Abstract
The ubiquitin-proteasome system (UPS) plays a major role in the homeostasis of cellular protein. We demonstrate that each of the major hematologic diseases (AML, ALL, and MDS) has a specific and different plasma profile of UPS protein and enzymatic activities. While high levels of proteasome and ubiquitin proteins and enzymatic activities are detected in the plasma samples from patients, normalizing enzymatic activities, show that each proteasome has lower enzymatic activities in these diseases as compared with normal controls. Proteasome protein levels in AML are strong predictor of survival independently of cytogenetics, performance status and age. The Ch-L activity when normalized to the level of proteasome protein show significant negative correlation with survival in ALL.
Keywords: acute myeloid leukemia; acute lymphoblastic leukemia; myelodysplastic syndrome; proteasome, ubiquitin; chymotrypsin-like activity; survival; response
Introduction
The ubiquitin-proteasomes system (UPS) is a major non-lysosomal proteolytic system in cells that plays a major role in regulating cellular functions including the cell cycle, apoptosis, differentiation, DNA repair, and many other activities.(1-6) The UPS works through a complex interaction of its component parts. (6) Ubiquitin is activated at the appropriate time, then binds the protein to be hydrolyzed and transfers it to the 26S proteasome structure for degradation. (3) The 26S proteasome itself consists of two 19S regulatory structures and a central 20S core with three different active sites (b1, b2, and b5); each site is responsible for the enzymatic activities of caspase-like (Cas-L), trypsin-like (Tr-L), and chymotrypsin-like (Ch-L), respectively. (5) These enzymes are responsible for the digestion of proteins that are to be degraded. Recent studies demonstrated that proteasomes from different tissue or cell types have different enzymatic activity patterns as well as molecular compositions. (7,8) In fact, erythrocytes, monocytes, T-cells, and platelets can be distinguished based on the structure of the 20S proteasomes in each cell type. (7) Furthermore, isoelectric focusing showed different compositions between cardiac muscle and liver with significant differences in their enzymatic activities. (8)
We have reported that proteasome enzymatic activities can be measured in plasma.(9,10) We also demonstrated that plasma samples from patients with various leukemias have significantly higher proteasome enzymatic acitivities when compared with plasma samples from healthy controls. Plasma proteasome activities from patients with leukemia behave similarly to those from cellular proteasomes and most likely reflect the enzymatic activities in the leukemic cells.(10) These proteasome enzymatic activities may originate from the free circulating proteasomes that are reported to be detectable at higher levels in patients with various autoimmune diseases and various types of cancer.(7, 11-14) Zoeger et al. reported that the circulating proteasomes in patients with autoimmune diseases are functionally and structurally different from those seen in erythrocytes, which are the dominant circulating cells, supporting the concept that the circulating proteasomes are generated from pathologic processes in the body.(7) Additional evidence supporting this concept includes work by Jacob et al. that correlated circulating proteasome levels with outcome in multiple myeloma.(11) We have also reported that proteasome activity correlates with clinical behavior in patients with chronic lymphocytic leukemia (CLL), acute myeloid leukaemia (AML), and myelodysplastic syndrome (MDS). (9.10)
Here we studied the levels of circulating proteasome and ubiquitin as well as their enzymatic activities in patients with AML, acute lymphoblastic leukemia (ALL), and MDS in an effort to build a profile for the UPS in these diseases. We also correlated these profiles with clinical outcome and various clinical and laboratory data. In addition, as a part of this profiling, we developed a method to determine the enzymatic activity of the proteasome in plasma to distinguish between tumor-load driven values and actual enzymatic activities of each proteasome in each disease. We demonstrate that creating this profile is helpful as a panel of biomarkers in predicting clinical behavior and stratifying patients.
Methods
Patients and Samples
All samples from patients and healthy volunteers were collected under an internal review board-approved protocol with written informed consent. Patient samples were collected during the period 2001 thru 2003 without selection prior to initiating therapy at MD Anderson Cancer Center (MD Anderson). All patients were newly diagnosed, but the majority were referred after diagnosis by their local physician within a few days of their diagnosis. Diagnosis of AML and advanced MDS was made at MD Anderson based on blood counts, flow cytometry, and molecular studies performed on peripheral blood and bone marrow samples. Plasma was separated from EDTA peripheral blood tubes by centrifuging at 1500 × g for 10 minutes at 4°C. Plasma samples obtained from apparently healthy volunteers were used as controls for each study. Plasma samples were stored at −70°C .until analysis Both AML and MDS patients were treated at MD Anderson with standard therapy based on idarubicine+ ara-C with minor variations (± topotecan or fludarabine). All patients with MDS had advanced disease and were candidates for chemotherapy. Advanced MDS disease is defined by the presence of severe anemia (hemoglobin <8 g/dL), thrombocytopenia (<50 ×109/L platelet count), or >10% blasts.
Measurement of Proteasome Protein Levels
For proteasome quantitation in human plasma, a sandwich immunoassay based on electro-chemiluminescence technology (MSD, Gaithersburg, Maryland) was used. Monoclonal antibody (MCP20, Biomol cat#pw8100, Plymouth, PA) was coated on MSD goat anti-mouse (GAM) plates to capture proteasome alpha subunit. Proteasome standards (range, 0.1-400ng/mL) (Biomol International, Cat# PW8720), controls, and patient plasma samples were all diluted1:20 in MSD lysis buffer and added to each well. After incubation at room temperature (RT) for 2 hours, the plate was washed. After the wash, detection antibody (polyclonal, anti-core subunit, Biomol International, Cat# PW8155-0100) was added to each well and incubated for an hour. After another wash, the Sulfo-tag labeled goat anti-rabbit (GAR) antibody was added to each well and incubated for another hour. After a final wash, The MSD read buffer was added and signal was detected by MSD Sector2400. The proteasome level (ng/mL) was calculated using the proteasome standard curve.
