar]
- 29. Dinesh-Kumar SP, Tham W-H, Baker BJ (2000) Structure–function analysis of the tobacco mosaic virus resistance gene N . Proc Natl Acad Sci USA 97: 14789–14794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Halterman DA, Wei F, Wise RP (2003) Powdery mildew-induced Mla mRNAs are alternatively spliced and contain multiple upstream open reading frames. Plant Physiol 131: 558–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Borhan MH, Holub EB, Beynon JL, Rozwadowski K, Rimmer SR (2004) The Arabidopsis TIR-NB-LRR gene RAC1 confers resistance to Albugo candida (White rust) and is dependent on EDS1 but not PAD4 . Mol Plant-Microbe Interact 17: 711–719. [DOI] [PubMed] [Google Scholar]
- 32. Schornack S, Ballvora A, Gürlebeck D, Peart J, Ganal M, et al. (2004) The tomato resistance protein Bs4 is a predicted non-nuclear TIR-NB-LRR protein that mediates defense responses to severely truncated derivatives of AvrBs4 and overexpressed AvrBs3. Plant J 37: 46–60. [DOI] [PubMed] [Google Scholar]
- 33. Zhang X, Gassmann W (2003) RPS4-mediated disease resistance requires the combined presence of RPS4 transcripts with full-length and truncated open reading frames. Plant Cell 15: 2333–2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gassmann W (2008) Alternative splicing in plant defense. In Current Topics in Microbiology and Immunology, Vol. 326 , Nuclear Pre-mRNA Processing in Plants. Eds. Reddy ASN, Golovkin M, Springer-Verlag, Berlin, Heidelberg, pp 219–233. [DOI] [PubMed] [Google Scholar]
- 35. Sekine K-T, Ishihara T, Hase S, Kusano T, Shah J, et al. (2006) Single amino acid alterations in Arabidopsis thaliana RCY1 compromise resistance to Cucumber mosaic virus, but differentially suppress hypersensitive response-like cell death. Plant Mo. Biol 62: 669–682. [DOI] [PubMed] [Google Scholar]
- 36. Takahashi H, Miller J, Nozaki Y, Sukamto, Takeda M, et al. (2002) RCY1, an Arabidopsis thaliana RPP8/HRT family resistance gene, conferring resistance to cucumber mosaic virus requires salicylic acid, ethylene and a novel signal transduction mechanism. Plant J 32: 655–667. [DOI] [PubMed] [Google Scholar]
- 37. Takahashi H, Kanayama Y, Zheng MS, Kusano T, Hase S, et al. (2004) Antagonistic interactions between the SA and JA signaling pathways in Arabidopsis modulate expression of defense genes and gene-for-gene resistance to Cucumber mosaic virus . Plant Cell Physiol 45: 803–809. [DOI] [PubMed] [Google Scholar]
- 38. Ishihara T, Sekine K-T, Hase S, Kanayama Y, Seo S, et al. (2008) Overexpression of the Arabidopsis thaliana EDS5 gene enhances resistance to viruses. Plant Biol 10: 451–461. [DOI] [PubMed] [Google Scholar]
- 39.Farrell Jr RE (2007) The regulation of gene expression in plants and animals. In Regulation of Gene Expression in Plants: The Role of Transcript Structure and Processing. Ed. Bassett CL. Springer, New York, USA, pp 1–38.
- 40. Rose A, Elfersi T, Parra G, Korf I (2008) Promoter-proximal introns in Arabidopsis thaliana are enriched in dispersed signals that elevate gene expression. Plant Cell 20: 543–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Jeong YM, Mun JH, Lee I, Woo JC, Hong CB, et al. (2006) Distinct roles of the first introns on the expression of Arabidopsis profilin gene family members. Plant Physiol 140: 196–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Bourdon V, Harvey A, Lonsdale D (2001) Introns and their positions affect the translational activity of mRNA in plant cells. EMBO Rep 2: 394–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Callis J, Fromm M, Walbot V (1987) Introns increase gene expression in cultured maize cells. Genes Dev 1: 1183–1200. [DOI] [PubMed] [Google Scholar]
- 44. Dean C, Favreau M, Bond-Nutter D, Bedbrook J, Dunsmuir P (1989) Sequences downstream of translation start regulate quantitative expression of two petunia rbcS genes. Plant Cell 1: 201–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Xu Y, Yu H, Hall TC (1994) Rice triosephosphate isomerase gene 59 sequence directs β-glucuronidase activity in transgenic tobacco but requires an intron for expression in rice. Plant Physiol 106: 459–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Rethmeier N, Seurinck J, Van Montagu M, Cornelissen M (1997) Intron-mediated enhancement of transgene expression in maize is a nuclear, gene-dependent process. Plant J 12: 895–899. [DOI] [PubMed] [Google Scholar]
- 47. Rose AB, Last RL (1997) Introns act post-transcriptionally to increase expression of the Arabidopsis thaliana tryptophan pathway gene PAT1. Plant J 11: 455–464. [DOI] [PubMed] [Google Scholar]
