Abstract
Aims/hypothesis
Glucokinase (GCK) plays a role in glucose metabolism and glucose-stimulated insulin secretion. Rare mutations in GCK cause MODY. We investigated whether common variation (minor allele frequency ≥0.01) in GCK is associated with metabolic traits and type 2 diabetes.
Methods
Four exonic single-nucleotide polymorphisms (SNPs) and three SNPs predicted to cause loss of promoter function were identified in whole-genome sequence data from 234 Pima Indians. These seven tag SNPs and rs4607517, a type 2 diabetes variant established in other studies, were analysed in 415 full-heritage non-diabetic Pima Indians characterised for metabolic traits, and 7,667 American Indians who had data on type 2 diabetes and BMI.
Results
A novel 3′ untranslated region (3′UTR) SNP, chr7:44184184-G/A, was associated with the rate of carbohydrate oxidation post-absorptively (β = 0.22 mg [kg estimated metabolic body size (EMBS)]−1 min−1, p = 0.005) and during a hyperinsulinaemic–euglycaemic clamp (β = 0.24 mg [kg EMBS]−1 min−1, p = 0.0002), the rate of carbohydrate oxidation in a respiratory chamber (β = 311 kJ/day, p = 0.03) and 24 h energy expenditure, which was attributable to the thermic effect of food (β = 520 kJ/day, p = 3.39 × 10−6). This 3′UTR SNP was also associated with diabetes (OR 1.36, 95% CI 1.11, 1.65, p = 0.002), where the A allele (allele frequency 0.05) was associated with a lower rate of carbohydrate oxidation, lower 24 h energy expenditure and higher risk for diabetes. In a Cox proportional hazards model, a rate of insulin-stimulated carbohydrate oxidation lower than the mean rate at baseline predicted a higher risk for developing diabetes than for those above the mean (hazard rate ratio 2.2, 95% CI 1.3, 3.6, p = 0.002).
Conclusions/interpretation
Common variation in GCK influences the rate of carbohydrate oxidation, 24 h energy expenditure and diabetes risk in Pima Indians.
Electronic supplementary material
The online version of this article (doi:10.1007/s00125-014-3234-8) contains peer-reviewed but unedited supplementary material, which is available to authorised users.
Keywords: Carbohydrate oxidation, Energy expenditure, GCK, Thermic effect of food, Type 2 diabetes
Introduction
Glucokinase (GCK) is a hexokinase isozyme (hexokinase IV) that catalyses glucose to glucose-6-phosphate (G6P) and is involved in the first step of both glycolysis and glycogen synthesis. GCK is predominantly expressed in hepatocytes and pancreatic beta cells, with isoforms distinct in the N terminus. The pancreatic beta cell isoform is a key enzyme in regulating glucose-stimulated insulin secretion and is considered to be a glucose sensor. The liver isoform plays a central role in regulating glucose homeostasis and is a major component of the hepatic glucose-sensing system involved in glucose synthesis, breakdown and storage [1–3]. Rare heterozygous inactivating mutations in GCK cause MODY, mainly due to a reduced glucose-stimulated insulin secretion [4]. While rare mutations in GCK cause MODY, common variants have been associated with HbA1c levels, fasting glucose concentrations and type 2 diabetes in white and other populations [5–7]. No rare coding variants in GCK were identified in 234 Pima Indians with whole-genome sequence data (unpublished data, Y. L. Muller). Thus, in the current study, we investigated the effects of common and low-frequency GCK variants with a minor allele frequency (mAF) ≥0.01 on metabolic traits and type 2 diabetes risk in Pima Indians.
Methods
Participants with outpatient longitudinal data on type 2 diabetes and BMI
Electronic supplementary material (ESM) Fig. 1 shows a flow chart depicting the study design and selection of participants. All individuals in this study are participants of a longitudinal study of the aetiology of type 2 diabetes among the Gila River Indian Community in Arizona, where most of the residents are Pima Indians or Tohono O’odham (a closely related tribe) [8]. Diabetes was determined by prior clinical diagnosis or an oral glucose tolerance test according to the criteria of the American Diabetes Association [9]. A population-based sample of full-heritage Pima Indians (n = 3,604, including 736 sibships [sibship is defined as sibs ≥2], Table 1) was initially used to assess associations with type 2 diabetes. A non-overlapping sample of mixed-heritage American Indians from the same longitudinal study (n = 4,063, including 739 sibships; reported heritage, on average, was one-half Pima and three-quarters American Indian, Table 1) was used to assess replication. Among these samples, BMI was measured at biennial examinations and maximum BMI observed in the longitudinal study was analysed in 3,391 full-heritage Pima Indians and 3,406 mixed-heritage American Indians (Table 1) who were examined when aged ≥15 years. Fasting serum glucose concentrations were measured in 2,542 full-heritage Pima Indians and 2,887 mixed-heritage American Indians that were non-diabetic, including individuals who subsequently developed diabetes and those who remained non-diabetic (Table 1).
Table 1.
Characteristics of full-heritage Pima Indians and mixed-heritage American Indians analysed in the population-based association studies
| Characteristic | n | Male sex (%) | Age (years) | BMI (kg/m2) |
|---|---|---|---|---|
| Full-heritage Pima Indians | ||||
| Type 2 diabetes study | 3,604 | |||
| Diabetic (46%) | 1,658 | 37 | 49.1 ± 14.1 | 38.7 ± 8.6 |
| Non-diabetic | 1,946 | 48 | 32.1 ± 14.6 | 36.1 ± 8.5 |
| BMI study | 3,391 | 42 | 36.1 ± 13.3 | 37.4 ± 8.7 |
| Fasting glucose concentration study | 2,542 | 43 | 39.0 ± 14.5 | 35.7 ± 8.3 |
| Mixed-heritage American Indians | ||||
| Type 2 diabetes study | 4,063 | |||
| Diabetic (21%) | 853 | 41 | 41.1 ± 14.2 | 38.7 ± 8.7 |
| Non-diabetic | 3,210 | 47 | 24.9 ± 11.9 | 33.5 ± 8.4 |
| BMI study | 3,406 | 45 | 29.2 ± 12.0 | 34.8 ± 8.8 |
| Fasting glucose concentration study | 2,887 | 44 | 30.4 ± 12.3 | 33.5 ± 8.3 |
Age and BMI data are shown as means ± SD
Subset of participants with additional inpatient data on quantifiable metabolic traits
Among the full-heritage Pima Indians described above, 415 non-diabetic individuals (including 99 sibships; male sex 58%, age 27 ± 6 years and BMI 34 ± 8 kg/m2 at the time of metabolic testing) had undergone detailed studies of metabolic and anthropometric phenotypes for risk factors related to type 2 diabetes and obesity. Body composition, including percentage body fat, fat mass and fat-free mass, was estimated by underwater weighing until 1996 and by dual energy x-ray absorptiometry (DPX-1; Lunar Radiation Corp., Madison, WI, USA ) thereafter [10]. Glucose tolerance was determined by a 75 g OGTT, with measurements of fasting, 30, 60, 120 and 180 min plasma glucose and insulin concentrations [11]. A hyperinsulinaemic–euglycaemic clamp (insulin infusion rate of 40 mU m−2 min−1 with simultaneous glucose tracers) was used to measure rates of post-absorptive (basal) and insulin-stimulated glucose disappearance as previously described [11]. Indirect calorimetry measurements using a ventilated hood system were performed before and during the insulin infusion to assess rates of energy expenditure and substrate oxidation [12, 13]. Pancreatic beta cell function was assessed by the acute insulin response (AIR) after a 25 g intravenous glucose bolus and calculated as the mean increment in plasma insulin concentrations from 3 to 5 min [11].