Measurement of Ubiquitin Protein
The level of ubiquitin protein (poly-ubiquitin) in plasma was quantitated by a sandwich immunoassay also using electro-chemiluminescence technology. Briefly, after 2 hours blockage of the MSD GAM plate, an anti-ubiquitin monoclonal antibody (clone FK1, Biomol, cat#PW8805) was coated overnight onto the MSD GAM single spot plate at 4°C on a shaker. Ubiquitin positive control (Cat# 89899, Pierce, Rockford, IL) was used as a calibrator to create a 7 point standard curve using Hel cell lysate. Plasma samples were diluted 1:2 using MSD lysis buffer. Calibrator, reference standards, and plasma samples were added to the plate and incubated for 3 hours on a shaker at room temperature. In this incubation, ubiquitin (poly-ubiquitin) was specifically captured with the anti-polyubiquitin. After washing, Sulfo-tag labeled anti-ubiquitin was added to the plate and incubated for 1 hour. After a final wash, the MSD read buffer was added to the plate and signal was read on the MSD Sector2400. The ubiquitin levels were determined by reading against a standard curve and converting to ng ubiquitin/mL plasma.
Measurement of Proteasmome enzymatic activities
Assays have been previously described. (9,10) Briefly, Ch-L, Cas-L, and Tr-L activities were assayed by continuously monitoring the production of 7-amino-4-methylcoumarin (AMC) from 3 separate fluorogenic peptides. Plasma was first mixed with 10% SDS at room temperature for 15 min to activate the plasma. The reaction wells contained 30 μL assay buffer (0.05% SDS in 25 mM HEPES), 10 μL activated plasma, and 10 μL of the fluorogenic peptide-AMC substrate. To measure the release of free AMC with time, the SpectraMax Gemini EM (Molecular Devices Corporation, Sunnyvale, CA) instrument was used with the following parameters: excitation, 380 nm; emission, 460 nm; read interval, 1min; read length, 30 min at 37 °C. Enzymatic activities were quantitated by generating a standard curve of AMC (range, 0–8 μM). The slope of the AMC standard curve was used as a conversion factor to calculate the 3 enzymatic activities for each sample as pmol AMC/sec/mL plasma.
Determining the specific enzymatic activities of Proteasomes
In order to measure the specific enzymatic activity of the individual proteasome, we divided the level of the enzymatic activity in the sample by the level of proteasome, which are measured in the same quantity of plasma sample. This allowed us to derive three new values as follows: Ch-L specific activity (Ch-L/p) = Ch-L/proteasome level, Cas-L specific activity (Cas-L/P) = Cas-L/proteasome level, and Tr-L specific activity (Tr-L/p) = Tr-L/proteasome level.
Statistical Methods
Clinical and biological characteristics were analyzed for their association with response and survival using log-rank test and multivariate Cox proportional hazards models.(16) Estimates of survival curves from the time of initiating therapy were calculated according to the Kaplan-Meier product-limit method. (15) Univariate and multivariate Cox proportional hazard models were developed; predictive variables with P values of less than 0.10 for the univariate Cox proportional hazards model were included in a multivariate model.
Results
High levels of circulating proteasome and ubiquitin in patients with acute leukaemia
Complete clinical data for AML, MDS, and ALL patients were recorded at the time of diagnosis prior to initiating therapy (Table 1). Patients with advanced MDS were treated with AML therapy. Few AML patients had good cytogenetics [inv16, t(8;21), or t(15;17)] and about one-third had unfavorable cytogenetics (−5, −7, and complex abnormalities); the majority of the AML and MDS patients had intermediate cytogenetics (diploid and other cytogenetics). Most of the MDS patients (70%) had refractory anemia with excess blasts in transformation (RAEB-T) (Table 1) and can be classified as acute leukemia based on the WHO classification. However, since these patients are biologically different from those with AML based on the data presented here (see below), we elected to keep them separate from the AML patients and not include them in any of the survival or response analysis. Due to the low numbers of patients with MDS, no survival analysis on this group of patients was performed.
Table 1. Characteristics of Patients with Acute Myeloid Leukemia (AML), Myelodysplastic Syndrome (MDS), or Acute Lymphoblastic Leukemia (ALL).
| Characteristic | AML, n = 147 | MDS, n = 27 | ALL, n= 34 |
|---|---|---|---|
| Median age, years (range) |
64 (17-84) | 63 (24-75) | 37 (18-78) |
| Performance Status | |||
| 0-1 | 112 | 1 | 28 |
| 2-4 | 35 | 26 | 6 |
| Cytogenetics, | |||
| Favorable | 8 | 0 | |
| Unfavorable | 51 | 11 | |
| Intermediate | 88 | 16 | |
| Hyperdiploid | 2 | ||
| Hypodiploid | 2 | ||
| Ph+ | 3 | ||
| Other | 27 | ||
| Median white blood cell count (range) ×109/L |
3.8 (0.4-161.0) | 2.6 (0.8-23.9) | 7.8 (0.9-74) |
| Median Hemoglobin, g/dL (range) |
7.8 (3.4-13.1) | 7.4 (2.2-11.0) | 8 (4.0-15) |
| Median Platelets ×109/L (range) |
54 (6-377) | 36 (10-270) | 84 (11-485) |
| LDH (U/L) | 772 (289- 11064) |
474 (254- 2322) |
1020 (352- 15113) |
| FAB classification | |||
| M0-2 | 78 | ||
| M3 | 2 | ||
| M4-5 | 38 | ||
| M6/M7 | 5 | ||
| RARS | 1 | ||
| RAEB | 5 | ||
| RAEB-T | 19 | ||
| CMML | 1 | ||
| L1-L2 | 27 | ||
| L3 | 7 |
Abbreviations: RARS, refractory anemia with ring sideroblasts; RAEB, refractory anemia with excess blasts; RAEB-T, refractory anemia with excess blasts in transformation; CMML, chronic myelomonocytic leukemia.
