Abstract
It has been proposed that differential kinetics of CD4/CD8 co-receptors regulate fate choice of selected thymocytes. Sustained signals by interaction between MHC class II and TCR/CD4 is required for commitment to the CD4 helper lineage. While prematurely terminated MHC-TCR/CD4 interaction in transgenic mouse models results in lineage redirection, it is unclear if CD4 expression is actively maintained by endogenous cis-elements to facilitate prolonged signaling under physiological conditions. Here we show that sustained CD4 expression in post-selection thymocytes requires an intronic sequence containing an uncharacterized DNase I hypersensitivity site (DHS) located 3’ to the silencer. Despite normal CD4 expression before selection, thymocytes lacking a 1.5 kb sequence in intron 1 including the 0.4 kb silencer and the DHS, but not the 0.4 kb silencer alone, failed to maintain CD4 expression upon positive selection and are redirected to the CD8 lineage following MHC class II-restricted selection. Furthermore, CpG dinucleotides adjacent to the DHS are hypermethylated in CD8+ T cells. These results indicate that the 1.5 kb cis-element is required in post-selection thymocytes for helper lineage commitment, presumably mediating the maintenance of CD4 expression, and suggest that inactivation of the cis-element by DNA methylation may contribute to epigenetic Cd4 silencing.
Introduction
CD4 and CD8 co-receptor expression defines subsets of mature T cells with distinct functions. Both CD4+ Th and CD8+ CTLare derived from a common precursor pool of CD4+ CD8+ (double positive, DP) thymocytes. As a consequence of random TCR-alpha chain rearrangements and interaction with MHC-peptide complexes presented on thymic epithelial cells, thymocytes expressing MHC class II (MHCII)- or class I (MHCI)-restricted TCR with appropriate avidity differentiate into the Th or CTL lineage, respectively (1–3). However, it is not understood how positive selection signals regulate fates of selected thymocytes.
It has been proposed that distinct expression kinetics of CD4 and CD8 co-receptors during positive selection determine the Th versus CTL lineage choice (2). Among several models that have been proposed and experimentally tested, the kinetic signaling model by Singer and colleague (4) has been supported by several lines of experimental evidence using mouse genetics. Cd8a/Cd8b1 genes, which encode two subunits of the CD8 co-receptor, are regulated by multiple stage-specific enhancers (5). During positive selection, CD8 co-receptor is transiently down-regulated due to reduced transcription of Cd8a/Cd8b1 regardless of TCR-MHC specificity, while CD4 expression remains constant or is promptly restored after transient down-regulation at the DPdull stage. These distinct expression kinetics of the two co-receptors cause differential duration of positive selection signals. MHCII-restricted thymocytes receive prolonged signal due to continued interaction between TCR/CD4 and MHCII, whereas MHCI-restricted thymocytes lose signals as surface CD8 is down-regulated. This model was further tested in transgenic mice or knock-in models, in which Cd4 was only transiently expressed under the control of Cd8a/Cd8b1 regulatory elements (6, 7). These experiments demonstrated that sustained CD4 expression is required for induction of ThPOK and commitment to the Th lineage following MHCII-restricted selection. Transient signaling through CD4/TCR interacting with MHCII-peptide complexes results in redirection to the CTL lineage. However, these experiments were performed exclusively in genetically modified mouse models to drive Cd4 expression under the control of heterologous regulatory elements and it therefore remains unknown whether Cd4 expression is actively maintained through activation of endogenous regulatory elements to permit prolonged signals.
Cis-regulatory elements that are required or sufficient for Cd4 expression have been identified and validated by genetic experiments (8, 9). Cd4 expression in developing thymocytes and T cells are predominantly regulated by a 0.35 kb proximal enhancer (Ep4) located 12 kb 5’ to the promoter and a 0.4 kb silencer in the first intron. Deletion of the silencer results in ectopic expression of CD4 in double-negative (DN) thymocytes and MHCI-restricted CD8+ T cells (10, 11). Transgenic constructs under the control of the Cd4 promoter (P4) and Ep4 are expressed in all thymocyte subsets and mature T cells, indicating that Ep4 is constitutively active during T cell development and that the silencer activity defines stage- and lineage-specific Cd4 expression (10, 11). Targeted deletion of Ep4 abolishes CD4 expression in DP thymocytes (8). Despite lack of CD4 expression in DP thymocytes, MHCII-restricted positive selection does occur in Ep4-deficient mice (8). These MHCII-restricted thymocytes turn on CD4, albeit at a lower level and frequency, and become ThPOK-expressing mature T cells, suggesting that Cd4 is driven by an unidentified enhancer, which becomes active following positive selection. This possibility is also supported by a previous study showing that an Ep4-P4 transgenic reporter is silenced in activated T cells without a change in endogenous CD4 expression (12).
In this study, we analyzed epigenetic marks in thymocytes and mature T cells and examined the requirement of an intronic sequence containing a peak of DNase I hypersensitivity site (DHS) located immediately 3’ to the 0.4 kb silencer and 3 kb downstream of the Cd4 promoter (DHS+3). While deletion of the 0.4 kb silencer alone showed little impact on T cell lineage commitment, mice lacking a 1.5 kb intronic sequence encompassing both the silencer and DHS+3 failed to sustain CD4 expression in positively selected thymocytes, which resulted in lineage redirection of MHCII-restricted thymocytes to the CD8+ T cell lineage. Our results suggest that the Cd4 locus utilizes distinct cis-elements between pre-selection and post-selection thymocytes and that the sustained CD4 expression in post-selection thymocytes, which requires the 1.5 kb sequence, or possibly the 1.1 kb sequence 3’ to the silencer, is essential for the lineage commitment to the helper lineage.
Material and Methods
Mice
Cd4tm2Litt (Cd4(0.4k)−, Δ0.4kb) and Cd4tm1Yzo (Cd4(1.5k)F) alleles were described previously (11, 13). These alleles were backcrossed 12 generations to the C57BL6 (B6) background before intercrossing or crossing to Cre transgenic lines. Cd4tm1Yzo was bred to Cd4-cre (Taconic) (14) to delete the 1.5 kb genomic fragment encompassing the silencer and DHS+3 (Δ1.5kb), or Vav1-cre (Jackson Laboratory) (15) for data shown in Supplementary Fig. 3. Mice homozygous for the Cd4tm2Litt or Cd4tm1Yzo were used all experiments. C57BL6 mice (National Cancer Institutes, Frederic, MD) were used as wild-type control. Bone marrow chimeras were generated by intravenous transfer of bone marrow cells (5 × 106 cells) into lethally irradiated B2m−/− mice (16) purchased from Jackson Laboratory. The recipient mice were treated with Trimethoprim and Sulfamethoxazole for four weeks and analyzed twelve weeks after transplantation. All mice were housed in a specific pathogen free animal facility at Washington University School of Medicine and experiments were performed according to a protocol approved by the Animal Studies Committee at Washington University in St. Louis.
Epigenetic Analysis
Data from deep sequencing analyses were previously published (17, 18), or have been deposited in NCBI Gene Expression Omnibus (GEO) by the ENCODE Project. Data from the ENCODE project were used in accordance with ENCODE Consortia Data Release, Data Use, and Publication Policies. FASTQ files or BigWig files were downloaded from GEO. FASTQ files were mapped to a mouse genome assembly mm9 using bowtie2 (19), and converted to TDF files. BigWig and TDF files were visualized in IGV browser (ver 2.3, Broad Institute).