- 48. Parra G, Bradnam K, Rose AB, Korf I (2011) Comparative and functional analysis of intron-mediated enhancement signals reveals conserved features among plants. Nucleic Acids Res 39: 5328–5337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Glisovic T, Bachorik JL, Yong J, Dreyfuss G (2008) RNA-binding proteins and post-transcriptional gene regulation. FEBS Lett 582: 1977–1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Diem MD, Chan CC, Younis I, Dreyfuss G (2007) PYM binds the cytoplasmic exon-junction complex and ribosomes to enhance translation of spliced mRNAs. Nature Struct Mol Biol 14: 1173–1179. [DOI] [PubMed] [Google Scholar]
- 51. Park N-I, Muench DG (2007) Biochemical and cellular characterization of the plant ortholog of PYM, a protein that interacts with the exon junction complex core proteins Mago and Y14. Planta 225: 625–639. [DOI] [PubMed] [Google Scholar]
- 52. Koroleva OA, Calder G, Pendle AF, Kim SH, Lewandowska D, et al. (2009) Dynamic behavior of Arabidopsis eIF4A-III, putative core protein of exon junction complex: Fast relocation to nucleolus and splicing speckles under hypoxia. Plant Cell 21: 1592–1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Mufarrege EF, Gonzalez DH, Curi GC (2011) Functional interconnections of Arabidopsis exon junction complex proteins and genes at multiple steps of gene expression. J Exp Botany 62: 5025–5036. [DOI] [PubMed] [Google Scholar]
- 54. Tomaru K, Hidaka J (1960) Strains of cucumber mosaic virus isolated from tobacco plants. III. A yellow strain. Bull Hatano Tobacco Exp Station 46: 143–149. [Google Scholar]
- 55. Takahashi H, Ehara Y (1993) Severe chlorotic spot symptoms in cucumber mosaic virus strain Y-infected tobaccos are induced by a combination of the virus coat protein gene and two host recessive genes. Mol Plant-Microbe Interact 6: 182–189. [DOI] [PubMed] [Google Scholar]
- 56.Green MR, Sambrook J (2012) Molecular Cloning. A Laboratory Manual, 4th edn. Cold Spring Harbor,NY, USA: Cold Spring Harbor Laboratory Press.
- 57. Hondo D, Hase S, Kanayama Y, Yoshikawa N, Takenaka S, et al. (2007) Up-regulation of LeATL6 that encodes a fungal elicitor-responsive ubiquitin ligase induces jasmonic acid-dependent proteinase inhibitor gene expression in tomato. Mol Plant-Microbe Interact 20: 72–81. [DOI] [PubMed] [Google Scholar]
- 58. Baker SS, Wilhelm KS, Thomashow MF (1994) The 59-region of Arabidopsis thaliana cor15a has cis-acting elements that confer cold-, drought-, and ABA-regulated gene expression. Plant Mol Biol 24: 701–713. [DOI] [PubMed] [Google Scholar]
- 59. Murray MG, Thompson WF (1980) Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res 8: 4321–4325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bechtold N (1998) In planta Agrobacterium-mediated transformation of adult Arabidopsis thaliana plants by vacuum infiltration. In Methods in Molecular Biology. Eds. J Martinez-Zapater, J Salinas, Humana Press, Totowa, NJ, USA, pp 259–266. [DOI] [PubMed]
- 61. Murashige T, Skoog F (1962) A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant 15: 473–497. [Google Scholar]
- 62. Takahashi H, Goto N, Ehara Y (1994) Hypersensitive response in cucumber mosaic virus-inoculated Arabidopsis thaliana. Plant J 6: 369–377. [Google Scholar]
- 63. Takahashi H, Kai A, Yamashita M, Ando S, Sekine K-T, et al. (2012b) Cyclic nucleotide-gated ion channel-mediated cell death may not be critical for R gene-conferred resistance to Cucumber mosaic virus in Arabidopsis thaliana . Physiol Mol Plant Pathol 79: 40–48. [Google Scholar]
- 64. Takahashi H, Nakaho K, Ishihara T, Ando S, Wada T, et al. (2014) Transcriptional profile of tomato roots exhibiting Bacillus thuringiensis-induced resistance to Ralstonia solanacearum . Plant Cell Rep 33: 99–110. [DOI] [PubMed] [Google Scholar]
- 65. Bowling SA, Guo A, Cao H, Gordon AS, Klessig DF, et al. (1994) A mutation in Arabidopsis that leads to constitutive expression of systemic acquired resistance. Plant Cell 6: 1845–1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Kim M, Ahn JW, Jin UH, Choi D, Paek KH, et al. (2003) Activation of the programmed cell death pathway by inhibition of proteasome function in plants. J Biol Chem 278: 19406–19415. [DOI] [PubMed] [Google Scholar]
- 67. Hakata M, Nakamura H, Iida-Okada K, Miyao A, Kajikawa M, et al. (2010) Production and characterization of a large population of cDNA-overexpressing transgenic rice plants using Gateway-based full-length cDNA expression libraries. Breeding Sci 60: 575–585. [Google Scholar]
- 68. Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248–254. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Detection of RCY1 transcripts in N. benthamiana leaves transiently expressing P35S-cRCY1-HA and P35S-gRCY1-HA . RCY1 transcripts in N. benthamiana leaves agro-infiltrated with the intron-containing genomic RCY1 coding region (P35S-gRCY1-HA), RCY1 cDNA without introns (P35S-cRCY1-HA), or pRI201-AN (Vector) as an empty vector control were detected by northern hybridization. Full-length RCY1 transcripts are indicated as bands marked by the arrow. RNA was extracted from three independent plants (1, 2, and 3) per vector-infiltrated plants.