The ALL patients were adult with median age of 37 and included 7 patients classified as having Burkitt-type ALL. Only 3 of these patients were positive for Philadelphia chromosome. These patients were treated with Hyper-CVAD (cyclophosphamide, vincristine, adriamycin, and dexamethasone).
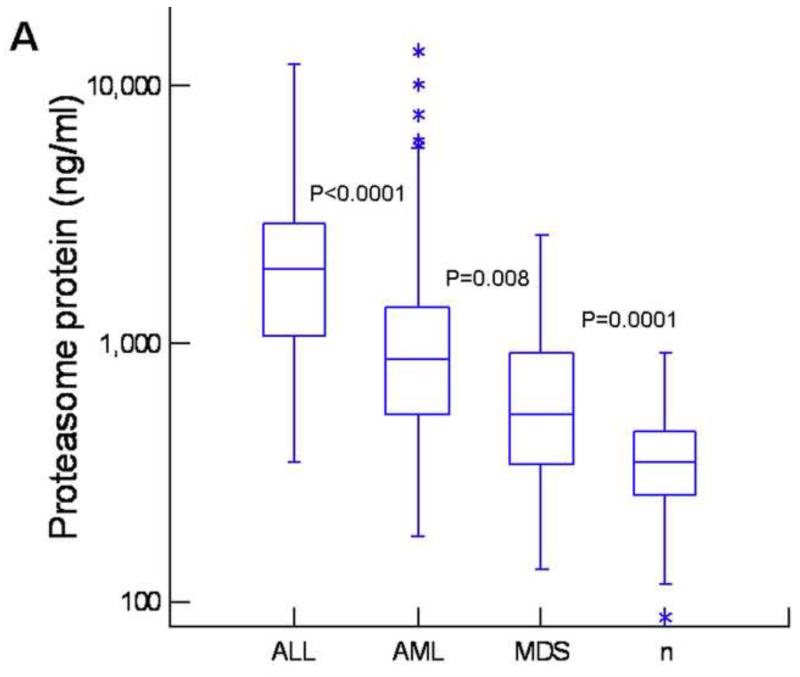
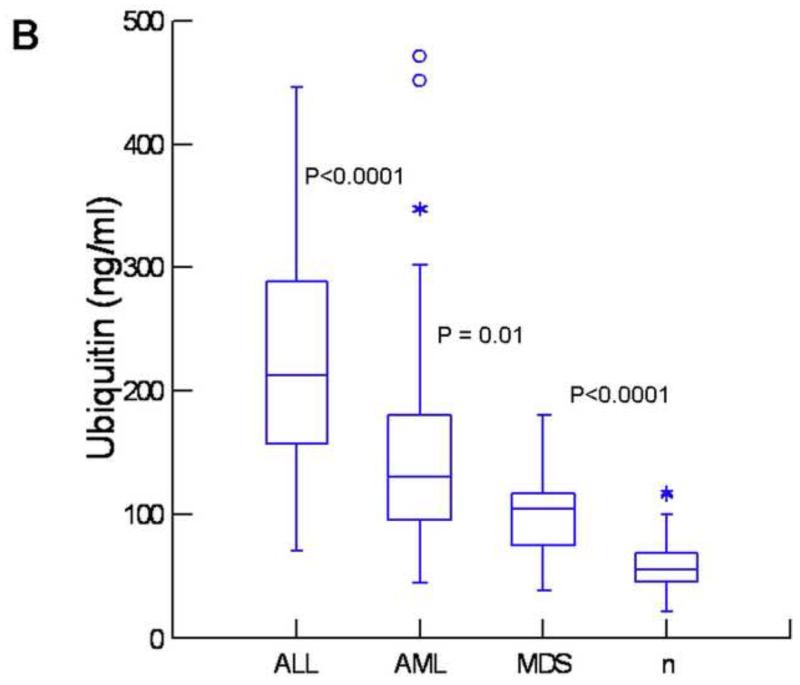
Frozen plasma samples from these patients as well as from healthy controls were analyzed for the levels of ubiquitin and proteasome using our newly developed chemiluminescent immunoassays, which have higher sensitivity than ELISA tests. (17) For the quantitation of ubiquitin, we used anti-polyubiquitin antibodies, therefore, only polyubiquitin was measured in our assay. Both assays were sensitive (100 pg/mL for proteasome and 2 ng/mL for ubiquitin), highly accurate (<15% recovery for both), and highly reproducible (<15% CV, inter-assay). As shown in Figure 1A, patients with leukemia or MDS had significantly higher levels of proteasome as compared with healthy controls (P<0.0001). Patients with ALL had the highest levels, significantly higher than in AML and MDS. A similar pattern was observed with ubiquitin (Fig1B) with highest levels in patients with ALL and lowest level in MDS. These patterns were somewhat different from the results of proteasome enzyme activities. While activities of Ch-L, Tr-L, and Cas-L were all elevated in AML, MDS, and ALL (P<0.0001) as compared with healthy controls (Fig 1,C, D, E), Ch-L activity was not significantly different between AML, MDS, and ALL. Tr-L activity was significantly higher in ALL as compared with AML and MDS. In contrast, Cas-L activity was not different between AML and ALL, but significantly higher in AML and ALL than in MDS (Fig1D).