Chromatin Immunoprecipitation (ChIP)
Native chromatin immunoprecipitation was performed by following a previously published protocol (20). In brief, native chromatin prepared from 5 − 10 × 106 cells was digested with micrococcal nuclease (Worthington) and subjected to immunoprecipitation using antibodies against H3K27ac (ab4729, Abcam) or H3K4me2 (ab7760, Abcam) coupled to Dynabeads protein G (Life Technologies). Precipitated DNA was purified and enrichment of the Cd4 regulatory regions was quantitated by real-time PCR with the following primers using a Roche LC480II and Luminaris Color SYBR green PCR reagent (Thermofisher):
Cd4Sil-F: TACGAAGCTAGGCAACAGAGGAAG,
Cd4Sil-R: TGTGGTCCCGAATGCGTTT,
Tcrb-enhancer (Eβ)-F: GCTCCATCTCCAGGAGTCAC,
Eβ-R: AAGTGTGGTTCCCAAAATGC,
DHS+3-F: AAGGAGGAAGAGCCCAATAGAG,
DHS+3-R: TGTGTCAGTCCCTGGTGAGTAG.
T cell culture and retroviral transduction
T cells were purified by sorting (FACS AriaII, BD) or magnetic enrichment using FlowComp CD4 and CD8 Dynabeads (Life Technologies) and cultured in the presence of 0.1 µg/ml of anti-CD3 (145-2C11, Biolegend) and 1 µg/ml of anti-CD28 (37.51, BioXcell) in multi-well plastic plates pre-coated with rabbit anti-hamster IgG antiserum (MP Biomedicals). Retroviral transduction was performed as previously described (21). GFP+ cells were purified by cell sorting and deletion of the targeted sequence was assessed by real-time PCR using the primers described above. 5-AZA treatment was done as previously described (13). Dead cells were excluded by DAPI staining.
Bisulfite sequencing
Genomic DNA was prepared using a Quick-gDNA MiniPrep kit (Zymo Research) and treated with bisulfite using an EZ DNA Methylation-Gold kit (Zymo Research). Treated genomic DNA was PCR amplified, cloned by TA cloning into pCR2.1-TOPO (Life Technologies) and sequenced. Following primers were used for PCR amplification of the treated genomic DNA:
CD4DHS3_methyl_F1: ggatTTtgtgaagggtggttgttg,
CD4DHS3_methyl_R1: ggTaTagttgaTttgggTTag,
CD4DHS3_methyl_F2: gggTTatgtgagggtggTag,
CD4DHS3_methyl_R2: aagTatttaagggaagggtgtg,
CD4DHS3_methyl_F3: TTTttgaggTttTtgtggttg,
CD4DHS3_methyl_R3: gtgataagagttgaaggagTag.
Luciferase reporter assay
The Cd4 intronic fragments were PCR amplified and cloned into KpnI and XhoI sites of a pGL3-Promoter plasmid expressing the firefly luciferase (FFL) under the control of the SV40 promoter (Promega). Jurkat cells were co-transfected with each SV40-FFL plasmid and a CMV-RL plasmid (Promega) using a TransIT-Jurkat transfection reagent (Mirus Bio) according to the manufacturer’s instruction. Luciferase activities were measured 16 hours after transfection.
Statistical Analysis
Statistical differences were tested using GraphPad Prism 6.0 by two-tailed unpaired Student’s t-test for two group comparisons or one-way ANOVA with the Tukey post-hoc analysis for comparison between more than three groups. P-values smaller than 0.05 were considered significant. P-values were abbreviated as follows. *: P < 0.05, **: P < 0.01, ***: P < 0.001, ****: P < 0.0001.
Results
The 1.5 kb intronic sequence containing the silencer and DHS+3 regulates CD4 expression in post-selection thymocytes
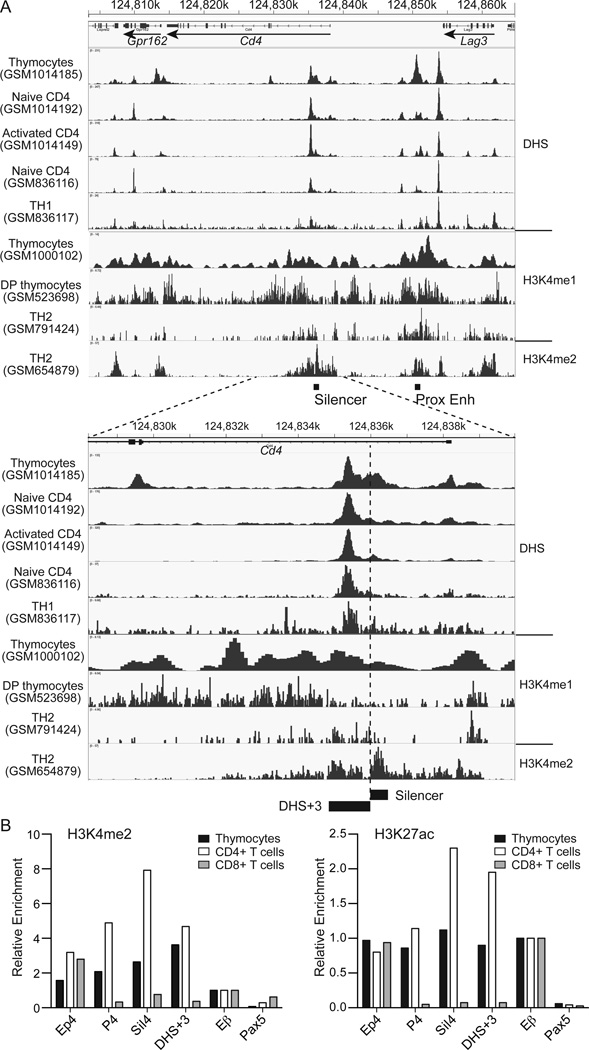
To locate a potential regulatory element, we analyzed publicly available deep sequencing data for DHS and ChIP against permissive histone modifications in DP thymocytes and mature T cells (Fig. 1A). In total thymocytes, there were multiple DHS peaks in a genomic region encompassing the Cd4 locus, including two strong peaks corresponding to Ep4 and the promoter region for Lag3. A slightly weaker peak was found in the first intron of Cd4 immediately 3’ to the 0.4 kb silencer region (designated as DHS+3). In mature T cells, while the Lag3 promoter peak remained high, the peak in intron 1 became dominant compared to the peak at Ep4, suggesting that the Cd4 locus is regulated by distinct elements between thymocytes and mature CD4+ T cells. Consistent with active chromatin states in ChIPseq data, both Cd4 silencer and DHS+3 were enriched for the H3K4me2 and H3K27ac marks in thymocytes and mature CD4+ T cells, but devoid of these marks in CD8+ T cells (Fig. 1B). Sequence analysis between mouse and human genomes using the ECR browser (http://ecrbrowser.dcode.org/) showed that DHS+3 is highly conserved between mouse and human to a similar degree to the 0.4 kb silencer and contained a cluster of predicted binding sites for the HNF family of nuclear receptors, TCF1-LEF1 complexes, STAT3, SZF1, c-REL, RUNX protein, WT1 and CACD factors (Supplementary Fig. 1). These results suggest a potential regulatory role of DHS+3 in regulation of Cd4 gene expression in developing thymocytes and T cells.