To measure 24 h energy expenditure, study participants entered a respiratory chamber for 23 h and 15 min after an overnight fast and after at least 3 days of a weight-maintaining diet [14]. Four meals were provided at 08:00, 11:00, 16:00 and 19:00 hours. Fresh air was drawn through the chamber, and CO2 production and O2 consumption were measured and calculated every 15 min and extrapolated to the 24 h period [15]. Spontaneous physical activity (SPA) was detected by radar sensors and expressed as percentage of time in motion per 15 min interval. The energy cost of SPA was calculated as the product of average SPA over 24 h and the slope of the regression line between energy expenditure and SPA between 08:00 and 23:00 hours [15]. Sleeping metabolic rate was defined as the average energy expenditure of all 15 min periods between 1:00 and 5:00 hours during which SPA was <1.5%, and was extrapolated to 24 h [16]. The 24 h respiratory quotient (RQ) was calculated as the ratio of 24 h to 24 h . Carbohydrate and lipid oxidation rates were derived from the 24 h RQ after accounting for protein oxidation, which was estimated from the 24 h urinary nitrogen excretion [15].
Identification and genotyping of single-nucleotide polymorphisms
Single-nucleotide polymorphisms (SNPs) in exons and the putative promoter region (∼1.4 kb upstream of the translational start site) of GCK were obtained from whole-genome sequence data (30–40× coverage) of 234 individuals who were predominantly full-heritage Pima Indians (Complete Genomics, Mountain View, CA, USA; Illumina, San Diego, CA, USA). Individuals had been characterised for metabolic traits in our Clinical Research Center and were selected from different nuclear families to maximise identification of genetic variation. Genome sequence data were compared with the reference sequence GRCh37/hg19. SNPs not reported in NCBI dbSNP/1000 genomes (http://www.ncbi.nlm.nih.gov/SNP/; or http://browser.1000genomes.org/) were classified as ‘novel’. Linkage disequilibrium (LD) was determined using Haploview (version 4.2, Broad Institute, Cambridge, MA, USA). Tag SNPs were selected using the Tagger algorithm with a pairwise r 2 ≥ 0.8 taken as indicative of redundancy.
SNPs were genotyped for association analyses using the TaqMan Allelic Discrimination Assay on an ABI Prism 7900 (Applied Biosystems, Carlsbad, CA, USA) or BeadXpress System (Illumina).
Allelic specific gene expression
Total RNA was isolated from percutaneous abdominal adipose tissue biopsies from 14 individuals who were heterozygous for the 3′ untranslated region (3′UTR) SNP chr7:44184184-G/A. RNA was reverse-transcribed to cDNA and allelic specific expression was performed on an ABI Prism 7900 using a TaqMan probe (Applied Biosystems). The ratio of allele G/A expression was normalised to the genomic DNA control.
Statistical analyses
Statistical analyses were performed using the software of the SAS Institute (version 9.2, Cary, NC, USA). A logistic regression analysis was used to assess the association of genotypes with type 2 diabetes and was adjusted for age, sex, birth year and heritage as covariates. The model was fit with the generalised estimating equation (GEE) to account for dependence among siblings. Genotype was analysed as a numeric variable representing 0, 1 or 2 copies of a given allele. To estimate the proportion of European ancestry, 45 informative markers with large differences in allele frequency between populations [17] were used as a covariate in these analyses. The association of quantifiable traits with genotypes was analysed by linear regression using the GEE procedure to account for correlation among siblings. Results were adjusted for covariates as indicated. The rate of carbohydrate oxidation, as a metabolic predictor of diabetes, was assessed in individuals with normal glucose tolerance (NGT) who had measures of carbohydrate oxidation rate at baseline during a hyperinsulinaemic–euglycaemic clamp. These individuals also had biennial follow-up OGTTs to determine diabetes status. The Cox proportional hazards model was used to determine the hazard rate ratio (HRR) for developing type 2 diabetes associated with the rate of insulin-stimulated carbohydrate oxidation including age, sex, percentage body fat, AIR and non-oxidative glucose disposal rate as covariates. Follow-up time was defined as the time from the measure of carbohydrate oxidation rate during an insulin clamp to either type 2 diabetes onset or the last evaluation when an individual remained non-diabetic. To analyse the effect of genotype on the energy expenditure trajectory, a mixed model analysis was used including time, time2 and time3 as fixed effects to model the non-linearity of the trajectory. Results were adjusted for age, sex, fat mass, fat-free mass and SPA.
Results
Association of GCK SNPs with carbohydrate oxidation rate
Whole-genome sequence data from 234 Pima Indians were used to identify common SNPs (mAF ≥ 0.01). Five SNPs were identified in exon regions: two synonymous amino acid substitutions, GCK-G193G and GCK-Y215Y (rs144723656), and three 3′-UTR SNPs, rs13306388, rs55714218 and a novel SNP chr7:44184184-G/A. Four SNPs were identified in the putative promoter region and were predicted to cause loss of function by the Ingenuity Variant Analysis (https://variants.ingenuity.com): rs1799831, rs1799884, rs193226243 and rs1476891. These nine SNPs and rs4607517 (which has been associated with type 2 diabetes in other studies [5–7] and maps 6.6 kb upstream of GCK) were analysed for pairwise LD. Eight tag SNPs capture all ten SNPs (r 2 ≥ 0.8, ESM Fig. 2).
