Abstract
Dedifferentiated chondrosarcomas are cartilaginous tumors that consist of two distinguishable components, a lowgrade chondrosarcoma (chondrogenic) component and a highgrade dedifferentiated (anaplastic) component. The tumor cells in both components seem to originate from a single precursor, but there are a substantial number of genetic alterations in the anaplastic component. The underlying mechanism of dedifferentiation is unknown, but cell cycle regulators p16, p53 and retinoblastoma appear to have important roles in tumor development and dedifferentiation. In this article, molecular pathogenesis of dedifferentiated chondrosarcomas is reviewed.
Keywords: Dedifferentiated chondrosarcoma, dedifferentiation, pathogenesis
INTRODUCTION
Chondrosarcomas, the second most common form of primary bone cancer, primarily affect the cartilaginous cells of the arms, shoulders, legs, spine and pelvis.1 Approximately, 10% of chondrosarcomas undergo dedifferentiation, a process in which a portion of the tumor undergoes an anaplastic transformation.1 These dedifferentiated tumors are comprised of two distinguishable, juxtaposed components: A chondrogenic component and a highgrade noncartilaginous sarcoma (anaplastic component) [Figure 1].2
Figure 1.
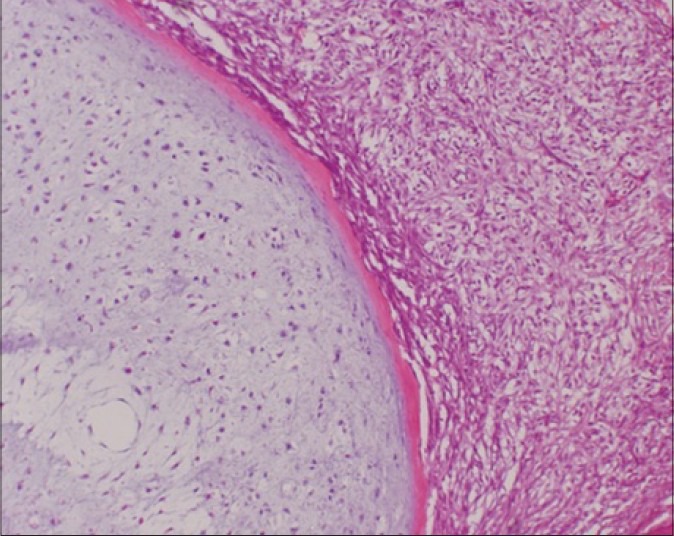
Dedifferentiated chondrosarcoma is characterized by the coexistence of low-grade chondrosarcoma components (left) and highgrade dedifferentiated components with features of undifferentiated sarcoma (right) with a distinct border (H and E, ×150)
CLINICAL FINDINGS
Femur is the most frequently involved bone (30%), followed by the pelvis (20%), humerus (16%), ribs (7%) and scapula (7%) respectively. Majority of tumors occur in patients >50 years of age, with a mean age of approximately 60 years.1 Most lesions occur centrally in the medullary cavities, although there are reports of dedifferentiation in juxtacortical chondrosarcomas or from a preexisting osteochondroma.3
Dedifferentiated chondrosarcomas are highly malignant tumors with a very poor prognosis. Median survival has been as short as 6 months and 5-year rates of survival range from as low as 10% to 13%. Patients rarely survive more than 2 years.4,5,6 Surgery is the primary form of treatment. The effectiveness of chemotherapy has not been proven, but it has been used for select cases.5,7,8 Metastasis, especially to the lungs, is the most common problem encountered during treatment.6,8,9 This article is a review of molecular studies of dedifferentiated chondrosarcoma, with an emphasis on the process and characteristics of dedifferentiation.
MATERIALS AND METHODS
Search strategy
We searched PubMed for relevant studies in the English language literature using the search term “dedifferentiated chondrosarcoma” for articles published after the original report.2 This search retrieved 224 articles, of which 92 case reports were excluded. The contents of the remaining 132 were examined by abstract or the entire paper. Six articles were selected in addition to one text book1 for the clinical review and 27 articles were selected for discussing the pathogenesis of dedifferentiated chondrosarcoma, particularly the mechanism for “dedifferentiation” in chondrosarcoma.