Figure 1. High levels of proteasome and ubiquitin in patients with leukemia and myelodysplastic syndrome.
Box plots showing levels of proteasome (A), ubiquitin (B), Ch-L activity (C), cas-L activity (D), and Tr-L activity (E) in acute lymphoblastic leukemia (ALL), acute myeloid leukemia (AML), myelodysplastic syndrome (MDS), and healthy controls (n). The P-values between adjacent groups is shown. Asterisks and circles indicate outliers and extreme values, respectively.
To explore this possibility further, we normalized the levels of activities to the levels of proteasome protein by dividing the proteasome enzymatic activities by the protein concentration. These levels were then compared between AML, ALL, and MDS and the healthy control group (Fig 2). The circulating proteasomes had Ch-L specific activity (Ch-L/p) that was significantly lower in ALL and AML than in healthy controls, whereas the proteasomes in MDS patients had similar specific activity to those in healthy controls (Fig 2A). Furthermore, proteasomes in patients with ALL had significantly lower levels of Ch-L/p than in patients with AML (Fig 2A). In contrast, Cas-L specific activity (Cas-L/P) was similar in AML and healthy controls, but was significantly higher in patients with MDS than controls and AML (Fig 2B). Patients with ALL had significantly lower levels of Cas-L/p than healthy controls or AML (Fig 2B). The circulating proteasomes had Tr-L specific activity that varied between diseases (Fig 2C). ALL had the lowest levels of Tr-L/p, significantly lower than AML and healthy controls. Patients with MDS had significantly higher levels of Tr-L/p than healthy controls and AML. There was no significant difference in Tr-L/p levels between AML patients and healthy controls (Fig 2C).
Figure 2. Relatively low enzymatic activity of proteasomes in acute leukemias despite high number of proteasomes.
Box plots showing levels of proteasome activities after normalization to the levels of proteasome protein. Normalized Ch-L (Ch-L/p), normalized Cas-L (Cas-L/p), and normalized Tr-L (Tr-L/p) are shown in panels A, B, and C, respectively. Abbreviations: ALL, acute lymphoblastic leukemia; AML, acute myeloid leukemia; MDS, myelodysplastic syndrome; and n, healthy control.The P-values between adjacent groups is shown. Asterisks and circles indicate outliers and extreme values, respectively.
This data suggest that the specific activities of each proteasome are in general lower in leukemia than in normal cells, but more proteasomes are present in leukemic cells leading to more net enzymatic activity in leukemic cells.
Clinical and laboratory correlations
Correlation data is tabulated in Table 2 for all three diseases. Positive correlation was seen between ubiquitin and proteasome levels, especially in patients with ALL, and also ubiquitin and WBC count in AML and ALL. Proteasome, but not ubiquitin levels correlated with WBC in MDS. LDH correlated with proteasome levels in AML and especially ALL, but not MDS. LDH correlated with ubiquitin only in ALL. In MDS, proteasome levels correlated with blast count in both peripheral blood and bone marrow.
Table 2. Spearman correlations between proteasome and ubiquitin with various clinical and laboratory data.
| AML | MDS | ALL | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||||||||
| Protea somes |
Ubiqui tin |
Ch-L/P | Tr-L/P | Cas-L/P | Proteas omes |
Ubiq uitin |
Ch- L/P |
Tr- L/P |
Cas- L/P |
Protea somes |
Ubiq uitin |
Ch- L/P |
Tr- L/P |
Cas- L/P |
|
| Age | −0.03 | −0.07 | −0.04 | 0.02 | −0.03 | −0.11 | −0.16 | 0.00 | 0.02 | 0.07 | −0.08 | −0.27 | 0.19 | −0.09 | 0.17 |
| B2M | 0.31 | 0.23 | −0.13 | −0.28 | −0.03 | 0.32 | −0.07 | −0.06 | −0.23 | −0.10 | 0.54 | 0.37 | −0.29 | −0.45 | −0.31 |
| WBC | 0.36 | 0.46 | −0.18 | −0.22 | −0.02 | 0.40 | 0.15 | −0.32 | −0.35 | −0.29 | 0.46 | 0.41 | −0.61 | −0.21 | −0.55 |
| Blood blasts (%) |
0.30 | 0.25 | −0.16 | −0.19 | 0.01 | 0.38 | −0.07 | −0.39 | −0.41 | −0.35 | 0.62 | 0.48 | −0.57 | −0.47 | −0.68 |
| Platelets | 0.05 | 0.07 | −0.16 | 0.10 | −0.09 | −0.02 | 0.09 | 0.01 | 0.14 | −0.14 | −0.23 | −0.28 | −0.02 | 0.29 | 0.24 |
| HGB | −0.06 | 0.08 | −0.05 | 0.07 | −0.10 | −0.32 | −0.22 | −0.01 | 0.15 | 0.01 | −0.22 | −0.19 | 0.00 | 0.35 | 0.08 |
| Marrow Blasts (%) |
0.07 | 0.26 | 0.01 | −0.06 | 0.05 | 0.51 | 0.20 | −0.12 | −0.43 | −0.22 | −0.11 | 0.03 | −0.09 | 0.00 | 0.19 |
| BUN | 0.06 | 0.09 | 0.03 | −0.14 | 0.05 | −0.09 | −0.22 | −0.05 | −0.04 | −0.05 | −0.11 | −0.03 | 0.09 | −0.05 | −0.07 |
| Creatinine | 0.10 | 0.06 | −0.12 | −0.21 | 0.01 | −0.08 | 0.00 | −0.10 | −0.17 | −0.20 | −0.10 | −0.07 | 0.15 | −0.04 | −0.10 |
| LDH | 0.56 | 0.32 | −0.17 | −0.45 | −0.04 | 0.31 | 0.11 | −0.01 | −0.13 | −0.15 | 0.74 | 0.69 | −0.52 | −0.58 | −0.57 |
