Abstract
AIM: To investigate the role of Na+/K+/2Cl- cotransporter 1 (NKCC1) in the regulation of genes involved in cell cycle progression and the clinicopathological significance of its expression in esophageal squamous cell carcinoma (ESCC).
METHODS: An immunohistochemical analysis was performed on 68 primary tumor samples obtained from ESCC patients that underwent esophagectomy. NKCC1 expression in human ESCC cell lines was analyzed by Western blotting. Knockdown experiments were conducted using NKCC1 small interfering RNA, and the effects on cell cycle progression were analyzed. The gene expression profiles of cells were analyzed by microarray analysis.
RESULTS: Immunohistochemical staining showed that NKCC1 was primarily found in the cytoplasm of carcinoma cells and that its expression was related to the histological degree of differentiation of SCC. NKCC1 was highly expressed in KYSE170 cells. Depletion of NKCC1 in these cells inhibited cell proliferation via G2/M phase arrest. Microarray analysis identified 2527 genes with altered expression levels in NKCC1depleted KYSE170. Pathway analysis showed that the top-ranked canonical pathway was the G2/M DNA damage checkpoint regulation pathway, which involves MAD2L1, DTL, BLM, CDC20, BRCA1, and E2F5.
CONCLUSION: These results suggest that the expression of NKCC1 in ESCC may affect the G2/M checkpoint and may be related to the degree of histological differentiation of SCCs. We have provided a deeper understanding of the role of NKCC1 as a mediator and/or a biomarker in ESCC.
Keywords: Na+/K+/2Cl- cotransporter 1, Esophageal cancer, Cell cycle
Core tip: The objectives of the present study were to investigate the role of Na+/K+/2Cl- cotransporter 1 (NKCC1) in the regulation of genes involved in cell cycle progression and the clinicopathological significance of its expression in esophageal squamous cell carcinoma (ESCC). An immunohistochemical analysis revealed that the expression of NKCC1 in ESCC samples was related to the histological type. Microarray results suggested that NKCC1 exhibits marked effects on the expression of genes related to G2/M cell cycle progression. A deeper understanding of the role of NKCC1 may lead to its use as an important biomarker and/or a novel therapeutic target for ESCC treatment.
INTRODUCTION
Several studies have recently shown that ion channels and transporters play important roles in fundamental cellular functions. Their physiological roles in cell proliferation have been studied in more detail because ion transport across the cell membrane is involved in the regulation of cell volume, which is indispensable for cell cycle progression. Several reports have demonstrated the important roles of Cl- channels/transporters, such as Ca2+- activated 2Cl- channels and Cl-/HCO3- exchangers, in gastrointestinal cancer cells[1,2]. These studies indicated that transepithelial Cl- transport plays an important role in the proliferation of gastrointestinal cancer cells.
The Na+/K+/2Cl- cotransporter (NKCC) is a member of the cation-chloride cotransporter family. NKCC transports one sodium ion, one potassium ion, and two chloride ions across the plasma membrane and is sensitive to loop diuretics, such as furosemide and bumetanide. There are two isoforms of NKCC, and NKCC1 is ubiquitously expressed in various types of cells including epithelial cells[3,4]. We previously examined transepithelial Cl- transport in various types of cancer cells[5-7] and showed that NKCC1 plays an important role in the proliferation of gastric and prostate cancer cells[8,9]. However, the role of NKCC1 in the proliferation of esophageal squamous cell carcinoma (ESCC) cells and its detailed regulatory mechanisms have not been fully investigated. Furthermore, the clinicopathological meaning of NKCC1 expression in ESCCs remains uncertain.
The objectives of the present study were to investigate the role of NKCC1 in the regulation of genes involved in cell cycle progression and the clinicopathological significance of its expression in ESCC. We analyzed the expression of NKCC1 in human ESCC samples and determined its relationship with the degree of histological differentiation of SCC samples. Furthermore, microarray analyses showed that depletion of NKCC1 with small interfering RNA (siRNA) changed the expression levels of many genes involved in G2/M cell cycle progression. Our results indicate that NKCC1 plays an important role in the tumor progression of ESCCs.
MATERIALS AND METHODS
Cell lines, antibodies, and other reagents
The human ESCC cell lines TE2, TE5, TE9, and TE13 were obtained from the Cell Resource Center for Biomedical Research at the Institute of Development, Aging, and Cancer (Tohoku University, Sendai, Japan)[10]. The human ESCC cell lines KYSE70 and KYSE170 were obtained from Kyoto University (Kyoto, Japan)[11]. These cells were grown in RPMI-1640 medium (Nacalai Tesque, Kyoto, Japan) supplemented with 100 U/mL of penicillin, 100 μg/mL of streptomycin, and 10% fetal bovine serum. Cells were cultured in flasks or dishes in a humidified incubator at 37 °C under 5% CO2 in air.
The anti-NKCC1 antibody used for immunohistochemical analysis and the protein assay were obtained from Sigma-Aldrich (St. Louis, MO). The anti-Ki-67 antibody was purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Horseradish peroxidase (HRP)-conjugated anti-rabbit secondary antibodies were purchased from Cell Signaling Technology (Beverly, MA), and the antibody for glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was obtained from Santa Cruz Biotechnology. Furosemide was purchased from Nacalai Tesque, Inc. (Kyoto, Japan).
Patients and primary tissue samples
ESCC tumor samples were obtained from 68 patients with a histologically confirmed primary ESCC who underwent esophagectomy at Kyoto Prefectural University of Medicine between 1998 and 2007 and were embedded in paraffin after 12 h of formalin fixation. Patient eligibility criteria were as follows: no synchronous or metachronous cancers (in addition to ESCC) and no preoperative chemotherapy or radiation therapy. We excluded patients with non-curative resected tumors or non-consecutive data. All patients provided written informed consent. Relevant clinicopathological and survival data were obtained from the hospital database. Staging was principally based on the International Union Against Cancer/tumor node metastasis Classification of Malignant Tumors (7th edition)[12].
Immunohistochemistry
Paraffin sections (4 μm thick) of tumor tissues were subjected to immunohistochemical staining for the NKCC1 protein using the avidin-biotin-peroxidase method. Briefly, paraffin sections were dewaxed with xylene and dehydrated with a graded series of alcohols. Antigen retrieval was performed by heating the samples in Dako REAL Target Retrieval Solution (Glostrup, Denmark) for 40 min at 98 °C. Endogenous peroxidases were quenched by incubating the sections for 30 min in 0.3% H2O2. Sections were then treated with protein blocker and incubated overnight at 4 °C with anti-NKCC1 or anti-Ki-67 antibody. The avidin-biotin-peroxidase complex (Vectastain ABC Elite kit; Vector laboratories, Burlingame, CA) was visualized with diaminobenzidine tetrahydrochloride. Sections were counterstained with hematoxylin, dehydrated with a graded series of alcohols, cleared in xylene, and mounted.
Immunohistochemical samples stained with NKCC1 were graded semi-quantitatively by considering both the staining intensity and the percentage of positive tumor cells using an immunoreactive score (IRS)[13]. Staining intensity was scored as 0 (no staining), 1 (weak staining), 2 (moderate staining), or 3 (strong staining). The proportion of positive tumor cells was scored as 1 (1%-10%), 2 (11%-50%), 3 (51%-80%), or 4 (81% or more). Each sample’s score was calculated as the maximum multiplied product of the intensity and proportion scores. Scores of 6 or more and scores of less than 6 were defined as high grade and low grade NKCC1 expression, respectively.
Tumor cells with nuclei containing brown immunoreactive products were considered Ki-67 positive (Figure 1). To evaluate the positive staining rate, the number of Ki-67 labeled cells was quantified in five randomly selected fields at a magnification of × 400. The positive staining rate in each case was calculated as the number of positive cells divided by the total number of examined cells in all examined fields. The mean Ki-67 labeling index was 29.4% (range, 2.9%-55.9%) in 68 primary tumor samples.
Figure 1.
Immunohistochemical staining of a primary tumor sample of human esophageal squamous cell carcinomas with a Ki-67 antibody. The expression of Ki-67 was clearly identified in the nucleus of ESCCs (Magnification × 200).
Western blotting
Cells were harvested in M-PER lysis buffer (Pierce, Rockford, IL) supplemented with protease inhibitors (Pierce, Rockford, IL). The protein concentration was measured with a modified Bradford assay (Bio-Rad, Hercules, CA). Cell lysates containing equal amounts of total protein were separated by SDS-PAGE and then transferred onto PVDF membranes (GE Healthcare, Piscataway, NJ). These membranes were then probed with the indicated antibodies, and proteins were detected using an ECL Plus Western Blotting Detection System (GE Healthcare, Piscataway, NJ).
Small interfering RNA transfection
Cells were transfected with 10 nmol/L NKCC1 Small interfering RNA (siRNA) (Stealth RNAi™ siRNA No.HSS109914; Invitrogen, Carlsbad, CA) using the Lipofectamine RNAiMAX reagent (Invitrogen), according to the manufacturer’s instructions. The medium containing siRNA was replaced with fresh medium after 24 h. The control siRNA provided (Stealth RNAi™ siRNA Negative Control; Invitrogen) was used as a negative control.