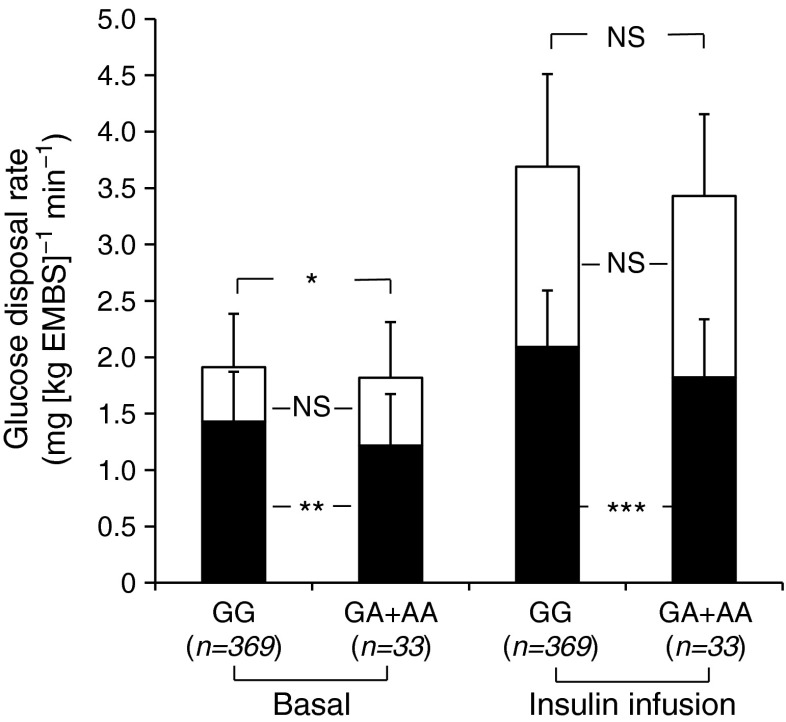
These eight tag SNPs (mAF ≥ 0.01) were genotyped in a population-based sample of 3,604 full-heritage Pima Indians of which 415 non-diabetic individuals had detailed measures of metabolic and anthropometric phenotypes for risk factors related to type 2 diabetes and obesity. Associations of these eight tag SNPs with metabolic traits were analysed in these 415 individuals. For the novel 3′UTR SNP chr7:44184184-G/A (mAF 0.05), only two of the 415 individuals were homozygous for the minor A allele, so their data were combined with those of the G/A heterozygotes for statistical analyses. Individuals with the A allele had a lower mean rate of basal carbohydrate oxidation (Fig. 1, β = 0.22 mg [kg estimated metabolic body size (EMBS)]−1 min−1 per risk allele, p = 0.005, adjusted for age, sex and percentage body fat) and lower rate of insulin-stimulated carbohydrate oxidation (β = 0.24 mg [kg EMBS]−1 min−1, adjusted p = 0.0002) when compared with individuals with the G allele. However, the non-oxidative glucose disposal rate at baseline and during insulin stimulation was not different between the two groups (adjusted p = 0.18 at basal state, p = 0.92 during insulin stimulation).
Fig. 1.
Oxidative and non-oxidative glucose disposal rates post-absorptively and during insulin infusion at 40 mU m−2 min−1 based on genotypes for the 3′UTR SNP chr7:44184184-G/A. Black bar, oxidative glucose disposal rate; white bar, non-oxidative glucose disposal rate. Error bar: SD. *p < 0.05; **p < 0.01; ***p < 0.001
Furthermore, individuals with the A allele had a higher lipid oxidation rate at baseline by 0.08 mg [kg EMBS]−1 min−1 (Table 2, p = 0.007, adjusted for age, sex and percentage body fat) and during insulin infusion by 0.09 mg [kg EMBS]−1 min−1 (adjusted p = 0.01) when compared with individuals with the G allele. These changes in lipid oxidation rate may be secondary to the changes in carbohydrate oxidation rate. Individuals with the A allele also had a lower basal rate of endogenous glucose production (β = 0.08 mg [kg EMBS]−1 min−1, adjusted p = 0.03), but no difference in endogenous glucose production rate during insulin infusion (adjusted p = 0.31). This 3′UTR SNP was not associated with the whole-body insulin-stimulated glucose disposal rate (adjusted p = 0.19). The resting metabolic rate also did not differ (p = 0.94, adjusted for age, sex, fat mass and fat-free mass) between those with the A allele and those with the G allele.
Table 2.
Metabolic characteristics of non-diabetic full-heritage individuals by genotypes of the 3′UTR SNP chr7:44184184-G/A
| Characteristic | chr7:44184184 (mean ± SD) | p valuea | ||
|---|---|---|---|---|
| G/G | G/A + AA | Estimate | ||
| Participants (n) | 369 | 33 | ||
| Body fat (%) | 32.9 ± 8.6 | 34.0 ± 7.9 | −0.82 | 0.54 |
| Oral glucose tolerance test | ||||
| Fasting plasma glucose (mmol/l) | 4.4 ± 0.5 | 5.0 ± 0.8 | −0.05 | 0.68 |
| 2 h plasma glucose (mmol/l) | 6.9 ± 1.7 | 6.9 ± 1.8 | −0.15 | 0.56 |
| Log10 fasting plasma insulin (pmol/l) | 3.0 ± 1.1 | 3.0 ± 1.1 | 0.87 | 0.67 |
| Log10 2 h plasma insulin (pmol/l) | 2.4 ± 1.0 | 2.4 ± 1.0 | 0.86 | 0.93 |
| Hyperinsulinaemic– euglycaemic clamp (mg [kg EMBS]−1 min−1)b | ||||
| Log10 glucose disposal rate | 0.55 ± 0.1 | 0.51 ± 0.1 | 0.02 | 0.19 |
| Carbohydrate oxidation | 2.10 ± 0.5 | 1.76 ± 0.5 | 0.24 | 0.0002 |
| Lipid oxidation | 0.41 ± 0.3 | 0.55 ± 0.3 | −0.09 | 0.01 |
| Endogenous glucose output | 0.38 ± 0.4 | 0.33 ± 0.3 | 0.05 | 0.31 |
| Basal glucose output (mg [kg EMBS]−1 min−1) | 1.91 ± 0.2 | 1.83 ± 0.2 | 0.08 | 0.03 |
| Basal carbohydrate oxidation (mg [kg EMBS]−1 min−1) | 1.42 ± 0.4 | 1.20 ± 0.5 | 0.22 | 0.005 |
| Basal lipid oxidation (mg [kg EMBS]−1 min−1) | 0.71 ± 0.3 | 0.80 ± 0.2 | −0.08 | 0.007 |
| Resting metabolic rate (kJ/day) | 7,314 ± 1,298 | 7,574 ± 1,583 | −8.08 | 0.94 |
| Participants, NGT (n) | 268 | 23 | ||
| Log10 AIR (pmol/l) | 3.2 ± 1.1 | 3.2 ± 1.2 | 0.91 | 0.46 |
| Log10 30-min plasma insulin (pmol/l) | 3.2 ± 1.1 | 3.2 ± 1.1 | 0.89 | 0.43 |
| Participants in metabolic chamber study (n) | 277 | 23 | ||