DISCUSSION
HISTOLOGICAL FEATURES
The histological features of the chondrogenic component of dedifferentiated chondrosarcomas are similar to those of conventional chondrosarcomas, including the expression of collagen type II and aggrecan in the matrix and S100 protein in the cells. These cartilaginous molecules are absent in the anaplastic components, with the exception of areas with chondroblastic osteosarcoma like features.10 The anaplastic component has increased proliferative activity, as shown by the expression of Ki-67 and proliferating cell nuclear antigen and a higher malignant potential than the chondrogenic component.11 Histological features of the anaplastic component can vary and include undifferentiated sarcomas, osteosarcomas, angiosarcomas, fibrosarcomas, rhabdomyosarcomas, leiomyosarcomas or giant cell tumors.7 However, clinical outcome is not affected by the type of high grade tumor in the dedifferentiated component.10
The anaplastic component is characterized by multiple differentiations, corresponding to various histological features.10 Both intramembranous (direct) and endochondral (indirect) ossification is observed. Intramembranous ossification occurs when pluripotent mesenchymal cells directly enter osteoblast lineage, whereas in endochondral ossification; chondrocytes differentiate and are replaced by osteoblasts. The potential for multiple differentiations of precursor cells is supported by the fact that a mutated human embryonic muscle cell line exhibits features of dedifferentiated chondrosarcomas.12
The plasminogen activator system is a key regulator of invasion and tumor angiogenesis. Plasminogen activator inhibitor 1 (PAI-1) acts as an inhibitor of tissue-type plasminogen activator and urokinase-type plasminogen activator.13 In dedifferentiated chondrosarcoma, the anaplastic component displays diffuse coexpression of t-PA, u-PA and PAI-1.14 Interestingly, the expression of PAI-1 in the chondrogenic component of dedifferentiated chondrosarcomas is associated with good prognosis.15 Matrix metalloproteinases (MMPs) are a family of zinc-dependent endopeptidases that are principally involved in the breakdown of the extra cellular matrix, as well as tumor angiogenesis. MMPs are regulated by the tissue inhibitors of MMPs (TIMPs).16 Upregulation of MMP2, MT1-MMP and TIMP2 has been reported in high grade malignant cartilaginous tumors, as well as in the anaplastic component of dedifferentiated chondrosarcoma.17 MMPs interact with the plasminogen activator system and upregulation of both the plasminogen activator system and MMPs apparently represent the malignant potential of the anaplastic component.
Oncogenesis in cartilaginous lesions
Heterozygous mutations of isocitrate dehydrogenase 1 and 2 (IDH1 and IDH2, respectively) near or in the active sites of the enzyme that lead to the accumulation of δ-2-hydroxyglutarate have been associated with oncogenesis. Mutations occur early in oncogenesis in gliomas/glioblastomas and acute myelogenous leukemia and represent early genetic abnormalities in all cartilaginous tumors.18 These IDH1 and IDH2 mutations account for more than half of mutations in benign and malignant cartilaginous tumors, including dedifferentiated chondrosarcomas.19
The c-Myc gene, located on chromosome 8, encodes a transcription factor that acts in the nucleus to stimulate cell growth and division.20 c-Myc amplification is frequently observed in highgrade malignant cartilaginous tumors including dedifferentiated chondrosarcomas, while no amplification is found in benign and lowgrade cartilaginous tumors.21 In the absence of c-Myc amplification, polysomy 8 is found frequently in low to highgrade malignant cartilaginous tumors.21 c-Myc abnormalities appear to correspond to early oncogenesis in all chondrosarcomas, particularly with highgrade malignant cartilaginous tumors including dedifferentiated chondrosarcomas.