| Ch-L | 0.80 | 0.37 | 0.05 | −0.55 | 0.06 | 0.76 | 0.64 | 0.12 | −0.54 | −0.13 | 0.47 | 0.34 | 0.12 | −0.38 | −0.02 |
| Tr-L | 0.20 | 0.15 | −0.07 | 0.47 | −0.05 | −0.20 | 0.06 | 0.36 | 0.71 | 0.46 | 0.01 | 0.15 | 0.02 | 0.54 | −0.05 |
| Cas-L | 0.80 | 0.45 | −0.07 | −0.56 | 0.19 | 0.73 | 0.54 | 0.06 | −0.39 | −0.03 | 0.44 | 0.25 | −0.07 | −0.44 | 0.22 |
| Proteasome | 0.42 | −0.49 | −0.72 | −0.37 | 0.43 | −0.48 | −0.77 | −0.64 | 0.77 | −0.75 | −0.80 | −0.74 | |||
| Ubiquitin | 0.42 | −0.16 | −0.29 | 0.00 | 0.43 | 0.23 | −0.20 | −0.07 | 0.77 | −0.50 | −0.47 | −0.65 | |||
| Ch-L/P | −0.49 | −0.16 | 0.39 | 0.75 | −0.48 | 0.23 | 0.56 | 0.83 | −0.75 | −0.50 | 0.61 | 0.75 | |||
| Tr-L/P | −0.72 | −0.29 | 0.39 | 0.32 | −0.77 | −0.20 | 0.56 | 0.72 | −0.80 | −0.47 | 0.61 | 0.52 | |||
| Cas-L/P | −0.37 | 0.00 | 0.75 | 0.32 | −0.64 | −0.07 | 0.83 | 0.72 | −0.74 | −0.65 | 0.75 | 0.52 | |||
We were unable to demonstrate a difference in proteasome or ubiquitin levels between the three cytogenetic groups (unfavorable, intermediate or favorable) in the AML group using Kruskal-Wallis ANOVA statistic. However, there was significant correlation between higher levels of proteasome and ubiquitin and poor performance status (P=0.03 and P=0.02, respectively).
Predicting Clinical behaviour
We explored the ability of proteasome protein levels or its derived (normalized) enzymatic activities as well as ubiquitin in predicting response to therapy, relapse, and survival. Proteasome level as a continuous variable predicted response to therapy in AML (P=0.04), but not in ALL (data not shown). Ubiquitin levels did not predict response in AML nor ALL. Using the median proteasome level (875 ng/mL) as a cut-off, AML patients with proteasome levels less than median had significantly better response rate than those with higher levels (P=0.01). As for survival, Table 3 lists the statistically significant predictors of survival in AML and ALL. Patients with MDS were too few for survival analysis. The correlation with survival for the enzymatic activities in AML has previously been published(10); therefore, we focused here on proteasome protein level and the normalized values of its enzymatic activities.
Table 3. Cox regression model for predicting survival and complete remission duration (CRD) in Patients with Acute Myeloid Leukemia and Acute lymphoblastic leukemia.
| Beta | Standard Error |
t-value | Wald Statistic |
P-value | |
|---|---|---|---|---|---|
| Univariate Survival as predicted by proteasome protein in AML |
|||||
| In all patients | 0.0003 | 0.0001 | 4.4383 | 19.6988 | <0.00001 |
| In Intermediate cytogenetic group | 0.000190 | 0.000086 | 2.222028 | 4.937406 | 0.026 |
| In poor Cytogenetic group | 0.000298 | 0.000087 | 3.418611 | 11.68690 | 0.0006 |
|
| |||||
| Multivariate in AML patients | |||||
| Proteasome | 0.0002 | 0.0001 | 3.0434 | 9.2622 | 0.002 |
| Cytogenetic grouping | 0.7860 | 0.1859 | 4.2286 | 17.8812 | <0.00001 |
| Age grouping (<70 vs >70) | 0.6489 | 0.1986 | 3.2673 | 10.6751 | 0.001 |
| Performance status (<2 vs >2) | 0.6099 | 0.2130 | 2.8631 | 8.1974 | 0.004 |
|
| |||||
| CRD in AML | |||||
| Ch-L/P | 0.2956 | 0.1678 | 1.7619 | 3.1041 | 0.08 |
| Cas-L/P | 0.1460 | 0.0821 | 1.7774 | 3.1591 | 0.08 |
|
| |||||
| Univariate, survival in ALL | |||||
| Proteasome | 0.00001 | 0.0000 | 1.6713 | 2.7934 | 0.09 |
| Ch-L/P | −1.3914 | 0.6323 | −2.2005 | 4.8422 | 0.03 |
| Cas-L/P | −0.7028 | 0.3980 | −1.7661 | 3.1191 | 0.08 |
|
| |||||
| Univariate, CRD in ALL | |||||
| Ubiquitin | 0.0051 | 0.0027 | 1.9299 | 3.7247 | 0.05 |
| Ch-L/P | −1.1975 | 0.6623 | −1.8083 | 3.2699 | 0.07 |
| Cas-L/P | −1.8295 | 0.7336 | −2.4939 | 6.2195 | 0.01 |
As shown in Table 3, using Cox proportional hazard model, proteasome, but not ubiquitin levels, were strong predictors of survival when used as continuous variable (P<0.0001) (Table 3). Even when we considered AML patients in the unfavorable cytogenetic group (n = 51), proteasome levels were strong predictors of survival in this group of patients (P=0.0006). The proteasome level was also predictive of survival in the intermediate cytogenetic group (P=0.03) (Table 3). In addition to proteasome levels, multivariate analysis was performed incorporating the major factors that are known to be predictors of survival in AML including cytogenetic grouping, age grouping, and performance status. Proteasome levels remained a strong predictor of survival independent of all other factors (P=0.002) (Table 3). Furthermore, when the median was used as a cut-off, patients with levels higher than the median had significantly shorter survival (P=0.04) (Fig 3). Restricting the analysis to only patients with unfavorable cytogenetics also showed that patients with proteasome levels >875 ng/mL had significantly shorter survival (P=0.04) (Fig 4).