| Body fat (%) | 33.2 ± 8.2 | 33.9 ± 7.6 | 0.08 | 0.99 |
| 24 h RQ | 0.85 ± 0.02 | 0.85 ± 0.02 | 0.004 | 0.53 |
| Carbohydrate oxidation (kJ/day) | 4,589 ± 988 | 4,363 ± 959 | 311 | 0.03 |
| Lipid oxidation (kJ/day) | 4,007 ± 1,269 | 3,866 ± 1,034 | 134 | 0.61 |
| Protein oxidation (kJ/day) | 1,231 ± 536 | 1,281 ± 477 | −60 | 0.53 |
| 24 h energy expenditure (kJ/day) | 9,927 ± 1,700 | 9,734 ± 1,486 | 520 | 3.39 × 10−6 |
| Sleeping metabolic rate (kJ/day) | 7,046 ± 1,214 | 7,138 ± 1,017 | 136 | 0.27 |
| Energy cost of SPA (kJ/day) | 1,562 ± 565 | 1,206 ± 494 | 164 | 0.16 |
Values for mean ± SD were unadjusted. The effect size estimates were calculated as the differences in least square means and were adjusted for covariates
Rate of glucose disappearance during insulin stimulation, fasting, 30 min and 2 h plasma insulin concentrations and AIR were log10-transformed before analyses to approximate a normal distribution
aThe p value for percentage body fat was adjusted for age and sex. The p value for AIR was adjusted for age, sex, percentage body fat and rate of glucose disappearance during insulin stimulation. The p values for resting metabolic rate and sleeping metabolic rate were adjusted for age, sex, fat mass and fat-free mass. The p value for 24 h energy expenditure was adjusted for age, sex, fat mass, fat-free mass and SPA. The p values for 24 h RQ and macronutrient oxidation were adjusted for age, sex, percentage body fat and energy balance. All remaining p values were adjusted for age, sex and percentage body fat
bEMBS is equivalent to fat-free mass + 17.7 kg
Association of GCK SNPs with 24 h energy expenditure
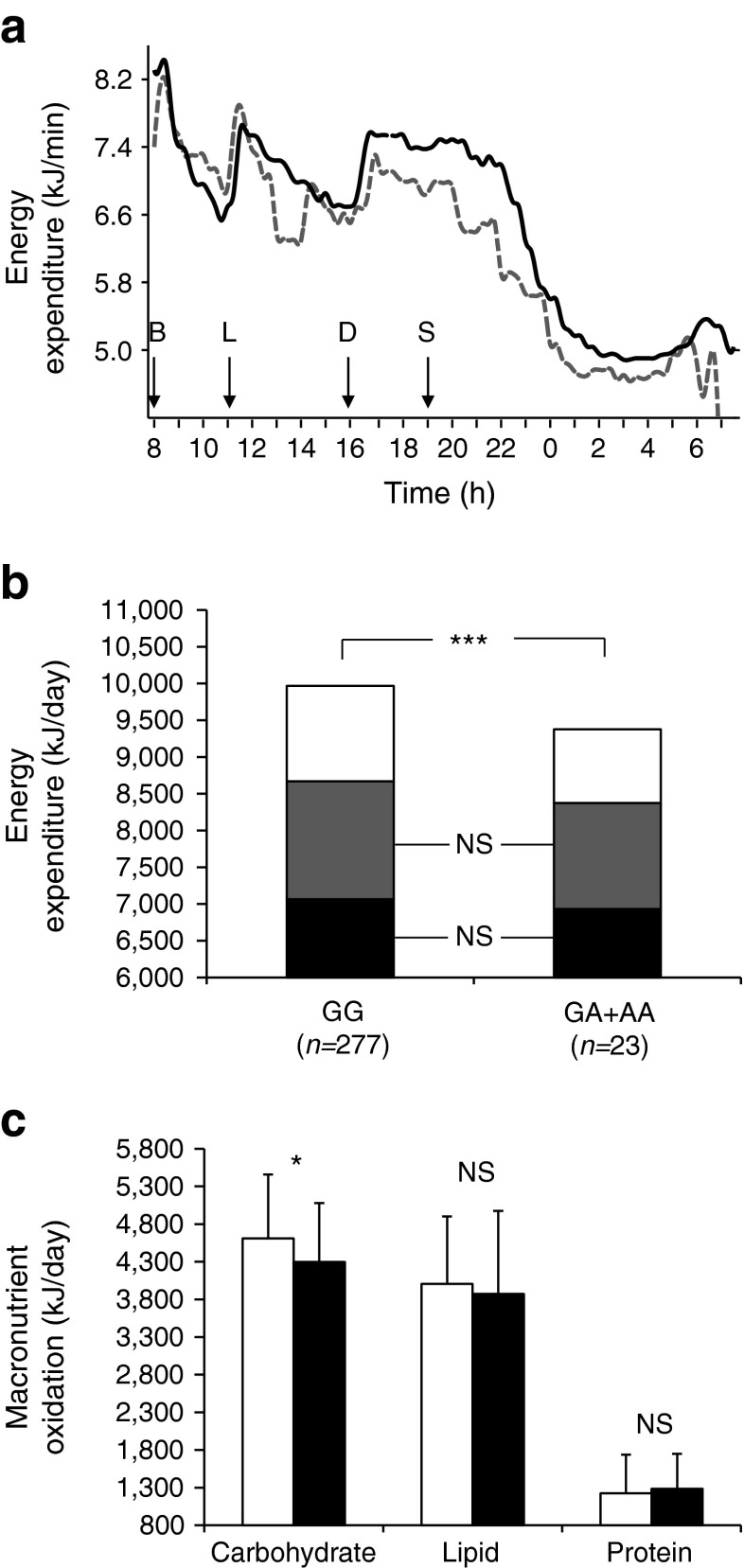
In the respiratory chamber study, participants with the A allele for the 3′UTR SNP chr7:44184184-G/A had a lower 24 h energy expenditure (by 520 kJ/day) than those with the G allele (as the difference in least square means); this was more evident in the postprandial state, as shown by the energy expenditure trajectory over the day (Fig. 2a).
Fig. 2.
(a) Time course of 24 h energy expenditure in the respiratory chamber based on genotypes for the 3′UTR SNP chr7:44184184-G/A. Arrows indicate when meals were provided (B, breakfast; L, lunch; D, dinner; S, snack); solid line, homozygous GG (n = 251); dashed line, GA + AA (n = 22). (b) Components of 24 h energy expenditure based on genotypes for the 3′UTR SNP. White bar, the thermic effect of food; grey bar, SPA; black bar, sleeping metabolic rate. ***p < 0.001. (c) 24 h macronutrient oxidation in the respiratory chamber based on genotype for the 3′UTR SNP. White bar, GG (n = 277); black bar, GA + AA (n = 23). Error bar: SD. *p < 0.05
Three components of 24 h energy expenditure, including the sleeping metabolic rate (60–70% of total energy expenditure), energy cost of SPA (20–30%) and thermic effect of food (awake and fed thermogenesis, 10%) were described previously [15, 18]. The difference in 24 h energy expenditure between genotypes of the 3′UTR SNP is attributable to the difference in the thermic effect of food (Fig. 2b, β = 520 kJ/day, p = 3.39 × 10−6 for 24 h energy expenditure, adjusted for age, sex, fat mass, fat-free mass and SPA), but not to SPA (p = 0.16, adjusted for age and sex) or sleeping metabolic rate (adjusted for age, sex, fat mass and fat-free mass, p = 0.27). To confirm this observation, we further analysed the effects of genotype on the energy expenditure trajectory during the postprandial state (daytime) vs fasting state (night-time) using a mixed model analysis. After accounting for age, sex, fat mass, fat-free mass and SPA, individuals with the A allele had a lower rate of energy expenditure during the day (β = −0.46 kJ/min, p = 0.0001, from 8:00 hours on one day to 01:00 hours on the next day), compared with those with the G allele, but no difference was observed during the night (β = −0.07 kJ/min, p = 0.50, from 01:00 hours to 05:00 hours). This result demonstrates that the difference in 24 h energy expenditure between genotypes is driven by the thermic effect of food in the postprandial state.