Cell cycle analysis
The cell cycle phase was evaluated 48 h after siRNA transfection by fluorescence-activated cell scoring (FACS). Briefly, cells were treated with Triton X-100 and RNase, and nuclei were stained with propidium iodide (PI) prior to DNA content measurement using a Becton Dickinson FACS Calibur instrument (Becton Dickinson, Mountain view, CA). At least 10000 cells were analyzed, and ModFit LT software (Verity Software House, Topsham, ME) was used to analyze cell cycle distribution.
Cell proliferation
Cells were seeded in 6-well plates at a density of 1.0 × 105 cells per well and incubated at 37 °C with 5% CO2. siRNA was transfected 24 h after the cells seeded. Cells were detached from the flasks with trypsin-EDTA 72 h after siRNA transfection and were counted using a hemocytometer.
Real time reverse transcription-polymerase chain reaction
Total RNA was extracted using an RNeasy kit (Qiagen, Valencia, CA). Messenger RNA (mRNA) expression was measured by quantitative real-time PCR (7300 Real-Time PCR System; Applied Biosystems, Foster City, CA) with TaqMan Gene Expression Assays (Applied Biosystems), according to the manufacturer’s instructions. Expression levels were measured for the following genes: NKCC1 (Hs00169032_m1), MAD2L1 (Hs01554513_g1), DTL (Hs00978565_m1), BLM (Mm00476150_m1), CDC20 (Hs00426680_mH), BRCA1 (Hs01556193_m1), and E2F5 (Hs00231092_m1) (Applied Biosystems). Expression was normalized for each gene to the housekeeping gene beta-actin (ACTB, Hs01060665_g1; Applied Biosystems). Assays were performed in triplicate.
Microarray sample preparation and hybridization
Total RNA was extracted using an RNeasy kit (Qiagen). RNA quality was monitored with an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA). Cyanine-3 (Cy3)-labeled cRNA was prepared from 0.1 μg of total RNA using a Low Input Quick Amp Labeling Kit (Agilent), according to the manufacturer’s instructions. Samples were purified using RNeasy columns (Qiagen). A total of 0.60 μg of Cy3-labelled cRNA was fragmented and hybridized to an Agilent SurePrint G3 Human Gene Expression 8 × 60K Microarray for 17 h. Slides were washed and scanned immediately on an Agilent DNA Microarray Scanner (G2565CA) using the one color scan setting for 8 × 60K array slides.
Processing of microarray data
Scanned images were analyzed with Feature Extraction Software 10.10 (Agilent) using default parameters to obtain background-subtracted and spatially detrended Processed Signal intensities. Signal transduction networks were analyzed with Ingenuity Pathway Analysis (IPA) software (Ingenuity Systems, Inc., Redwood City, CA).
Statistical analysis
Fisher’s exact test was used to evaluate the differences between proportions, and Student’s t tests (for comparisons between two groups) and Tukey-Kramer HSD tests (for multiple comparisons) were used to evaluate continuous variables. Survival curves were constructed by the Kaplan-Meier method, and differences in survival were examined using the log-rank test. Differences were considered significant when the relevant P value was < 0.05.
These analyses were performed using the statistical software JMP (version 8, SAS Institute Inc., Cary, NC). Correlation analysis was performed by creating Fit Y by X plots using JMP.
RESULTS
NKCC1 protein expression in human ESCCs
An immunohistochemical examination of non-cancerous esophageal epithelia performed with the NKCC1 antibody demonstrated that cells with NKCC1 expression were chiefly confined to the lower and middle layer of the squamous epithelium but were absent from the basal and parabasal cell layers (Figure 2A). Photographs of well differentiated, moderately differentiated, or poorly differentiated ESCC tumor samples with high or low NKCC1 expression are shown in Figure 2B. NKCC1 expression was observed in the cytoplasm of ESCC cells in all groups. NKCC1 staining scores were significantly increased as histological differentiation decreased (Figure 2C).
Figure 2.
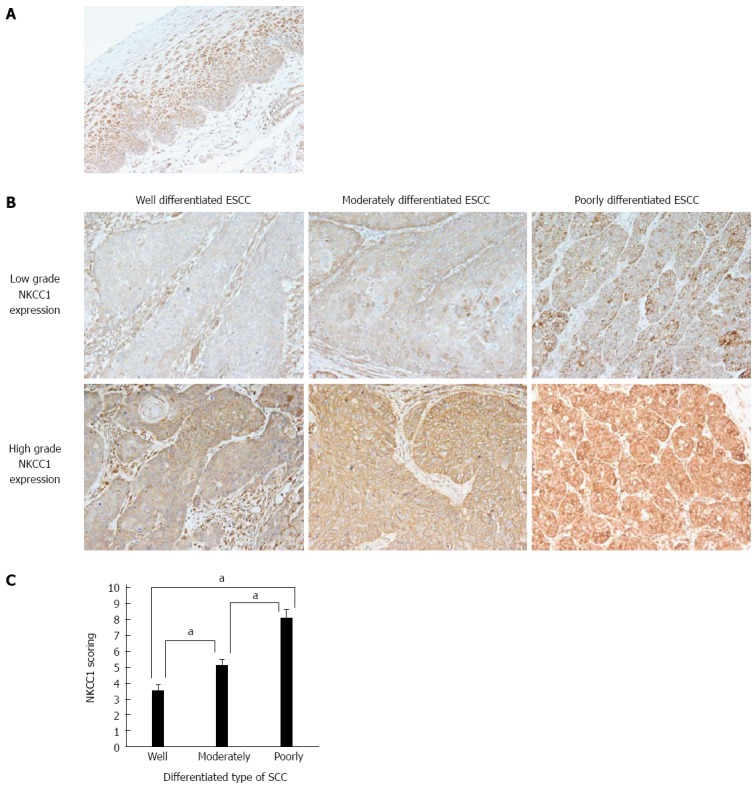
Na+/K+/2Cl- cotransporter 1 protein expression in human esophageal squamous cell carcinomas. A: Immunohistochemical staining of human esophageal epithelia with an Na+/K+/2Cl- cotransporter 1 (NKCC1) antibody. Cells with NKCC1 expression were primarily confined to the lower and middle layers of the squamous epithelium with the exception of the basal and parabasal cell layers; B: Immunohistochemical staining of well differentiated, moderately differentiated, or poorly differentiated esophageal squamous cell carcinoma (ESCC) tumor samples with high or low grade NKCC1 expression (magnification: × 200); C: NKCC1 staining scores according to the differentiation type of SCC. Mean ± SEM. Well differentiated ESCC; n = 15. Moderately differentiated ESCC; n = 31. Poorly differentiated ESCC; n = 22. aP < 0.05 vs control, Tukey-Kramer HSD test.
We divided ESCC patients into 2 groups, a low grade NKCC1 expression group with staining scores < 6, n = 28, and a high grade NKCC1 expression group with staining scores ≥ 6, n = 40, and compared their clinicopathological features. We found that the percentage of poorly differentiated SCC samples was significantly higher in the high grade group (47.5%) when compared to the low grade group (10.7%) (Table 1). No correlation was found between NKCC1 expression and any other clinicopathological parameter. No correlation was found between NKCC1 expression and the Ki-67 labeling index (Table 1). Furthermore, the 5-year survival rate did not differ between the high grade group (69.9 %) and the low grade group (63.5 %) (P = 0.501, the log-rank test). Subgroup analysis of pStage I patients showed that the 5-year survival rate of the high grade group (86.5%) tended to be lower than that of the low grade group (100.0 %), although no significant difference was observed (P = 0.403, the log-rank test). These results suggest that NKCC1 plays an important role in the differentiation of ESCC cells, although a significant prognostic impact could not be determined.
Table 1.
Correlations between clinicopathological parameters and Na+/K+/2Cl- cotransporter 1 expression
| Variable |
NKCC1 expression |
P value | ||
| Low grade | High grade | |||
| Age (yr) | < 60 | 12 | 10 | 0.1874 |
| ≥ 60 | 16 | 30 | ||
| Gender | Male | 25 | 32 | 0.5049 |
| Female | 3 | 8 | ||
| Location of tumor | Ce/Ut | 4 | 3 | 0.4346 |
| Mt/Lt/Ae | 24 | 37 | ||
| Tumor size (mm) | < 50 | 18 | 30 | 0.4206 |
| ≥ 50 | 10 | 10 | ||
| Histological type | Differentiated type SCC | 25 | 21 | 0.0015a |
| Poorly differentiated type SCC | 3 | 19 | ||
| pT | pT1 | 10 | 21 | 0.2191 |
| pT2-3 | 18 | 19 | ||
| pN | negative | 13 | 20 | 0.8095 |
| positive | 15 | 20 | ||
| pStage | I | 6 | 16 | 0.1231 |
| II-III | 22 | 24 | ||
| Ki-67 labeling index | 28.7 ± 2.3 | 29.9 ± 2.0 | 0.6834 | |
Ce: Cervical esophagus; Ut: Upper thoracic esophagus; Mt: Middle thoracic esophagus; Lt: Lower thoracic esophagus; Ae: Abdominal esophagus; pT: Pathological T stage; pN: Pathological N stage; pStage: Pathological stage; SCC: Squamous cell carcinoma;
P < 0.05 vs control, Fisher’s exact test.
