Abstract
The mammary gland is a highly regenerative organ that can undergo multiple cycles of proliferation, lactation and involution, a process controlled by stem cells. The last decade much progress has been made in the identification of signaling pathways that function in these stem cells to control self-renewal, lineage commitment and epithelial differentiation in the normal mammary gland. The same signaling pathways that control physiological mammary development and homeostasis are also often found deregulated in breast cancer. Here we provide an overview on the functional and molecular identification of mammary stem cells in the context of both normal breast development and breast cancer. We discuss the contribution of some key signaling pathways with an emphasis on Notch receptor signaling, a cell fate determination pathway often deregulated in breast cancer. A further understanding of the biological roles of the Notch pathway in mammary stem cell behavior and carcinogenesis might be relevant for the development of future therapies.
Keywords: Breast cancer, mammary stem cells, notch signaling
INTRODUCTION
Stem cells in adult tissues are of key importance for physiological tissue renewal and regeneration after injury. Understanding the molecular pathways that govern normal stem cell function may aid in the development of tissue-specific cell replacement therapies whereby stem cells adopt specific cell fates and functions in any desired organ. Although still in its infancy, cell replacement therapy using autologous stem cells holds great promise for overcoming genetic disorders and tissue regeneration after damage. Under pathological conditions such as uncontrolled proliferation and metastases formation in cancer, populations of tumor cells have been identified that are collectively referred to as “tumor-initiating cells (TIC) or cancer stem cells (CSC)” controlling cancer cell maintenance and growth. These cells are thought to harbor many properties of normal stem cells in that they exhibit self-renewing capacity, multipotency and their ability to initiate and sustain neoplastic growth. Increasing evidence indicates that these CSC are responsible for cancer cell maintenance that underlies malignant progression and recurrence after treatment failure.
Breast cancer affects almost 1 in 8 women in the Western world with a total of about one million new cases per year worldwide of which 35% will eventually die. Breast cancer is a complex and heterogeneous disease with several histological and molecular manifestations within tumors and between patients. A comprehensive understanding of the etiology of breast cancer is paramount to the identification of novel therapies and improving existing strategies for treatment and prevention of the disease. The mammary gland is a dynamic organ that goes through significant changes during the menstrual cycle, development, pregnancy, lactation, and involution. Normal mammary gland development and homeostasis is a stem cell driven process and key signaling pathways have been identified that control these processes. Mounting evidence indicates that the same genes that control physiological organ development and function are often deregulated in cancer.
Here we provide an overview on the functional and molecular characterization and identification of mammary stem cells in the context of normal breast development and their role in breast cancer. We highlight the key molecular pathways controlling these processes with a special emphasis on the role of Notch receptor signaling, a highly conserved cell fate determination pathway frequently altered in human breast cancer.
STEM CELLS
Stem cells can be categorized into two types: pluripotent and multipotent. Pluripotent stem cells are cells that can give rise to the three germ layers, endoderm, mesoderm and ectoderm and propagate by symmetric cell division; i.e. the capacity to give rise to identical daughter cells with identical fates. Examples are embryonic stem cells and embryonic germ cells, which will not be discussed further here. Unlike embryonic stem cells, adult stem cells are multipotent and lineage-restricted and divide by asymmetric cell divisions producing a daughter and a mother at every division with distinct cell fates.
Multipotent stem cells are present in many if not all adult tissues and are rare immature cells characterized by the ability to undergo unlimited self-renewal and differentiate into all cells of a given lineage [1]. Until recently it was assumed that stem cells are quiescent or slow-dividing so-called label retaining cells (LRCs) [2], but that dogma has recently been challenged by the identification of rapidly dividing populations of stem cells in the epidermis and gut where tissue replenishment is a continuous process [3-5]. It has become clear that these organs have both slow and fast dividing stem cell populations that perform different and overlapping functions in regulating tissue homeostasis [6].
The enormous interest in stem cell research over the last decade has led to the identification of many different markers that allow the identification and purification of cell types with long term repopulating activity in various tissues and organs. So far the most well characterized stem cells are those which form blood, namely Hematopoietic Stem Cells (HSCs) [1]. Currently, there is a good understanding of hierarchical organization of hematopoietic lineage commitment [1]. A similar organization has been observed in other self-renewing tissues such as the intestinal epithelium [7]. Approaches analogous to the ones applied for the hematopoietic system and intestinal stem cells have been employed for the identification of mammary stem cells. Not surprisingly the continuous cell renewal required during each menstrual cycle and for lactation as well as regression in the mammary gland is also controlled by mammary stem cells [8]. The existence of a slow (LRCS) and a fast proliferating stem/progenitor populations within the mammary gland has been postulated [9].
Cancer Stem Cells
Stem cells are obvious targets of accumulating oncogenic mutations due to their longevity and capacity for indefinite proliferation [10]. However, it was only in nineties until John Dick and colleagues established the existence of tumor initiating cells or CSCs from myeloid leukemia. By careful immunophenotyping and fractionation of subpopulations of tumor cells, they established that only a fraction of the bulk of the myeloid tumor had the capacity to self-renew in vitro and give rise to transplantable leukemias in vivo [11, 12]. Whereas all cancer cells harbored the initiating mutation, only a fraction was able to initiate and maintain neoplastic growth. From these studies the concept was put forward that leukemia’s are composed of a hierarchy of undifferentiated immature cells to more differentiated cells with limited potential for self-renewal. Soon hereafter colon [13], brain [14], breast [15], pancreas [16], prostate [17], and melanoma [18] CSCs have been postulated. According to this hypothesis, normal stem cells that acquire mutations during tumor evolution continue to exist within tumors and are responsible for the initiation and maintenance of neoplastic growth. TICs retain key stem cell properties such as self-renewal and the capacity to generate progenitor cells, in contrast with the bulk of tumor cells. There is increasing evidence that TICs are enriched in breast cancer patients after conventional treatment, indicating their intrinsic therapeutic resistance [19]. Thus, the first step towards understanding breast carcinogenesis is to identify the pathways that regulate normal breast development and homeostasis. This understanding may lead to insight into the pathways that drive cancer formation, progression, maintenance and resistance to therapy.
MAMMARY GLAND DEVELOPMENT
In mammals the first step in mammary morphogenesis is a thickening of the ventral ectoderm also referred to as the milk or mammary line. This structure gives rise to placodes: the precursor to the mammary bud that will give rise to a ductal branching network rooted within the fat pad and attached to the nipple. Whereas humans only have one pair of placodes that develop into two breasts, mice have 5 pair symmetrically distributed along the rostral-caudal axis between the upper- and hind limbs developing in 10 functional mammary glands. Early mammary gland morphogenesis relies on coordinated signaling between the epithelium and the underlying mesenchyme similar to the development of other epithelial appendages (e.g. limbs, hair follicles). There are important differences however between murine and human mammary gland development. A brief overview of the key steps in mouse mammary gland development is given below and indicated where human development differs significantly.
Mouse Mammary Gland Development
Mouse mammary gland development starts around embryonic day 10.5 (E10.5) and is complete just before birth at E19 day. Just around the time of milk line development Wnt10b expression marks the epithelial and mesenchymal cells destined to form the future mammary gland [20]. Canonical Wnt signaling promotes mammary placode development and is essential for initiation of mammary gland morphogenesis and maintained in the ducts until E15.5. Activation of Wnt signaling induces placode formation and size [21, 22]. Canonical Wnt signaling is mediated by the transcription factor Lef1 and epithelial Wnt10b expression is driven by Fgf10 produced by mesenchymal cells from the somites, which is essential for midline and placode formation [23]. Together with Lef1, Tbx3 is also expressed during early mammary gland development. The combination of signaling pathways Tbx3, Fgf and Wnt regulates epithelial-mesenchymal interactions during this time. Both Fgf and Wnt signaling seem to maintain Tbx3 expression while this leads to the expression of Lef1 [24]. It is important to note that due to the spatial distribution of placodes along the rostro-caudal axis, each pair is also exposed to unique signaling cues [25]. During the embryonic development, the mammary gland remains quiescent until E15.5-E16.5 when ductal growth is stimulated by steroid hormones [26]. A combination of steroid and locally acting growth hormones like Insulin-like growth factor [27], estrogen, progesterone, and somatotropin [28] mediate developmental signals and work synergistically in the transmission of these signals to the stromal and epithelial components of the mammary gland [29]. The lumen is generated by apoptosis of central cells in the multilayered epithelium while ducts expand into the fatpads by growth centers with high mitotic rates at the tip of ducts called terminal end buds (TEBs) [30] where stem cells are thought to reside [31].
After birth, ductal growth and branching from TEBs give rise to fully developed mammary outgrowths between 3-12 weeks of age. Mammary gland goes through changes during pregnancy such as lobulo-alveolar outgrowth (pregnancy), differentiation-secretion (lactation), and apoptotic regression (involution) as part of the normal reproductive cycle [32]. Peptide and steroid hormones together with the interactions of extracellular matrix are responsible of regulating these different phases. Estrogen, progesterone, and prolactin and their cognate receptors are well known regulators required for the successful completion of this developmental cycle [33].
The adult mammary gland is composed of a highly branched system of ducts that terminate in a lobulus. Lobules are composed of alveoli, secretory cells that produce milk. Ducts are composed of two major cell types: an inner layer of luminal epithelial cells and an outer layer of contractile myoepithelial cells which are responsible for contraction in response to oxytocin during lactation [34]. The rapid proliferation and differentiation of mammary gland stem cells at the onset of lactation is necessary to provide secretory activity for milk production [35]. After weaning milk production ceases and the gland involutes [36].
Morphological and Functional Differences Between Mouse and Human
Mammary development in humans starts as a primary ectodermal outgrowth in 4-6 months old embryo [37]. At this stage, the primary bud contains a central and a peripheral-basal cell population which will give rise to different cell layers [38]. The existence and organization of these distinct cell layers is important in both mice and humans for the correct functioning of the gland and a unique feature of the mammary gland. The epithelial buds start to form from the primary bud in the 21-25 weeks of embryonic age [38] (Fig. 1). Breast development differs between individuals of mice at the level of birth. While some individuals have a few branched ducts at that point of development, highly structured ductal tree together with regular lobules as it is observed in adults are also observed [39-41]. After birth, the effect of maternal hormones lessens and the newborn’s breast involutes. In females, the ductal tree development and the stromal enlargement continue further only during puberty [42].
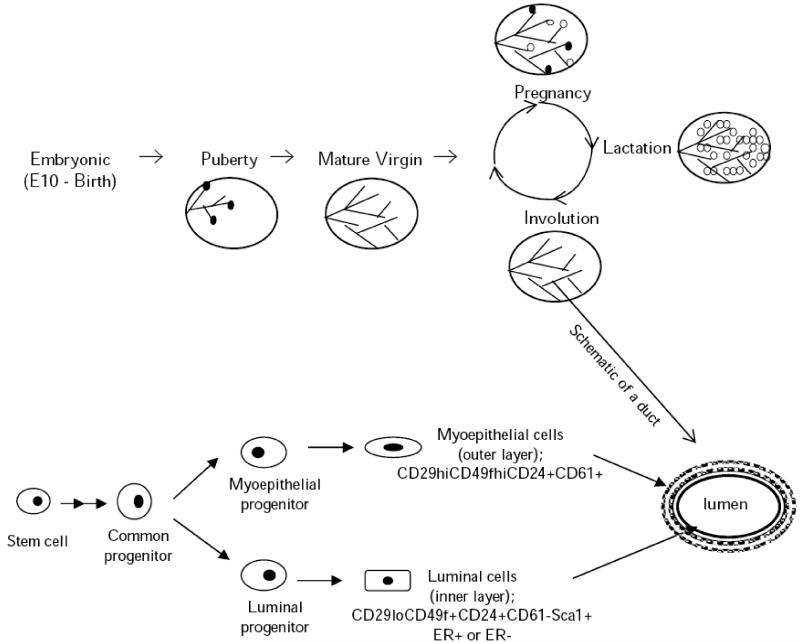
Fig. (1). A schematic representation of mouse mammary gland development and a duct.
The closed circular structures appearing at the ductal tips during puberty represent terminal end buds and during pregnancy, also alveolar buds. Mature alveoli are symbolized by the open circles.
Mouse and human mammary gland also differ in terms of morphology. In humans, branching ducts connect to groups of small terminal ductal and alveolar structures, which are collectively called terminal ductal lobular units (TDLUs). On the contrary, TDULs are not a feature of mouse mammary epithelial tree. Instead, in the mouse, once the mammary gland is stimulated by hormones, ducts branch and elongate from TEBs. Despite these differences, there is ample evidence supporting overall similar morphology and function including the existence of mammary epithelial stem cells [43].