In the chamber, individuals with the A allele also had a decreased rate of carbohydrate oxidation (by 311 kJ/day) compared with those with the G allele (Fig. 2c, p = 0.03, adjusted for age, sex, percentage body fat and energy balance). This was comparable with the difference in the rate of carbohydrate oxidation during a hyperinsulinaemic–euglycaemic clamp, in which a ∼70 kg (EMBS) individual with the A allele was estimated to have a lower rate of insulin-stimulated carbohydrate oxidation (by ∼406 kJ/day). However, neither the rate of lipid oxidation nor protein oxidation differed between genotypes (p = 0.61 and p = 0.53, respectively, adjusted for age, sex, percentage body fat and energy balance), indicating that the 3′UTR SNP affected energy expenditure primarily via carbohydrate oxidation.
All eight tag SNPs were analysed for associations with metabolic traits in 415 full-heritage non-diabetic individuals (data not shown). Only the 3′UTR SNP was associated with the rate of carbohydrate oxidation and 24 h energy expenditure (Figs 1 and 2, Table 2).
Association of GCK SNPs with type 2 diabetes and BMI
The eight tag SNPs were genotyped in a population-based sample of 3,604 full-heritage Pima Indians and a replication sample of 4,063 mixed-heritage American Indians for association analyses of type 2 diabetes and BMI. The 3′UTR SNP chr7:44184184-G/A had a nominal association with type 2 diabetes in full-heritage Pima Indians (Table 3; OR 1.37, 95% CI 1.06, 1.78, p = 0.015, adjusted for age, sex, birth year and heritage). This SNP also had a borderline association with type 2 diabetes in a replication sample of mixed-heritage American Indians (OR 1.32, 95% CI 0.97, 1.79, adjusted p = 0.075). A lower p value was observed in a combined analysis of all individuals (OR 1.36, 95% CI 1.11, 1.65, adjusted p = 0.002, n = 7,667). Due to the low allele frequency (mAF 0.05), hence limited statistical power, this SNP only achieved a type 2 diabetes association at p = 0.002 despite a considerable effect size (OR 1.36). The A allele, associated with a lower rate of carbohydrate oxidation rate and 24 h energy expenditure, was associated with a higher prevalence of type 2 diabetes. In addition, the promoter SNP rs1476891 (D′ = 0.88, r 2 = 0 with the 3′UTR SNP, ESM Fig. 2) had a nominal association with type 2 diabetes in a combined analysis (OR 1.15, 95% CI 1.02, 1.31, adjusted p = 0.03).
Table 3.
Associations of eight tag SNPs in GCK with type 2 diabetes and fasting glucose concentrations in American Indians
| SNP | Risk/Non | Full-heritage, type 2 diabetes (n = 3,604) | Mixed-heritage, type 2 diabetes (n = 4,063) | Combined, type 2 diabetes (n = 7,667) | Combined, fasting glucose (n = 5,429) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| RAF | OR (95% CI) | p value | RAF | OR (95% CI) | p value | OR (95% CI) | p value | β value | p value | ||
| rs4607517a 5′-upstream | T/C | 0.34 | 0.97 (0.86, 1.09) | 0.616 | 0.29 | 1.20 (1.04, 1.38) | 0.010 | 1.07 (0.97, 1.17) | 0.158 | 0.06 | 8.7 × 10−7 |
| rs1476891 promoter | A/G | 0.10 | 1.26 (1.04, 1.53) | 0.016 | 0.16 | 1.08 (0.91, 1.29) | 0.380 | 1.15 (1.02,1.31) | 0.029 | 0.03 | 0.053 |
| rs193226243 promoter | A/G | 0.09 | 1.11 (0.91, 1.36) | 0.316 | 0.06 | 0.97 (0.72, 1.30) | 0.837 | 1.07 (0.90, 1.26) | 0.445 | 0.08 | 0.0003 |
| rs1799831 promoter | T/C | 0.94 | 1.02 (0.79, 1.31) | 0.894 | 0.89 | 0.82 (0.68, 1.00) | 0.056 | 0.91 (0.77, 1.06) | 0.215 | 0.01 | 0.476 |
| GCK-G193G Gly193Gly | C/T | 0.01 | 1.17 (0.68, 2.01) | 0.581 | 0.01 | 1.98 (0.97, 4.08) | 0.061 | 1.38 (0.86, 2.20) | 0.177 | 0.25 | 4.3 × 10−5 |
| rs13306388 3′UTR | C/T | 0.74 | 1.04 (0.91, 1.19) | 0.603 | 0.76 | 1.04 (0.89, 1.22) | 0.605 | 1.04 (0.94, 1.15) | 0.470 | 0.01 | 0.516 |
| chr7:44184184 3′UTR | A/G | 0.05 | 1.37 (1.06, 1.78) | 0.015 | 0.03 | 1.32 (0.97, 1.79) | 0.075 | 1.36 (1.11, 1.65) | 0.002 | −0.02 | 0.473 |
| rs55714218 3′UTR | G/- | 0.78 | 1.05 (0.92, 1.20) | 0.462 | 0.71 | 0.84 (0.73, 0.97) | 0.014 | 0.95 (0.86, 1.05) | 0.292 | −0.01 | 0.515 |
The analysis for ‘combined’ is conducted in full-heritage Pima Indians and mixed-heritage American Indians. The risk allele (given first) for rs4607517 is defined as the observed risk allele in European studies, while for other SNPs it is defined as the allele with a higher risk of diabetes in full-heritage Pima Indians; ORs are given per copy of this allele. RAF is the frequency of the risk allele. Beta for fasting glucose concentrations represents the effect in mmol/l per copy of the risk allele. The p values were adjusted for age, sex, birth year and heritage
aEstablished type 2 diabetes variant in European populations
SNP rs4607517, which was reproducibly associated with fasting glucose concentrations in other populations [5–7], also had strong associations with fasting glucose concentrations in a combined analysis of 5,429 American Indians (β = 0.06 mmol/l per risk allele, p = 8.7 × 10−7, adjusted for age, sex, birth year and heritage) (Table 3). However, this SNP was not associated with type 2 diabetes (adjusted p = 0.16, n = 7,667). In addition, the synonymous SNP Gly193Gly and rs193226243 were also associated with fasting glucose concentrations (β = 0.25 mmol/l, adjusted p = 4.3 × 10−5; β = 0.08 mmol/l, adjusted p = 0.0003, respectively).
Beta cell function was assessed by AIR in 298 full-heritage Pima Indians with NGT. SNP rs4607517, associated with fasting glucose concentrations, was nominally associated with AIR (β = 0.89 [log10 scale], p = 0.07, adjusted for age, sex, percentage body fat and rate of glucose disappearance during insulin stimulation). The promoter SNP rs193226243 was also associated with AIR (β = 0.92 [log10 scale], adjusted p = 0.02). However, the 3′UTR SNP was not associated with AIR (adjusted p = 0.46).