(TIFF)
Assay for the enhancement of promoter activity by the RCY1 intron sequence. Schematic of the β-glucronidase (gusA)-coding vector constructs: P35S-GUS with gusA under control of the CaMV 35S promoter (P35S) and nopaline synthase terminator (TNOS); RCY1intI+II-P35S-GUS in which RCY1 intron sequences shown by the black fold lines, were inserted into SbfI site upstream of P35S of P35S-GUS (A). The position of insertion of the intron–containing fragment from P35S-gRCY1-HA into RCY1intI+II-P35S-GUS is indicated by dotted lines. Relative GUS protein quantities in N. benthamiana leaves transiently expressing P35S-GUS or RCY1intI+II-P35S-GUS were measured by ELISA at 0, 24, 48, and 72 h after agro-infiltration (B). Four independent plants transiently expressing each vector construct were analyzed. The averages of relative GUS protein amounts ±SE are shown in B.
(TIFF)
Schematic structure of the COR15a and PRF3 vector constructs under control of the CaMV 35S promoter. COR15a or PRF3 cDNA without introns but with HA-epitope tags (HA) at their 3′-ends were cloned between the CaMV 35S promoter with the 5′-UTR sequence of the Arabidopsis Alcohol Dehydrogenase gene (ADH5′-UTR) and Heat Shock Protein gene terminator (THSP) in the pRI201-AN binary vector. The resulting constructs were named P35S-cCOR15a-HA and P35S-cPRF3-HA, respectively. The COR15a- or PRF3-coding regions are indicated by gray boxes and the two splice junction sites are indicated by vertical lines in the boxes. Genomic COR15a or PRF3 tagged with HA at its 3′-end under control of the CaMV 35S promoter (P35S-gCOR15a-HA and P35S-gPRF3-HA) contains intron sequences indicated by the black dashed lines.
(TIFF)
Comparison of HA-epitope-tagged COR15a and PRF3 transcript and protein levels among N. benthamiana leaf tissues transiently expressing intron-containing genomic COR15a or PRF3 or COR15a or PRF3 cDNAs without introns. COR15a (A) or PRF3 (B) transcripts in N. benthamiana leaf tissues transiently expressing either the intron-containing genomic COR15a (P35S-gCOR15a-HA) or PRF3 (P35S-gPRF3-HA), or the cDNAs for COR15a (P35S-cCOR15a-HA) or PRF3 (P35S-cPRF3-HA) without introns, were detected by northern hybridization. pRI201-AN (Vector) was used as an empty-vector control. As an internal control for RNA sample quantities, 18S rRNA is shown. The size of each band and the position of 18S rRNA were shown at right side of the panels. COR15a (C) or PRF3 (D) protein amounts in each line were quantified by band intensity using Quantity One software. Four independent plants transiently expressing each vector construct were analyzed. The averages of relative COR15a-HA and PRF3-HA protein amounts ±SE are shown. The COR15a-HA and PRF3-HA proteins in leaf tissues of each line were also detected by immunoblotting (E). As controls, pRI201-AN (Vector), P35S-gRCY1-HA, and P35S-cRCY1-HA were agro-infiltrated into N. benthamiana leaves. As an internal control for protein sample quantities, the large subunit of RuBisCO was visualized by staining with CBB. In this experiment, 1/50 volume of total protein sample of leaf accumulating COR15a-HA against that of RCY1-HA and PRF3-HA was applied on the gel, since the level of COR15a-HA accumulation was essentially much higher than others. The size of each band and the position of RuBisCO large subunit were shown at right side of the panel.
(TIFF)
(XLSX)
(XLSX)
(XLSX)
Data Availability Statement
The authors confirm that all data underlying the findings are fully available without restriction. All data are included within the manuscript and Supporting Information files.