Figure 3. Kaplan-Meier estimates of patient survival grouped by plasma proteasome protein levels in all patients with acute myeloid leukemia.
Patients with proteasome levels higher than the median 875 ng/mL show significantly shorter survival (P=0.04) N:E indicates the total number of patients and the number of patients with an event (death).
Figure 4. Kaplan-Meier estimates of patient survival grouped by plasma proteasome protein levels in patients with acute myeloid leukemia and unfavorable cytogenetic abnormalities.
Patients with proteasome levels higher than 875 ng/ml show significantly shorter survival. N:E indicates the total number of patients and the number of patients with an event (death).
None of the normalized enzymatic activities (Ch-L/p, Cas-L/p, Tr-L/p) were statistically adequate for the prediction of survival in AML, which is significantly different from the predictive value of absolute enzymatic activities that we have previously reported.(10) However, Ch-L/p and Cas-L/p showed a marginal correlation with survival in AML (P=0.08 for both) (Table 3).
As for patients with ALL, proteasome levels, ubiquitin levels, and proteasome activities were not predictors of survival. However, there was a significant negative correlation between Ch-L/p as a continuous variable and survival (P=0.03) (Table 3). The Cas-L/p specific activity showed a slight, but not significant, negative correlation with survival in ALL (P=0.08). When we used the median as cut-off point, ALL patients with Ch-L/p level < 0.74 (pMol AMC/sec/pg proteasome) had significant longer survival (P=0.0015) (Fig 5). Using the median for Cas-L/p, we also found a negative correlation with survival (P=0.03) in ALL. Patients with lower than the median Cas-L/p (0.88 pMol AMC/sec/pg proteasome) had significantly longer survival. Complete remission duration (CRD) in AML did not correlate with any of the proteasome or ubiquitin parameters, but in ALL, ubiquitin levels as continuous variable correlated significantly with CRD (P=0.05) (Table 3). Unlike with survival, Cas-L/p correlated negatively better than Ch-L/p with CRD (P=0.01 vs. P=0.07) as continuous variables (Table 3).
Figure 5. Kaplan-Meier estimates of patient survival grouped by plasma normalized Ch-L activity (Ch-L/p) levels in patients with acute lymphoblastic leukemia.
Patients with Ch-L/p level higher than the median 0.88 pMol AMC/Sec/pg proteasome show significantly better survival.
Discussion
The UPS plays a major role in the most important processes that control cell homeostasis in normal and neoplastic states. Therefore, it is not surprising that this system is variable dependent on the biology of specific cancer cells in a specific patient. Analyzing the various aspects of the UPS in tumor cells may provide a profile that can be used for classifying and stratifying cancer patients for diagnosis, therapy, and prediction of clinical behavior.
It is frequently difficult to have access to the cancer cells even in hematologic diseases. This difficulty is compounded by the problem of dilution effects by normal cells. The advantages of using plasma is that plasma, at least in hematologic diseases, reflects the tumor rather than the normal cells in its content of cellular proteins and nucleic acids.(18-25) By studying the levels of proteasome, ubiquitin, and proteasome enzymatic activities in the plasma, the data suggest that we are most likely studying the UPS profile in the leukemic blasts. In our previous paper, we have demonstrated that the plasma proteasome measurement was specific for proteasomes and only proteasome inhibitors can inhibit the detected activities in plasma in a fashion identical to that seen in cells.(9) We also showed that this inhibition is dose dependent in the plasma. (9) More importantly, we cannot rule out the possibility that the plasma proteasomes may have different enzymatic activities as compared with cellular due to the presence of inhibitors, which has been reported in alveolar fluid.(26) Sixt et al reported that proteasomal activity and albumin degradation rate in the alveolar fluid from patients with acute respiratory distress syndrome (ARDS) were approximately 17-fold lower than in healthy subjects.(26) In addition, the proteasomal activity in normal alveolar fluid was inhibited by fluid from patients with ARD.(26) Multiple mechanisms have been reported for the inhibition of the enzymatic activities of proteasomes including the PA28 protein and the S19 capping complex.(27) The possibility that these inhibitors are present in the plasma and not in the cytoplasm of cells cannot be ruled out. However, our preliminary data from analyzing cell lysates from bone marrow from patients with various types of leukemias suggests a pattern similar to that seen in plasma with few exceptions (data not shown).