Developmental Patterning and Breast Cancer
Pathways involved in patterning and morphogenesis during mammary gland development are also implicated in breast cancer formation. Among these are neuregulin3 (NRG3) (an epidermal growth factor receptor (EGFR) ligand [44] involved in placode induction), Wnt signaling (essential for midline specification [20]), fibroblast growth factor (FGF) (critical for inductive signaling to the placode [45]) and Notch signaling [46] important in luminal cell fate commitment which are all frequently activated/mutated in human breast cancer. Importantly, the same pathways have also been identified by insertion mutagenesis as common proviral insertions in mouse mammary tumor virus (MMTV) induced breast cancer [47] indicating also a causal role for these signaling cascades in breast cancer in mice. Similar insertions by a complete proviral sequence, 95% to 99% homologous to MMTV, were observed also in human breast cancer tissue. This virus was named as Human Mammary Tumor Virus (HMTV) [48]. The percentage of HMTV occurrence is shown to reach up to 42% in breast cancers occurring in Europe, North America [49] and Australia [50] compared to the healthy breast tissues, which do have 1 to 2 % HMTV prevalence. High percentage of insertions seems to play a role in some human breast cancers.
EVIDENCE FOR MAMMARY STEM CELLS
The postnatal development of the mammary gland makes it possible to surgically remove the rudimentary gland leading to an epithelium free (cleared) fat pad and to study mammary development by transplantation from donor mice. The first functional evidence for the presence of murine mammary epithelial stem cells comes from transplantation experiments of DeOme [51]. This study showed that small tissue fragments isolated from a randomly selected portion of the mammary gland were capable of regenerating a functional ductal tree upon transplantation into a cleared mammary fat-pad. Moreover, explants taken from a regenerated gland could be serially transplanted to other fat-pads [52]. While these experiments strongly pointed to a long-lived pool of epithelial progenitors, whether these contained “true” stem cells was questioned since transplant ability and ductal outgrowth could only be maintained for up to 7 generations [53-55]. More conclusive evidence for a mammary stem cell was obtained by clonal analysis using insertion mutagenesis with MMTV. These elegant studies demonstrated for the first time that an entire mammary gland composed of luminal and myoepithelial cells was a clonal derivative of a single cell. This “stem cell” was also capable of forming restricted lineages that produced only lobules or only ductal outgrowths without lobules that cannot expand upon impregnation [56]. Accordingly, it appears that mammary epithelial cell populations generated by a single stem cell comprise distinct and multipotent epithelial cells which are able to proliferate. Moreover, MMTV tagged progenitors survived multiple rounds of pregnancy and involution demonstrating that these cell had the capacity for robust proliferation and differentiation and were protected from apoptosis. It is important to note that such a stem-cell population would be exquisitely sensitive to oncogenic transformation.
It took almost another decade for researchers to identify markers that could be used to enrich for this multi-potent mammary stem cell (MaSC) and corroborate these earlier findings [57, 58].
Molecular Characterization of Mammary Stem Cells (MaSC)
Researchers have been using several complementary methods to isolate and study mammary stem/progenitor cells. The isolation of stem cells based on the expression of cell surface markers has been the most commonly used technique. The knowledge obtained in many different systems such as neuronal, hematopoietic and epidermal lineage has been an advantage for the researchers working on the characterization of mammary epithelial stem cells. The following part will provide an overview of the techniques used to identify mammary stem cells.
The side population technique; The Side Population (SP) is a population of cells that are characterized for their ability to actively exclude vital dyes from being taken up by the cell such as Hoechst and Rhodamine 123 while maintaining other stem cell-like markers [59, 60]. Dye uptake is an active process mediated by ATP-binding cassette family of multi drug resistance proteins e.g. MDR1/P-glycoprotein) and SP staining can be inhibited by drugs such as verapamil. Drug efflux is restricted to normal stem cells and CSC and not observed in differentiated cells. Expression of ABC is closely correlated with stem cells from a wide variety of tissues sources ranging from embryonic stem cells to bone marrow [61]. Mammary gland SP isolated from reduction mammoplasty [62], normal human [63, 64] and mouse mammary gland [65, 66] respectively have been shown to be enriched by stem/progenitor cells based on their ability to generate lobuloalveolar and ductal outgrowths upon transplantation.
Label retention; Label retaining cell (LRC) is another approach to identify stem cells. This approach is based on differential retention of nucleotide analogs (3H-Thymidine (3HTdR) or 5-BromodeoxyUridine (BrdU)) that are incorporated during S-phase between slowly proliferating versus fast proliferating cells. The LRC hypothesis has been very popular for many years particularly for epidermal and intestinal stem cells [2, 67] however recent work has challenged this hypothesis by the isolation of fast-cycling stem cells [6]. LRCs have also been reported in the mammary gland [66, 68, 69]. The undifferentiated nature of LCRs in mammary gland has been identified by the lack of differentiation markers [9]. This population of cells has been shown to divide asymmetrically and is able to repopulate stem cell niches. Although asymmetric cell divisions of stem cells have been proposed before, in a more recent study, Smith et al. tried to answer the question by labeling mice first with 3HTdR and then with BrdU. 3HTdR label was retained by LRCs in the mammary gland. Following the pulse of BrdU as a second label, most of 3HTdR labeled LRCs were shown to have the BrdU label as well. As a next step, it was shown that these cells were able to undergo asymmetric divisions and pass the newly synthesized BrdU labeled strand to their progeny while retaining the 3HTdR label [69].
Hormone receptor status; steroid hormones have a huge impact on mammary development and the majority of breast cancers are estrogen receptor (ER) positive and are an important target for anti-hormonal therapy [70]. In view of this, it is not surprising that researchers have been trying to characterize MaSCs with respect to steroid receptor expression. Previously, studies of Clarke et al. demonstrated the existence of steroid receptor positive cells within LRC and SP with full self-renewal and differentiation capacity [71]. On the other hand, more recent studies revealed that both mouse and human MaSCs show a ER, PR and ErbB2 negative phenotype [72]; but yet they respond to ovarian hormone signaling [73].
Mammosphere culture; this approach has been used to functionally identify MaSC and TICs based on their ability to form mammospheres in culture that give rise to tumors containing all the differentiated cell types present in the original tumors. [64]. Using this technique, the differentiation capacity (based on the markers specific for different lineages of the mammary gland) and clonality (i.e. by retroviral tagging) of cells enriched in the mammospheres was demonstrated. Cells from mammospheres can be serially passaged and retain their multipotency for multiple passages.
ALDEFLUOR assay; this is another promising approach to identify stem cells such as hematopoietic and neural stem cells which have high aldehyde dehydrogenase 1 (ALDH1) activity [74]. ALDH1 is a detoxifying enzyme responsible for the oxidation of retinol to retinoic acid and it may have a role in the early differentiation of stem cells [75, 76]. ALDH1+ cells were shown to possess functional and phenotypic characteristics of mammary stem cells even though they seem to be restricted luminal epithelial layer [77]. It has been shown that ALDH1+ cells can survive and proliferate under anchorage independent conditions and they are capable of self-renewal. Furthermore clonogenic assays demonstrated that these cells do have the ability to give rise to mixed colonies of myoepithelial and luminal cells. ALDH1 expressing cells have been demonstrated in the normal human mammary gland and human breast cancers [78].
Cell surface markers; to date, different combinations of cell surface antigens have been used for the isolation of human and mouse mammary epithelial stem/progenitor cells. Although several research groups reached to an agreement on the cell surface expression profiles of stem and certain progenitor cell groups, it is important to keep in mind that the interpretation of the FACS data obtained so far mostly depends on the use of antibodies conjugated to different fluorochromes, antibody titrations and good controls. In the following part, we will review markers used to identify mouse or human mammary stem cells that are summarized in Table 1.
Table 1. Commonly Used Surface Markers to Identify Mouse and Human Mammary Stem Cells.
| Mammary Gland Stem Cells | Marker |
|---|---|
| Mouse | CD24, CD29, CD49f, CD61, Sca-1. |
| Human | ALDH1, c-KIT, CD10, CD24, CD44, CD49f, CD90, CD133, EpCAM, MUC-1 |
Mouse Mammary Stem Cells
Defined subsets of mouse mammary epithelial cells have been used to show stem cell capacity by being able to reconstitute mammary glands when transplanted into cleared fat pads [43]. So far, the most useful markers for mouse MaSCs have been CD49 ( 6 integrin), CD29 ( 1 integrin), CD61 ( 3 integrin), CD24, and Sca-1 [79]. Mouse MaSCs could be purified based on CD24+CD29hiCD49fhiSca1- profile following the removal of stromal and hematopoietic cells [57, 58, 80].
A single genetically tagged cell from this population could give rise to an entire epithelial tree upon transplantation [57]. The self renewal and multi-lineage capacity of these cells was demonstrated by serial transplantations into cleared fat pads leading to mature functional glands with the formation of milk producing alveolar units upon impregnation. Moreover, when these genetically tagged cells were cultured together with wild type cells, they were able to produce chimeric structures [57, 58]. By these experiments, researchers were able to show that the purified cells based on the markers mentioned above did indeed contain stem cells. It is important to note that only a small group of MaSCs is characterized by the CD24+CD29hiCD49fhiSca1− population, others have identified other combinations of markers that indicate stemness (e.g. CD45−Ter119−CD31−CD49fhiCD24med. Such populations are not pure since committed progenitors and differentiated cells are also present that express these markers as well [58].
Some of the markers used to enrich MaSC populations are interesting due to their regulatory roles. For example high expression levels of integrin 6 (CD49f) and 1 (CD29) both important for adhesion and migration indicates an important role for stemness and microenvironment interactions [81]. It has been previously shown that both 6 and 1 integrin expression decreases during breast cancer progression [82, 83]. Reduced levels of integrins might give rise to reduced interaction of MaSCs with the microenvironment and a failure in the self-renewal and proliferation. Deletion of 1 integrins from Keratin-5 expressing basal cells led to defects in mammary morphogenesis, proliferation and survival of mammary epithelial cells. 1 integrin deficient transplants efficiently regenerated an entire new gland albeit slower than control transplants and were characterized by disorganized ductal outgrowth indicating a lack of regenerative potential in the absence of integrin 1 [84]. A comprehensive review on the role of integrins and extracellular matrix in mammary gland development has recently appeared [85].
Human Mammary Stem Cells
Common patterns of X chromosome-inactivation and loss of heterozygosity in adjacent areas of breast epithelium (so-called “Field effect”) have provided proof for the clonality of lineages within the normal and neoplastic human mammary gland [86, 87]. Further evidence for a hierarchical model of human breast epithelial development has been obtained by in vitro clonogenic assays where the human mammary stem cells have been shown to reside within the ducts since it was possible to serially passage ductal fragments [64, 88, 89]. The attempts to characterize human mammary stem cells have resulted in identification of a combination of markers such as epithelial cell adhesion molecule (EpCAM), ALDH1 and CD49f [88-92].
Eirew et al. developed a xenotransplantation approach to study the in vivo characteristics of human mammary epithelial cells. A population of cells having EpCAMlowCD49fhi profile was sorted by FACS from reduction mammoplasty samples and these cells were subsequently transplanted into immunodeficient mice together with irradiated fibroblasts. The in vivo experiments of Eirew et al. and Lim et al. revealed that EpCAMlowCD49fhi population are positioned basally within the mammary gland ducts and posses stem cell characteristics [90, 93]. Even though these two markers do identify different cell types when used alone, together they define a subset of human mammary cells, which have regenerative capacity in a xenograft mouse model. The self-renewal and regenerative capacity of EpCAMlowCD49fhicells was further supported by transplantation into humanized mouse mammary fat pads -Human in Mouse model (HIM)-[93, 94]. In this model, the mouse mammary fat pads are humanized by pre-injection of immortalized human fibroblasts. Using the same method, Ginestier identified another subpopulation of human stem/progenitor cell population expressing high levels of ALDH1, with high engraftment capability into NOD/SCID mice [77]. It should be noted that not only MaSCs but also luminal lineage restricted cells express ALDH1 therefore it is necessary to use a combination of different markers together with ALDH1 such as EPCAM, CD49f, MUC1 and others to obtain the highest enriched fraction of cells with MaSC ability.