Figure 1. DNase I hypersensitivity and permissive histone modifications at the Cd4 locus in thymocytes and mature T cells.
(A) DNase I hypersensitivity sites (DHS) and permissive histone modifications from publicly available datasets were analyzed and shown in the IGV browser. Nucleotide positions on chromosome 6 of the mm9 assembly of mouse genome and locations and orientations of Cd4, Lag3 and Gpr162 genes are shown. (B) Chromatin immunoprecipitation analysis of H3K4me2 and H3K27ac modifications at Ep4, Cd4 promoter, Cd4 silencer and DHS+3. Primers for Tcrb enhancer (Eβ) and Pax5 promoter were used as positive and negative control, respectively. Values were calculated relative to input DNA and normalized against enrichment at Eβ. Data represent three experiments with similar results.
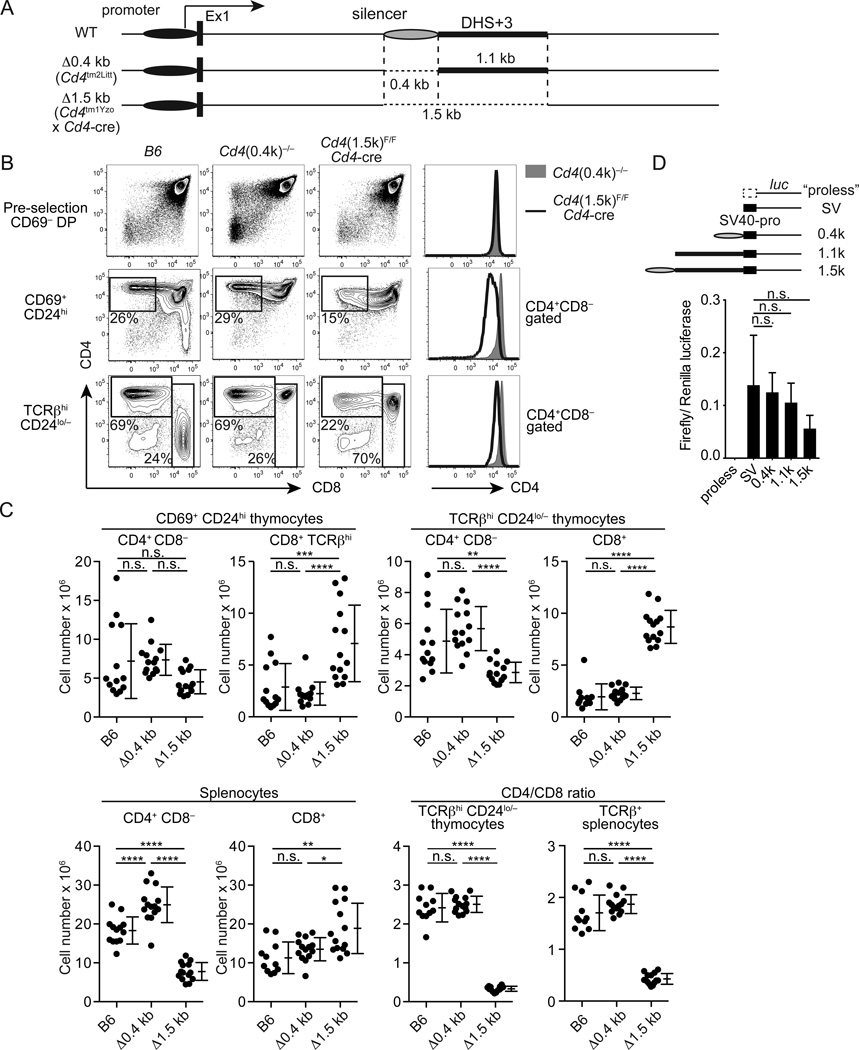
To determine whether the intronic region in the Cd4 locus containing DHS+3has the regulatory role in Cd4 gene regulation in vivo, we examined CD4 expression and T cell development in mice lacking a 1.5kb fragment (Δ1.5kb, Cd4tm1Yzo crossed to Cd4-cre) encompassing the 0.4 kb silencer and an additional 1.1 kb region containing DHS+3and mice lacking only the 0.4 kb silencer (Δ0.4kb) (Fig. 2A). We did not observe differences in total thymus or spleen cellularity between age-matched wild-type B6, Δ0.4kb and Δ1.5kb mice (Supplementary Fig. 2A). CD4 expression in pre-selection CD69− TCRβlo/− CD24hi DP thymocytes, CD69+ CD24hi CD8− post-selection intermediate and TCRβhi CD24lo/− CD8− mature thymocytes was grossly comparable between B6 and Δ0.4kb mice (Fig. 2B, Supplementary Fig. 2B). These results indicate that the 0.4kb silencer is dispensable for normal CD4 expression in developing thymocytes in vivo. In contrast, despite normal expression in pre-selection thymocytes, the CD4 level was reduced in post-selection intermediate and mature thymocytes in Δ1.5kb mice (Fig. 2B, Supplementary Fig. 2B). Besides reduced CD4 expression, the absolute number of TCRβhi CD24lo/− CD4SP cells was reduced in Δ1.5kb mice by two-fold and TCRβhi CD24lo/− CD8+ mature thymocytes increased by three-fold compared to B6 or Δ0.4kb mice (Fig. 2C). A similar decrease in CD4+ CD8− T cells and increase in CD8+ T cells were observed in the spleen of Δ1.5kb mice (Fig. 2C). Total numbers of TCRβhi CD24lo/− thymocytes were also increased by 1.5-fold in Δ1.5kb mice compared to B6 or Δ0.4kb mice (Supplementary Fig. 2A). We observed nearly identical phenotypes when the 1.5 kb sequence was deleted with the Vav1-cre deleter (Supplementary Fig. 3), indicating that these phenotypes were not caused by the presence of multiple copies of Cd4 regulatory elements in the Cd4-cre transgenic mice. Unlike CD4+ T cells in ThPOK/Zbtb7b hypomorphic mutant (ThPOKFN) mice (22), the remaining CD4+ T cells in Δ1.5kb mice expressed ThPOK protein at a comparable level to B6 CD4+ T cells and did not express Runx3 (Supplementary Fig. 2C). They also expressed IFNγ and IL-2at comparable frequencies to control Δ0.4kb CD4+ CD8− or CD8+ T cells under non-polarizing conditions (Supplementary Fig. 2D). These results indicate that the 1.5 kb intronic sequence containing is required for sustained CD4 expression in post-selection thymocytes and mature T cells, and that diminished CD4 expression after positive selection leads to reduced Th lineage differentiation.
Figure 2. A 1.5 kb intronic sequence containing the silencer and DHS+3 is essential for sustained CD4 expression in post-selection thymocytes and mature CD4+ T cells.