NKCC1 controls cell cycle progression in ESCC cells
We examined six ESCC cell lines, TE2, TE5, TE9 TE13, KYSE70, and KYSE170, to determine NKCC1 protein expression levels. Western blotting analysis revealed that NKCC1 was highly expressed in the KYSE170 cell line, and lower levels of expression were observed in the TE2 and TE5 cell lines (Figure 3A). We conducted knockdown experiments using NKCC1 siRNA in KYSE170 cells and analyzed the effects of NKCC1 depletion on cell cycle progression. NKCC1 siRNA effectively reduced NKCC1 protein levels (Figure 3B) and NKCC1 mRNA levels (Figure 3C) in the KYSE170 cell line. The downregulation of NKCC1 induced G2/M phase arrest in KYSE170 cells (Figure 3D). The cell counts of NKCC1 depleted cells were significantly lower when compared to those of control siRNA transfected cells 72 h after siRNA transfection (Figure 3E). Furthermore, the NKCC blocker furosemide significantly inhibited the proliferation of KYSE170 cells (Figure 3F). Similar trends were found in several cell lines, including TE9, TE13 and KYSE 70, which expressed NKCC1 (Figure 4). These results suggest that NKCC1 plays an important role in regulating cell cycle progression and cell proliferation in ESCC cells.
Figure 3.
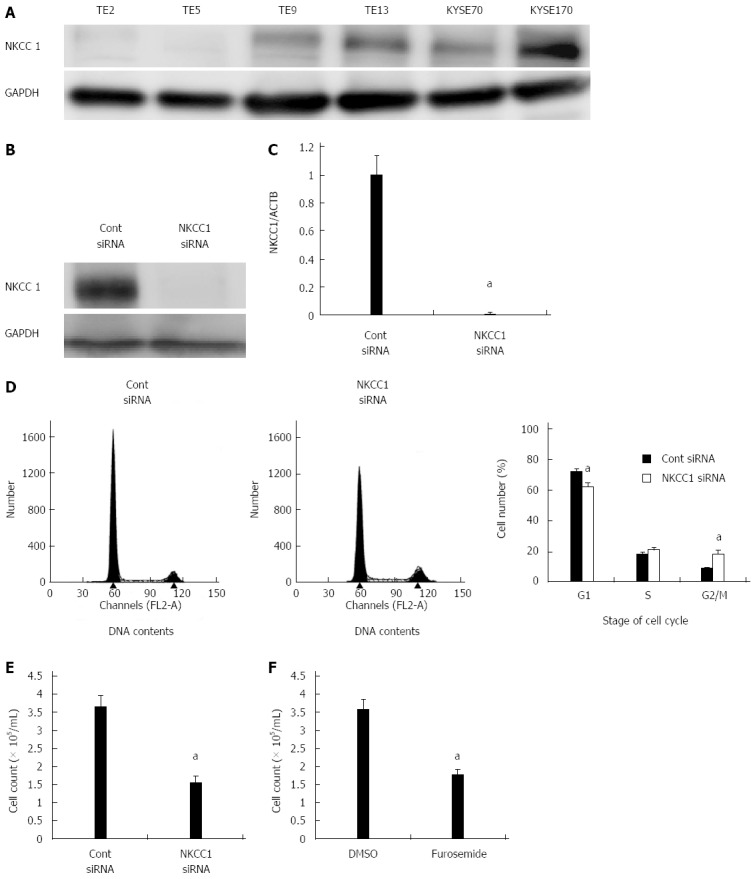
Na+/K+/2Cl- cotransporter 1 controls cell cycle progression in esophageal squamous cell carcinoma cells. A: Na+/K+/2Cl- cotransporter 1 (NKCC1) protein expression was analyzed in 6 esophageal squamous cell carcinoma (ESCC) cell lines. Western blotting revealed that NKCC1 was highly expressed in the KYSE170 cell line, and lower levels of expression were observed in TE2 and TE5 cells. B: Western blotting revealed that NKCC1 small interfering RNA (siRNA) effectively reduced the protein levels of NKCC1 in KYSE170 cells; C: NKCC1 siRNA effectively reduced the mRNA levels of NKCC1 in KYSE170 cells. The mean ± SEM. n = 4. aP < 0.05 vs the control siRNA group; D: The depletion of NKCC1 induced G2/M phase arrest in KYSE170 cells. Cells transfected with control or NKCC1 siRNA were stained with propidium iodide (PI) and analyzed by flow cytometry. The mean ± SEM. n = 5. aP < 0.05 vs control siRNA; E: The depletion of NKCC1 inhibited the proliferation of KYSE170 cells. Cell number was counted 72 h after siRNA transfection. The mean ± SEM. n = 5. aP < 0.05 (significantly different from control siRNA); F: The NKCC blocker furosemide inhibited the proliferation of KYSE170 cells. Cell number was counted 72 h after drug stimulation (500 μmol/L furosemide). The mean ± SEM. n = 5. aP < 0.05 vs control (significantly different from 500 μmol/L DMSO). GAPDH: Glyceraldehyde-3-phosphate dehydrogenase.
Figure 4.
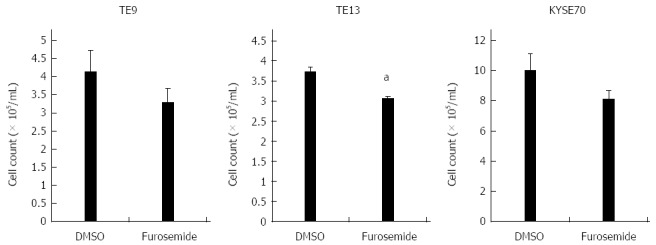
Effects of the Na+/K+/2Cl- cotransporter blocker furosemide on the proliferation of TE9, TE13 and KYSE70 cells. Cell number was counted 72 h after drug stimulation (500 μmol/L furosemide). The mean ± SEM. n = 3. aP < 0.05 vs control (significantly different from 500 μmol/L DMSO).
Gene expression profiles of NKCC1 depleted cells
We analyzed the gene expression profiles of NKCC1 depleted KYSE170 cells in microarray and bioinformatics studies. Microarray analysis showed that the expression levels of 2527 genes displayed fold changes of > 2.0 in KYSE170 cells upon depletion of NKCC1. Of these genes, 1157 were upregulated and 1370 were downregulated in NKCC1 siRNA depleted KYSE170 cells. A list of 20 genes with expression levels that were the most strongly up- or downregulated in NKCC1 depleted KYSE170 cells is shown in Table 2. NKCC1 (SLC12A2) expression was downregulated in NKCC1 depleted KYSE170 cells (fold change: -28.92; Table 2). Ingenuity Pathway Analysis showed that “Cancer” was the top-ranked disease and that “Cell Cycle” was the top-ranked biological function related to NKCC1 depletion. Furthermore, “Cell Cycle: G2/M DNA Damage Checkpoint Regulation” was one of the top-ranked canonical pathways related to NKCC1 depletion (Table 3), and this result was in agreement with the results obtained via cell cycle analysis. Among the 2527 genes with expression levels that were altered by NKCC1 depletion, 267 genes exhibited cell proliferation-related functions (Table 4). Among these genes, 82 genes were upregulated, and the other 185 genes were downregulated. We then examined the signal transduction networks induced by NKCC1 depletion (Table 3). All of the top 3 ranked signal networks were related to the cell cycle (Figure 5). These results indicate that the expression level of NKCC1 influences genes related to cellular growth and cell cycle progression.
Table 2.