Evidence for Tumor Initiating Cells in Breast Cancer
A large body of evidence has demonstrated the similarities between stem cells and cancer cells. It is well established that stem and progenitor cells are likely targets of genetic mutations necessary for carcinogenesis. The stem cell thory of cancer proposes that there is a small group of cells–cancer stem cells (CSCs) or tumor initiating cells (TICs)- within tumors that foster tumor initiation and maintenance [95, 96]. The first evidence for TICs was based on studies which showed human acute myeloid leukemia originated from a small proportion of cancer cells [12]. Since then, many groups have investigated and demonstrated the existence of tumor cells capable of self-renewal from hematopoietic to solid cancers by using different techniques [26]. A frequently applied approach has been the transplantation of tumor cells classified by cell surface markers into NOD/SCID mice to investigate the ability of subpopulations of tumor cells to give rise to new tumors. Similar approaches have been used to identify TICs in breast cancer, which will be discussed below. The risk of developing breast cancer is 1 in every 8 women in the western world and it increases with age. This is thought to be related to the accumulation of multiple mutations in cells of the mammary gland over lifetime of women. Damaged stem cells with unlimited potential of proliferation can give rise to delayed cancers such as breast cancer after having a normal phenotype for decades. This is important because once it becomes possible to identify such cells; approaches to eliminate them would decrease the risk of breast cancer incidence and recurrence. The first prospective identification of breast cancer stem cells from human breast cancer specimens reported that within lineage - (tumor cells depleted with expression of normal antigens CD2, CD3, CD10, CD16, CD18, CD31, CD64, and CD140b) population up to 35 % of cells displayed a CD44+CD24-/low marker profile that was able to efficiently generate transplantable tumors in mice [15, 97]. Interestingly, ALDH1+ phenotype within a number of human breast tumor samples defined a subpopulation of cancer stem cells not overlapping with the EpCAM+CD44+CD24− phenotype [62]. When the cells were isolated based on ALDH1+EpCAM+CD44+CD24− phenotype, this population was highly enriched with TIC. Another study showed that ALDH1 expression did not differ between BRCA1 (breast cancer 1) mutation carriers (who are at increased risk of developing breast cancer) and non-carriers [78]. Others however have shown a close correlation between Brca1-deficiency and ALDH1 FLUOR positivity in clinical samples and experimental models of Brca1-related cancer [98]. Even though numerous studies [99-102] have proposed the existence of breast TICs, to date the exact nature of these cells remain undefined and far from clinical utility.
SIGNALING CUES REGULATING MAMMARY STEM CELL RENEWAL AND BREAST CANCER
Normal mammary gland development is tightly controlled by TGF-β , Wnt, FGF Hedgehog, EGF, Estrogen and Notch signaling pathways [103]. It has been demonstrated that deregulation of these pathways in the mammary gland can give rise to tumor development. In the following part, we will discuss the role of these pathways with a focus on Notch signaling.
Wnt/β-Catenin
Wnt proteins are highly conserved secreted signaling molecules that regulate cell fate throughout metazoan development and homeostasis. Canonical Wnt proteins transduce signals through frizzled (FZD) transmembrane receptors which lead to inactivation of the tumor suppressor protein APC and stabilizes β-catenin which is the intracellular effector of the Wnt pathway [104]. The importance of Wnt pathway activation in breast cancer was demonstrated by identification of Wnt a common insertion site of MMTV [105]. Besides promoting maintenance and proliferation of stem cells, Wnt signaling is also shown to be crucial in pluripotency. The expression of Nanog, a gene known to be associated with pluripotency and self-renewal of stem cells, is suppressed by the Wnt-responsive transcription factor T cell factor 3 (TCF3) [106]. Wnt involvement in pluripotency is further supported by the effect of upregulation of Oct-4, a transcription factor mostly known through its involvement in the inhibition self-renewal of undifferentiated embryonic stem cells [107]. Aberrant activation of the canonical Wnt pathway is also associated with cancer development in early-onset familial adenomatous polyposis (FAP) and late-onset spontaneous forms of colon cancer [108-109]. Upregulated Wnt activity due to the loss of the Wnt inhibitor SFRP1 (Secreted Frizzled-related protein 1) and high levels of β-catenin is correlated with poor prognosis in breast cancer [110, 111]. Moreover, β-catenin expression has been shown to be associated with survival of mammary epithelial stem cells and more efficient self-renewal [112, 113]. Transplantation of mammary epithelial cells overexpressing constitutively active β-catenin into cleared mammary fat pads gave rise to hyperplasia’s [112]. These results illustrate that upregulated canonical Wnt signaling increases the mammary stem cell activity in vivo. Conclusive evidence for a role of Wnts in mammary stem cell renewal was recently obtained by the Nusse lab that demonstrated that purified Wnts are sufficient to promote clonogenic growth of mammary stem cells in culture and support their long-term repopulation in vivo [114]. It is likely that Wnt induced mitogenic effects in the mammary stem cell niche are causative to tumor initiation.
Hedgehog
The Hedgehog (Hh) pathway is another highly conserved pathway essential for early embryonic patterning and cell fate determination and in the self-renewal and maintenance adult tissues including mammary gland [115-117]. In mammalian cells, signaling takes place between the three secreted ligands; Sonic Hedgehog (Shh), Desert Hegdehog (Dhh) and Indian Hedgehog (Ihh) and cells that express the transmembrane receptor for Hedgehog ligand Patched-1 (PTCH1) and Patched-2 (PTCH2). In the absence of ligands, Ptch forms a complex with another transmembrane protein called Smoothened (Smo) inhibiting the binding of Smo to Gli: a transcription factor. In the presence of Hh, Patched binds to Hh, enabling Smo to activate Gli proteins (Gli1, Gli2 and Gli3) that activate gene transcription. Deregulated Hh signaling has been shown to be associated with a range of malignancies. Mutations found in the Hh pathway genes leading to ligand independent activation have been shown to be associated with several malignancies such as medulloblastoma, sarcoma and basal cell carcinoma [118, 119]. Overexpression of Hh ligands is frequently observed in gastrointestinal tract and lung carcinomas [120].
The evidence supporting Hh involvement in breast carcinogenesis is increasing. In one of the first studies, Lewis and colleagues showed that heterozygous disruption of Ptch1 and Gli-2, impairs ductal morphogenesis resulting in ductal hyperplasias and dysplasias [121]. Constitutive activation of Smo also leads to aberrant proliferation and ductal dysplasia [122]. The Hh pathway was first implicated in breast cancer by Kubo and colleagues who showed a correlation between high levels of Shh, Ptch and Gli1 with growth inhibition by the Hh inhibitor cyclopamine [123]. Although activating mutations in Hh pathway are common in many cancers, they are not frequently associated with breast cancer [124]. Instead, epigenetic events may be more important in Hh pathway activity in breast cancer as demonstrated by the finding that demethylating and deactylating agents can upregulate Ptch expression in breast cancer cell lines [125]. Finally, Patched polymorphisms have been shown to be associated with the risk of oral contraceptive use on breast cancer risk [126]. This suggests that Hh signaling pathway might an important role in hormone induced development of breast cancer.
Furthermore, in an in vitro study, addition of Shh ligand or overexpression of Gli2 into mammosphere cultures increased primary and secondary mammosphere formation and size, which was reversed by cyclopamine, a specific inhibitor of the pathway. Finally, Hh signaling has been associated with the tumorigenic phenotype of CD24+/Cd44-/low/Lin− TICs [127]. Altogether, the Hh pathway plays pivotal roles during normal mammary gland development and likely also plays unrecognized roles in breast cancer as well.
Transforming Growth Factor–Beta TGFβ
The transforming growth factor beta (TGFβ) family of polypeptide growth factors comprises secreted proteins of which there are three isoforms: TGFβ1, TGFβ2, and TGFβ3. TGFβ proteins bind to Type II receptors that heterodimerize to Type I receptors which triggers serine/threonine phosphorylation and activation of receptor SMAD proteins that in turn heterodimerize with other SMAD proteins which enter the nucleus and activate gene-transcription. TGFβ signaling controls many cellular processes during development and in adult tissues including cell proliferation, differentiation, and apoptosis, in species from worms to mammals. Defects in TGFβ signaling occur in many inherited and acquired diseases including cancer [128]. The function of TGFβ in mammary morphogenesis appears to be cell and context dependent. Several groups demonstrated the importance of different local concentrations of TGFβ within ductal epithelium as an inhibitory factor in the ductal growth and lateral branching [129, 130] TGFβ within luminal epithelial cells controls alveolar development [131, 132]. The role of TGFβ in breast cancer is complex as TGFβ has both growth suppressive as well growth promoting activities. In general TGFβ promotes tumor progression in several ways including suppression of immune responses, stimulation of angiogenesis and promotion of epithelial-to-mesenchymal transition [133], [134].
Estrogens and Progestagens
The steroid hormones estrogen and progesterone have key roles during normal mammary gland physiology from puberty to menopause. Deregulation of this hormonal regulation also markedly influences breast cancer risk forming the basis for anti-hormonal therapies in breast cancer treatment. Although the role of hormones in cancer risk has already been known for a long time it was only recently that two groups reported the underlying mechanism [73], [135]. The thought that TICs would directly respond to hormones is attractive however these cells do not express the receptors for either hormone. It appears that proliferative effect of hormones on breast tissue occurs during the normal reproductive cycle and pregnancy when hormones rise causing significant increases in MaSC numbers. The proliferative effect is indirectly mediated by the response of the Niche (basal and luminal cells) surrounding the stem cells. These produce a Wnt4a signal that stimulates MaSC proliferation. These findings pave the way to understanding why hormones can modulate growth and proliferation of the normal mammary gland and may sustain malignant growth. They also suggest that during such hormonal surges stem cells may be particularly vulnerable for accumulating mutations that lay dormant until later in life when women develop breast cancer. Such breast cancers may ultimately be unresponsive to hormonal therapies if their origin is driven by TICs. In hindsight the relationship between estrogens, MaSC and breast cancer may not be totally unexpected given the interplay between transcriptional regulation of estrogen/progesterone and cyclinD1; a gene frequently upregulated in breast cancer [136, 137]. Mice lacking the cyclinD1 gene have defects in hormone-induced proliferation during pregnancy [138] and are protected from breast cancer [139]. Paradoxically these findings do not explain why pregnancy and breast-feeding in the long term protects against breast cancer in humans [140].
Notch
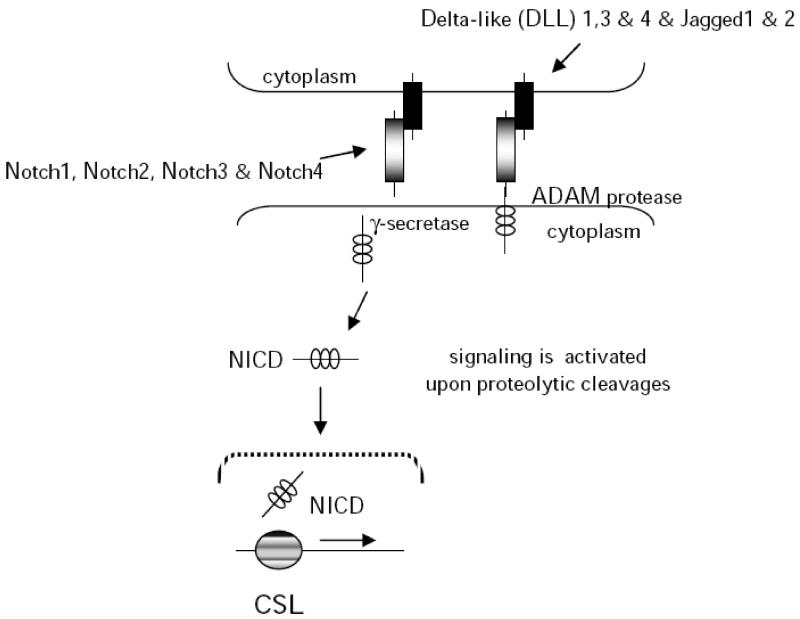
Notch signaling is a short-range cell-cell communication pathway that controls virtually every aspect of metazoan development and cellular responses to maintain tissue homeostasis in adults. Notch signaling occurs between transmembrane bound ligands and receptors on adjacent cells and is conserved from flies to mammals. Flies have a single Notch receptor and two ligands (Delta and Serrate/Jagged) whereas mammalian cells have four receptors and at least five Delta/Jagged type ligands (Fig. 2). Notch proteins are single pass type I transmembrane receptors, with an extracellular domain involved in ligand binding, and a cytoplasmic domain involved in signal transduction [141]. During maturation in the trans-Golgi network, Notch precursors are first cleaved at Site-1 (S1) by Furin-like convertase producing a heterodimeric type I receptor with the Notch extracellular domain (NECD) non-covalently bound to a transmembrane/intracellular fragment (TMIC). The extracellular domain of Notch proteins is composed of 29-36 epidermal growth factor (EGF) like repeats involved in ligand interactions, three cysteïne rich LIN12/Notch (LNR) repeats and a heterodimerization domain (HD). The intracellular domain of Notch (NICD) contains nuclear localization signals together with a transcriptional activation domain (TAD) and a PEST domain [142]. The canonical Notch signaling cascade is regulated by proteolysis. In the absence of ligand, mature Notch receptors are held into an inactive “proteolysis resistant” state because the Negative Regulatory Region (NRR) composed of the HD domain and the Lin12/Notch repeats (LNR) inhibits Notch activation. Ligand binding to Notch receptors unfolds the NRR permitting cleavage by the metalloproteases ADAM10/Kuzbanian at a site close to the membrane termed site-2 [142] leading to shedding of the Notch extracellular domain [142, 143]. Extracellular cleavage of Notch triggers the intramembranous cleavage by the multi-subunit protein complex termed γ-secretase containing the aspartyl protease Presenilin. This leads to the release and translocation of Notch intracellular domain (NICD) to the nucleus where it interacts with the transcription factor CSL (CBF1 in humans; RBP-Jk in mice) to activate Hes/Hey family genes, which are involved in growth and proliferation, differentiation, and survival [144]. In the absence of Notch signaling, CSL inhibits the transcription of target genes.