(A) A genomic configuration of Δ0.4kb and Δ1.5kb mutant alleles of Cd4. (B) CD4 and CD8 expression in pre-selection thymocytes (CD69−), post-selection intermediate (CD69+ CD24hi) and mature (TCRβhi CD24lo/−) thymocytes from control B6, Δ0.4kb and Δ1.5kb mice. Histogram overlays for CD4 expression at each developmental stage are shown on right. (C) Statistical analysis of absolute numbers of CD4+ CD8− and CD8+ cells and CD4/CD8 ratios in thymi and spleens of control B6, Δ0.4kb and Δ1.5kb mice. N=9–12. (D) Luciferase reporter assays to test enhancer activities of the 0.4 kb silencer and DHS+3 containing sequences. SV40 promoter-driven firefly luciferase expression constructs with the 0.4 kb, 1.1 kb or 1.5 kb intronic sequence were transfected into Jurkat cells. A construct lacking the SV40 promoter (“proless”) was used as negative control. Firefly luciferase activities were normalized against CMV promoter-driven Renilla luciferase activities used as internal control. Experiments were performed in triplicate and data from two independent experiments were combined. Data are shown with means and SD. Statistical difference was assessed by one-way ANOVA and the Tukey test.
To determine whether the intronic sequence deleted in Δ1.5kb mice is sufficient to mediate an enhancer activity, we inserted the 1.5 kb or 1.1 kb genomic sequence to an SV40 promoter-driven luciferase expression plasmid and transfected into Jurkat cells. Neither the 1.5 kb nor 1.1 kb sequence showed an enhancement in luciferase expression (Fig. 2D), suggesting that the 1.5 kb sequence by itself is insufficient to provide an enhancer activity and that other unknown sequences in the endogenous Cd4 locus cooperate with the deleted region to drive gene expression.
Deletion of the 1.5 kb sequence containing DHS+3 results in lineage redirection
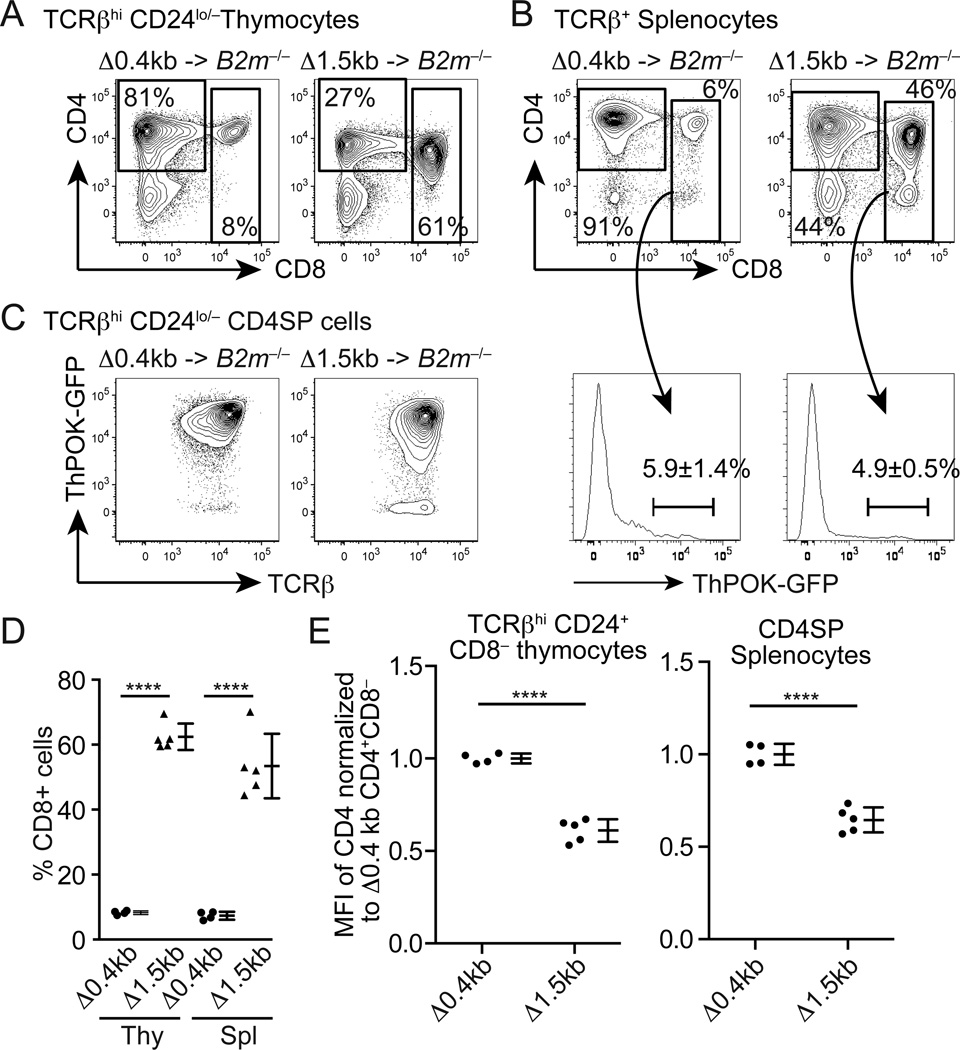
Reduced numbers of CD4SP thymocytes and CD4+ CD8− T cells and increased numbers of CD8+ mature thymocytes and splenic CD8+ T cells in Δ1.5kb mice suggest lineage redirection of MHCII-restricted thymocytes as observed in ThPOK-deficient mice (22–25) or transgenic mice, in which Cd4 is expressed under the control of Cd8a/Cd8b1 regulatory elements (6, 7). To determine whether the 1.5 kb genomic region containing the silencer and DHS+3 is required for commitment to the CD4+ Th lineage, we generated bone marrow chimeras using B2m−/− mice as recipients. In these chimeras, thymocytes are selected predominantly by MHCII. In chimeras receiving Δ0.4kb bone marrow cells (in ThPOKGFP/+ background), the majority of mature thymocytes and splenic T cells were CD4+ CD8− (Fig. 3A, B). These CD4SP cells uniformly expressed the ThPOK-GFP reporter (Fig. 3C). In contrast, we observed diminished CD4SP and significantly increased CD8+ mature thymocytes and T cells in the chimera receiving Δ1.5kb bone marrow (Fig. 3A, B, D), indicating that MHCII-restricted thymocytes were redirected to the CD8+ CTL lineage in the absence of the 1.5 kb sequence. While we ThPOK expression was slightly reduced or largely intact (Fig. 3C), CD4 expression was significantly reduced in CD4SP cells lacking the 1.5 kb sequence compared to those lacking only the 0.4kb silencer (Fig. 3E). While MHCII-restricted CD8+ T cells in ThPOK-deficient mice still turn on the ThPOK-GFP reporter (22), redirected CD8+ T cells in Δ1.5kb mice were mostly GFPlo/− (Fig. 3B), suggesting that Δ1.5kb cells failed to upregulate ThPOK following MHCII-restricted selection and were subsequently redirected to the CD8 lineage. These results indicate that the 1.5 kb sequence containing DHS+3 is required for CD4+ Th lineage commitment following MHCII-restricted selection, presumably by sustaining CD4 expression.
Figure 3. Deletion of the 1.5 kb intronic sequence results in lineage redirection of MHCII-restricted thymocytes.
(A and B) CD4 and CD8 expression in TCRβhi CD24lo/− mature thymocytes (A) and TCRβ+ splenic T cells (B) from B2m−/− mice reconstituted with bone marrow cells from Δ0.4kb × ThPOKGFP/+ (N=4) or Δ1.5kb × ThPOKGFP/+(N=5) mice. Frequencies of ThPOK-GFP+ CD8+ T cells are shown with means and SD. (C) ThPOK-GFP reporter expression in CD4+ CD8− thymocytes in the B2m−/− chimeras. (D) Statistical analysis of percentages of CD8+ cells in TCRβhi CD24lo/− thymocytes (Thy) and TCRβ+ splenic T cells (Spl) shown as mean and SD. (E) Comparison of CD4 expression between Δ0.4kb and Δ1.5kb CD4+ CD8− cells as defined in (A) and (B). Mean florescence intensity (MFI) of CD4 staining of each sample was normalized against average MFI of Δ0.4kb cells and shown as means and SD. Statistical significance was tested by unpaired two–tailed Student’s t-test.