Our data show that levels of proteasomes and ubiquitin increase significantly in the plasma of patients with acute leukemias. This leads to increased proteasome enzyme activity in the plasma. However, this increase is most likely driven by the increase in proteasome due to the tumor (leukemia) load. The data also suggest that leukemic cells have higher number of proteasomes, but the specific enzymatic activities are, in general, lower than the specific activity of mature hematopoietic cells. More importantly, lymphoblasts have a different UPS profile than that of AML and both have a different profile from MDS. Our measurements of normalized enzymatic activities are expressed as activities per pg of plasma proteasome. Therefore, we are not exactly measuring activities per proteasome molecule, but rather per specific quantity of proteasomes. Most of our patients with MDS had RAEB-T and proteasome profiling showed that these patients are different from AML patients.
In addition, correlation with clinical behavior is different depending on the disease. Clearly proteasome levels and their activity is major player in AML disease. In contrast, the relative proteasome activity, especially Ch-L and Cas-L, play a major role in the clinical behavior of ALL.
The ability of proteasome to predict survival within the intermediate cytogenetic group, as well as within the unfavorable cytogenetic group, has particular clinical importance. These patient groups are difficult to manage, and profiling proteasome protein activity using plasma may be useful. In addition, testing for Cas-L/p may be useful to identify patients with ALL who will have short remission duration.
Although we studied here only acute leukemia, similar data from chronic leukemia also demonstrate the importance of profiling UPS in plasma to predict clinical behavior (unpublished data). All this suggests that this profiling might be important in other types of cancer and raise the possibility of stratifying patients with solid tumors, since high proteasome levels have been reported in various solid tumors. The concept of proteasome profiling is new and should be explored further.
Acknowledgements
All authors have seen and approved the manuscript, and declare there are no conflicts of interest or financial interests.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Authors’ Contributions
W.M. developed assays, analyzed data, performed assays, helped writing paper; H.K., Z.E. and S.O’B. developed concept, interpreted data, provided materials; X.Z. and X.W. performed testing and M.A. developed the concept, designed the study, analyzed data, interpreted data and approved paper.
Conflict of Interest
All authors declare there are no conflicts of interest or financial interests.
References
- 1.Reinstein E, Ciechanover A. Narrative review: protein degradation and human diseases: the ubiquitin connection. Ann Intern Med. 2006;145:676–84. doi: 10.7326/0003-4819-145-9-200611070-00010. [DOI] [PubMed] [Google Scholar]
- 2.Maki CG, Huibregtse JM, Howley PM. In vivo ubiquitination and proteasome-mediated degradation of p53. Cancer Res. 1996;56:2649–54. [PubMed] [Google Scholar]
- 3.Coux O, Tanaka K, Goldberg AL. Structure and functions of the 20S and 26S proteasomes. Annu Rev Biochem. 1996;65:801–47. doi: 10.1146/annurev.bi.65.070196.004101. [DOI] [PubMed] [Google Scholar]
- 4.Hershko A, Ciechanover A. The ubiquitin system for protein degradation. Annu Rev Biochem. 1992;61:761–807. doi: 10.1146/annurev.bi.61.070192.003553. [DOI] [PubMed] [Google Scholar]
- 5.Ciechanover A. Intracellular protein degradation: from a vague idea thru the lysosome and the ubiquitin-proteasome system and onto human diseases and drug targeting. Cell Death Differ. 2005;12:1178–90. doi: 10.1038/sj.cdd.4401692. [DOI] [PubMed] [Google Scholar]
- 6.Goldberg AL. Functions of the proteasome: from protein degradation and immune surveillance to cancer therapy. Biochem Soc Trans. 2007;35:12–7. doi: 10.1042/BST0350012. [DOI] [PubMed] [Google Scholar]
- 7.Zoeger A, Blau M, Egerer K, Feist E, Dahlmann B. Circulating proteasomes are functional and have a subtype pattern distinct from 20S proteasomes in major blood cells. Clin Chem. 2006;52:2079–86. doi: 10.1373/clinchem.2006.072496. [DOI] [PubMed] [Google Scholar]
- 8.Drews O, Wildgruber R, Zong C, Sukop U, Nissum M, Weber G, Gomes AV, Ping P. Mammalian proteasome subpopulations with distinct molecular compositions and proteolytic activities. Mol Cell Proteomics. 2007;6:2021–31. doi: 10.1074/mcp.M700187-MCP200. [DOI] [PubMed] [Google Scholar]
- 9.Ma W, Kantarjian H, O’Brien S, Jilani I, Zhang X, Estrov Z, Ferrajoli A, et al. Enzymatic activity of circulating proteasomes correlates with clinical behavior in patients with chronic lymphocytic leukemia. Cancer. 2008;112:1306–12. doi: 10.1002/cncr.23301. [DOI] [PubMed] [Google Scholar]