Fig. (2). A schematic of activation of the Notch pathway.
Notch signaling is activated by proteolytic cleavages followed by the release and translocation of Notch intracellular domain to the nucleus where it acts on downstream targets by binding the transcription factor CSL to activate Hes/Hey family genes which are involved in cell growth, differentiation, and survival.
Most if not all Notch signaling requires metalloprotease and γ-secretase cleavage to release the NICD and induce transcriptional activation by binding to CSL [145]. Notch activity is frequently deregulated in human cancer by overexpression or mutation [146]. Mutations are found in the extracellular HD and intracellular PEST domain and induce ligand-independency and increased stability of Notch [147]. Mutations that affect the activity of Notch are also found in negative regulators such as the ubiquitin ligase Fbw-7 [148, 149]. Given the frequent involvement of Notch in human malignancies, targeting Notch cleavage appears an attractive drug target. At the same time this provides challenges for drug development since such drugs would also target physiological Notch activation. Several clinical trials are underway that evaluate the efficacy of γ-secretase inhibitors (GSIs) as anti-cancer drugs [150]. While targeting γ-secretase has shown encouraging results in targeting tumors with activated Notch it has many pitfalls as well. Among these is the lack of specificity and the mechanism based toxicity caused by attenuating physiological Notch function. For example, the gastrointestinal toxicity caused by precocious secretory differentiation of intestinal epithelial cells prevents long-term use because of intestinal stem cell depletion [151-153]. Real et al. showed that such side-effects may be overcome by combination treatment with glucocorticoid and GSIs which suppressed gut toxicity through inhibition of KLF4 [154]. Since γ-secretase has many different substrates, pleiotropic effects are unavoidable when targeting this enzyme, although modulators have been identified that may show selectivity for specific substrates [155]. Despite the fact that GSIs have been studied intensely for the past decade, a safe-drug has yet to enter clinical practice [150].
Notch in Mammary Development
Members of Notch family have been shown to play important roles in normal breast development by several studies (Fig. 3). In one of the earliest reports, Uyttendaele et al. demonstrated that normal breast epithelial cells failed to differentiate upon overexpression of a constitutively active form of Notch4 in vitro [156]. This was supported by other reports where a constitutively active form of Notch4 was expressed in transgenic mice and lead to a failure in mammary development followed by the progression of mammary tumors [157-159]. The increase in mammosphere numbers in mammosphere culture systems upon activation of Notch signaling via an exogenous peptide showed that Notch signaling has a promoting role in self-renewal in human primary mammary epithelial cells [160]. In addition, Notch activation facilitated proliferation and branching morphogenesis which could be inhibited by using Notch antagonists. Altogether, these reports demonstrate the importance of Notch signaling in the regulation of ductal branching in normal development where aberrant Notch signaling seems to disrupt differentiation and causes excessive proliferation.
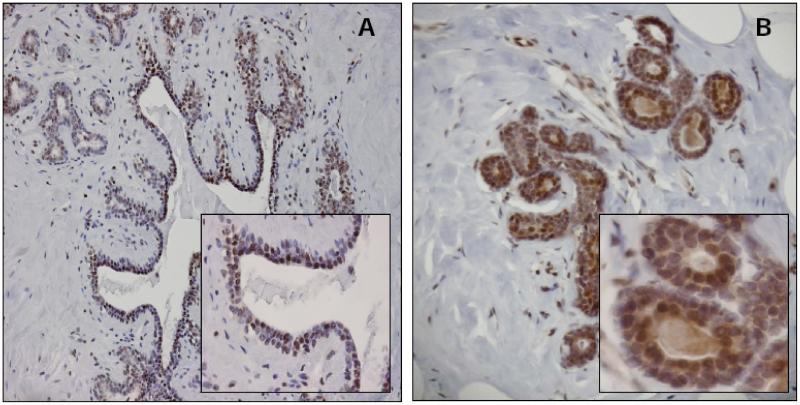
Fig. (3). Notch1 Signaling in Normal Human Breast.
We have observed high Notch1 activity in normal human breast epithelium, indicating the activation of this pathway during normal breast development. Normal human breast tissue probed with an antibody against (A) cleaved Notch1, representing Notch1 activity (10× and 40× magnification), and (B) Hes1 (a downstream target of Notch pathway) (10× and 40× magnification). Similar patterns of expression are evident.
Notch3, another member of the family, was shown to be upregulated in mammospheres formed by normal breast tissue [160] suggesting its role in self-renewal. Notch3 has also been shown to be upregulated in breast cancer cells [161]. Causality between deregulated Notch signaling and mammary carcinogenesis was exemplified by the identification of Notch1 and Notch4 loci as common insertions in MMTV induced tumors [162, 163]. The integration of MMTV into the Notch4 locus results in a Notch protein lacking most of the extracellular domain creating a ligand-independent protein [164-167]. WAP (whey acidic protein) or MMTV (mouse mammary tumor virus)-driven expression of Notch4 in transgenic mice also leads to developmental defects and formation of mammary tumors [158]. The failure in development leads to differentiation defects and hyper-proliferation of immature ductal cells. A possible explanation can be that specified cells within the mammary epithelium govern a proliferating stem cell fate that makes the cells vulnerable to mutational events driving tumorigenesis [158].
Notch activation is likely to be important for alveolar development since Notch4 overexpression leads to aberrant alveolar growth and lactation in both WAP and MMTV-Int3 female mice [157-159]. This observation was supported by the observation that loss of RBP-Jk and Pofut1, a fucoslytransferases necessary for the activity of Notch proteins, leads to a disturbed balance in basal cells at the expense of luminal cell fates [168].
Notch and Mammary Stem Cells
Dontu et al., were among the first who demonstrated the involvement of Notch signaling in mammary stem cell renewal [160]. By utilizing the mammosphere system, they were able to show that human mammary stem/progenitor cells formed an increasing number of mammospheres when Notch signaling was activated, and that sphere formation was inhibited when Notch signaling was blocked. In the same study, they demonstrated the role of Notch4 in myoepithelial lineage commitment and branching morphogenesis of human mammary stem/progenitor cells in three dimensional matrigel cultures. More genetic approaches have also been used to block the Notch pathway, such as the targeted disruption of RBP-jk in the mouse mammary gland [168]. Even though no aberrant changes were observed in virgin animals, alveolar cell maintenance and basal cell proliferation increased dramatically by the loss of RBP-jk upon pregnancy. Thus Notch appears to be regulating alveolar development during pregnancy by controlling the luminal cell fate since the luminal cells fail to differentiate upon loss of Notch signaling. The role of Notch signaling in luminal cell fate commitment was identified by Bouras who demonstrated high Notch1 activity in luminal progenitor cells in vivo [169]. Moreover, constitutive activation of Notch1 in CD29hiCD24+cells was shown to stimulate luminal cell fate commitment leading to excessive proliferation and eventually tumorigenesis. Conversely, knockdown of RBP-Jk led to an increase in number, size and clonogenic capacity of CD29hiCD24+ mammary epithelial cells. Notch pathway inhibition induced aberrant luminal cell differentiation and expansion of basal cells. In contrast, Notch4 was shown to be down-regulated in luminal restricted stem/progenitor cells whereas Notch3, was shown to be upregulated [169]. Functional studies supported this further where Notch3 signaling was blocked and this retarded the luminal cell fate commitment where it stimulated myoepithelial cell differentiation. This was further confirmed by Rouf who performed a transcriptome analysis and showed that upregulation of Notch3 and down-regulation of Notch4 characterized the luminal cell lineage [91].
Notch in Mammary Carcinogenesis
MMTV insertions in the Notch1 locus were first identified in MMTV-Neu mammary tumors [162] as a collaborating oncogene in ErbB2/Neu driven mammary tumorigenesis. Notch1 was shown to be rearranged in 2 out of 24 MMTV-Neu mammary tumors investigated. These insertions caused expression of constitutive active truncated Notch1 proteins in a similar manner to that observed in the case of Notch4. Wnt and Notch signaling also collaborate in the transformation of human primary mammary epithelial cells (HMECs) [170]. In this study, ectopic expression of Wnt1 was shown to induce overexpression of Notch ligands (Dll1, Dll3, and Dll4). Further, it induced Notch receptor cleavage, which leads to Notch activity resulting in mammary epithelial transformation. HMECs with constitutive activation of Notch1 detached from culture plates and began to form spherical structures. However these spheres failed to proliferate, indicating that Notch1 activation alone was not sufficient to induce transformation of these cells. Interestingly, Notch1 activation in 3 dimensional cultures of MCF-10A, an immortalized human mammary epithelial cell line, yields heterogeneous phenotypes such as large and hyper-proliferative structures or small and growth arrested structures [171]. Based on these results, it is postulated that different phenotypes are related with dose and context dependence of Notch pathway [172].
There is increasing evidence that activation of Notch signaling is a frequent event in human breast cancers. In one of the earliest studies, immunohistochemical analysis of Notch1 was observed in high percentage (~57%) of breast cancers in concert with high Ha-RAS expression [173]. Further in vitro experiments revealed that oncogenic Ha-RAS acts at a post-transcriptional level to increase Notch1 activity and that Ha-RAS induced transformation requires Notch1 activation. High Notch1 levels have been correlated mainly with poorly differentiated breast tumors suggesting a possible defect in cell fate decisions, whereas Notch2 expression is inversely correlated to poor prognosis [174]. These data support the notion that different members of the Notch family might have different roles depending on cellular context. Analysis of several human breast cancer cases showed high JAG1 levels besides high Notch1 and Notch3 expression in association to poor overall survival [100, 161, 175]. These findings are in line with research demonstrating the tumor-promoting role of Notch1 and Notch3 in mice [100, 176]. Similarly, Pece et al. showed that loss of NUMB expression (a negative regulator of the Notch pathway) is commonly observed in primary human breast cancer [177].
Notch Signaling as a Prognostic Tool and a Therapeutic Drug Target
To date, it is clear that the Notch pathway plays a crucial role in breast cancer development. Therefore, targeting Notch activity in cancer may provide a bona fide approach for the development of novel therapeutic intervention strategies. Strategies include: (i) preventing ligand-receptor interaction using soluble ligand [178, 179], (ii) ectodomain blocking antibodies [180-183] (iii) targeting metalloprotease cleavage [143, 184] and (iv) targeting γ-secretase cleavage [173]. Crosstalk between Notch signaling and other pathways frequently altered in breast cancer are exploited to sensitize breast cancers to common cancer therapeutics drugs by combined Notch inhibition [185, 186]. Finally, approaches to block transcriptional activation have also been successful and may be implemented for future treatment regimes [187].
Inhibition of γ-secretase activity has been so far the most well developed approach to prevent the activation of Notch signaling. γ-secretase inhibitors (GSIs) have been heavily investigated since the gamma secretase complex also cleaves Aβ peptide which plays an important role in Alzheimer’s disease [188]. These inhibitors can also block the Notch pathway, generating increasing interest for their potential usage as new cancer therapeutics. In case of breast cancer treatment, GSIs seem to have potential in combination with other therapies for individualized therapy. The optimal use of Notch inhibitors was demonstrated to be dependent on ER status of the tumor. It has been suggested that GSIs might be more efficient in combination with chemotherapy for the treatment of ERα/PR negative tumors [189] which do not overexpress Her2/Neu. Accordingly, another study showed that inhibition of Her2 overexpression by trastuzumab or tyrosine kinase inhibitors (TKI) increases Notch1 activity, which then sensitized breast cancer cell lines [185] for GSI treatment.
Expression patterns of Notch pathway genes are also likely to have a prognostic relevance. Recent studies stressed the importance of the detection of Notch expression as a potential prognostic marker in breast cancer. Yao et al. demonstrated the correlation of the expression of Notch1, Notch4 and Jag1 with known prognostic factors such as ER status, tumor grade, Ki67, lymphovascular invasion, lymph node status and tumor size [190].