The 1.5 kb sequence is required for establishment of stable CD4 expression but not for maintenance in mature T cells
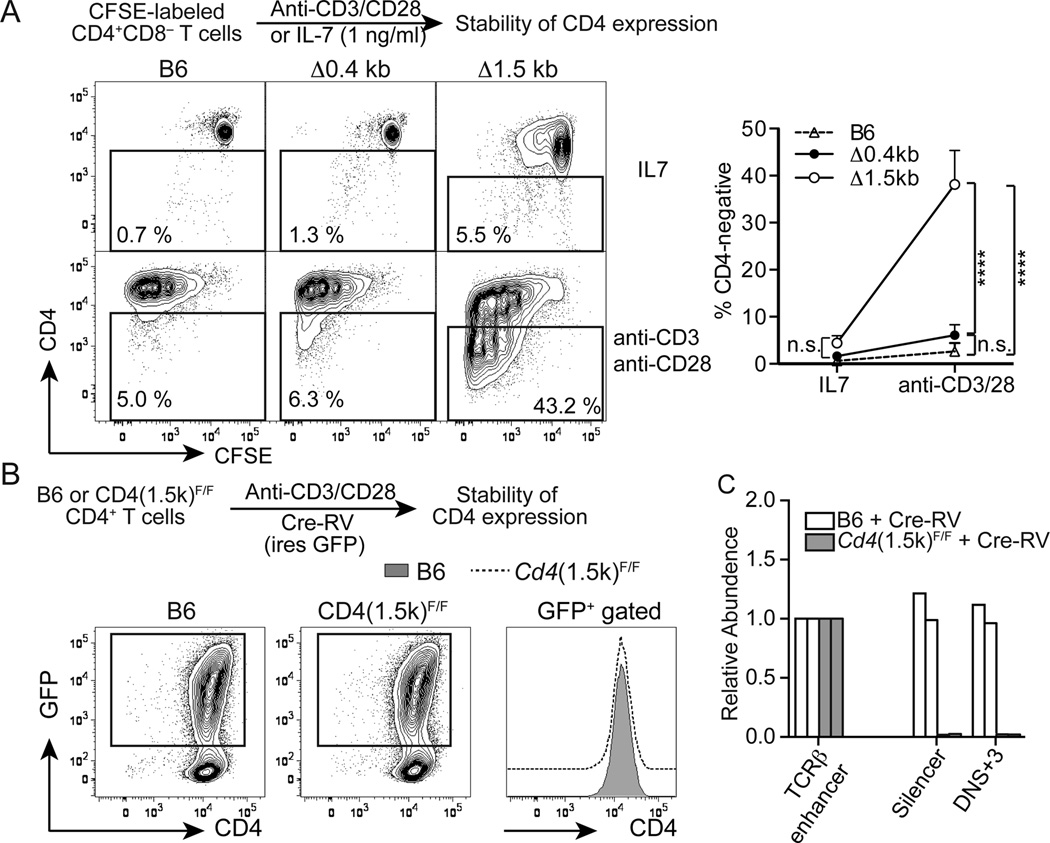
Mice lacking Ep4 develop CD4lo CD8− T cells but the CD4 expression is unstable (8). Since CD4 expression in Δ1.5kb CD4+ T cells was similarly low, we tested whether CD4 expression could be maintained during mitosis. CFSE-labeled CD4+ T cells from B6 or Δ0.4kb mice maintained their CD4 expression during rapid proliferation in the presence of anti-CD3 and anti-CD28 stimulation as well as under a culture condition without rapid cell divisions (1 ng/ml IL-7) (Fig. 4A). In contrast, a large proportion of Δ1.5kb CD4+ T cells failed to maintain CD4 following cell divisions (Fig. 4A). This instability of CD4 expression could result from a constitutive requirement for the 1.5 kb sequence in maintenance of CD4 expression in activated T cells or from a defective establishment of stable CD4 expression during development in the absence of the 1.5 kb sequence. To test these possibilities, we used a retrovirus expressing Cre recombinase to delete the 1.5 kb sequence in mature CD4+ T cells and tracked CD4 expression in dividing cells. CD4 expression was comparable between control B6 CD4+ T cells and Cd4(1.5k)F/F CD4+ T cells following Cre-mediated deletion of the 1.5 kb sequence in activated CD4+ T cells (Fig.4B and 4C), indicating that the 1.5 kb sequence is dispensable for the maintenance of CD4 expression in normally developed CD4+ T cells. These results also suggest that stable CD4 expression is established in Ep4 and the 1.5 kb sequence-dependent manners during thymocyte differentiation.
Figure 4. The 1.5 kb intronic sequence is essential for establishment of stable CD4 expression during development but dispensable for maintenance of CD4 expression in activated T cells.
(A) CD4 expression in CFSE-labeled CD4+ CD8− T cells from control B6, Δ0.4kb and Δ1.5kb under non/slowly dividing (IL-7) and rapidly dividing (anti-CD3 and anti-CD28) conditions. Percentages of CD4− cells under either condition after 4 days of culture are shown with means and SD. Statistical differences were assessed by two-way ANOVA and the Tukey test. N=4.(B) CD4 and GFP expression in Cre-expressing retrovirus (Cre-RV)-transduced B6 and Cd4(1.5k)F/F CD4+ CD8− T cells on day 4 after transduction and a histogram overlay of CD4 expression in GFP+ gated cells are shown. N=3. (C) Efficiency of Cre-mediated deletion of DHS+3 was assessed by real-time PCR using the same primers as those used in Fig. 1 and shown as relative abundance to the Eβ sequence. N=4.