Association of the eight tag SNPs with maximum BMI were also analysed (Table 4). SNP rs193226243 had a nominal association with BMI in a combined analyses of 6,797 American Indians (β = 0.02 (loge scale), p = 0.008, adjusted for age, sex, birth year and heritage). None of other SNPs was consistently associated with BMI.
Table 4.
Associations of eight tag SNPs in GCK with maximal BMI in full-heritage Pima Indians, mixed-heritage American Indians and the combined samples
| SNP | Risk/Non | Full-heritage Pima Indian (n = 3,391), mean BMI (kg/m2) | Mixed-heritage American Indian (n = 3,406), mean BMI (kg/m2) | Combined (n = 6,797) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| RAF | Risk/Risk | Risk/Non | Non/Non | β value | p value | RAF | Risk/Risk | Risk/Non | Non/Non | β value | p value | β value | p value | ||
| rs4607517 | T/C | 0.34 | 38.2 | 37.5 | 37.2 | 0.013 | 0.028 | 0.29 | 34.8 | 35.1 | 34.7 | 0.003 | 0.608 | 0.008 | 0.065 |
| rs1476891 | A/G | 0.10 | 37.7 | 36.9 | 37.6 | −0.006 | 0.512 | 0.16 | 34.3 | 34.4 | 35.0 | −0.002 | 0.781 | −0.005 | 0.433 |
| rs193226243 | A/G | 0.09 | 39.2 | 37.9 | 37.4 | 0.012 | 0.216 | 0.06 | 30.7 | 36.4 | 34.6 | 0.029 | 0.020 | 0.020 | 0.008 |
| rs1799831 | T/C | 0.94 | 37.5 | 36.9 | 38.1 | 0.012 | 0.307 | 0.89 | 34.9 | 34.8 | 30.4 | 0.005 | 0.555 | 0.009 | 0.238 |
| GCK-G193G | C/T | 0.01 | 38.7 | 37.4 | 0.046 | 0.085 | 0.01 | 35.1 | 34.8 | −0.028 | 0.548 | 0.020 | 0.432 | ||
| rs13306388 | C/T | 0.74 | 37.3 | 37.7 | 38.2 | −0.006 | 0.342 | 0.76 | 34.8 | 34.7 | 34.6 | −0.002 | 0.720 | −0.005 | 0.321 |
| Chr7:44184184 | A/G | 0.05 | 41.4 | 37.9 | 37.4 | 0.008 | 0.571 | 0.03 | 30.8 | 36.6 | 34.7 | 0.018 | 0.268 | 0.012 | 0.254 |
| rs55714218 | G/− | 0.78 | 37.8 | 37.7 | 37.1 | 0.002 | 0.822 | 0.71 | 34.7 | 34.8 | 34.7 | 0.002 | 0.728 | 0.003 | 0.555 |
BMI is the maximum value observed in the longitudinal study from all examinations after age 15 years. The risk allele is defined as the allele with a higher risk of diabetes in full-heritage Pima Indians; the regression coefficient (B) represents the effect on the logarithmic scale (loge) per copy of the risk allele. Mean BMI was unadjusted. The p values were adjusted for age, sex, birth year and heritage
The predictive effect of carbohydrate oxidation rate on development of type 2 diabetes
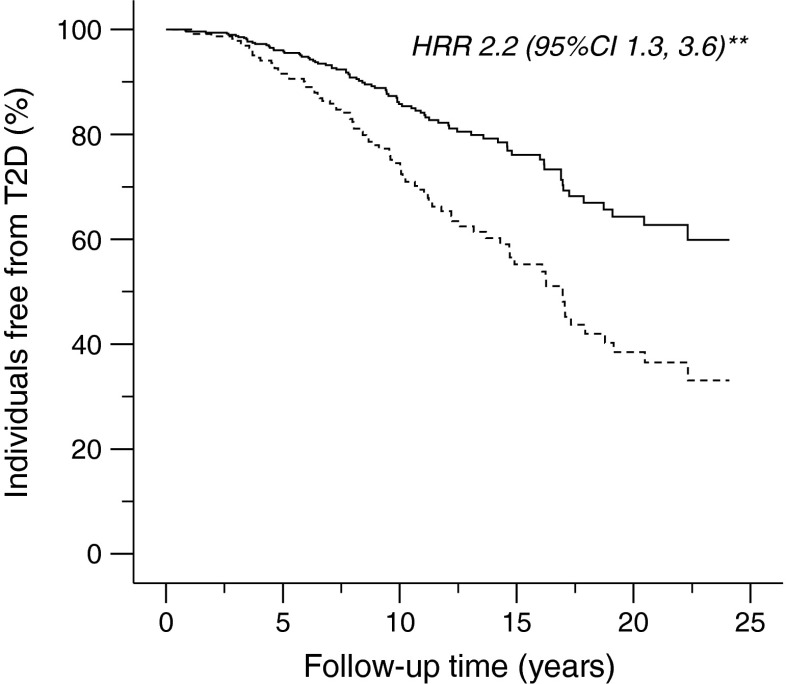
We further evaluated the relationship between the rate of insulin-stimulated carbohydrate oxidation and the risk of developing type 2 diabetes in 287 full-heritage Pima Indians. Of these 287 individuals with NGT who had measures of carbohydrate oxidation rate during insulin stimulation at baseline (baseline age 26.4 ± 6.0 years) and also had follow-up data for development of diabetes (follow-up time 7.8 ± 8.2 years), 99 (34%) developed type 2 diabetes. Figure 3 shows the Kaplan–Meier survival curve for time to type 2 diabetes onset with participants categorised as those with a higher or lower rate of insulin-stimulated carbohydrate oxidation than the mean of 1.63 mg (kg EMBS)−1 min−1. Individuals with a lower rate of insulin-stimulated carbohydrate oxidation (n = 150; carbohydrate oxidation rate 1.35 ± 0.23 mg [kg EMBS]−1 min−1; age 26.4 ± 5.8 years) had a shorter period to type 2 diabetes onset, hence a higher risk for developing type 2 diabetes as compared with those with a higher rate of carbohydrate oxidation (n = 137; carbohydrate oxidation rate 1.94 ± 0.26 mg [kg EMBS]−1 min−1; age 26.5 ± 6.3 years) (HRR 2.2, 95% CI 1.3, 3.6, p = 0.002, adjusted for age, sex, percentage body fat, AIR and non-oxidative glucose disposal rate). The same result was observed with individuals categorised as those with a higher or lower median rate of insulin-stimulated carbohydrate oxidation (1.61 mg [kg EMBS]−1 min−1). In a Cox proportional hazards analysis, a lower than the mean rate of insulin-stimulated non-oxidative glucose disposal at baseline also predicted a higher risk for developing type 2 diabetes than a rate above the mean (HRR 2.5, 95% CI 1.4, 4.5, p = 0.002, adjusted for age, sex, percentage body fat, AIR and glucose oxidation rate; data not shown).