CONCLUSION
The existence of MaSCs was already demonstrated more than 50 years ago but we are only beginning to understand the identity of these cells, the signaling pathways that regulate their homeostasis and their role in diseases such as breast cancer. Currently, there are a number of important questions remaining to be answered; what are the functional differences between human and murine stem cells in the mammary gland? Where do these stem cells reside within their organs and in what micro-environment (niche)? This knowledge is needed to improve our understanding of breast stem cells and their role in cancer formation and progression. Notch signaling has emerged as an important cell fate determinant in mammary gland morphogenesis where it plays seemingly opposite roles in cell renewal and differentiation. Notch is also frequently deregulated in breast cancer and in some cases expression of Notch or its ligands has shown prognostic significance. The γ-secretase inhibitors are emerging as potent drugs that attenuate Notch signaling and are currently being evaluated in clinical trials including those with breast cancer. The coming years will reveal if and how breast cancer patients may benefit from treatments with Notch inhibition.
ACKNOWLEDGEMENTS
The authors acknowledge Patrick Derksen, Jan Theys and Elsken van der Wall for helpful suggestions and critical input. This work is supported by the European Research Council under the European Community Seventh Framework Programme (FP7/2007-2013)/ERC Grant 208259 (to MV) and the Dutch Cancer Society Grant KWF UU2006-3623 (to MV).
REFERENCES
- [1].Weissman IL. Stem cells: units of development, units of regeneration, and units in evolution. Cell. 2000;100:157–68. doi: 10.1016/s0092-8674(00)81692-x. [DOI] [PubMed] [Google Scholar]
- [2].Cotsarelis G, Sun TT, Lavker RM. Label-retaining cells reside in the bulge area of pilosebaceous unit: implications for follicular stem cells, hair cycle, and skin carcinogenesis. Cell. 1990;61:1329–37. doi: 10.1016/0092-8674(90)90696-c. [DOI] [PubMed] [Google Scholar]
- [3].Barker N, Clevers H. Tracking down the stem cells of the intestine: strategies to identify adult stem cells. Gastroenterology. 2007;133:1755–60. doi: 10.1053/j.gastro.2007.10.029. [DOI] [PubMed] [Google Scholar]
- [4].Jaks V, Barker N, Kasper M, et al. Lgr5 marks cycling, yet long-lived, hair follicle stem cells. Nat Genet. 2008;40:1291–9. doi: 10.1038/ng.239. [DOI] [PubMed] [Google Scholar]
- [5].Snippert HJ, van Es JH, van den Born M, et al. Prominin-1/CD133 marks stem cells and early progenitors in mouse small intestine. Gastroenterology. 2009;136:2187–94 e1. doi: 10.1053/j.gastro.2009.03.002. [DOI] [PubMed] [Google Scholar]
- [6].Li L, Clevers H. Coexistence of quiescent and active adult stem cells in mammals: Science. 2010;327:542–5. doi: 10.1126/science.1180794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].van der Flier LG, Clevers H. Stem cells, self-renewal, and differentiation in the intestinal epithelium. Annu Rev Physiol. 2009;71:241–60. doi: 10.1146/annurev.physiol.010908.163145. [DOI] [PubMed] [Google Scholar]
- [8].Lydon JP. Stem cells: Cues from steroid hormones. Nature. 2010;465:695–6. doi: 10.1038/465695a. [DOI] [PubMed] [Google Scholar]
- [9].Woodward WA, Chen MS, Behbod F, Rosen JM. On mammary stem cells. J Cell Sci. 2005;118:3585–94. doi: 10.1242/jcs.02532. [DOI] [PubMed] [Google Scholar]
- [10].Sell S. Stem cell origin of cancer and differentiation therapy. Crit Rev Oncol Hematol. 2004;51:1–28. doi: 10.1016/j.critrevonc.2004.04.007. [DOI] [PubMed] [Google Scholar]
- [11].Lapidot T, Sirard C, Vormoor J, et al. A cell initiating human acute myeloid leukaemia after transplantation into SCID mice. Nature. 1994;367:645–8. doi: 10.1038/367645a0. [DOI] [PubMed] [Google Scholar]
- [12].Bonnet D, Dick JE. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat Med. 1997;3:730–7. doi: 10.1038/nm0797-730. [DOI] [PubMed] [Google Scholar]
- [13].O’Brien CA, Pollett A, Gallinger S, Dick JE. A human colon cancer cell capable of initiating tumour growth in immunodeficient mice. Nature. 2007;445:106–10. doi: 10.1038/nature05372. [DOI] [PubMed] [Google Scholar]
- [14].Singh SK, Hawkins C, Clarke ID, et al. Identification of human brain tumour initiating cells. Nature. 2004;432:396–401. doi: 10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- [15].Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci USA. 2003;100:3983–8. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Li C, Heidt DG, Dalerba P, et al. Identification of pancreatic cancer stem cells. Cancer Res. 2007;67:1030–7. doi: 10.1158/0008-5472.CAN-06-2030. [DOI] [PubMed] [Google Scholar]
- [17].Collins AT, Berry PA, Hyde C, Stower MJ, Maitland NJ. Prospective identification of tumorigenic prostate cancer stem cells. Cancer Res. 2005;65:10946–51. doi: 10.1158/0008-5472.CAN-05-2018. [DOI] [PubMed] [Google Scholar]
- [18].Schatton T, Frank MH. Cancer stem cells and human malignant melanoma. Pigment Cell Melanoma Res. 2008;21:39–55. doi: 10.1111/j.1755-148X.2007.00427.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Li X, Lewis MT, Huang J, et al. Intrinsic resistance of tumorigenic breast cancer cells to chemotherapy. J Natl Cancer Inst. 2008;100:672–9. doi: 10.1093/jnci/djn123. [DOI] [PubMed] [Google Scholar]
- [20].Veltmaat JM, Van Veelen W, Thiery JP, Bellusci S. Identification of the mammary line in mouse by Wnt10b expression. Dev Dyn. 2004;229:349–56. doi: 10.1002/dvdy.10441. [DOI] [PubMed] [Google Scholar]
- [21].Chu EY, Hens J, Andl T, et al. Canonical WNT signaling promotes mammary placode development and is essential for initiation of mammary gland morphogenesis. Development. 2004;131:4819–29. doi: 10.1242/dev.01347. [DOI] [PubMed] [Google Scholar]
- [22].DasGupta R, Fuchs E. Multiple roles for activated LEF/TCF transcription complexes during hair follicle development and differentiation. Development. 1999;126:4557–68. doi: 10.1242/dev.126.20.4557. [DOI] [PubMed] [Google Scholar]
- [23].Veltmaat JM, Relaix F, Le LT, et al. Gli3-mediated somitic Fgf10 expression gradients are required for the induction and patterning of mammary epithelium along the embryonic axes. Development. 2006;133:2325–35. doi: 10.1242/dev.02394. [DOI] [PubMed] [Google Scholar]
- [24].Eblaghie MC, Song SJ, Kim JY, et al. Interactions between FGF and Wnt signals and Tbx3 gene expression in mammary gland initiation in mouse embryos. J Anat. 2004;205:1–13. doi: 10.1111/j.0021-8782.2004.00309.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Robinson GW. Cooperation of signalling pathways in embryonic mammary gland development. Nat Rev Genet. 2007;8:963–72. doi: 10.1038/nrg2227. [DOI] [PubMed] [Google Scholar]
- [26].Farnie G, Clarke RB. Mammary stem cells and breast cancer--role of Notch signalling. Stem Cell Rev. 2007;3:169–75. doi: 10.1007/s12015-007-0023-5. [DOI] [PubMed] [Google Scholar]
- [27].Jones JI, Clemmons DR. Insulin-like growth factors and their binding proteins: biological actions. Endocr Rev. 1995;16:3–34. doi: 10.1210/edrv-16-1-3. [DOI] [PubMed] [Google Scholar]
- [28].Cowie AT. The relative growth of the mammary gland in normal, gonadectomized and adrenalectomized rats. J Endocrinol. 1949;6:145–57. doi: 10.1677/joe.0.0060145. [DOI] [PubMed] [Google Scholar]
- [29].Humphreys RC, Lydon J, O’Malley BW, Rosen JM. Mammary gland development is mediated by both stromal and epithelial progesterone receptors. Mol Endocrinol. 1997;11:801–11. doi: 10.1210/mend.11.6.9891. [DOI] [PubMed] [Google Scholar]
- [30].Molyneux G, Regan J, Smalley MJ. Mammary stem cells and breast cancer. Cell Mol Life Sci. 2007;64:3248–60. doi: 10.1007/s00018-007-7391-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Dontu G, El-Ashry D, Wicha MS. Breast cancer, stem/progenitor cells and the estrogen receptor. Trends Endocrinol Metab. 2004;15:193–7. doi: 10.1016/j.tem.2004.05.011. [DOI] [PubMed] [Google Scholar]
- [32].Wagner KU, Boulanger CA, Henry MD, et al. An adjunct mammary epithelial cell population in parous females: its role in functional adaptation and tissue renewal. Development. 2002;129:1377–86. doi: 10.1242/dev.129.6.1377. [DOI] [PubMed] [Google Scholar]
- [33].Holland MS, Holland RE. The cellular perspective on mammary gland development: stem/progenitor cells and beyond. J Dairy Sci. 2005;88(Suppl 1):E1–8. doi: 10.3168/jds.S0022-0302(05)73132-5. [DOI] [PubMed] [Google Scholar]
- [34].Daniel CW, Smith GH. The mammary gland: a model for development. J Mammary Gland Biol Neoplasia. 1999;4:3–8. doi: 10.1023/a:1018796301609. [DOI] [PubMed] [Google Scholar]
- [35].Tucker HA. Physiological control of mammary growth, lactogenesis, and lactation. J Dairy Sci. 1981;64:1403–21. doi: 10.3168/jds.S0022-0302(81)82711-7. [DOI] [PubMed] [Google Scholar]
- [36].Capuco AV, Wood DL, Baldwin R, McLeod K, Paape MJ. Mammary cell number, proliferation, and apoptosis during a bovine lactation: relation to milk production and effect of bST. J Dairy Sci. 2001;84:2177–87. doi: 10.3168/jds.S0022-0302(01)74664-4. [DOI] [PubMed] [Google Scholar]
- [37].Howard BA, Gusterson BA. Human breast development. J Mammary Gland Biol Neoplasia. 2000;5:119–37. doi: 10.1023/a:1026487120779. [DOI] [PubMed] [Google Scholar]
- [38].Jolicoeur F, Gaboury LA, Oligny LL. Basal cells of second trimester fetal breasts: immunohistochemical study of myoepithelial precursors. Pediatr Dev Pathol. 2003;6:398–413. doi: 10.1007/s10024-003-1125-y. [DOI] [PubMed] [Google Scholar]
- [39].Rabi T, Ramachandran C, Fonseca HB, et al. Novel drug amooranin induces apoptosis through caspase activity in human breast carcinoma cell lines. Breast Cancer Res Treat. 2003;80:321–30. doi: 10.1023/A:1024911925623. [DOI] [PubMed] [Google Scholar]
- [40].Rudland PS. Histochemical organization and cellular composition of ductal buds in developing human breast: evidence of cytochemical intermediates between epithelial and myoepithelial cells. J Histochem Cytochem. 1991;39:1471–84. doi: 10.1177/39.11.1918925. [DOI] [PubMed] [Google Scholar]
- [41].Monaghan P, Perusinghe NP, Cowen P, Gusterson BA. Peripubertal human breast development. Anat Rec. 1990;226:501–8. doi: 10.1002/ar.1092260412. [DOI] [PubMed] [Google Scholar]
- [42].Wellings SR, Jensen HM, Marcum RG. An atlas of subgross pathology of the human breast with special reference to possible precancerous lesions. J Natl Cancer Inst. 1975;55:231–73. [PubMed] [Google Scholar]
- [43].Visvader JE. Keeping abreast of the mammary epithelial hierarchy and breast tumorigenesis. Genes Dev. 2009;23:2563–77. doi: 10.1101/gad.1849509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Lin SY, Xia W, Wang JC, et al. Beta-catenin, a novel prognostic marker for breast cancer: its roles in cyclin D1 expression and cancer progression. Proc Natl Acad Sci USA. 2000;97:4262–6. doi: 10.1073/pnas.060025397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Reis-Filho JS, Simpson PT, Turner NC, et al. FGFR1 emerges as a potential therapeutic target for lobular breast carcinomas. Clin Cancer Res. 2006;12:6652–62. doi: 10.1158/1078-0432.CCR-06-1164. [DOI] [PubMed] [Google Scholar]
- [46].Stylianou S, Clarke RB, Brennan K. Aberrant activation of notch signaling in human breast cancer. Cancer Res. 2006;66:1517–25. doi: 10.1158/0008-5472.CAN-05-3054. [DOI] [PubMed] [Google Scholar]