Twenty genes displaying the highest change in expression levels in Na+/K+/2Cl- cotransporter 1 depleted KYSE170 cells
| Gene Symbol | Gene ID | Gene Name | Fold Change |
| Upregulated Genes | |||
| C18orf34 | NM_001105528 | Chromosome 18 open reading frame 34 | 155.49 |
| KCNA6 | NM_002235 | Potassium voltage-gated channel, shaker-related subfamily, member 6 | 140.4 |
| CCDC147 | NM_001008723 | Coiled-coil domain containing 147 | 105.98 |
| C20orf202 | NM_001009612 | Chromosome 20 open reading frame 202 | 86.17 |
| A1CF | NM_138933 | APOBEC1 complementation factor | 70.98 |
| SH3GL2 | NM_003026 | SH3-domain GRB2-like 2 | 70.93 |
| PTGFR | NM_001039585 | Prostaglandin F receptor (FP) | 66.99 |
| NDN | NM_002487 | Necdin homolog (mouse) | 66.45 |
| INPP5D | NM_001017915 | Inositol polyphosphate-5-phosphatase, 145 kDa | 52.83 |
| CYP2E1 | NM_000773 | Cytochrome P450, family 2, subfamily E, polypeptide 1 | 52.44 |
| AGBL3 | NM_178563 | ATP/GTP binding protein-like 3 | 50.88 |
| UBTFL1 | NM_001143975 | Upstream binding transcription factor, RNA polymerase I-like 1 | 47.88 |
| PADI2 | NM_007365 | Peptidyl arginine deiminase, type II | 46.83 |
| CCR1 | NM_001295 | Chemokine (C-C motif) receptor 1 | 44.86 |
| ARC | NM_015193 | Activity-regulated cytoskeleton-associated protein | 44.41 |
| COLEC10 | NM_006438 | Homo sapiens collectin sub-family member 10 (C-type lectin) | 44.28 |
| DNAH6 | NM_001370 | Dynein, axonemal, heavy chain 6 | 41.96 |
| BOLL | NM_033030 | Bol, boule-like (Drosophila) | 41.31 |
| CORO2B | NM_006091 | Coronin, actin binding protein, 2B | 41.04 |
| MUC7 | NM_152291 | Mucin 7, secreted | 36.97 |
| Downregulated Genes | |||
| NPFFR1 | NM_022146 | Neuropeptide FF receptor 1 | -54.97 |
| LRRFIP1 | NM_001137550 | Leucine rich repeat (in FLII) interacting protein 1 | -44.72 |
| PPIL6 | NM_173672 | Peptidylprolyl isomerase (cyclophilin)-like 6 | -44.46 |
| CRHR2 | NM_001883 | Corticotropin releasing hormone receptor 2 | -39.78 |
| CMTM2 | NM_144673 | CKLF-like MARVEL transmembrane domain containing 2 | -39.62 |
| C5 | NM_001735 | Complement component 5 | -39.13 |
| KCNMA1 | NM_001014797 | Potassium large conductance calcium-activated channel, subfamily M, alpha member 1 | -38.59 |
| HESX1 | NM_003865 | HESX homeobox 1 | -33.03 |
| SLC22A2 | NM_003058 | Solute carrier family 22 (organic cation transporter), member 2 | -32.49 |
| WNT8B | NM_003393 | Wingless-type MMTV integration site family, member 8B | -32.17 |
| GRIA1 | NM_000827 | Glutamate receptor, ionotropic, AMPA 1 | -31.27 |
| ZNF367 | NM_153695 | Zinc finger protein 367 | -30.04 |
| GPR128 | NM_032787 | G protein-coupled receptor 128 | -29.88 |
| SLC12A2 | NM_001046 | Solute carrier family 12 (sodium/potassium/chloride transporters), member 2 | -28.92 |
| KCNG2 | NM_012283 | Potassium voltage-gated channel, subfamily G, member 2 | -28.3 |
| ECT2L | NM_001077706 | Epithelial cell transforming sequence 2 oncogene-like | -27 |
| ERMN | NM_020711 | Ermin, ERM-like protein | -26.61 |
| DPP10 | NM_020868 | Dipeptidyl-peptidase 10 (non-functional) | -26.58 |
| TSPAN7 | NM_004615 | Tetraspanin 7 | -25.54 |
| APOA1 | NM_000039 | Apolipoprotein A-I | -25.21 |
Table 3.
Top biological functions, canonical pathways, and networks of Na+/K+/2Cl- cotransporter 1 according to Ingenuity Pathway Analysis
| Top Biological Functions | ||
| Diseases and disorders | ||
| Name | P value | Number of molecules |
| Cancer | 2.08E-12 - 1.59E-02 | 277 |
| Gastrointestinal disease | 8.03E-12 - 1.60E-02 | 149 |
| Reproductive system disease | 2.25E-09 - 1.57E-02 | 138 |
| Hematological disease | 1.72E-06 - 1.22E-02 | 70 |
| Hereditary disorder | 2.10E-06 - 1.57E-02 | 120 |
| Molecular and cellular functions | ||
| Name | P value | Number of molecules |
| Cell cycle | 1.06E-20 - 1.60E-02 | 158 |
| Cellular assembly and organization | 1.06E-20 - 1.36E-02 | 111 |
| DNA replication, recombination, and repair | 1.06E-20 - 1.16E-02 | 133 |
| Cellular movement | 8.36E-11 - 1.51E-02 | 82 |
| Cell death | 2.98E-06 - 1.56E-02 | 216 |
| Top canonical pathways | ||
| Name | P value | Ratio |
| Role of BRCA1 in the DNA damage response | 9.79E-6 | 12/65 (0.185) |
| Mitotic roles of Polo-Like kinase | 7.37E-5 | 11/69 (0.159) |
| Estrogen-mediated S-phase entry | 4.91E-4 | 6/28 (0.214) |
| Cell Cycle: G2/M DNA damage checkpoint regulation | 5.07E-4 | 8/49 (0.163) |
| Role of CHK proteins in cell cycle checkpoint control | 5.37E-4 | 9/56 (0.161) |
| Top networks | ||
| Associated network functions | Score | |
| Cellular assembly and organization; DNA replication, recombination, and repair; Cell cycle | 47 | |
| Cellular assembly and organization, Cell cycle, DNA replication, recombination, and repair | 43 | |
| Cell cycle; DNA replication, recombination, and repair; Cancer | 37 | |
| Digestive system development and function, organismal injury and abnormalities, cellular function and maintenance | 37 | |
| Cellular assembly and organization; DNA replication, recombination, and repair; Cardiovascular disease | 35 | |
Table 4.
Cell growth-related genes with expression levels in KYSE170 cells that were changed by the depletion of Na+/K+/2Cl- cotransporter 1
| Gene symbol | Gene ID |
Biological functions |
Fold change | |
| Cell growth and proliferation | Cell cycle | |||
| Upregulated genes | ||||
| NDN | NM_002487 | ● | 66.45 | |
| INPP5D | NM_001017915 | ● | ● | 52.83 |
| CCR1 | NM_001295 | ● | 44.86 | |
| COL1A2 | NM_000089 | ● | 36.72 | |
| EDAR | NM_022336 | ● | 35.86 | |
| RBP4 | NM_006744 | ● | 30.40 | |
| DCLK1 | NM_004734 | ● | 28.95 | |
| RARRES1 | NM_002888 | ● | 22.92 | |
| FMOD | NM_002023 | ● | 22.85 | |
| BARX1 | NM_021570 | ● | 21.47 | |
| MAPK10 | NM_138980 | ● | 20.00 | |
| ADORA2A | NM_000675 | ● | 18.78 | |
| CHRNA7 | NM_001190455 | ● | ● | 18.47 |
| SOX10 | NM_006941 | ● | 18.03 | |
| FGF20 | NM_019851 | ● | 16.73 | |