Hypermethylation of CpG dinucleotides adjacent to DHS+3in CD8+ T cells
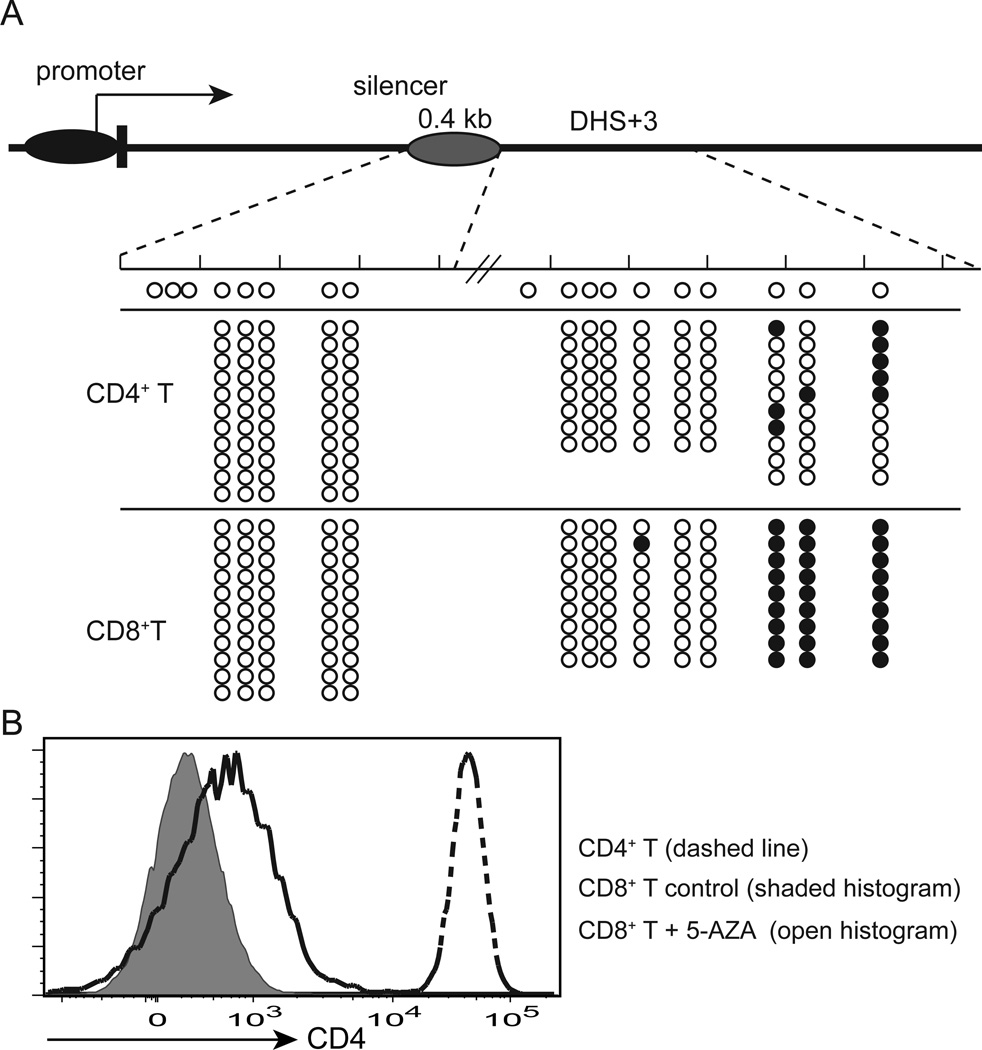
During normal T cell development, CD8+ T cells epigenetically shutoff Cd4 in a silencer-dependent manner (11, 13). Although epigenetic silencing is linked to DNA methylation (26), a previous study showed that there is no difference in CpG methylation in the Cd4 promoter region between CD4+ and CD8+ T cells (13). We hypothesized that the silencer or DHS+3 could be a target of CD8+ T cell-specific DNA methylation, which may contribute to epigenetic Cd4 silencing. To test this hypothesis, we mapped CpG methylation in the 1.5 kb region containing the 0.4 kb silencer and DHS+3 by bisulfite sequencing. While the silencer was hypomethylated both in CD4+ and CD8+ T cells, a few CpG dinucleotides located immediately 3’ to the DHS+3 peak were hypermethylated in CD8+ T cells (Fig. 5A). In contrast, CpG methylation at the same dinucletides was substantially reduced in CD4+ T cells. To further test whether maintenance of Cd4 silencing requires DNA methylation, we treated CD8+ T cells from B6 mice with the DNA methyltransferase inhibitor, 5-AZA, and examined CD4 expression. We observed CD4 upregulation in CD8+ T cells treated with 5-AZA, even though the level of CD4 expression was substantially lower than control CD4+ T cells (Fig. 5B). These results suggest that DNA methylation may be required for maintenance of Cd4 silencing and that DHS+3 could be a target of CD8+ T cell-specific DNA methylation.
Figure 5. CpG dinucleotides adjacent to DHS+3 are hypermethylated in CD8+ T cells.
(A) Bisulfite sequencing analysis of CpG methylation in the Cd4 silencer and DHS+3 in CD4+ and CD8+ T cells from B6 mice. Data are pooled from three experiments. Open and closed circles indicate unmethylated (converted) and methylated CpG dinucleotides, respectively. (B) Epigenetic Cd4 silencing in CD8+ T cells was partially reversed by inhibition of DNA methyltransferase activity. CD4 expression in 5-AZA treated CD8+ T cells and control5-AZA-treated CD4+ and untreated CD8+ T cells is shown as a histogram overlay. N=4.
Discussion
In this study, we characterized the requirement for the silencer and its neighboring DHS in the Cd4 gene regulation. Our data showed that a 1.5 kb intronic region containing DHS+3 and the previously established 0.4 kb silencer is required for sustained CD4 expression in post-selection thymocytes and commitment of MHCII-restricted thymocytes to the helper lineage. Despite normal expression in pre-selection DP thymocytes, CD4 expression was diminished in CD69+ CD24hi post-selection thymocytes in the absence of the 1.5 kb region and a large proportion of MHCII-restricted thymocytes were redirected to the CD8lineage. Because we did not observe CD4 down-regulation in mice lacking the 0.4 kb silencer alone, sustained CD4 expression in post-selection thymocytes is possibly mediated by the 1.1 kb sequence located immediate 3’ to the 0.4 kb silencer. The 1.1 kb genomic region contains a DHS peak mapped to an evolutionarily conserved sequence and is marked by H3K27ac and H3K4me2, histone modifications known to be enriched in active enhancer regions. Although we could not detect an enhancer activity mediated by the 1.5 kb or 1.1 kb sequence in a luciferase reporter assay in vitro, our data suggest that the 1.5 kb intronic sequence is required, though not sufficient, for enhancer activity essential for sustained CD4 expression. This region itself may not function as a classical enhancer, which serves as a dock for transcriptional activator or co-activator binding. It may rather be necessary to establish a permissive chromatin conformation via functioning as a flexible hinge to place two or more distally located elements in a close proximity. Although we speculate that reduced CD4 expression in Δ1.5kb mice causes lineage redirection, we do not rule out the possibility that it acts in trans on other loci encoding molecules involved in helper versus cytotoxic lineage decisions. A previous study suggests that the 0.4 kb silencer and Runx proteins that bind to the silencer mediate association of Cd4 and Cd8a/Cd8b1 loci in thymocytes (27). While a deletion of only the 0.4 kb silencer did not alter CD8 expression or cause marked lineage redirection, intra- or inter-chromosomal associations mediated by the 1.5 kb sequence, which contains the silencer element, may play a role in the helper lineage commitment.
In the kinetic signaling model, sustained positive selection signals instruct cells to differentiate into the CD4+ Th lineage, while an artificial interruption of MHCII-restricted positive selection signals by transgenic expression of CD4 results in lineage redirection (2, 6, 7). In cells lacking the 1.5 kb sequence, MHCII-restricted positive selection signals may prematurely be interrupted due to CD4 down-regulation after selection is initiated, thus leading to insufficient ThPOK induction and the development of MHCII-restricted CD8+ cells. Our results suggest that the Cd4 locus may undergo enhancer switching in part similar to Cd8a/Cd8b1 loci, and that CD4 expression may actively be maintained by a post-selection stage-specific enhancer that requires the 1.5 kb sequence. Although transcription factor requirements for CD4 versus CD8 lineage decision have been explained in large part by Gata3, ThPOK, and Runx3/CBFβ (3, 28–30), there may be additional factors which also play key roles in the cell fate decision process presumably through regulation of sustained Cd4 expression. The putative post-selection enhancer appears likely to account for Ep4-independent CD4 expression following MHCII-restricted selection (8). Because CD4 expression was not completely inactivated in Δ1.5kb cells, the post-selection enhancer activity may not be completely lost by the deletion, or other enhancers, including Ep4, may be at least partially redundant in post-selection thymocytes. Interestingly, while CD4+ T cells from Cd4(Ep4)−/− or Δ1.5kb mice failed to establish stable CD4 expression, the requirements for the 1.5 kb sequence and Ep4 are transient, and they are not required for maintenance of CD4 expression. These findings suggest that both Ep4 and the silencer/DHS+3-containing cis-element have stage-specific functions during thymocytes development.