Fig. 3.
Survival curve for time to type 2 diabetes (T2D) onset in 287 full-heritage Pima Indians with NGT at baseline. Solid line, carbohydrate oxidation rate > a mean of 1.63 mg (kg EMBS)−1 min−1 (n = 137); dashed line, carbohydrate oxidation rate <1.63 mg (kg EMBS)−1 min−1 (n = 150). Data are plotted up to the follow-up time of ∼23 years, and omitted at longer follow-up time when only 1% of participants were involved. HRR 2.2, 95% CI 1.3, 3.6; **p < 0.01
Allelic specific GCK expression
To investigate whether the alleles of the 3′UTR SNP chr7:44184184-G/A differentially influence gene expression, allelic specific expression of GCK was assessed in adipose tissue biopsies from individuals heterozygous for this SNP. No difference in the allelic expression of GCK was observed in this tissue (G/A = 0.993 vs expected ratio of 1, p = 0.2, data not shown). Since liver and pancreas tissue biopsies from Pima Indians were not available for study, the allelic specific expression of GCK in these tissues is not known.
Discussion
GCK is the main glucose-phosphorylating enzyme in the liver and pancreatic beta cells. It converts glucose to G6P as a first and rate-limiting step in glycolysis, which plays a part in the process of glucose oxidation [1–3]. Our study indicates that a novel variant in the 3′UTR of GCK, with a risk allele frequency of 0.05, is associated with a lower rate of glucose oxidation post-absorptively, during insulin stimulation and after a diet of mixed consumption, which is in agreement with the role of GCK in glycolysis. This variant was not associated with non-oxidative glucose disposal, suggesting that glucose storage (glycogen synthesis) was not affected. It is known that rare mutations in GCK occurring in MODY result from a reduced glucose-stimulated insulin secretion. Although rs193226243 and rs4607517 had borderline associations with AIR, none of the other common variants were associated with AIR. Therefore, our data suggest that common variation in GCK predominantly influences glycolysis and the rate of glucose oxidation in hepatocytes. These data are consistent with the observations that overexpression of GCK in mouse liver or rat isolated hepatocytes enhances glucose oxidation [19, 20]. Nevertheless, a subtle effect of common GCK variants on beta cell function cannot be ruled out. Since the GCK variants affect the threshold for glucose sensing, the effect size and/or sample size may be too small to render a statistical difference in insulin secretion. It is also possible that the 25 g intravenous glucose bolus used in the AIR measurement may be above the threshold at which GCK exerts its effect, thus limiting a positive detection.
Hepatic GCK serves as a major component of the hepatic glucose-sensing system involved in glucose synthesis, breakdown and storage. While glycolysis and glycogen synthesis pathways are activated during the postprandial state, gluconeogenesis and glycogen breakdown are involved in hepatic glucose production in the post-absorptive state. Our data indicate that the association of the 3′UTR GCK variant with the rate of basal hepatic glucose production is likely due to an association with the rate of basal glucose oxidation. However, this variant was not associated with hepatic glucose production during insulin stimulation, despite previous findings demonstrating that a GCK variant was associated with hepatic insulin resistance [21].
In addition to its pivotal role in glucose metabolism, a new role for hepatic GCK in energy metabolism has emerged in recent studies. Tsukita and co-workers reported that upregulation of hepatic GCK by high-fat diet feeding in mice suppresses brown adipose tissue (BAT) thermogenesis via leptin-mediated neural signals and downregulation of uncoupling protein-1 [22, 23]. This GCK-mediated liver-to-BAT neuronal relay system provides a novel mechanism in modulating obesity predisposition in mice. In this study, we report that the novel 3′UTR variant in GCK had a significant effect on 24 h energy expenditure through a change in the thermic effect of food. This effect resulted from a change in the rate of carbohydrate oxidation rather than from any apparent effect on BAT thermogenesis.
Measures of energy expenditure, thermic effect of food and substrate oxidation are predictors of weight change. In Pima Indians, individuals with lower than expected energy expenditure are at higher risk for future long-term increases in weight and fat mass [16]. The thermic effect of food is also reduced in obese individuals and predicts their future weight gain [18]. A higher rate of insulin-stimulated carbohydrate oxidation during a hyperinsulinaemic–euglycaemic clamp predicts a future weight again [24]. The rate of carbohydrate oxidation in a respiratory chamber also predicts short-term changes in body weight [14], but not long-term changes [16]. In the present study, the 3′UTR SNP, associated with rates of energy expenditure and carbohydrate oxidation, was not associated with BMI. Nevertheless, this 3′UTR SNP was associated with risk of type 2 diabetes in Pima Indians. This most likely results from the effect on carbohydrate oxidation since we found that a lower carbohydrate oxidation rate during insulin stimulation was associated with a higher risk of type 2 diabetes, independent of age, sex, percentage body fat, AIR and non-oxidative glucose disposal rate. Thus, while rare mutations in GCK cause MODY and neonatal diabetes, our data indicate that common variation in GCK with a modest effect on the rate of carbohydrate oxidation contributes to risk of type 2 diabetes.
In summary, our study in individuals who had been extensively characterised for metabolic traits provides cohesive evidence to support a hepatic effect of a novel 3′UTR variant in GCK on influencing carbohydrate oxidation, energy expenditure and type 2 diabetes risk; this is consistent with the role of GCK in hepatic glycolysis and energy metabolism. However, our functional analysis of this 3′UTR SNP in adipose tissue did not support a role in allelic imbalance of GCK expression in this particular tissue. Interpretation of this negative result is unclear since this SNP could potentially affect transcriptional regulation or mRNA stability in a tissue-specific manner, and we do not have access to liver or pancreatic beta cells from Pima individuals. Alternatively, this 3′UTR SNP might alter GCK translation via an effect on microRNA binding, or perhaps this SNP is in LD with an undiscovered functional variant. Future studies in liver or pancreatic biopsy tissues would clarify some of these possible mechanisms.
Electronic supplementary material
Below is the link to the electronic supplementary material.
(PDF 249 kb)
(PDF 85 kb)
Acknowledgements
We thank the clinical staff of the Phoenix Epidemiology and Clinical Research Branch for conducting the study. We also thank all the participants from the Gila River Indian Community.
Funding
This work was supported by the intramural research programme of the National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health.
Duality of interest
The authors declare that there is no duality of interest associated with this manuscript.
Contribution statement
CB is responsible for the integrity of the work as a whole. YLM, PP, LJB and CB contributed to the study design. YLM, DH, KH, BG, WCK, RLH, LJB and CB contributed to the data acquisition and PP, SK MST, WCK and CB contributed to the data analysis. All authors contributed to data interpretation and manuscript drafting, and approved the final version.