- [47].Callahan R, Smith GH. Common integration sites for MMTV in viral induced mouse mammary tumors. J Mammary Gland Biol Neoplasia. 2008;13:309–21. doi: 10.1007/s10911-008-9092-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Wang Y, Holland JF, Bleiweiss IJ, et al. Detection of mammary tumor virus env gene-like sequences in human breast cancer. Cancer Res. 1995;55:5173–9. [PubMed] [Google Scholar]
- [49].Wang Y, Jiang JD, Xu D, et al. A mouse mammary tumor virus-like long terminal repeat superantigen in human breast cancer. Cancer Res. 2004;64:4105–11. doi: 10.1158/0008-5472.CAN-03-3880. [DOI] [PubMed] [Google Scholar]
- [50].Ford CE, Faedo M, Rawlinson WD. Mouse mammary tumor virus-like RNA transcripts and DNA are found in affected cells of human breast cancer. Clin Cancer Res. 2004;10:7284–9. doi: 10.1158/1078-0432.CCR-04-0767. [DOI] [PubMed] [Google Scholar]
- [51].Deome KB, Faulkin LJ, Jr., Bern HA, Blair PB. Development of mammary tumors from hyperplastic alveolar nodules transplanted into gland-free mammary fat pads of female C3H mice. Cancer Res. 1959;19:515–20. [PubMed] [Google Scholar]
- [52].Hoshino K, Gardner WU. Transplantability and life span of mammary gland during serial transplantation in mice. Nature. 1967;213:193–4. doi: 10.1038/213193a0. [DOI] [PubMed] [Google Scholar]
- [53].Daniel CW, De Ome KB, Young JT, Blair PB, Faulkin LJ., Jr. The in vivo life span of normal and preneoplastic mouse mammary glands: a serial transplantation study. Proc Natl Acad Sci USA. 1968;61:53–60. doi: 10.1073/pnas.61.1.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Daniel CW, Young LJ, Medina D, DeOme KB. The influence of mammogenic hormones on serially transplanted mouse mammary gland. Exp Gerontol. 1971;6:95–101. doi: 10.1016/0531-5565(71)90053-2. [DOI] [PubMed] [Google Scholar]
- [55].Smith GH, Medina D. A morphologically distinct candidate for an epithelial stem cell in mouse mammary gland. J Cell Sci. 1988;90(Pt 1):173–83. doi: 10.1242/jcs.90.1.173. [DOI] [PubMed] [Google Scholar]
- [56].Kordon EC, Smith GH. An entire functional mammary gland may comprise the progeny from a single cell. Development. 1998;125:1921–30. doi: 10.1242/dev.125.10.1921. [DOI] [PubMed] [Google Scholar]
- [57].Shackleton M, Vaillant F, Simpson KJ, et al. Generation of a functional mammary gland from a single stem cell. Nature. 2006;439:84–8. doi: 10.1038/nature04372. [DOI] [PubMed] [Google Scholar]
- [58].Stingl J, Eirew P, Ricketson I, et al. Purification and unique properties of mammary epithelial stem cells. Nature. 2006;439:993–7. doi: 10.1038/nature04496. [DOI] [PubMed] [Google Scholar]
- [59].Hirschmann-Jax C, Foster AE, Wulf GG, et al. A distinct “side population” of cells with high drug efflux capacity in human tumor cells. Proc Natl Acad Sci USA. 2004;101:14228–33. doi: 10.1073/pnas.0400067101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Montanaro F, Liadaki K, Schienda J, et al. Demystifying SP cell purification: viability, yield, and phenotype are defined by isolation parameters. Exp Cell Res. 2004;298:144–54. doi: 10.1016/j.yexcr.2004.04.010. [DOI] [PubMed] [Google Scholar]
- [61].Zhou S, Schuetz JD, Bunting KD, et al. The ABC transporter Bcrp1/ABCG2 is expressed in a wide variety of stem cells and is a molecular determinant of the side-population phenotype. Nat Med. 2001;7:1028–34. doi: 10.1038/nm0901-1028. [DOI] [PubMed] [Google Scholar]
- [62].Ginestier C, Wicha MS. Mammary stem cell number as a determinate of breast cancer risk. Breast Cancer Res. 2007;9:109. doi: 10.1186/bcr1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Clayton H, Titley I, Vivanco M. Growth and differentiation of progenitor/stem cells derived from the human mammary gland. Exp Cell Res. 2004;297:444–60. doi: 10.1016/j.yexcr.2004.03.029. [DOI] [PubMed] [Google Scholar]
- [64].Dontu G, Abdallah WM, Foley JM, et al. In vitro propagation and transcriptional profiling of human mammary stem/progenitor cells. Genes Dev. 2003;17:1253–70. doi: 10.1101/gad.1061803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Alvi AJ, Clayton H, Joshi C, et al. Functional and molecular characterisation of mammary side population cells. Breast Cancer Res. 2003;5:R1–8. doi: 10.1186/bcr547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Welm BE, Tepera SB, Venezia T, et al. Sca-1(pos) cells in the mouse mammary gland represent an enriched progenitor cell population. Dev Biol. 2002;245:42–56. doi: 10.1006/dbio.2002.0625. [DOI] [PubMed] [Google Scholar]
- [67].Potten C, Owen G, Booth D. Intestinal stem cells protect their genome by selective segregation of template DNA strands. Journal of cell science. 2002;115:2381–8. doi: 10.1242/jcs.115.11.2381. [DOI] [PubMed] [Google Scholar]
- [68].Zeps N, Dawkins HJ, Papadimitriou JM, Redmond SL, Walters MI. Detection of a population of long-lived cells in mammary epithelium of the mouse. Cell Tissue Res. 1996;286:525–36. doi: 10.1007/s004410050722. [DOI] [PubMed] [Google Scholar]
- [69].Smith GH. Label-retaining epithelial cells in mouse mammary gland divide asymmetrically and retain their template DNA strands. Development. 2005;132:681–7. doi: 10.1242/dev.01609. [DOI] [PubMed] [Google Scholar]
- [70].Allred DC, Brown P, Medina D. The origins of estrogen receptor alpha-positive and estrogen receptor alpha-negative human breast cancer. Breast Cancer Res. 2004;6:240–5. doi: 10.1186/bcr938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Clarke RB, Spence K, Anderson E, et al. A putative human breast stem cell population is enriched for steroid receptor-positive cells. Dev Biol. 2005;277:443–56. doi: 10.1016/j.ydbio.2004.07.044. [DOI] [PubMed] [Google Scholar]
- [72].Asselin-Labat ML. Mammary stem and progenitor cells: critical role of the transcription factor Gata-3. Med Sci (Paris) 2007;23:1077–80. doi: 10.1051/medsci/200723121077. [DOI] [PubMed] [Google Scholar]
- [73].Asselin-Labat ML, Vaillant F, Sheridan JM, et al. Control of mammary stem cell function by steroid hormone signalling. Nature. 2010;465:798–802. doi: 10.1038/nature09027. [DOI] [PubMed] [Google Scholar]
- [74].Armstrong L, Stojkovic M, Dimmick I, et al. Phenotypic characterization of murine primitive hematopoietic progenitor cells isolated on basis of aldehyde dehydrogenase activity. Stem Cells. 2004;22:1142–51. doi: 10.1634/stemcells.2004-0170. [DOI] [PubMed] [Google Scholar]
- [75].Duester G. Families of retinoid dehydrogenases regulating vitamin A function: production of visual pigment and retinoic acid. Eur J Biochem. 2000;267:4315–24. doi: 10.1046/j.1432-1327.2000.01497.x. [DOI] [PubMed] [Google Scholar]
- [76].Sophos NA, Vasiliou V. Aldehyde dehydrogenase gene superfamily: the 2002 update. Chem Biol Interact. 2003;143-144:5–22. doi: 10.1016/s0009-2797(02)00163-1. [DOI] [PubMed] [Google Scholar]
- [77].Ginestier C, Hur MH, Charafe-Jauffret E, et al. ALDH1 is a marker of normal and malignant human mammary stem cells and a predictor of poor clinical outcome. Cell Stem Cell. 2007;1:555–67. doi: 10.1016/j.stem.2007.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Heerma van Voss MR, van der Groep P, Bart J, van der Wall E, van Diest PJ. Expression of the stem cell marker ALDH1 in the normal breast of BRCA1 mutation carriers. Breast Cancer Res Treat. 2010;123:611–2. doi: 10.1007/s10549-010-1005-x. [DOI] [PubMed] [Google Scholar]
- [79].Stingl J. Detection and analysis of mammary gland stem cells. J Pathol. 2009;217:229–41. doi: 10.1002/path.2457. [DOI] [PubMed] [Google Scholar]
- [80].Sleeman KE, Kendrick H, Ashworth A, Isacke CM, Smalley MJ. CD24 staining of mouse mammary gland cells defines luminal epithelial, myoepithelial/basal and non-epithelial cells. Breast Cancer Res. 2006;8:R7. doi: 10.1186/bcr1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Asselin-Labat ML, Vaillant F, Shackleton M, et al. Delineating the epithelial hierarchy in the mouse mammary gland. Cold Spring Harb Symp Quant Biol. 2008;73:469–78. doi: 10.1101/sqb.2008.73.020. [DOI] [PubMed] [Google Scholar]
- [82].Vaillant F, Asselin-Labat ML, Shackleton M, et al. The mammary progenitor marker CD61/beta3 integrin identifies cancer stem cells in mouse models of mammary tumorigenesis. Cancer Res. 2008;68:7711–7. doi: 10.1158/0008-5472.CAN-08-1949. [DOI] [PubMed] [Google Scholar]
- [83].Abel RM, Bryan RT, Rafaat F, et al. Axillary lipoblastoma--tumor recurrence in the right atrium. J Pediatr Surg. 2003;38:1246–7. doi: 10.1016/s0022-3468(03)00279-3. [DOI] [PubMed] [Google Scholar]
- [84].Taddei I, Deugnier MA, Faraldo MM, et al. Beta1 integrin deletion from the basal compartment of the mammary epithelium affects stem cells. Nat Cell Biol. 2008;10:716–22. doi: 10.1038/ncb1734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Muschler J, Streuli CH. Cell-matrix interactions in mammary gland development and breast cancer. Cold Spring Harb Perspect Biol. 2010;2:a003202. doi: 10.1101/cshperspect.a003202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Deng G, Lu Y, Zlotnikov G, Thor AD, Smith HS. Loss of heterozygosity in normal tissue adjacent to breast carcinomas. Science. 1996;274:2057–9. doi: 10.1126/science.274.5295.2057. [DOI] [PubMed] [Google Scholar]
- [87].Lakhani SR, Slack DN, Hamoudi RA, et al. Detection of allelic imbalance indicates that a proportion of mammary hyperplasia of usual type are clonal, neoplastic proliferations. Lab Invest. 1996;74:129–35. [PubMed] [Google Scholar]
- [88].Stingl J, Eaves CJ, Zandieh I, Emerman JT. Characterization of bipotent mammary epithelial progenitor cells in normal adult human breast tissue. Breast Cancer Res Treat. 2001;67:93–109. doi: 10.1023/a:1010615124301. [DOI] [PubMed] [Google Scholar]
- [89].Villadsen R, Fridriksdottir AJ, Ronnov-Jessen L, et al. Evidence for a stem cell hierarchy in the adult human breast. J Cell Biol. 2007;177:87–101. doi: 10.1083/jcb.200611114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Eirew P, Stingl J, Raouf A, et al. A method for quantifying normal human mammary epithelial stem cells with in vivo regenerative ability. Nat Med. 2008;14:1384–9. doi: 10.1038/nm.1791. [DOI] [PubMed] [Google Scholar]
- [91].Raouf A, Zhao Y, To K, et al. Transcriptome analysis of the normal human mammary cell commitment and differentiation process. Cell Stem Cell. 2008;3:109–18. doi: 10.1016/j.stem.2008.05.018. [DOI] [PubMed] [Google Scholar]
- [92].Stingl J, Eaves CJ, Kuusk U, Emerman JT. Phenotypic and functional characterization in vitro of a multipotent epithelial cell present in the normal adult human breast. Differentiation. 1998;63:201–13. doi: 10.1111/j.1432-0436.1998.00201.x. [DOI] [PubMed] [Google Scholar]
- [93].Lim E, Vaillant F, Wu D, et al. Aberrant luminal progenitors as the candidate target population for basal tumor development in BRCA1 mutation carriers. Nat Med. 2009;15:907–13. doi: 10.1038/nm.2000. [DOI] [PubMed] [Google Scholar]
- [94].Kuperwasser C, Chavarria T, Wu M, et al. Reconstruction of functionally normal and malignant human breast tissues in mice. Proc Natl Acad Sci USA. 2004;101:4966–71. doi: 10.1073/pnas.0401064101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001;414:105–11. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- [96].Kelly PN, Dakic A, Adams JM, Nutt SL, Strasser A. Tumor growth need not be driven by rare cancer stem cells. Science. 2007;317:337. doi: 10.1126/science.1142596. [DOI] [PubMed] [Google Scholar]