| FAM5C | NM_199051 | ● | ● | 14.35 |
| MBD2 | NM_015832 | ● | 12.13 | |
| EGF | NM_001963 | ● | ● | 11.50 |
| TLR5 | NM_003268 | ● | 11.35 | |
| TNN | NM_022093 | ● | 10.98 | |
| SLC1A2 | NM_004171 | ● | 10.74 | |
| CD36 | NM_001001547 | ● | 10.43 | |
| CD52 | NM_001803 | ● | 10.34 | |
| NR2E3 | NM_016346 | ● | 10.31 | |
| PLCB1 | NM_182734 | ● | 10.26 | |
| MYCN | NM_005378 | ● | ● | 10.23 |
| ZNF365 | NM_199451 | ● | 10.12 | |
| ERG | NM_004449 | ● | 10.04 | |
| MSH4 | NM_002440 | ● | 10.03 | |
| DMRT1 | NM_021951 | ● | 9.61 | |
| RNF128 | NM_194463 | ● | 9.37 | |
| CD69 | NM_001781 | ● | 8.89 | |
| PDE3A | NM_000921 | ● | ● | 8.58 |
| ACVR1C | NM_145259 | ● | 8.57 | |
| SPI1 | NM_001080547 | ● | ● | 8.55 |
| SH2D3C | NM_170600 | ● | 8.53 | |
| IFNG | NM_000619 | ● | ● | 8.22 |
| MRAS | NM_012219 | ● | 8.20 | |
| MCF2L | NM_024979 | ● | 7.43 | |
| RRAD | NM_004165 | ● | ● | 7.42 |
| E2F5 | NM_001951 | ● | ● | 7.32 |
| BGN | NM_001711 | ● | 7.13 | |
| KIFC1 | NM_002263 | ● | 7.02 | |
| ABCC6 | NM_001171 | 6.98 | ||
| SERPINE1 | NM_000602 | ● | 6.74 | |
| CIITA | NM_000246 | ● | 6.74 | |
| GJB6 | NM_006783 | ● | 6.45 | |
| TP53INP1 | NM_033285 | ● | ● | 6.38 |
| GHRL | NM_016362 | ● | 6.30 | |
| CCNG2 | NM_004354 | ● | ● | 6.29 |
| RORC | NM_005060 | ● | 6.24 | |
| NCF1 | NM_000265 | ● | 6.24 | |
| NFATC4 | NM_001136022 | ● | 6.15 | |
| CHRM5 | NM_012125 | ● | 6.01 | |
| HMOX1 | NM_002133 | ● | ● | 6.00 |
| IL18RAP | NM_003853 | ● | 5.98 | |
| C8orf4 | NM_020130 | ● | 5.95 | |
| L1CAM | NM_024003 | ● | 5.87 | |
| TNFSF8 | NM_001244 | ● | 5.85 | |
| MSMB | NM_002443 | ● | 5.77 | |
| ITPR1 | NM_002222 | ● | 5.77 | |
| ITGAL | NM_002209 | ● | ● | 5.73 |
| INHBA | NM_002192 | ● | ● | 5.53 |
| HEYL | NM_014571 | ● | 5.38 | |
| JAK3 | NM_000215 | ● | ● | 5.30 |
| MMP13 | NM_002427 | ● | 5.23 | |
| NNMT | NM_006169 | ● | 5.06 | |
| BNIPL | NM_138278 | ● | 4.86 | |
| LTC4S | NM_145867 | ● | 4.74 | |
| MMP24 | NM_006690 | ● | 4.72 | |
| MMP1 | NM_002421 | ● | 4.66 | |
| CD19 | NM_001770 | ● | ● | 4.52 |
| ADC | NM_052998 | ● | 4.46 | |
| TGFBR1 | NM_004612 | ● | ● | 4.33 |
| RHOB | NM_004040 | ● | 4.32 | |
| CDKN1C | NM_000076 | ● | ● | 4.30 |
| HOXB13 | NM_006361 | ● | ● | 4.20 |
| IPMK | NM_152230 | ● | 4.07 | |
| BMF | NM_001003940 | ● | 4.06 | |
| VTCN1 | NM_024626 | ● | ● | 4.05 |
| CEACAM1 | NM_001712 | ● | 2.97 | |
| TSSK3 | NM_052841 | ● | 2.00 | |
| Downregulated genes | ||||
| CRHR2 | NM_001883 | ● | -39.78 | |
| C5 | NM_001735 | ● | ● | -39.13 |
| KCNMA1 | NM_001014797 | ● | -38.59 | |
| GRIA1 | NM_000827 | ● | -31.27 | |
| SLC12A2 | NM_001046 | ● | -28.92 | |
| APOA1 | NM_000039 | ● | -25.21 | |
| PRKAR2B | NM_002736 | ● | ● | -24.25 |
| TF | NM_001063 | ● | ● | -22.85 |
| BRCA2 | NM_000059 | ● | ● | -22.04 |
| AURKC | NM_001015878 | ● | -21.69 | |
| PLXNA4 | NM_181775 | ● | -20.54 | |
| TYR | NM_000372 | ● | -20.39 | |
| BACH2 | NM_021813 | ● | -20.01 | |
| KIF14 | NM_014875 | ● | -18.06 | |
| HEY2 | NM_012259 | ● | -18.01 | |
| TMPO | NM_003276 | ● | ● | -15.97 |
| FCGR3A | NM_000569 | ● | -15.18 | |
| ARF6 | NM_001663 | -15.09 | ||
| MYBL1 | NM_001080416 | ● | ● | -14.60 |
| CCNA1 | NM_003914 | ● | ● | -14.29 |
| ESCO2 | NM_001017420 | ● | -14.05 | |
| TOP2A | NM_001067 | ● | ● | -13.91 |
| CENPI | NM_006733 | ● | -13.87 | |
| ATAD2 | NM_014109 | ● | -13.41 | |
| POSTN | NM_006475 | ● | -12.91 | |
| MKI67 | NM_002417 | ● | ● | -12.41 |
| ABCB1 | NM_000927 | ● | ● | -12.15 |
| KIF20B | NM_016195 | ● | ● | -11.53 |
| SPN | NM_001030288 | ● | -11.41 | |
| MAD2L1 | NM_002358 | ● | ● | -11.16 |
| HLA-DPB1 | NM_002121 | ● | -11.12 | |
| SGOL1 | NM_001012410 | ● | -10.91 | |
| RRM2 | NM_001034 | ● | -10.71 | |
| FANCD2 | NM_033084 | ● | ● | -10.55 |
| FANCA | NM_001018112 | ● | ● | -10.31 |
| HDAC2 | NM_001527 | ● | ● | -9.94 |
| NUF2 | NM_145697 | ● | -9.76 | |
| CLSPN | NM_022111 | ● | ● | -9.57 |
| RAD54L | NM_003579 | ● | -9.47 | |
| KLHL13 | NM_033495 | ● | -9.32 | |
| CCNA2 | NM_001237 | ● | ● | -9.13 |
| MCM10 | NM_182751 | ● | ● | -9.11 |
| MCTS1 | NM_014060 | ● | -9.02 | |
| ANLN | NM_018685 | ● | -9.00 | |
| HMGB2 | NM_002129 | ● | ● | -8.86 |
| VPREB1 | NM_007128 | ● | -8.81 | |
| KIF4A | NM_012310 | ● | -8.78 | |
| SPC25 | NM_020675 | ● | -8.75 | |
| ALOX5 | NM_000698 | ● | ● | -8.55 |
| PBK | NM_018492 | ● | -8.20 | |
| TNFRSF11B | NM_002546 | ● | -8.20 | |
| CIT | NM_001206999 | ● | -8.17 | |
| HELLS | NM_018063 | ● | ● | -8.1 |
| CDC45 | NM_003504 | ● | ● | -8.07 |
| DTL | NM_016448 | ● | ● | -8.00 |
| RGS3 | NM_017790 | -7.93 | ||
| TYMS | NM_001071 | ● | ● | -7.87 |
| NDC80 | NM_006101 | ● | -7.86 | |
| ERCC6L | NM_017669 | ● | -7.82 | |
| CENPE | NM_001813 | ● | -7.75 | |
| TTK | NM_003318 | ● | ● | -7.74 |
| SIM2 | NM_009586 | ● | -7.61 | |
| KRT4 | NM_002272 | ● | -7.55 | |
| RAD51AP1 | NM_006479 | ● | -7.55 | |
| LTA | NM_000595 | ● | -7.51 | |
| PAK2 | NM_002577 | ● | ● | -7.50 |
| SLC5A8 | NM_145913 | ● | -7.41 | |
| BLM | NM_000057 | ● | ● | -7.40 |
| NUSAP1 | NM_016359 | ● | -7.36 | |
| JDP2 | NM_130469 | ● | ● | -7.19 |
| CASP3 | NM_004346 | ● | ● | -7.17 |
| NEIL3 | NM_018248 | ● | -7.17 | |
| POLH | NM_006502 | ● | ● | -7.11 |
| KIF20A | NM_005733 | ● | ● | -7.08 |
| MYO7A | NM_000260 | -6.93 | ||
| NRGN | NM_006176 | ● | -6.82 | |
| NCAPG | NM_022346 | ● | ● | -6.78 |
| CDCA8 | NM_018101 | ● | ● | -6.72 |
| CEP55 | NM_018131 | ● | -6.65 | |
| DLGAP5 | NM_014750 | ● | ● | -6.60 |
| CDC25C | NM_001790 | ● | ● | -6.59 |
| ARL2BP | NM_012106 | ● | -6.58 | |
| IL12A | NM_000882 | ● | ● | -6.53 |
| MYH14 | NM_001077186 | ● | -6.52 | |
| SKA1 | NM_001039535 | ● | -6.46 | |
| CASC1 | NM_018272 | ● | -6.44 | |
| HJURP | NM_018410 | ● | -6.42 | |
| TACC3 | NM_006342 | ● | ● | -6.33 |
| ENPP3 | NM_005021 | ● | -6.30 | |
| STIL | NM_001048166 | ● | ● | -6.27 |
| KNTC1 | NM_014708 | ● | -6.26 | |