In addition to lineage redirection of MHCII-selected thymocytes to the CTL lineage, we also observed an approximately1.5-fold increase in the number of total mature thymocytes with a relatively mild reduction of CD4SP thymocytes in Δ1.5kb mice compared to WT B6 or Δ0.4kb mice. A recent study using Nur-77 GFP reporter and Bim−/− mice demonstrated that more than a half of MHCII-selected thymocytes that differentiate to CD4SP cells are negatively selected during normal thymocyte development (31). In Bim−/− mice, which are defective in negative selection, both CD4SP and CD8SP thymocytes substantially increase with an enrichment of Nur77hi cells, which are normally eliminated by negative selection. Therefore, the increased number of mature thymocytes may reflect reduced negative selection resulting from CD4 down-regulation in post-selection thymocytes in Δ1.5kb mice, but not in Δ0.4kb or B6 mice.
Previously, Leung et al. generated a mouse strain, in which a larger proportion of Cd4 intron 1 was deleted (32). Because the 0.4 kb silencer was also deleted in this strain, CD8+ T cells expressed CD4 similarly to the two lines of mice we used in this study. In addition to loss of Cd4 silencing in CD8+ T cells, the study reported phenotypes similar to those we observed in Δ1.5 kb mice, including diminished CD4 expression, reduced CD4+ T cells and increased CD8+ T cells due to redirection of MHCII-restricted T cells to the CD8 lineage. The paper was concluded in favor of the stochastic model of CD4 versus CD8 lineage decisions based on their interpretation that MHCII-restricted thymocytes that would have chosen to express CD8 and terminate CD4 were “rescued”, resulting in the development of MHCII-restricted CD8+ T cells. However, we argue against their interpretation because deletion of the 0.4 kb silencer alone, which allows both MHCI-restricted cells and MHCII-restricted cells to express high levels of CD4, resulted in the development of substantially reduced CD8+ T cells compared to mice with the 1.5 kb deletion. Based on these data, we concluded that the development of MHCII-restricted CD8+ T cells in our Δ1.5kb mice resulted primarily from reduced CD4 expression after positive selection, which compromised “prolonged” positive selection signals required for CD4 lineage commitment, rather than a rescue of “CD8-fated” MHCII-restricted thymocytes from cell death.
CpG dinucleotides adjacent to DHS+3were hypermethylated in CD8+ T cells. This result could be interpreted in two different ways. In one possibility, inactivation of the putative post-selection enhancer in CD8+ T cells may be important for epigenetic silencing. This is consistent with our observation that CD8+ T cells treated with 5-AZA de-repressed CD4, although we are unable to rule out the possibility that 5-AZA treatment induced global gene expression changes that might indirectly cause CD4 de-repression. TheCD4 de-repression occurred in the presence of the silencer, which may restrict CD4 de-repression to only a modest extent. Synergistic effects by the silencer-dependent repression and inactivation of DHS+3 by DNA methylation may completely shut off the locus activity in CD8+ T cells. An alternative but not necessarily mutual exclusive possibility is that DHS+3 is demethylated specifically in CD4+ T cells, which allows full activation of the Cd4 locus activity. Future analysis examining de novo DNA methyltransferase or Tet family demethylase protein recruitment to the Cd4 locus may address this possibility.
In summary, our study provides genetic evidence that the 1.5 kb intronic sequence containing the silencer and DHS+3 serves as an essential cis-element for sustained CD4 expression in post-selection thymocytes and lineage commitment to the CD4 Th lineage.
Supplementary Material
Acknowledgements
We thank Dan R. Littman (NYU School of Medicine, NY), Yong-Rui Zou (Feinstein Institute, NY), Ichiro Taniuchi (RIKEN, Japan), and William E Paul (NIAID/NIH) for materials, Sunnie Hsiung for technical assistance, and Eugene Oltz and Chyi-Song Hsieh for critical reading of the manuscript.
This study was supported by grants from the National Institutes of Health (R01AI097244 to T.E.) and the Edward Mallinckrodt Jr. Foundation (T.E.) and by the Lucille P. Markey Pathway Program (C.C.).
Abbreviations used in the manuscript
- ChIP
chromatin immunoprecipitation
- DHS
DNase I hypersensitivity
- DP
double positive
- H3K4me2
di-methylated histone H3 lysine 4
- H3K27ac
acetylated histone H3 lysine 27
Footnotes
Disclosure
The authors have no financial conflicts of interest.
References
- 1.Kappes DJ, He X, He X. CD4-CD8 lineage commitment: an inside view. Nature immunology. 2005;6:761–766. doi: 10.1038/ni1230. [DOI] [PubMed] [Google Scholar]
- 2.Singer A, Adoro S, Park JH. Lineage fate and intense debate: myths, models and mechanisms of CD4- versus CD8-lineage choice. Nature reviews. Immunology. 2008;8:788–801. doi: 10.1038/nri2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Collins A, Littman DR, Taniuchi I. RUNX proteins in transcription factor networks that regulate T-cell lineage choice. Nature reviews. Immunology. 2009;9:106–115. doi: 10.1038/nri2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brugnera E, Bhandoola A, Cibotti R, Yu Q, Guinter TI, Yamashita Y, Sharrow SO, Singer A. Coreceptor reversal in the thymus: signaled CD4+8+ thymocytes initially terminate CD8 transcription even when differentiating into CD8+ T cells. Immunity. 2000;13:59–71. doi: 10.1016/s1074-7613(00)00008-x. [DOI] [PubMed] [Google Scholar]
- 5.Kioussis D, Ellmeier W. Chromatin and CD4, CD8A and CD8B gene expression during thymic differentiation. Nature reviews. Immunology. 2002;2:909–919. doi: 10.1038/nri952. [DOI] [PubMed] [Google Scholar]
- 6.Adoro S, McCaughtry T, Erman B, Alag A, Van Laethem F, Park JH, Tai X, Kimura M, Wang L, Grinberg A, Kubo M, Bosselut R, Love P, Singer A. Coreceptor gene imprinting governs thymocyte lineage fate. The EMBO journal. 2012;31:366–377. doi: 10.1038/emboj.2011.388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sarafova SD, Erman B, Yu Q, Van Laethem F, Guinter T, Sharrow SO, Feigenbaum L, Wildt KF, Ellmeier W, Singer A. Modulation of coreceptor transcription during positive selection dictates lineage fate independently of TCR/coreceptor specificity. Immunity. 2005;23:75–87. doi: 10.1016/j.immuni.2005.05.011. [DOI] [PubMed] [Google Scholar]