- [97].Ponti D, Costa A, Zaffaroni N, et al. Isolation and in vitro propagation of tumorigenic breast cancer cells with stem/progenitor cell properties. Cancer Res. 2005;65:5506–11. doi: 10.1158/0008-5472.CAN-05-0626. [DOI] [PubMed] [Google Scholar]
- [98].Liu S, Ginestier C, Charafe-Jauffret E, et al. BRCA1 regulates human mammary stem/progenitor cell fate. Proc Natl Acad Sci USA. 2008;105:1680–5. doi: 10.1073/pnas.0711613105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Fillmore CM, Kuperwasser C. Human breast cancer cell lines contain stem-like cells that self-renew, give rise to phenotypically diverse progeny and survive chemotherapy. Breast Cancer Res. 2008;10:R25. doi: 10.1186/bcr1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Sansone P, Storci G, Giovannini C, et al. p66Shc/Notch-3 interplay controls self-renewal and hypoxia survival in human stem/progenitor cells of the mammary gland expanded in vitro as mammospheres. Stem Cells. 2007;25:807–15. doi: 10.1634/stemcells.2006-0442. [DOI] [PubMed] [Google Scholar]
- [101].Horwitz KB, Dye WW, Harrell JC, Kabos P, Sartorius CA. Rare steroid receptor-negative basal-like tumorigenic cells in luminal subtype human breast cancer xenografts. Proc Natl Acad Sci USA. 2008;105:5774–9. doi: 10.1073/pnas.0706216105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Yu F, Yao H, Zhu P, et al. let-7 regulates self renewal and tumorigenicity of breast cancer cells. Cell. 2007;131:1109–23. doi: 10.1016/j.cell.2007.10.054. [DOI] [PubMed] [Google Scholar]
- [103].Hennighausen L, Robinson GW. Information networks in the mammary gland. Nat Rev Mol Cell Biol. 2005;6:715–25. doi: 10.1038/nrm1714. [DOI] [PubMed] [Google Scholar]
- [104].Clevers H. Wnt/beta-catenin signaling in development and disease. Cell. 2006;127:469–80. doi: 10.1016/j.cell.2006.10.018. [DOI] [PubMed] [Google Scholar]
- [105].Nusse R. Wnt signaling and stem cell control. Cell Res. 2008;18:523–7. doi: 10.1038/cr.2008.47. [DOI] [PubMed] [Google Scholar]
- [106].Pereira L, Yi F, Merrill BJ. Repression of Nanog gene transcription by Tcf3 limits embryonic stem cell self-renewal. Mol Cell Biol. 2006;26:7479–91. doi: 10.1128/MCB.00368-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Hochedlinger K, Yamada Y, Beard C, Jaenisch R. Ectopic expression of Oct-4 blocks progenitor-cell differentiation and causes dysplasia in epithelial tissues. Cell. 2005;121:465–77. doi: 10.1016/j.cell.2005.02.018. [DOI] [PubMed] [Google Scholar]
- [108].Miki Y, Nishisho I, Horii A, et al. Disruption of the APC gene by a retrotransposal insertion of L1 sequence in a colon cancer. Cancer Res. 1992;52:643–5. [PubMed] [Google Scholar]
- [109].Barker N, Clevers H. Mining the Wnt pathway for cancer therapeutics. Nat Rev Drug Discov. 2006;5:997–1014. doi: 10.1038/nrd2154. [DOI] [PubMed] [Google Scholar]
- [110].Klopocki E, Kristiansen G, Wild PJ, et al. Loss of SFRP1 is associated with breast cancer progression and poor prognosis in early stage tumors. Int J Oncol. 2004;25:641–9. [PubMed] [Google Scholar]
- [111].Ugolini F, Charafe-Jauffret E, Bardou VJ, et al. WNT pathway and mammary carcinogenesis: loss of expression of candidate tumor suppressor gene SFRP1 in most invasive carcinomas except of the medullary type. Oncogene. 2001;20:5810–7. doi: 10.1038/sj.onc.1204706. [DOI] [PubMed] [Google Scholar]
- [112].Liu BY, McDermott SP, Khwaja SS, Alexander CM. The transforming activity of Wnt effectors correlates with their ability to induce the accumulation of mammary progenitor cells. Proc Natl Acad Sci USA. 2004;101:4158–63. doi: 10.1073/pnas.0400699101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Tepera SB, McCrea PD, Rosen JM. A beta-catenin survival signal is required for normal lobular development in the mammary gland. J Cell Sci. 2003;116:1137–49. doi: 10.1242/jcs.00334. [DOI] [PubMed] [Google Scholar]
- [114].Zeng YA, Nusse R. Wnt proteins are self-renewal factors for mammary stem cells and promote their long-term expansion in culture. Cell Stem Cell. 2010;6:568–77. doi: 10.1016/j.stem.2010.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Bailey EC, Scott MP, Johnson RL. Hedgehog signaling in animal development and human disease. Ernst Schering Res Found Workshop. 2000:211–35. doi: 10.1007/978-3-662-04264-9_12. [DOI] [PubMed] [Google Scholar]
- [116].Berman DM, Karhadkar SS, Maitra A, et al. Widespread requirement for Hedgehog ligand stimulation in growth of digestive tract tumours. Nature. 2003;425:846–51. doi: 10.1038/nature01972. [DOI] [PubMed] [Google Scholar]
- [117].Cohen MM., Jr. The hedgehog signaling network. Am J Med Genet A. 2003;123A:5–28. doi: 10.1002/ajmg.a.20495. [DOI] [PubMed] [Google Scholar]
- [118].Unden AB, Zaphiropoulos PG, Bruce K, Toftgard R, Stahle-Backdahl M. Human patched (PTCH) mRNA is overexpressed consistently in tumor cells of both familial and sporadic basal cell carcinoma. Cancer Res. 1997;57:2336–40. [PubMed] [Google Scholar]
- [119].Hahn H, Wicking C, Zaphiropoulous PG, et al. Mutations of the human homolog of Drosophila patched in the nevoid basal cell carcinoma syndrome. Cell. 1996;85:841–51. doi: 10.1016/s0092-8674(00)81268-4. [DOI] [PubMed] [Google Scholar]
- [120].Merchant AA, Matsui W. Targeting Hedgehog--a cancer stem cell pathway. Clin Cancer Res. 2010;16:3130–40. doi: 10.1158/1078-0432.CCR-09-2846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Lewis MT, Ross S, Strickland PA, et al. Defects in mouse mammary gland development caused by conditional haploinsufficiency of Patched-1. Development. 1999;126:5181–93. doi: 10.1242/dev.126.22.5181. [DOI] [PubMed] [Google Scholar]
- [122].Moraes RC, Zhang X, Harrington N, et al. Constitutive activation of smoothened (SMO) in mammary glands of transgenic mice leads to increased proliferation, altered differentiation and ductal dysplasia. Development. 2007;134:1231–42. doi: 10.1242/dev.02797. [DOI] [PubMed] [Google Scholar]
- [123].Kubo M, Nakamura M, Tasaki A, et al. Hedgehog signaling pathway is a new therapeutic target for patients with breast cancer. Cancer Res. 2004;64:6071–4. doi: 10.1158/0008-5472.CAN-04-0416. [DOI] [PubMed] [Google Scholar]
- [124].Vorechovsky I, Benediktsson KP, Toftgard R. The patched/hedgehog/smoothened signalling pathway in human breast cancer: no evidence for H133Y SHH, PTCH and SMO mutations. Eur J Cancer. 1999;35:711–3. doi: 10.1016/s0959-8049(99)00017-9. [DOI] [PubMed] [Google Scholar]
- [125].Wolf I, Bose S, Desmond JC, et al. Unmasking of epigenetically silenced genes reveals DNA promoter methylation and reduced expression of PTCH in breast cancer. Breast Cancer Res Treat. 2007;105:139–55. doi: 10.1007/s10549-006-9440-4. [DOI] [PubMed] [Google Scholar]
- [126].Chang-Claude J, Dunning A, Schnitzbauer U, et al. The patched polymorphism Pro1315Leu (C3944T) may modulate the association between use of oral contraceptives and breast cancer risk. Int J Cancer. 2003;103:779–83. doi: 10.1002/ijc.10889. [DOI] [PubMed] [Google Scholar]
- [127].Liu S, Dontu G, Mantle ID, et al. Hedgehog signaling and Bmi-1 regulate self-renewal of normal and malignant human mammary stem cells. Cancer Res. 2006;66:6063–71. doi: 10.1158/0008-5472.CAN-06-0054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].Shi Y, Massague J. Mechanisms of TGF-beta signaling from cell membrane to the nucleus. Cell. 2003;113:685–700. doi: 10.1016/s0092-8674(03)00432-x. [DOI] [PubMed] [Google Scholar]
- [129].Ewan KB, Shyamala G, Ravani SA, et al. Latent transforming growth factor-beta activation in mammary gland: regulation by ovarian hormones affects ductal and alveolar proliferation. Am J Pathol. 2002;160:2081–93. doi: 10.1016/s0002-9440(10)61158-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [130].Nelson CM, Vanduijn MM, Inman JL, Fletcher DA, Bissell MJ. Tissue geometry determines sites of mammary branching morphogenesis in organotypic cultures. Science. 2006;314:298–300. doi: 10.1126/science.1131000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [131].Gorska AE, Joseph H, Derynck R, Moses HL, Serra R. Dominant-negative interference of the transforming growth factor beta type II receptor in mammary gland epithelium results in alveolar hyperplasia and differentiation in virgin mice. Cell Growth Differ. 1998;9:229–38. [PubMed] [Google Scholar]
- [132].Joseph H, Gorska AE, Sohn P, Moses HL, Serra R. Overexpression of a kinase-deficient transforming growth factor-beta type II receptor in mouse mammary stroma results in increased epithelial branching. Mol Biol Cell. 1999;10:1221–34. doi: 10.1091/mbc.10.4.1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [133].Siegel PM, Shu W, Cardiff RD, Muller WJ, Massague J. Transforming growth factor beta signaling impairs Neu-induced mammary tumorigenesis while promoting pulmonary metastasis. Proc Natl Acad Sci USA. 2003;100:8430–5. doi: 10.1073/pnas.0932636100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [134].Giampieri S, Manning C, Hooper S, et al. Localized and reversible TGFbeta signalling switches breast cancer cells from cohesive to single cell motility. Nat Cell Biol. 2009;11:1287–96. doi: 10.1038/ncb1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [135].Joshi PA, Jackson HW, Beristain AG, et al. Progesterone induces adult mammary stem cell expansion. Nature. 2010;465:803–7. doi: 10.1038/nature09091. [DOI] [PubMed] [Google Scholar]
- [136].Zwijsen RM, Wientjens E, Klompmaker R, et al. CDK-independent activation of estrogen receptor by cyclin D1. Cell. 1997;88:405–15. doi: 10.1016/s0092-8674(00)81879-6. [DOI] [PubMed] [Google Scholar]
- [137].van Diest PJ, Michalides RJ, Jannink L, et al. Cyclin D1 expression in invasive breast cancer. Correlations and prognostic value. Am J Pathol. 1997;150:705–11. [PMC free article] [PubMed] [Google Scholar]
- [138].Sicinski P, Donaher JL, Parker SB, et al. Cyclin D1 provides a link between development and oncogenesis in the retina and breast. Cell. 1995;82:621–30. doi: 10.1016/0092-8674(95)90034-9. [DOI] [PubMed] [Google Scholar]
- [139].Yu Q, Geng Y, Sicinski P. Specific protection against breast cancers by cyclin D1 ablation. Nature. 2001;411:1017–21. doi: 10.1038/35082500. [DOI] [PubMed] [Google Scholar]
- [140].Kelsey J, Gammon M, John E. Reproductive factors and breast cancer. Epidemiologic Reviews. 1993;15:36. doi: 10.1093/oxfordjournals.epirev.a036115. [DOI] [PubMed] [Google Scholar]
- [141].Mumm JS, Kopan R. Notch signaling: from the outside in. Dev Biol. 2000;228:151–65. doi: 10.1006/dbio.2000.9960. [DOI] [PubMed] [Google Scholar]
- [142].Mumm JS, Schroeter EH, Saxena MT, et al. A ligand-induced extracellular cleavage regulates gamma-secretase-like proteolytic activation of Notch1. Mol Cell. 2000;5:197–206. doi: 10.1016/s1097-2765(00)80416-5. [DOI] [PubMed] [Google Scholar]
- [143].van Tetering G, van Diest P, Verlaan I, et al. Metalloprotease ADAM10 is required for Notch1 site 2 cleavage. J Biol Chem. 2009;284:31018–27. doi: 10.1074/jbc.M109.006775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [144].Bray SJ. Notch signalling: a simple pathway becomes complex. Nat Rev Mol Cell Biol. 2006;7:678–89. doi: 10.1038/nrm2009. [DOI] [PubMed] [Google Scholar]