Single point mutations in the ras genes play a role in tumor development by eliminating dependence on GTPase-activation of protein regulation. A small number of cases revealed H-ras mutations in dedifferentiated chondrosarcoma, but not in lowgrade conventional chondrosarcomas, suggesting that the mutation may be associated with the aggressive nature of the disease rather than dedifferentiation.22
Origin of dedifferentiation
The mechanism underlying dedifferentiation is controversial. There is debate as to whether the chondrogenic and anaplastic components derive from a common precursor cell.23 Molecular studies show that both tumor components share some genetic alterations and that the components derive from a single precursor. However, a substantial number of genetic alterations occur in anaplastic components after the division.24,25 Therefore, it is also possible, though less likely, that the highgrade dedifferentiated component is the result of a malignant transformation within the dense fibrotic material surrounding the necrotic areas at the margin of the lowgrade chondrosarcoma component, which is the same process by which sarcoma develops in bone infarcts and chronic osteomyelitis.26
Although the two components derive from a single precursor cell, the molecular mechanism, such as the timing of separation, has not been clarified. A possibility is that the anaplastic component originates from mature chondrosarcoma cells in the chondrogenic component as a result of direct transformation,2 or the cells of each component separate from a common single precursor cell early in tumorigenesis. The existence of a stable dedifferentiated chondrosarcoma cell line supports the premise that separation occurs early during tumor development.12
Chromosomal abnormalities
Chromosomal aberrations of increasing complexity develop during tumor progression in chondrosarcoma.27,28 Dedifferentiated chondrosarcoma has a hypodiploid to hypertetraploid chromosome number and presents a heterogeneous pattern of copy number imbalances.29 Trisomy 19 has been documented in half of the dedifferentiated chondrosarcomas.30 No structural or numerical chromosomal aberrations, highly specific for dedifferentiated chondrosarcomas, have been detected. However, there is some evidence for the clustering of breakpoints in specific regions of 6q13-22 and 9p21-24.30 Chromosomal aberrations in 6q13-21 are associated with locally aggressive behavior in benign and malignant cartilaginous tumors, including dedifferentiated chondrosarcoma.27,28 Aberrations in 5q have also been reported, but the abnormality is rather more frequent in highgrade conventional chondrosarcoma (35%) than dedifferentiated chondrosarcoma (13%).30 Array-based comparative genomic hybridization studies also show that the 5q14.2-q21.3, 6q16-q25.3, 9p24.2-q12 and 9p21.3 loci are specific for highgrade conventional chondrosarcoma and dedifferentiated chondrosarcoma.31 The breakpoints in 9p21-24 are seen in anaplastic components with osteosarcoma-like features. This observation raises the possibility that each highgrade subtype may be associated with a unique set of chromosomal changes.30
Molecular analysis of dedifferentiation
Defects in cell-cycle regulatory pathways play an important role in the oncogenesis of chondrosarcoma. p16 regulates cell cycle through the inhibition of cdk4 and cdk6.32 Analysis of a dedifferentiated chondrosarcoma cell line, MS0812, suggests that deletion of the p16 gene may play a major role in the malignant phenotype of dedifferentiated chondrosarcoma.12 Moreover, aberrant promoter methylation of the p16 gene has also been reported in both components of dedifferentiated chondrosarcoma.25 Interestingly, aberrant methylation of E-cadherin, a cell adhesion molecule, is also seen in both components. The aberrant methylation of p16 and E-cadherin may signal early oncogenesis of a dedifferentiated chondrosarcoma.25
p53 has many mechanisms of anticancer function and plays a role in cell cycle regulation, apoptosis, genomic stability and inhibition of angiogenesis. p53 mutations are the predominant mutations present in highgrade conventional chondrosarcomas and in dedifferentiated chondrosarcoma.33,34 In dedifferentiated chondrosarcoma, p53 mutation or loss of heterozygosity (LOH) is detected exclusively in the highgrade dedifferentiated component.11,24,25 The retinoblastoma (Rb) protein controls E2F-mediated transactivation of genes whose products are important for S phase entry and cell-cycle progression. Loss of Rb function is an essential step in oncogenesis. LOH of Rb is associated with decreased Rb expression and is significantly correlated with high malignancy in cartilaginous tumors. In addition, LOH of Rb has been found only in the anaplastic component.25,35 Aberrant promoter methylation in the fragile histidine triad (FHIT) gene is seen only in highgrade dedifferentiated components. Although FHIT is involved in the regulation of apoptosis and in cell cycle control, the molecular mechanism or functional pathway is still unknown. Irregularities in Rb, p53 and FHIT in the anaplastic component may be the key abnormalities in dedifferentiation of chondrosarcomas.11,24,25,35
CONCLUSIONS
Molecular studies in dedifferentiated chondrosarcomas have analyzed tumor development, as well as the mechanism of “dedifferentiation.” Among various abnormalities, cell-cycle regulation molecules p16, p53 and Rb may be important in the development of these tumors. Dedifferentiated chondrosarcomas are chemotherapy resistant, regardless of various histological features in the highgrade dedifferentiated components. Therefore, a comparative study of the anaplastic component with osteosarcoma-like features and conventional chemotherapy-sensitive osteosarcoma would be interesting, in order to identify the proteins involved in chemotherapy resistance.