| NR1I2 | NM_003889 | ● | ● | -6.24 |
| AKR1B10 | NM_020299 | ● | -6.22 | |
| E2F2 | NM_004091 | ● | ● | -6.20 |
| USP47 | NM_017944 | ● | -6.14 | |
| KIF11 | NM_004523 | ● | ● | -6.09 |
| E2F8 | NM_024680 | ● | ● | -6.05 |
| PLK1 | NM_005030 | ● | ● | -6.02 |
| CCDC6 | NM_005436 | ● | -6.00 | |
| ORC6 | NM_014321 | ● | -6.00 | |
| EXO1 | NM_003686 | ● | -5.95 | |
| GPC5 | NM_004466 | ● | -5.94 | |
| GSG2 | NM_031965 | ● | -5.93 | |
| PRC1 | NM_003981 | ● | -5.89 | |
| RAD51 | NM_002875 | ● | ● | -5.78 |
| KIF2C | NM_006845 | ● | ● | -5.71 |
| TNFRSF13C | NM_052945 | ● | -5.70 | |
| BLZF1 | NM_003666 | ● | -5.63 | |
| FEN1 | NM_004111 | ● | -5.51 | |
| PLK4 | NM_014264 | ● | ● | -5.49 |
| HAS2 | NM_005328 | ● | ● | -5.44 |
| PKMYT1 | NM_182687 | ● | -5.40 | |
| BUB1 | NM_001211 | ● | ● | -5.34 |
| BUB1B | NM_001211 | ● | ● | -5.34 |
| NEK2 | NM_002497 | ● | ● | -5.33 |
| IQGAP3 | NM_178229 | ● | -5.27 | |
| SKA3 | NM_145061 | ● | -5.23 | |
| PNN | NM_002687 | ● | ● | -5.20 |
| NTRK3 | NM_001007156 | ● | ● | -5.17 |
| IL25 | NM_022789 | ● | -5.09 | |
| UBE2C | NM_181803 | ● | ● | -5.09 |
| AURKB | NM_004217 | ● | ● | -5.08 |
| CDC6 | NM_001254 | ● | ● | -5.08 |
| CDKN2C | NM_078626 | ● | ● | -5.06 |
| EDN2 | NM_001956 | ● | -5.06 | |
| CDC20 | NM_001255 | ● | ● | -5.05 |
| RRM1 | NM_001033 | ● | ● | -5.05 |
| APC | NM_000038 | ● | ● | -5.04 |
| KIF15 | NM_020242 | ● | ● | -5.03 |
| LMNB1 | NM_005573 | ● | -5.02 | |
| NCAPG2 | NM_017760 | ● | -4.96 | |
| CCNE2 | NM_057749 | ● | ● | -4.94 |
| HMMR | NM_012484 | ● | -4.93 | |
| BRIP1 | NM_032043 | ● | -4.90 | |
| ECT2 | NM_018098 | ● | ● | -4.89 |
| CDT1 | NM_030928 | ● | ● | -4.87 |
| MCAM | NM_006500 | ● | -4.82 | |
| LAG3 | NM_002286 | ● | ● | -4.78 |
| ZWINT | NM_032997 | ● | -4.73 | |
| DCLK2 | NM_001040260 | ● | -4.72 | |
| TRAIP | NM_005879 | ● | -4.71 | |
| SSTR2 | NM_001050 | ● | ● | -4.69 |
| TXK | NM_003328 | ● | -4.65 | |
| TBC1D9 | NM_015130 | ● | -4.63 | |
| IL1RN | NM_173843 | ● | -4.61 | |
| CDCA7 | NM_031942 | ● | -4.56 | |
| STK38 | NM_007271 | ● | ● | -4.56 |
| CDCA5 | NM_080668 | ● | ● | -4.54 |
| E2F7 | NM_203394 | ● | -4.54 | |
| FIGNL1 | NM_001042762 | ● | -4.51 | |
| SMC4 | NM_005496 | ● | -4.50 | |
| CYCS | NM_018947 | ● | -4.48 | |
| FBN1 | NM_000138 | ● | -4.48 | |
| NCAPD3 | NM_015261 | ● | -4.46 | |
| IL16 | NM_172217 | ● | ● | -4.44 |
| PCNA | NM_002592 | ● | ● | -4.42 |
| FBXO5 | NM_001142522 | ● | -4.37 | |
| CKAP2 | NM_018204 | ● | -4.34 | |
| IL34 | NM_152456 | ● | -4.34 | |
| PSRC1 | NM_032636 | ● | ● | -4.33 |
| C11orf82 | NM_145018 | ● | -4.32 | |
| CHRDL1 | NM_145234 | ● | -4.31 | |
| RAD54B | NM_012415 | ● | -4.31 | |
| DIAPH3 | NM_001042517 | ● | -4.29 | |
| AKR1C1 | NM_001353 | ● | -4.26 | |
| INHBB | NM_002193 | ● | -4.25 | |
| MDM2 | NM_002392 | ● | ● | -4.25 |
| PRKAA1 | NM_206907 | ● | -4.25 | |
| MASTL | NM_032844 | ● | -4.23 | |
| MCM5 | NM_006739 | ● | -4.21 | |
| CD2AP | NM_012120 | ● | ● | -4.20 |
| BRCA1 | NM_007300 | ● | ● | -4.18 |
| TPX2 | NM_012112 | ● | ● | -4.15 |
| FGFBP1 | NM_005130 | ● | -4.14 | |
| EIF4G2 | NM_001172705 | ● | ● | -4.12 |
| AURKA | NM_198433 | ● | ● | -4.10 |
| PTTG1 | NM_004219 | ● | ● | -4.08 |
| ADRA1B | NM_000679 | ● | ● | -4.07 |
| RECQL4 | NM_004260 | ● | -4.02 | |
| GJB2 | NM_004004 | ● | -4.00 | |
| BIRC5 | NM_001012271 | ● | ● | -3.35 |
| TERF1 | NM_017489 | ● | ● | -3.18 |
| LRP1 | NM_032832 | ● | -2.43 | |
| CDK1 | NM_012395 | ● | ● | -2.21 |
| ARHGEF10 | NM_014629 | ● | -2.08 | |
Figure 5.
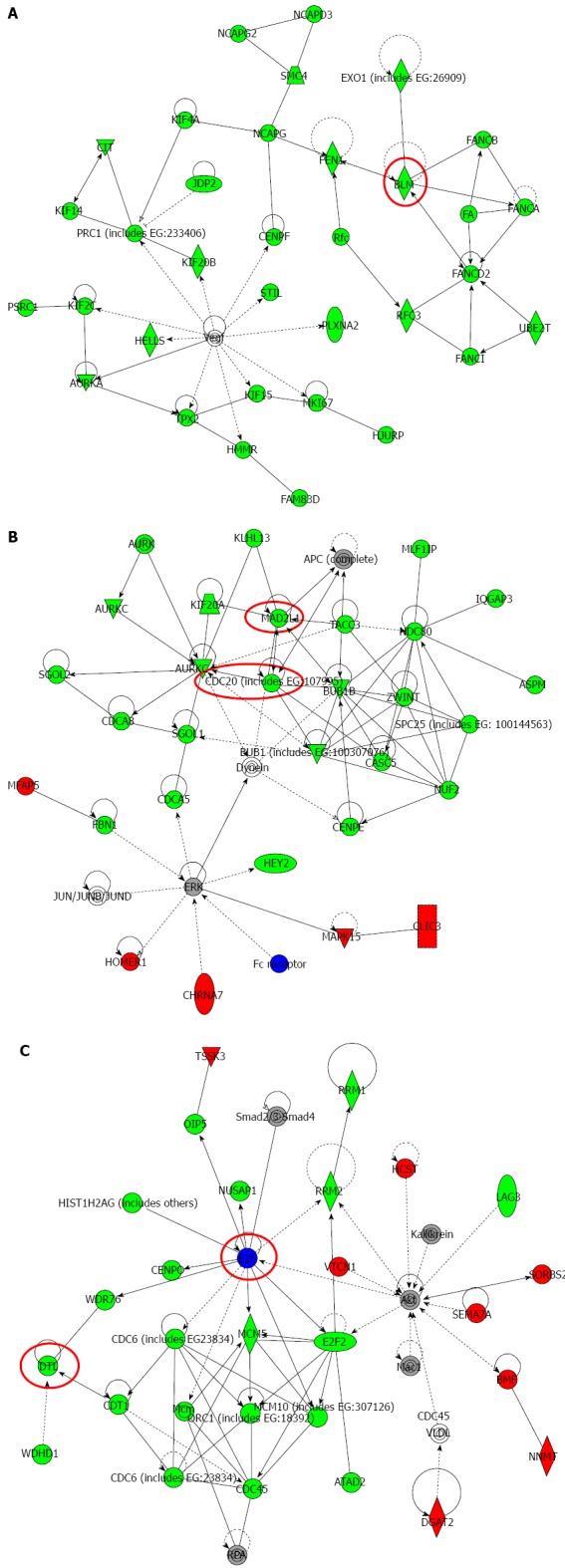
Top-ranked signaling networks related to Na+/K+/2Cl- cotransporter 1 downregulation according to ingenuity pathway analysis. A: This network is called “Cellular Assembly and Organization; DNA Replication, Recombination, and Repair; Cell Cycle”; B: This network is called “Cellular Assembly and Organization, Cell Cycle, DNA Replication, Recombination, and Repair”; C: This network is called “Cell Cycle; DNA Replication, Recombination, and Repair; Cancer”. Red and green indicate genes with expression levels that were higher or lower, respectively, than reference RNA levels. Genes analyzed for verification in Figure 6 were highlighted by red circles.
Verification of gene expression by real-time quantitative RT-PCR
Six genes (MAD2L1, DTL, BLM, CDC20, BRCA1, and E2F5) were further examined by quantitative Real time reverse transcription-polymerase chain reaction (RT-PCR). BLM was chosen from Network A, MAD2L1 and CDC20 from Network B, and DTL and E2F from Network C (Figure 5). BRCA1 was chosen because “Role of BRCA1 in DNA Damage Response” was the top-ranked canonical pathway related to NKCC1 (Table 3). All of these genes were related to the G2/M checkpoint according to IPA and are included in Table 4. The expression levels of MAD2L1, DTL, BLM, CDC20, and BRCA1 mRNA were significantly lower in NKCC1 depleted KYSE170 cells compared to control siRNA transfected cells (Figure 6). The expression levels of E2F5 mRNA were significantly higher in NKCC1 depleted KYSE170 cells compared to control siRNA transfected cells (Figure 6). Similar trends were found in several cell lines, including TE9, TE13 and KYSE 70 which expressed NKCC1 (Figure 7).These changes were in agreement with the microarray results and suggest that NKCC1 controls cell cycle progression via G2/M checkpoint regulation in ESCC cells.
Figure 6.
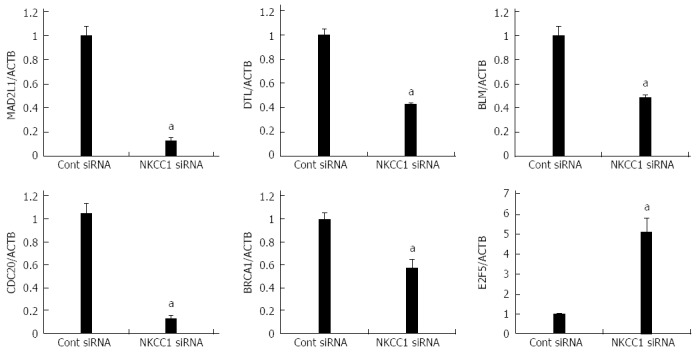
Verification of gene expression by real-time quantitative reverse transcription-polymerase chain reaction. The expression levels of six selected genes (MAD2L1, DTL, BLM, CDC20, BRCA1, and E2F5) in NKCC1 depleted KYSE170 cells were compared to those in control siRNA transfected cells using real-time quantitative reverse transcription-polymerase chain reaction. Gene expression levels were normalized to the level of ACTB. The mean ± SEM. n = 3. aP < 0.05 vs control siRNA.