- [145].Kopan R, Ilagan MX. The canonical Notch signaling pathway: unfolding the activation mechanism. Cell. 2009;137:216–33. doi: 10.1016/j.cell.2009.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [146].Radtke F, Raj K. The role of Notch in tumorigenesis: oncogene or tumour suppressor? Nat Rev Cancer. 2003;3:756–67. doi: 10.1038/nrc1186. [DOI] [PubMed] [Google Scholar]
- [147].Weng AP, Ferrando AA, Lee W, et al. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science. 2004;306:269–71. doi: 10.1126/science.1102160. [DOI] [PubMed] [Google Scholar]
- [148].O’Neil J, Grim J, Strack P, et al. FBW7 mutations in leukemic cells mediate NOTCH pathway activation and resistance to gamma-secretase inhibitors. J Exp Med. 2007;204:1813–24. doi: 10.1084/jem.20070876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [149].Thompson BJ, Buonamici S, Sulis ML, et al. The SCFFBW7 ubiquitin ligase complex as a tumor suppressor in T cell leukemia. J Exp Med. 2007;204:1825–35. doi: 10.1084/jem.20070872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [150].Bergmans BA, De Strooper B. gamma-secretases: from cell biology to therapeutic strategies. Lancet Neurol. 2010;9:215–26. doi: 10.1016/S1474-4422(09)70332-1. [DOI] [PubMed] [Google Scholar]
- [151].Milano J, McKay J, Dagenais C, et al. Modulation of notch processing by gamma-secretase inhibitors causes intestinal goblet cell metaplasia and induction of genes known to specify gut secretory lineage differentiation. Toxicol Sci. 2004;82:341–58. doi: 10.1093/toxsci/kfh254. [DOI] [PubMed] [Google Scholar]
- [152].Barten DM, Meredith JE, Jr., Zaczek R, Houston JG, Albright CF. Gamma-secretase inhibitors for Alzheimer’s disease: balancing efficacy and toxicity. Drugs R D. 2006;7:87–97. doi: 10.2165/00126839-200607020-00003. [DOI] [PubMed] [Google Scholar]
- [153].van Es JH, van Gijn ME, Riccio O, et al. Notch/gamma-secretase inhibition turns proliferative cells in intestinal crypts and adenomas into goblet cells. Nature. 2005;435:959–63. doi: 10.1038/nature03659. [DOI] [PubMed] [Google Scholar]
- [154].Real PJ, Tosello V, Palomero T, et al. Gamma-secretase inhibitors reverse glucocorticoid resistance in T cell acute lymphoblastic leukemia. Nat Med. 2009;15:50–8. doi: 10.1038/nm.1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [155].Weggen S, Eriksen JL, Das P, et al. A subset of NSAIDs lower amyloidogenic A beta 42 independently of cyclooxygenase activity. Nature. 2001;414:212–6. doi: 10.1038/35102591. [DOI] [PubMed] [Google Scholar]
- [156].Uyttendaele H, Soriano JV, Montesano R, Kitajewski J. Notch4 and Wnt-1 proteins function to regulate branching morphogenesis of mammary epithelial cells in an opposing fashion. Dev Biol. 1998;196:204–17. doi: 10.1006/dbio.1998.8863. [DOI] [PubMed] [Google Scholar]
- [157].Gallahan D, Jhappan C, Robinson G, et al. Expression of a truncated Int3 gene in developing secretory mammary epithelium specifically retards lobular differentiation resulting in tumorigenesis. Cancer Res. 1996;56:1775–85. [PubMed] [Google Scholar]
- [158].Smith GH, Gallahan D, Diella F, et al. Constitutive expression of a truncated INT3 gene in mouse mammary epithelium impairs differentiation and functional development. Cell Growth Differ. 1995;6:563–77. [PubMed] [Google Scholar]
- [159].Jhappan C, Gallahan D, Stahle C, et al. Expression of an activated Notch-related int-3 transgene interferes with cell differentiation and induces neoplastic transformation in mammary and salivary glands. Genes Dev. 1992;6:345–55. doi: 10.1101/gad.6.3.345. [DOI] [PubMed] [Google Scholar]
- [160].Dontu G, Jackson KW, McNicholas E, et al. Role of Notch signaling in cell-fate determination of human mammary stem/progenitor cells. Breast Cancer Res. 2004;6:R605–15. doi: 10.1186/bcr920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [161].Shipitsin M, Campbell LL, Argani P, et al. Molecular definition of breast tumor heterogeneity. Cancer Cell. 2007;11:259–73. doi: 10.1016/j.ccr.2007.01.013. [DOI] [PubMed] [Google Scholar]
- [162].Dievart A, Beaulieu N, Jolicoeur P. Involvement of Notch1 in the development of mouse mammary tumors. Oncogene. 1999;18:5973–81. doi: 10.1038/sj.onc.1202991. [DOI] [PubMed] [Google Scholar]
- [163].Gallahan D, Callahan R. Mammary tumorigenesis in feral mice: identification of a new int locus in mouse mammary tumor virus (Czech II)-induced mammary tumors. J Virol. 1987;61:66–74. doi: 10.1128/jvi.61.1.66-74.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [164].Robbins J, Blondel BJ, Gallahan D, Callahan R. Mouse mammary tumor gene int-3: a member of the notch gene family transforms mammary epithelial cells. J Virol. 1992;66:2594–9. doi: 10.1128/jvi.66.4.2594-2599.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [165].Kordon EC, Smith GH, Callahan R, Gallahan D. A novel non-mouse mammary tumor virus activation of the Int-3 gene in a spontaneous mouse mammary tumor. J Virol. 1995;69:8066–9. doi: 10.1128/jvi.69.12.8066-8069.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [166].Gallahan D, Callahan R. The mouse mammary tumor associated gene INT3 is a unique member of the NOTCH gene family (NOTCH4) Oncogene. 1997;14:1883–90. doi: 10.1038/sj.onc.1201035. [DOI] [PubMed] [Google Scholar]
- [167].Raafat A, Bargo S, Anver MR, Callahan R. Mammary development and tumorigenesis in mice expressing a truncated human Notch4/Int3 intracellular domain (h-Int3sh) Oncogene. 2004;23:9401–7. doi: 10.1038/sj.onc.1208187. [DOI] [PubMed] [Google Scholar]
- [168].Buono KD, Robinson GW, Martin C, et al. The canonical Notch/RBP-J signaling pathway controls the balance of cell lineages in mammary epithelium during pregnancy. Dev Biol. 2006;293:565–80. doi: 10.1016/j.ydbio.2006.02.043. [DOI] [PubMed] [Google Scholar]
- [169].Bouras T, Pal B, Vaillant F, et al. Notch signaling regulates mammary stem cell function and luminal cell-fate commitment. Cell Stem Cell. 2008;3:429–41. doi: 10.1016/j.stem.2008.08.001. [DOI] [PubMed] [Google Scholar]
- [170].Ayyanan A, Civenni G, Ciarloni L, et al. Increased Wnt signaling triggers oncogenic conversion of human breast epithelial cells by a Notch-dependent mechanism. Proc Natl Acad Sci USA. 2006;103:3799–804. doi: 10.1073/pnas.0600065103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [171].Mazzone M, Selfors LM, Albeck J, et al. Dose-dependent induction of distinct phenotypic responses to Notch pathway activation in mammary epithelial cells. Proc Natl Acad Sci USA. 2010;107:5012–7. doi: 10.1073/pnas.1000896107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [172].Vooijs M, Ong CT, Hadland B, et al. Mapping the consequence of Notch1 proteolysis in vivo with NIP-CRE. Development. 2007;134:535–44. doi: 10.1242/dev.02733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [173].Weijzen S, Rizzo P, Braid M, et al. Activation of Notch-1 signaling maintains the neoplastic phenotype in human Ras-transformed cells. Nat Med. 2002;8:979–86. doi: 10.1038/nm754. [DOI] [PubMed] [Google Scholar]
- [174].Parr C, Watkins G, Jiang WG. The possible correlation of Notch-1 and Notch-2 with clinical outcome and tumour clinicopathological parameters in human breast cancer. Int J Mol Med. 2004;14:779–86. doi: 10.3892/ijmm.14.5.779. [DOI] [PubMed] [Google Scholar]
- [175].Reedijk M, Odorcic S, Chang L, et al. High-level coexpression of JAG1 and NOTCH1 is observed in human breast cancer and is associated with poor overall survival. Cancer Res. 2005;65:8530–7. doi: 10.1158/0008-5472.CAN-05-1069. [DOI] [PubMed] [Google Scholar]
- [176].Hu C, Dievart A, Lupien M, et al. Overexpression of activated murine Notch1 and Notch3 in transgenic mice blocks mammary gland development and induces mammary tumors. Am J Pathol. 2006;168:973–90. doi: 10.2353/ajpath.2006.050416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [177].Pece S, Serresi M, Santolini E, et al. Loss of negative regulation by Numb over Notch is relevant to human breast carcinogenesis. J Cell Biol. 2004;167:215–21. doi: 10.1083/jcb.200406140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [178].Garces C, Ruiz-Hidalgo MJ, Font de Mora J, et al. Notch-1 controls the expression of fatty acid-activated transcription factors and is required for adipogenesis. J Biol Chem. 1997;272:29729–34. doi: 10.1074/jbc.272.47.29729. [DOI] [PubMed] [Google Scholar]
- [179].Hendrix MJ, Seftor RE, Seftor EA, et al. Transendothelial function of human metastatic melanoma cells: role of the microenvironment in cell-fate determination. Cancer Res. 2002;62:665–8. [PubMed] [Google Scholar]
- [180].Yasutomo K, Doyle C, Miele L, Fuchs C, Germain RN. The duration of antigen receptor signalling determines CD4+ versus CD8+ T-cell lineage fate. Nature. 2000;404:506–10. doi: 10.1038/35006664. [DOI] [PubMed] [Google Scholar]
- [181].Wu Y, Cain-Hom C, Choy L, et al. Therapeutic antibody targeting of individual Notch receptors. Nature. 2010;464:1052–7. doi: 10.1038/nature08878. [DOI] [PubMed] [Google Scholar]
- [182].Li K, Li Y, Wu W, et al. Modulation of Notch signaling by antibodies specific for the extracellular negative regulatory region of NOTCH3. J Biol Chem. 2008;283:8046–54. doi: 10.1074/jbc.M800170200. [DOI] [PubMed] [Google Scholar]
- [183].Aste-Amezaga M, Zhang N, Lineberger JE, et al. Characterization of Notch1 antibodies that inhibit signaling of both normal and mutated Notch1 receptors. PLoS One. 2010;5:e9094. doi: 10.1371/journal.pone.0009094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [184].Fridman JS, Caulder E, Hansbury M, et al. Selective inhibition of ADAM metalloproteases as a novel approach for modulating ErbB pathways in cancer. Clin Cancer Res. 2007;13:1892–902. doi: 10.1158/1078-0432.CCR-06-2116. [DOI] [PubMed] [Google Scholar]
- [185].Osipo C, Patel P, Rizzo P, et al. ErbB-2 inhibition activates Notch-1 and sensitizes breast cancer cells to a gamma-secretase inhibitor. Oncogene. 2008;27:5019–32. doi: 10.1038/onc.2008.149. [DOI] [PubMed] [Google Scholar]
- [186].Dong Y, Li A, Wang J, Weber JD, Michel LS. Synthetic lethality through combined Notch-epidermal growth factor receptor pathway inhibition in basal-like breast cancer. Cancer Res. 2010;70:5465–74. doi: 10.1158/0008-5472.CAN-10-0173. [DOI] [PubMed] [Google Scholar]
- [187].Moellering RE, Cornejo M, Davis TN, et al. Direct inhibition of the NOTCH transcription factor complex. Nature. 2009;462:182–8. doi: 10.1038/nature08543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [188].Panza F, Solfrizzi V, Frisardi V, et al. Beyond the neurotransmitter-focused approach in treating Alzheimer’s disease: drugs targeting beta-amyloid and tau protein. Aging Clin Exp Res. 2009;21:386–406. doi: 10.1007/BF03327445. [DOI] [PubMed] [Google Scholar]
- [189].Rizzo P, Miao H, D’Souza G, et al. Cross-talk between notch and the estrogen receptor in breast cancer suggests novel therapeutic approaches. Cancer Res. 2008;68:5226–35. doi: 10.1158/0008-5472.CAN-07-5744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [190].Yao K, Rizzo P, Rajan P, et al. Notch-1 and Notch-4 Receptors as Prognostic Markers in Breast Cancer. Int J Surg Pathol. 2011;19:607–613. doi: 10.1177/1066896910362080. [DOI] [PubMed] [Google Scholar]