Footnotes
Source of Support: Nil.
Conflict of Interest: None.
REFERENCES
- 1.Dorfman HD, Czerniak B. Year Book. Bone Tumors. St Louis, Missouri: Mosby; 1998. pp. 395–410. [Google Scholar]
- 2.Dahlin DC, Beabout JW. Dedifferentiation of low-grade chondrosarcomas. Cancer. 1971;28:461–6. doi: 10.1002/1097-0142(197108)28:2<461::aid-cncr2820280227>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- 3.Papagelopoulos PJ, Galanis EC, Mavrogenis AF, Savvidou OD, Bond JR, Unni KK, et al. Survivorship analysis in patients with periosteal chondrosarcoma. Clin Orthop Relat Res. 2006;448:199–207. doi: 10.1097/01.blo.0000194684.40624.a8. [DOI] [PubMed] [Google Scholar]
- 4.Frassica FJ, Unni KK, Beabout JW, Sim FH. Dedifferentiated chondrosarcoma. A report of the clinicopathological features and treatment of seventy-eight cases. J Bone Joint Surg Am. 1986;68:1197–205. [PubMed] [Google Scholar]
- 5.Mercuri M, Picci P, Campanacci L, Rulli E. Dedifferentiated chondrosarcoma. Skeletal Radiol. 1995;24:409–16. doi: 10.1007/BF00941235. [DOI] [PubMed] [Google Scholar]
- 6.Dickey ID, Rose PS, Fuchs B, Wold LE, Okuno SH, Sim FH, et al. Dedifferentiated chondrosarcoma: The role of chemotherapy with updated outcomes. J Bone Joint Surg Am. 2004;86-A:2412–8. [PubMed] [Google Scholar]
- 7.Bruns J, Fiedler W, Werner M, Delling G. Dedifferentiated chondrosarcoma – A fatal disease. J Cancer Res Clin Oncol. 2005;131:333–9. doi: 10.1007/s00432-004-0648-6. [DOI] [PubMed] [Google Scholar]
- 8.Grimer RJ, Gosheger G, Taminiau A, Biau D, Matejovsky Z, Kollender Y, et al. Dedifferentiated chondrosarcoma: Prognostic factors and outcome from a European group. Eur J Cancer. 2007;43:2060–5. doi: 10.1016/j.ejca.2007.06.016. [DOI] [PubMed] [Google Scholar]
- 9.Capanna R, Bertoni F, Bettelli G, Picci P, Bacchini P, Present D, et al. Dedifferentiated chondrosarcoma. J Bone Joint Surg Am. 1988;70:60–9. [PubMed] [Google Scholar]
- 10.Dornauer K, Söder S, Inwards CY, Bovee JV, Aigner T. Matrix biochemistry and cell biology of dedifferentiated chondrosarcomas. Pathol Int. 2010;60:365–72. doi: 10.1111/j.1440-1827.2010.02530.x. [DOI] [PubMed] [Google Scholar]
- 11.Simms WW, Ordóñez NG, Johnston D, Ayala AG, Czerniak B. p53 expression in dedifferentiated chondrosarcoma. Cancer. 1995;76:223–7. doi: 10.1002/1097-0142(19950715)76:2<223::aid-cncr2820760210>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- 12.Yang L, Chen Q, Zhang S, Wang X, Li W, Wen J, et al. A novel mutated cell line with characteristics of dedifferentiated chondrosarcoma. Int J Mol Med. 2009;24:427–35. doi: 10.3892/ijmm_00000249. [DOI] [PubMed] [Google Scholar]
- 13.Dellas C, Loskutoff DJ. Historical analysis of PAI-1 from its discovery to its potential role in cell motility and disease. Thromb Haemost. 2005;93:631–40. doi: 10.1160/TH05-01-0033. [DOI] [PubMed] [Google Scholar]
- 14.Häckel C, Czerniak B, Ayala AG, Radig K, Roessner A. Expression of plasminogen activators and plasminogen activator inhibitor 1 in dedifferentiated chondrosarcoma. Cancer. 1997;79:53–8. [PubMed] [Google Scholar]