Figure 7.

Expression levels of six selected genes (MAD2L1, DTL, BLM, CDC20, BRCA1, and E2F5) in Na+/K+/2Cl- cotransporter 1 depleted TE9, TE13 and KYSE70 cells. The expression levels of six selected genes (MAD2L1, DTL, BLM, CDC20, BRCA1, and E2F5) in Na+/K+/2Cl- cotransporter 1 (NKCC1) depleted TE9 (A), TE13 (B) and KYSE70 cells (C) were compared to those in control siRNA transfected cells using real-time quantitative RT-PCR. Gene expression levels were normalized to the level of ACTB. The mean ± SEM. n = 3. aP < 0.05 vs control siRNA.
DISCUSSION
The roles of ion transporters have recently been studied in cancer cells[14,15]. Some types of K+ channels have been reported to be expressed at high levels in colonic carcinoma[16,17]. The voltage-gated HERG channel has also exhibited cancer-specific expression in gastric cancer and its blocker diminished the G1 to S phase transition[18]. Increased mRNA levels of Ca2+ channels have also been reported in colorectal adenocarcinoma[19,20]. Furthermore, some reports have indicated that Cl- channels/transporters, such as Cl- channels, K+/Cl- cotransporters, and NKCC play important roles in the proliferation of colorectal, breast, lung, and prostate cancer cells[14,15]. To the best of our knowledge, the present study is the first report examining NKCC1 expression in ESCC tissue and the gene expression profile of NKCC1 depleted cancer cells.
We investigated the role of transepithelial Cl- transport in cancer cells[5-7]. In the present study, we found that the depletion of NKCC1 induced G2/M phase arrest in KYSE170 cells. We have previously shown that the blockage of NKCC inhibited G1/S cell cycle progression in gastric and prostate cancer cells[8,9], which suggests that the mechanism by which NKCC1 regulates cell cycle progression varies among cell types and their different genetic backgrounds. Microarray analysis showed that many of the genes that displayed changes in expression levels after NKCC1 depletion were well connected in the top-ranked signaling network related to the cell cycle, indicating that they are not only functionally related but are also regulated together at the level of expression by NKCC1-related signal transduction pathways.
With regard to signaling networks, we noted that the expression levels of several G2/M checkpoint-related genes were altered by the depletion of NKCC1. In the spindle checkpoint, the anaphase-promoting complex (APC) was activated by CDC20, which subsequently triggered anaphase. MAD2L1, a mitotic spindle assembly checkpoint protein, inhibited the activity of the APC by a direct physical interaction with a ternary complex containing CDC20[21,22]. DTL, BLM, BRCA1, and E2F5 are also known regulators of the G2/M checkpoint[23-26]. One possible mechanism by which NKCC1 changes the expression of these major G2/M checkpoint-related genes may be through the regulation of intracellular Cl- concentrations ([Cl-]i). Recent reports have indicated that [Cl-]i is a fundamental signal mediator for the regulation of various cellular functions[27-29]. For example, our study showed that [Cl-]i could act as a signal to regulate mRNA expression of the epithelial Na+ channel via a protein tyrosine kinase-dependent pathway in renal epithelial cells[29]. We have also previously shown that [Cl-]i regulated cell proliferation in gastric and prostate cancer cells[5-9]. We consider NKCC to be one of the important transporters that regulates [Cl-]i in the steady state and have previously shown that the blockage of NKCC decreased [Cl-]i[9]. Although the detailed mechanism should be verified by further studies, these observations suggest that the change in [Cl-]i induced by NKCC1 may be a critically important messenger that regulates the expression of these G2/M checkpoint-related genes in ESCC cells.
Our results demonstrate that no correlation was found between NKCC1 expression and the Ki-67 labeling index in immunohistochemical studies of ESCC expression. Ki-67 is commonly used to assess cell proliferation, and this factor reacts with a nuclear antigen present throughout the cell cycle (late G1, S, G2, and M phase) of proliferating cells but is absent from quiescent (G0) cells[30]. In the present study, we found that NKCC1 plays an important role in the G2/M phase of the cell cycle. The different rates of progression through each phase of the cell cycle may explain why no correlation was found between NKCC1 and Ki-67 expression, although further studies will be needed with a larger sample size to confirm these observations. Furthermore, in the present study NKCC1 expression was correlated with the degree of histological differentiation in SCC. Similarly, we previously found that mRNA levels and the functional expression levels of NKCC1 were higher in poorly differentiated type gastric adenocarcinoma cells compared to differentiated cells[8]. Furosemide (a NKCC blocker and a loop diuretic) is often used as a diuretic to maintain urine output and improve edema, ascites, or pleural effusion for the treatment of patients with terminal stage cancers. From this viewpoint, our observation that the blockage of NKCC1 diminished the proliferation of ESCC cells provides strong clinical evidence that furosemide can be used for ESCC patients with high NKCC1 expression, such as those with poorly differentiated SCC, and suggests the possibility of a novel tailor-made treatment.
In summary, we found that NKCC1 plays a role in the proliferation of ESCC cells. An immunohistochemical analysis revealed that the expression of NKCC1 in human ESCC samples was related to the histological type of ESCC. Our microarray results also suggest that NKCC1 exhibits marked effects on the expression of genes related to G2/M cell cycle progression. A deeper understanding of the role of NKCC1 may lead to its use as an important biomarker of tumor development and/or a novel therapeutic target for ESCC.
COMMENTS
Background
The roles of ion transporters have recently been studied in cancer cells, and several reports have demonstrated the important roles of Cl- channels/transporters in gastrointestinal cancer cells.
Research frontiers
Although previous reports showed that the Na+/K+/2Cl- cotransporter 1 (NKCC1) plays an important role in the proliferation of several types of cancer cells, its role in esophageal squamous cell carcinoma (ESCC) cells has not been fully investigated. Furthermore, the clinicopathological meaning of NKCC1 expression in ESCCs remains uncertain.
Innovations and breakthroughs
The authors analyzed the expression of NKCC1 in human ESCC samples and determined its relationship with the degree of histological differentiation of SCC samples. Depletion of NKCC1 in KYSE170 cells inhibited cell proliferation via G2/M phase arrest. The results of microarray showed that the top-ranked canonical pathway was the G2/M DNA damage checkpoint regulation pathway, which involves MAD2L1, DTL, BLM, CDC20, BRCA1, and E2F5.
Applications
The study results suggest that a deeper understanding of the role of NKCC1 may lead to its use as an important biomarker of tumor development and/or a novel therapeutic target for ESCC. The observation that the blockage of NKCC1 diminished the proliferation of ESCC cells provides clinical evidence that furosemide can be used for ESCC patients with high NKCC1 expression, and suggests the possibility of a novel tailor-made treatment.
Terminology
NKCC is a member of the cation-chloride cotransporter family. NKCC transports one sodium ion, one potassium ion, and two chloride ions across the plasma membrane and is sensitive to loop diuretics. There are two isoforms of NKCC, and NKCC1 is ubiquitously expressed in various types of cells including epithelial cells.
Peer review
This is a good descriptive study in which the authors analyzed the role of NKCC1 in the proliferation of ESCC. The authors showed NKCC1 was found in the cytoplasm and related to tumor differentiation in patients with ESCC. Depletion of NKCC1 lead to inhibition of cell proliferation, and microarray analysis showed that NKCC1 exhibits marked effects on the expression of genes related to G2/M cell cycle progression. The results are interesting and meaningful for further understand the role of NKCC1 on cancer development.