- 10.Ma W, Kantarjian H, Bekele B, Donahue A, Zhang X, Zhang Z, O’Brien S, Estey E, Estrov Z, Cortes J, Keating M, Giles F, Albitar M. Proteasome enzymatic activities in plasma as risk stratification of patients with acute myeloid leukemia and advanced-stage myelodysplastic syndrome. Clinical Cancer research. 2009;15:3820–6. doi: 10.1158/1078-0432.CCR-08-3034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jakob C, Egerer K, Liebisch P, Türkmen S, Zavrski I, Kuckelkorn U, Heider U, Kaiser M, Fleissner C, Sterz J, Kleeberg L, Feist E, Burmester GR, Kloetzel PM, Sezer O. Circulating proteasome levels are an independent prognostic factor for survival in multiple myeloma. Blood. 2007;109:2100–5. doi: 10.1182/blood-2006-04-016360. [DOI] [PubMed] [Google Scholar]
- 12.Henry L, Lavabre-Bertrand T, Vercambre L, Ramos J, Carillo S, Guiraud I, Pouderoux P, Bismuth M, Demattei C, Duny Y, Chaze I, Funakoshi N, Bureau JP, Daures JP, Blanc P. Plasma proteasome level is a reliable early marker of malignant transformation of liver cirrhosis. Gut. 2009;58(6):833–8. doi: 10.1136/gut.2008.157016. [DOI] [PubMed] [Google Scholar]
- 13.Stoebner PE, Lavabre-Bertrand T, Henry L, Guiraud I, Carillo S, Dandurand M, Joujoux JM, Bureau JP, Meunier L. High plasma proteasome levels are detected in patients with metastatic malignant melanoma. Br J Dermatol. 2005;152:948–53. doi: 10.1111/j.1365-2133.2005.06487.x. [DOI] [PubMed] [Google Scholar]
- 14.Lavabre-Bertrand T, Henry L, Carillo S, Guiraud I, Ouali A, Dutaud D, Aubry L, Rossi JF, Bureau JP. Plasma proteasome level is a potential marker in patients with solid tumors and hemopoietic malignancies. Cancer. 2001;92:2493–500. doi: 10.1002/1097-0142(20011115)92:10<2493::aid-cncr1599>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- 15.Kaplan EL, Meier P. Nonparametric estimation from incomplete observations. J. American Statistical Association. 1958;53:457–481. [Google Scholar]
- 16.Cox DR. Regression models and life tables. J Royal Stat Soc. 1972;34:187–220. [Google Scholar]
- 17.Yeh CH, Tseng R, Hannah A, Estrov Z, Estey E, Kantarjian H, Albitar M. Clinical correlation of circulating heat shock protein 70 in acute leukemia. Leuk Res. 2010;34:605–9. doi: 10.1016/j.leukres.2009.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Giles FJ, Albitar M. Plasma-based testing as a new paradigm for clinical testing in hematologic diseases. Expert Rev Mol Diagn. 2007;7:615–23. doi: 10.1586/14737159.7.5.615. [DOI] [PubMed] [Google Scholar]
- 19.Rogers A, Joe Y, Manshouri T, Dey A, Jilani I, Giles F, Estey E, Freireich E, Keating M, Kantarjian H, Albitar M. Relative increase in leukemia-specific DNA in peripheral blood plasma from patients with acute myeloid leukemia and myelodysplasia. Blood. 2004;103:801–818. doi: 10.1182/blood-2003-06-1840. [DOI] [PubMed] [Google Scholar]
- 20.Manshouri T, Do KA, Wang X, Giles FJ, O’Brien SM, Saffer H, Thomas D, Jilani I, Kantarjian HM, Keating MJ, Albitar M. Circulating CD20 is detectable in the plasma of patients with chronic lymphocytic leukemia and is of prognostic significance. Blood. 2003;101:2507–13. doi: 10.1182/blood-2002-06-1639. [DOI] [PubMed] [Google Scholar]
- 21.Ahmed M, Giles F, Joe Y, Weber DM, Jilani I, Manshouri T, Giralt S, De Lima M, Keating M, Albitar M. Use of plasma DNA in detection of loss of heterozygosity in patients with multiple myeloma. Eur J Haematol. 2003;71:174–8. doi: 10.1034/j.1600-0609.2003.00125.x. [DOI] [PubMed] [Google Scholar]
- 22.Jilani I, Estey E, Manshuri T, Caligiuri M, Keating M, Giles F, Thomas D, Kantarjian H, Albitar M. Better detection of FLT3 internal tandem duplication using peripheral blood plasma DNA. Leukemia. 2003;17:114–9. doi: 10.1038/sj.leu.2402743. [DOI] [PubMed] [Google Scholar]
- 23.Ma W, Kantarjian H, Jilani I, Gorre M, Bhalla K, Ottmann O, Giles F, Albitar M. Heterogeneity in detecting Abl kinase mutations and better sensitivity using circulating plasma RNA. Leukemia. 2006;20:1989–91. doi: 10.1038/sj.leu.2404355. [DOI] [PubMed] [Google Scholar]
- 24.Ma W, Kantarjian H, Zhang X, Sun W, Buller AM, Jilani I, Schwartz JG, Giles F, Albitar M. Higher detection rate of JAK2 mutation using plasma. Blood. 2008;111:3906–7. doi: 10.1182/blood-2008-02-139188. [DOI] [PubMed] [Google Scholar]
- 25.Jilani I, Kantarjian H, Faraji H, Gorre M, Cortes J, Ottmann O, Bhalla K, O’Brien S, Giles F, Albitar M. An immunological method for the detection of BCR-ABL fusion protein and monitoring its activation. Leuk Res. 2008;32:936–43. doi: 10.1016/j.leukres.2007.11.023. [DOI] [PubMed] [Google Scholar]
- 26.Sixt SU, Adamzik M, Spyrka D, Saul B, Hakenbeck J, Wohlschlaeger J, Costabel U, Kloss A, Giesebrecht J, Dahlmann B, Peters J. Alveolar extracellular 20S proteasome in patients with acute respiratory distress syndrome. Am J Respir Crit Care Med. 2009;179:1098–106. doi: 10.1164/rccm.200802-199OC. [DOI] [PubMed] [Google Scholar]
- 27.Shibatani T, Carlson EJ, Larabee F, McCormack AL, Früh K, Skach WR. Global organization and function of mammalian cytosolic proteasome pools: Implications for PA28 and 19S regulatory complexes. Mol Biol Cell. 2006;17:4962–71. doi: 10.1091/mbc.E06-04-0311. [DOI] [PMC free article] [PubMed] [Google Scholar]