- 15.Rozeman LB, de Bruijn IH, Bacchini P, Staals EL, Bertoni F, Bovée JV, et al. Dedifferentiated peripheral chondrosarcomas: Regulation of EXT-downstream molecules and differentiation-related genes. Mod Pathol. 2009;22:1489–98. doi: 10.1038/modpathol.2009.120. [DOI] [PubMed] [Google Scholar]
- 16.Birkedal-Hansen H, Moore WG, Bodden MK, Windsor LJ, Birkedal-Hansen B, DeCarlo A, et al. Matrix metalloproteinases: A review. Crit Rev Oral Biol Med. 1993;4:197–250. doi: 10.1177/10454411930040020401. [DOI] [PubMed] [Google Scholar]
- 17.Sakamoto A, Oda Y, Iwamoto Y, Tsuneyoshi M. Expression of membrane type 1 matrix metalloproteinase, matrix metalloproteinase 2 and tissue inhibitor of metalloproteinase 2 in human cartilaginous tumors with special emphasis on mesenchymal and dedifferentiated chondrosarcoma. J Cancer Res Clin Oncol. 1999;125:541–8. doi: 10.1007/s004320050314. [DOI] [PubMed] [Google Scholar]
- 18.van den Bent MJ, Dubbink HJ, Marie Y, Brandes AA, Taphoorn MJ, Wesseling P, et al. IDH1 and IDH2 mutations are prognostic but not predictive for outcome in anaplastic oligodendroglial tumors: A report of the European Organization for Research and Treatment of Cancer Brain Tumor Group. Clin Cancer Res. 2010;16:1597–604. doi: 10.1158/1078-0432.CCR-09-2902. [DOI] [PubMed] [Google Scholar]
- 19.Amary MF, Bacsi K, Maggiani F, Damato S, Halai D, Berisha F, et al. IDH1 and IDH2 mutations are frequent events in central chondrosarcoma and central and periosteal chondromas but not in other mesenchymal tumours. J Pathol. 2011;224:334–43. doi: 10.1002/path.2913. [DOI] [PubMed] [Google Scholar]
- 20.Broadhead ML, Clark JC, Myers DE, Dass CR, Choong PF. The molecular pathogenesis of osteosarcoma: A review. Sarcoma 2011. 2011 doi: 10.1155/2011/959248. 959248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Morrison C, Radmacher M, Mohammed N, Suster D, Auer H, Jones S, et al. MYC amplification and polysomy 8 in chondrosarcoma: Array comparative genomic hybridization, fluorescent in situ hybridization and association with outcome. J Clin Oncol. 2005;23:9369–76. doi: 10.1200/JCO.2005.03.7127. [DOI] [PubMed] [Google Scholar]
- 22.Sakamoto A, Oda Y, Adachi T, Oshiro Y, Tamiya S, Tanaka K, et al. H-ras oncogene mutation in dedifferentiated chondrosarcoma: Polymerase chain reaction-restriction fragment length polymorphism analysis. Mod Pathol. 2001;14:343–9. doi: 10.1038/modpathol.3880313. [DOI] [PubMed] [Google Scholar]
- 23.Bridge JA, DeBoer J, Travis J, Johansson SL, Elmberger G, Noel SM, et al. Simultaneous interphase cytogenetic analysis and fluorescence immunophenotyping of dedifferentiated chondrosarcoma. Implications for histopathogenesis. Am J Pathol. 1994;144:215–20. [PMC free article] [PubMed] [Google Scholar]