Footnotes
Supported by Grants-in-Aid for Young Scientists (B), NO. 22791295, NO. 23791557, and NO. 24791440; and a Grant-in-Aid for Scientific Research (C), NO.22591464 and NO. 24591957, from the Japan Society for the Promotion of Science
P- Reviewers: Kim MP, Zhao BS S- Editor: Qi Y L- Editor: A E- Editor: Liu XM
References
- 1.Bustin SA, Li SR, Dorudi S. Expression of the Ca2+-activated chloride channel genes CLCA1 and CLCA2 is downregulated in human colorectal cancer. DNA Cell Biol. 2001;20:331–338. doi: 10.1089/10445490152122442. [DOI] [PubMed] [Google Scholar]
- 2.Sarosi GA, Jaiswal K, Herndon E, Lopez-Guzman C, Spechler SJ, Souza RF. Acid increases MAPK-mediated proliferation in Barrett’s esophageal adenocarcinoma cells via intracellular acidification through a Cl-/HCO3- exchanger. Am J Physiol Gastrointest Liver Physiol. 2005;289:G991–G997. doi: 10.1152/ajpgi.00215.2005. [DOI] [PubMed] [Google Scholar]
- 3.Hebert SC, Mount DB, Gamba G. Molecular physiology of cation-coupled Cl- cotransport: the SLC12 family. Pflugers Arch. 2004;447:580–593. doi: 10.1007/s00424-003-1066-3. [DOI] [PubMed] [Google Scholar]
- 4.Mount DB, Mercado A, Song L, Xu J, George AL, Delpire E, Gamba G. Cloning and characterization of KCC3 and KCC4, new members of the cation-chloride cotransporter gene family. J Biol Chem. 1999;274:16355–16362. doi: 10.1074/jbc.274.23.16355. [DOI] [PubMed] [Google Scholar]
- 5.Shiozaki A, Otsuji E, Marunaka Y. Intracellular chloride regulates the G(1)/S cell cycle progression in gastric cancer cells. World J Gastrointest Oncol. 2011;3:119–122. doi: 10.4251/wjgo.v3.i8.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miyazaki H, Shiozaki A, Niisato N, Ohsawa R, Itoi H, Ueda Y, Otsuji E, Yamagishi H, Iwasaki Y, Nakano T, et al. Chloride ions control the G1/S cell-cycle checkpoint by regulating the expression of p21 through a p53-independent pathway in human gastric cancer cells. Biochem Biophys Res Commun. 2008;366:506–512. doi: 10.1016/j.bbrc.2007.11.144. [DOI] [PubMed] [Google Scholar]
- 7.Ohsawa R, Miyazaki H, Niisato N, Shiozaki A, Iwasaki Y, Otsuji E, Marunaka Y. Intracellular chloride regulates cell proliferation through the activation of stress-activated protein kinases in MKN28 human gastric cancer cells. J Cell Physiol. 2010;223:764–770. doi: 10.1002/jcp.22088. [DOI] [PubMed] [Google Scholar]
- 8.Shiozaki A, Miyazaki H, Niisato N, Nakahari T, Iwasaki Y, Itoi H, Ueda Y, Yamagishi H, Marunaka Y. Furosemide, a blocker of Na+/K+/2Cl- cotransporter, diminishes proliferation of poorly differentiated human gastric cancer cells by affecting G0/G1 state. J Physiol Sci. 2006;56:401–406. doi: 10.2170/physiolsci.RP010806. [DOI] [PubMed] [Google Scholar]
- 9.Hiraoka K, Miyazaki H, Niisato N, Iwasaki Y, Kawauchi A, Miki T, Marunaka Y. Chloride ion modulates cell proliferation of human androgen-independent prostatic cancer cell. Cell Physiol Biochem. 2010;25:379–388. doi: 10.1159/000303042. [DOI] [PubMed] [Google Scholar]
- 10.Nishihira T, Hashimoto Y, Katayama M, Mori S, Kuroki T. Molecular and cellular features of esophageal cancer cells. J Cancer Res Clin Oncol. 1993;119:441–449. doi: 10.1007/BF01215923. [DOI] [PubMed] [Google Scholar]
- 11.Shimada Y, Imamura M, Wagata T, Yamaguchi N, Tobe T. Characterization of 21 newly established esophageal cancer cell lines. Cancer. 1992;69:277–284. doi: 10.1002/1097-0142(19920115)69:2<277::aid-cncr2820690202>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- 12.Sobin L, Gospodarowicz M, Wittekind C, eds . TNM Classification of malignant tumors. 7th ed. Hoboken, NJ: John Wiley Sons, Inc; 2009. [Google Scholar]
- 13.Remmele W, Stegner HE. [Recommendation for uniform definition of an immunoreactive score (IRS) for immunohistochemical estrogen receptor detection (ER-ICA) in breast cancer tissue] Pathologe. 1987;8:138–140. [PubMed] [Google Scholar]
- 14.Kunzelmann K. Ion channels and cancer. J Membr Biol. 2005;205:159–173. doi: 10.1007/s00232-005-0781-4. [DOI] [PubMed] [Google Scholar]
- 15.Schönherr R. Clinical relevance of ion channels for diagnosis and therapy of cancer. J Membr Biol. 2005;205:175–184. doi: 10.1007/s00232-005-0782-3. [DOI] [PubMed] [Google Scholar]
- 16.Lastraioli E, Guasti L, Crociani O, Polvani S, Hofmann G, Witchel H, Bencini L, Calistri M, Messerini L, Scatizzi M, et al. herg1 gene and HERG1 protein are overexpressed in colorectal cancers and regulate cell invasion of tumor cells. Cancer Res. 2004;64:606–611. doi: 10.1158/0008-5472.can-03-2360. [DOI] [PubMed] [Google Scholar]
- 17.Kim CJ, Cho YG, Jeong SW, Kim YS, Kim SY, Nam SW, Lee SH, Yoo NJ, Lee JY, Park WS. Altered expression of KCNK9 in colorectal cancers. APMIS. 2004;112:588–594. doi: 10.1111/j.1600-0463.2004.apm1120905.x. [DOI] [PubMed] [Google Scholar]
- 18.Shao XD, Wu KC, Hao ZM, Hong L, Zhang J, Fan DM. The potent inhibitory effects of cisapride, a specific blocker for human ether-a-go-go-related gene (HERG) channel, on gastric cancer cells. Cancer Biol Ther. 2005;4:295–301. doi: 10.4161/cbt.4.3.1500. [DOI] [PubMed] [Google Scholar]
- 19.Wang XT, Nagaba Y, Cross HS, Wrba F, Zhang L, Guggino SE. The mRNA of L-type calcium channel elevated in colon cancer: protein distribution in normal and cancerous colon. Am J Pathol. 2000;157:1549–1562. doi: 10.1016/S0002-9440(10)64792-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tsavaler L, Shapero MH, Morkowski S, Laus R. Trp-p8, a novel prostate-specific gene, is up-regulated in prostate cancer and other malignancies and shares high homology with transient receptor potential calcium channel proteins. Cancer Res. 2001;61:3760–3769. [PubMed] [Google Scholar]
- 21.Luo X, Fang G, Coldiron M, Lin Y, Yu H, Kirschner MW, Wagner G. Structure of the Mad2 spindle assembly checkpoint protein and its interaction with Cdc20. Nat Struct Biol. 2000;7:224–229. doi: 10.1038/73338. [DOI] [PubMed] [Google Scholar]
- 22.Luo X, Tang Z, Xia G, Wassmann K, Matsumoto T, Rizo J, Yu H. The Mad2 spindle checkpoint protein has two distinct natively folded states. Nat Struct Mol Biol. 2004;11:338–345. doi: 10.1038/nsmb748. [DOI] [PubMed] [Google Scholar]
- 23.Sansam CL, Shepard JL, Lai K, Ianari A, Danielian PS, Amsterdam A, Hopkins N, Lees JA. DTL/CDT2 is essential for both CDT1 regulation and the early G2/M checkpoint. Genes Dev. 2006;20:3117–3129. doi: 10.1101/gad.1482106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ababou M, Dutertre S, Lécluse Y, Onclercq R, Chatton B, Amor-Guéret M. ATM-dependent phosphorylation and accumulation of endogenous BLM protein in response to ionizing radiation. Oncogene. 2000;19:5955–5963. doi: 10.1038/sj.onc.1204003. [DOI] [PubMed] [Google Scholar]
- 25.Shabbeer S, Omer D, Berneman D, Weitzman O, Alpaugh A, Pietraszkiewicz A, Metsuyanim S, Shainskaya A, Papa MZ, Yarden RI. BRCA1 targets G2/M cell cycle proteins for ubiquitination and proteasomal degradation. Oncogene. 2013;32:5005–5016. doi: 10.1038/onc.2012.522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wan Z, Zhi N, Wong S, Keyvanfar K, Liu D, Raghavachari N, Munson PJ, Su S, Malide D, Kajigaya S, et al. Human parvovirus B19 causes cell cycle arrest of human erythroid progenitors via deregulation of the E2F family of transcription factors. J Clin Invest. 2010;120:3530–3544. doi: 10.1172/JCI41805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jiang B, Hattori N, Liu B, Nakayama Y, Kitagawa K, Sumita K, Inagaki C. Expression and roles of Cl- channel ClC-5 in cell cycles of myeloid cells. Biochem Biophys Res Commun. 2004;317:192–197. doi: 10.1016/j.bbrc.2004.03.036. [DOI] [PubMed] [Google Scholar]
- 28.Menegazzi R, Busetto S, Dri P, Cramer R, Patriarca P. Chloride ion efflux regulates adherence, spreading, and respiratory burst of neutrophils stimulated by tumor necrosis factor-alpha (TNF) on biologic surfaces. J Cell Biol. 1996;135:511–522. doi: 10.1083/jcb.135.2.511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Niisato N, Eaton DC, Marunaka Y. Involvement of cytosolic Cl- in osmoregulation of alpha-ENaC gene expression. Am J Physiol Renal Physiol. 2004;287:F932–F939. doi: 10.1152/ajprenal.00131.2004. [DOI] [PubMed] [Google Scholar]
- 30.Gerdes J, Lemke H, Baisch H, Wacker HH, Schwab U, Stein H. Cell cycle analysis of a cell proliferation-associated human nuclear antigen defined by the monoclonal antibody Ki-67. J Immunol. 1984;133:1710–1715. [PubMed] [Google Scholar]