Abbreviations
- 3′UTR
3′ Untranslated region
- AIR
Acute insulin response
- BAT
Brown adipose tissue
- EMBS
Estimated metabolic body size
- G6P
Glucose-6-phosphate
- GCK
Glucokinase
- GEE
Generalised estimating equation
- HRR
Hazard rate ratio
- mAF
Minor allele frequency
- LD
Linkage disequilibrium
- NGT
Normal glucose tolerance
- RQ
Respiratory quotient
- SNP
Single-nucleotide polymorphism
- SPA
Spontaneous physical activity
Footnotes
Yunhua L. Muller and Paolo Piaggi contributed equally to this study.
References
- 1.Postic C, Shiota M, Magnuson MA. Cell-specific roles of glucokinase in glucose homeostasis. Recent Prog Horm Res. 2001;56:195–217. doi: 10.1210/rp.56.1.195. [DOI] [PubMed] [Google Scholar]
- 2.Matschinsky FM (2002) Regulation of pancreatic beta-cell glucokinase: from basics to therapeutics. Diabetes 51(Suppl. 3):S394-S404 [DOI] [PubMed]
- 3.Matschinsky FM, Glaser B, Magnuson MA. Pancreatic beta-cell glucokinase: closing the gap between theoretical concepts and experimental realities. Diabetes. 1998;47:307–315. doi: 10.2337/diabetes.47.3.307. [DOI] [PubMed] [Google Scholar]
- 4.Hussain K. Mutations in pancreatic β-cell glucokinase as a cause of hyperinsulinaemic hypoglycaemia and neonatal diabetes mellitus. Rev Endocr Metab Disord. 2010;11:179–183. doi: 10.1007/s11154-010-9147-z. [DOI] [PubMed] [Google Scholar]
- 5.Soranzo N, Sanna S, Wheeler E, et al. Common variants at 10 genomic loci influence hemoglobin A1(C) levels via glycemic and nonglycemic pathways. Diabetes. 2010;59:3229–3239. doi: 10.2337/db10-0502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weedon MN, Clark VJ, Qian Y, et al. A common haplotype of the glucokinase gene alters fasting glucose and birth weight: association in six studies and population-genetics analyses. Am J Hum Genet. 2006;79:991–1001. doi: 10.1086/509517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Holmkvist J, Almgren P, Lyssenko V, et al. Common variants in maturity-onset diabetes of the young genes and future risk of type 2 diabetes. Diabetes. 2008;57:1738–1744. doi: 10.2337/db06-1464. [DOI] [PubMed] [Google Scholar]
- 8.Knowler WC, Pettitt DJ, Saad MF, et al. Diabetes mellitus in the Pima Indians: incidence, risk factors and pathogenesis. Diabetes Metab Rev. 1990;6:1–27. doi: 10.1002/dmr.5610060101. [DOI] [PubMed] [Google Scholar]
- 9.The Expert Committee on the Diagnosis and Classification of Diabetes Mellitus Report of the Expert Committee on the Diagnosis and Classification of Diabetes Mellitus. Diabetes Care. 1997;20:1183–1197. doi: 10.2337/diacare.20.7.1183. [DOI] [PubMed] [Google Scholar]
- 10.Tataranni PA, Ravussin E. Use of dual-energy X-ray absorptiometry in obese individuals. Am J Clin Nutr. 1995;62:730–734. doi: 10.1093/ajcn/62.4.730. [DOI] [PubMed] [Google Scholar]
- 11.Lillioja S, Mott DM, Spraul M, et al. Insulin resistance and insulin secretory dysfunction as precursors of non-insulin-dependent diabetes mellitus. Prospective studies of Pima Indians. N Engl J Med. 1993;329:1988–1992. doi: 10.1056/NEJM199312303292703. [DOI] [PubMed] [Google Scholar]
- 12.Bogardus C, Lillioja S, Ravussin E, et al. Familial dependence of the resting metabolic rate. N Engl J Med. 1986;315:96–100. doi: 10.1056/NEJM198607103150205. [DOI] [PubMed] [Google Scholar]
- 13.Lillioja S, Bogardus C. Obesity and insulin resistance: lessons learned from the Pima Indians. Diabetes Metab Rev. 1988;4:517–540. doi: 10.1002/dmr.5610040508. [DOI] [PubMed] [Google Scholar]
- 14.Pannacciulli N, Salbe AD, Ortega E, Venti CA, Bogardus C, Krakoff J. The 24-h carbohydrate oxidation rate in a human respiratory chamber predicts ad libitum food intake. Am J Clin Nutr. 2007;86:625–632. doi: 10.1093/ajcn/86.3.625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ravussin E, Lillioja S, Anderson TE, Christin L, Bogardus C. Determinants of 24-hour energy expenditure in man. Methods and results using a respiratory chamber. J Clin Invest. 1986;78:1568–1578. doi: 10.1172/JCI112749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Piaggi P, Thearle MS, Bogardus C, Krakoff J. Lower energy expenditure predicts long-term increases in weight and fat mass. J Clin Endocrinol Metab. 2013;98:E703–E707. doi: 10.1210/jc.2012-3529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hanis CL, Chakraborty R, Ferrell RE, Schull WJ. Individual admixture estimates: disease associations and individual risk of diabetes and gallbladder disease among Mexican-Americans in Starr County, Texas. Am J Phys Anthropol. 1986;70:433–441. doi: 10.1002/ajpa.1330700404. [DOI] [PubMed] [Google Scholar]
- 18.Piaggi P, Krakoff J, Bogardus C, Thearle MS. Lower ‘awake and fed thermogenesis’ predicts future weight gain in subjects with abdominal adiposity. Diabetes. 2013;62:4043–4051. doi: 10.2337/db13-0785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ferre T, Riu E, Bosch F, Valera A. Evidence from transgenic mice that glucokinase is rate limiting for glucose utilization in the liver. FASEB J 10:1213-1218 [DOI] [PubMed]
- 20.Takeuchi H, Inoue Y, Ishihara H, Oka Y. Overexpression of either liver type or pancreatic beta cell type glucokinase via recombinant adenovirus enhances glucose oxidation in isolated rat hepatocytes. FEBS Lett. 1996;393:60–64. doi: 10.1016/0014-5793(96)00833-2. [DOI] [PubMed] [Google Scholar]
- 21.Chiu KC, Chuang LM, Yoon C, Saad MF. Hepatic glucokinase promoter polymorphism is associated with hepatic insulin resistance in Asian Indians. BMC Genet. 2000;1:2. doi: 10.1186/1471-2156-1-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tsukita S, Yamada T, Uno K. Hepatic glucokinase modulates obesity predisposition by regulating BAT thermogenesis via neural signals. Cell Metab. 2012;16:825–832. doi: 10.1016/j.cmet.2012.11.006. [DOI] [PubMed] [Google Scholar]
- 23.Yamada T, Tsukita S, Katagiri H. Identification of a novel interorgan mechanism favoring energy storage in overnutrition. Adipocyte. 2013;2:281–284. doi: 10.4161/adip.25499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Swinburn BA, Nyomba BL, Saad MF, et al. Insulin resistance associated with lower rates of weight gain in Pima Indians. J Clin Invest. 1991;88:168–173. doi: 10.1172/JCI115274. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF 249 kb)
(PDF 85 kb)