- 8.Chong MM, Simpson N, Ciofani M, Chen G, Collins A, Littman DR. Epigenetic propagation of CD4 expression is established by the Cd4 proximal enhancer in helper T cells. Genes & development. 2010;24:659–669. doi: 10.1101/gad.1901610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Taniuchi I, Ellmeier W, Littman DR. The CD4/CD8 lineage choice: new insights into epigenetic regulation during T cell development. Advances in immunology. 2004;83:55–89. doi: 10.1016/S0065-2776(04)83002-5. [DOI] [PubMed] [Google Scholar]
- 10.Sawada S, Scarborough JD, Killeen N, Littman DR. A lineage-specific transcriptional silencer regulates CD4 gene expression during T lymphocyte development. Cell. 1994;77:917–929. doi: 10.1016/0092-8674(94)90140-6. [DOI] [PubMed] [Google Scholar]
- 11.Taniuchi I, Sunshine MJ, Festenstein R, Littman DR. Evidence for distinct CD4 silencer functions at different stages of thymocyte differentiation. Molecular cell. 2002;10:1083–1096. doi: 10.1016/s1097-2765(02)00735-9. [DOI] [PubMed] [Google Scholar]
- 12.Manjunath N, Shankar P, Stockton B, Dubey PD, Lieberman J, von Andrian UH. A transgenic mouse model to analyze CD8(+) effector T cell differentiation in vivo. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:13932–13937. doi: 10.1073/pnas.96.24.13932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zou YR, Sunshine MJ, Taniuchi I, Hatam F, Killeen N, Littman DR. Epigenetic silencing of CD4 in T cells committed to the cytotoxic lineage. Nature genetics. 2001;29:332–336. doi: 10.1038/ng750. [DOI] [PubMed] [Google Scholar]
- 14.Lee PP, Fitzpatrick DR, Beard C, Jessup HK, Lehar S, Makar KW, Perez-Melgosa M, Sweetser MT, Schlissel MS, Nguyen S, Cherry SR, Tsai JH, Tucker SM, Weaver WM, Kelso A, Jaenisch R, Wilson CB. A critical role for Dnmt1 and DNA methylation in T cell development, function, and survival. Immunity. 2001;15:763–774. doi: 10.1016/s1074-7613(01)00227-8. [DOI] [PubMed] [Google Scholar]
- 15.de Boer J, Williams A, Skavdis G, Harker N, Coles M, Tolaini M, Norton T, Williams K, Roderick K, Potocnik AJ, Kioussis D. Transgenic mice with hematopoietic and lymphoid specific expression of Cre. European journal of immunology. 2003;33:314–325. doi: 10.1002/immu.200310005. [DOI] [PubMed] [Google Scholar]
- 16.Koller BH, Marrack P, Kappler JW, Smithies O. Normal development of mice deficient in beta 2M, MHC class I proteins, and CD8+ T cells. Science. 1990;248:1227–1230. doi: 10.1126/science.2112266. [DOI] [PubMed] [Google Scholar]
- 17.Nakayamada S, Kanno Y, Takahashi H, Jankovic D, Lu KT, Johnson TA, Sun HW, Vahedi G, Hakim O, Handon R, Schwartzberg PL, Hager GL, O'Shea JJ. Early Th1 cell differentiation is marked by a Tfh cell-like transition. Immunity. 2011;35:919–931. doi: 10.1016/j.immuni.2011.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wei G, Abraham BJ, Yagi R, Jothi R, Cui K, Sharma S, Narlikar L, Northrup DL, Tang Q, Paul WE, Zhu J, Zhao K. Genome-wide analyses of transcription factor GATA3-mediated gene regulation in distinct T cell types. Immunity. 2011;35:299–311. doi: 10.1016/j.immuni.2011.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nature methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ciofani M, Madar A, Galan C, Sellars M, Mace K, Pauli F, Agarwal A, Huang W, Parkurst CN, Muratet M, Newberry KM, Meadows S, Greenfield A, Yang Y, Jain P, Kirigin FK, Birchmeier C, Wagner EF, Murphy KM, Myers RM, Bonneau R, Littman DR. A validated regulatory network for Th17 cell specification. Cell. 2012;151:289–303. doi: 10.1016/j.cell.2012.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Egawa T, Littman DR. Transcription factor AP4 modulates reversible and epigenetic silencing of the Cd4 gene. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:14873–14878. doi: 10.1073/pnas.1112293108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Egawa T, Littman DR. ThPOK acts late in specification of the helper T cell lineage and suppresses Runx-mediated commitment to the cytotoxic T cell lineage. Nature immunology. 2008;9:1131–1139. doi: 10.1038/ni.1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.He X, He X, Dave VP, Zhang Y, Hua X, Nicolas E, Xu W, Roe BA, Kappes DJ. The zinc finger transcription factor Th-POK regulates CD4 versus CD8 T-cell lineage commitment. Nature. 2005;433:826–833. doi: 10.1038/nature03338. [DOI] [PubMed] [Google Scholar]
- 24.Muroi S, Naoe Y, Miyamoto C, Akiyama K, Ikawa T, Masuda K, Kawamoto H, Taniuchi I. Cascading suppression of transcriptional silencers by ThPOK seals helper T cell fate. Nature immunology. 2008;9:1113–1121. doi: 10.1038/ni.1650. [DOI] [PubMed] [Google Scholar]
- 25.Wang L, Wildt KF, Castro E, Xiong Y, Feigenbaum L, Tessarollo L, Bosselut R. The zinc finger transcription factor Zbtb7b represses CD8-lineage gene expression in peripheral CD4+ T cells. Immunity. 2008;29:876–887. doi: 10.1016/j.immuni.2008.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bird AP, Wolffe AP. Methylation-induced repression--belts, braces, and chromatin. Cell. 1999;99:451–454. doi: 10.1016/s0092-8674(00)81532-9. [DOI] [PubMed] [Google Scholar]
- 27.Collins A, Hewitt SL, Chaumeil J, Sellars M, Micsinai M, Allinne J, Parisi F, Nora EP, Bolland DJ, Corcoran AE, Kluger Y, Bosselut R, Ellmeier W, Chong MM, Littman DR, Skok JA. RUNX transcription factor-mediated association of Cd4 and Cd8 enables coordinate gene regulation. Immunity. 2011;34:303–314. doi: 10.1016/j.immuni.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kappes DJ. Expanding roles for ThPOK in thymic development. Immunological reviews. 2010;238:182–194. doi: 10.1111/j.1600-065X.2010.00958.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Taniuchi I, Ellmeier W. Transcriptional and epigenetic regulation of CD4/CD8 lineage choice. Advances in immunology. 2011;110:71–110. doi: 10.1016/B978-0-12-387663-8.00003-X. [DOI] [PubMed] [Google Scholar]
- 30.Xiong Y, Bosselut R. The enigma of CD4-lineage specification. European journal of immunology. 2011;41:568–574. doi: 10.1002/eji.201041098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stritesky GL, Xing Y, Erickson JR, Kalekar LA, Wang X, Mueller DL, Jameson SC, Hogquist KA. Murine thymic selection quantified using a unique method to capture deleted T cells. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:4679–4684. doi: 10.1073/pnas.1217532110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Leung RK, Thomson K, Gallimore A, Jones E, Van den Broek M, Sierro S, Alsheikhly AR, McMichael A, Rahemtulla A. Deletion of the CD4 silencer element supports a stochastic mechanism of thymocyte lineage commitment. Nature immunology. 2001;2:1167–1173. doi: 10.1038/ni733. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.