- 24.Bovée JV, Cleton-Jansen AM, Rosenberg C, Taminiau AH, Cornelisse CJ, Hogendoorn PC. Molecular genetic characterization of both components of a dedifferentiated chondrosarcoma, with implications for its histogenesis. J Pathol. 1999;189:454–62. doi: 10.1002/(SICI)1096-9896(199912)189:4<454::AID-PATH467>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 25.Röpke M, Boltze C, Neumann HW, Roessner A, Schneider-Stock R. Genetic and epigenetic alterations in tumor progression in a dedifferentiated chondrosarcoma. Pathol Res Pract. 2003;199:437–44. doi: 10.1078/0344-0338-00443. [DOI] [PubMed] [Google Scholar]
- 26.Sanerkin NG, Woods CG. Fibrosarcomata and malignant fibrous histiocytomata arising in relation to enchondromata. J Bone Joint Surg Br. 1979;61-B:366–72. doi: 10.1302/0301-620X.61B3.225333. [DOI] [PubMed] [Google Scholar]
- 27.Swarts SJ, Neff JR, Johansson SL, Bridge JA. Cytogenetic analysis of dedifferentiated chondrosarcoma. Cancer Genet Cytogenet. 1996;89:49–51. doi: 10.1016/0165-4608(96)00022-2. [DOI] [PubMed] [Google Scholar]
- 28.Sawyer JR, Swanson CM, Lukacs JL, Nicholas RW, North PE, Thomas JR. Evidence of an association between 6q13-21 chromosome aberrations and locally aggressive behavior in patients with cartilage tumors. Cancer. 1998;82:474–83. [PubMed] [Google Scholar]
- 29.Hallor KH, Staaf J, Bovée JV, Hogendoorn PC, Cleton-Jansen AM, Knuutila S, et al. Genomic profiling of chondrosarcoma: Chromosomal patterns in central and peripheral tumors. Clin Cancer Res. 2009;15:2685–94. doi: 10.1158/1078-0432.CCR-08-2330. [DOI] [PubMed] [Google Scholar]
- 30.O’Malley DP, Opheim KE, Barry TS, Chapman DB, Emond MJ, Conrad EU, et al. Chromosomal changes in a dedifferentiated chondrosarcoma: A case report and review of the literature. Cancer Genet Cytogenet. 2001;124:105–11. doi: 10.1016/s0165-4608(00)00335-6. [DOI] [PubMed] [Google Scholar]
- 31.Hameed M, Ulger C, Yasar D, Limaye N, Kurvathi R, Streck D, et al. Genome profiling of chondrosarcoma using oligonucleotide array-based comparative genomic hybridization. Cancer Genet Cytogenet. 2009;192:56–9. doi: 10.1016/j.cancergencyto.2009.03.009. [DOI] [PubMed] [Google Scholar]
- 32.Asp J, Sangiorgi L, Inerot SE, Lindahl A, Molendini L, Benassi MS, et al. Changes of the p16 gene but not the p53 gene in human chondrosarcoma tissues. Int J Cancer. 2000;85:782–6. doi: 10.1002/(sici)1097-0215(20000315)85:6<782::aid-ijc7>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- 33.Terek RM, Healey JH, Garin-Chesa P, Mak S, Huvos A, Albino AP. p53 mutations in chondrosarcoma. Diagn Mol Pathol. 1998;7:51–6. doi: 10.1097/00019606-199802000-00009. [DOI] [PubMed] [Google Scholar]
- 34.Dobashi Y, Sugimura H, Sato A, Hirabayashi T, Kanda H, Kitagawa T, et al. Possible association of p53 overexpression and mutation with high-grade chondrosarcoma. Diagn Mol Pathol. 1993;2:257–63. [PubMed] [Google Scholar]
- 35.Röpke M, Boltze C, Meyer B, Neumann HW, Roessner A, Schneider-Stock R. Rb-loss is associated with high malignancy in chondrosarcoma. Oncol Rep. 2006;15:89–95. doi: 10.3892/or.15.1.89. [DOI] [PubMed] [Google Scholar]


