Abstract
Bid is a proapopotic activator protein of the Bcl-2 family that plays a pivotal role in controlling mitochondrial outer membrane permeabilization during apoptosis. Here, we characterized the interaction of fluorescently labeled truncated Bid (tBid) with a mitochondria-like supported lipid bilayer at the single-molecule level. The proteins observed at the membrane exhibited a very wide range of mobility. Confocal images of the membrane displayed both diffraction-limited Gaussian spots and horizontal streaks, corresponding to immobile and mobile tBid species, respectively. We observed 1), fast-diffusing proteins corresponding to a loosely, probably electrostatically bound state; 2), slowly diffusing proteins, likely corresponding to a superficially inserted state; and 3), fully immobilized proteins, suggesting a fully inserted state. The stoichiometry of these proteins was determined by normalizing their fluorescence intensity by the brightness of a tBid monomer, measured separately using fluorescence fluctuation techniques. Strikingly, the immobile species were found to be mainly tetramers and higher, whereas the mobile species had on average a significantly lower stoichiometry. Taken together, these results show that as soluble Bid progresses toward a membrane-inserted state, it undergoes an oligomerization process similar to that observed for Bax.
Introduction
Apoptosis is a form of programmed cell death vital for development and homeostasis in multicellular organisms. In mammals, the initiation of apoptosis is tightly regulated through a network of protein-protein and protein-membrane interactions involving the Bcl-2 family of proteins (1,2). Bcl-2 proteins are functionally divided into two groups, anti- and proapoptotic. Antiapoptotic family members, such as Bcl-XL, share four conserved Bcl-2 homology (BH) regions, and they inhibit the onset of apoptosis. Proapoptotic members are further subdivided into two categories. Multiregion proapoptotic proteins, such as Bax, contain BH 1–3 regions and permeabilize organelle membranes by pore formation. Proapoptotic BH3 proteins contain only the BH3 region and function as inhibitors of the antiapoptotic proteins (3,4) or as activators that also promote pore formation by activating the multiregion proapoptotic proteins (5,6).
The BH3-interacting domain death agonist (Bid) is a 22 kDa protein that has been classified as a proapoptotic BH3 activator (5–8). Bid function is regulated by a number of posttranslational modifications. Of particular importance in apoptosis is the cleavage of cytosolic Bid by caspase-8 in response to apoptotic stimuli (9). The two fragments of cleaved Bid (cBid) remain bound together through hydrophobic interactions in solution, but in the presence of mitochondrial membranes they rapidly separate, and the larger, 15 kDa fragment, known as truncated Bid (tBid), inserts into the lipid bilayer (10,11). Membrane-bound tBid recruits cytosolic Bax to mitochondria, resulting in mitochondrial outer-membrane permeabilization followed by release of apoptotic factors in the cytoplasm, activation of cellular caspases, and, ultimately, cell death (12). Interestingly, tBid also recruits antiapoptotic Bcl-XL to bind membranes (13).
Evidence suggests that despite its classification as a BH3 protein, Bid resembles the multiregion Bcl-2 proteins Bax and Bcl-XL in many respects (14). Unlike other BH3 proteins, which are unstructured, Bid maintains a helical structure in solution when it is not membrane-associated, with a fold similar to that of Bax and Bcl-XL (15). The conformational changes that occur upon activation also exhibit similarities among the three proteins: hydrophobic helixes that are buried in the interior of the protein when in solution (helices 6 and 7 for Bid) become exposed and promote membrane binding (14). Furthermore, as we have recently shown, just like Bax, tBid may adopt multiple conformations at the membrane, and these conformations are not functionally equivalent (11). Finally, Bid contains a structurally conserved hydrophobic pocket in the core of the protein that resembles that of multiregion Bcl-2 proteins (16). These pockets play a central role in regulating the interactions between Bcl-2 family proteins. It is believed that the BH3 region of BH3 proteins can insert into the hydrophobic pocket of multiregion proteins (17). The presence of a homologous hydrophobic pocket in Bid suggests the possibility for interactions with other BH3 proteins. In particular, it raises the question of whether tBid might be able to form homooligomers when interacting with mitochondrial membranes, a hypothesis supported by previous cross-linking and fluorescence resonance energy transfer (FRET) experiments in apoptotic cells (18).
Investigations of the molecular mechanisms underlying Bid function have primarily used ensemble techniques, e.g., to characterize the interaction of Bid with other Bcl-2 family proteins and with membranes (10,11,18,19). Yet for proteins exhibiting different conformations and forming oligomers with multiple stoichiometries, the average data obtained from ensemble methods may not provide a full description of the system. We therefore used single-particle methods to observe the behavior of individual Bid complexes when bound to supported lipid bilayers (SLBs) with complex mitochondria-like lipid composition. Molecular complexes formed by tBid at the lipid bilayer were imaged with both confocal and total internal reflection fluorescence (TIRF) microscopy and additionally characterized by fluorescence correlation spectroscopy (FCS). We developed what we believe to be a new way of analyzing confocal images that allows simultaneous assessment of both the oligomeric state and the mobility of detected fluorescent objects. The first is accomplished by performing a calibration of the monomer specific brightness using fluorescence intensity distribution analysis (FIDA), the latter by recognizing that immobile and mobile objects have different signatures in confocal images, with mobile objects appearing as horizontal streaks (20). We find that membrane-associated tBid is present predominantly as monomers and dimers that have a low to high mobility, but that higher order oligomers with zero to low mobility are also present. We discuss the implications of these findings for the role of Bid in apoptosis.
Materials and Methods
Protein purification and labeling
Recombinant full-length murine Bid with an N-terminal His-tag was purified, fluorescently labeled at position 126, and cleaved as previously described (10,11,21). Labeling efficiency between 50 and 70% were achieved for Alexa Fluor 488 (Invitrogen Molecular Probes, now Life Technologies, Carlsbad, CA) and between 70 and 90% for the other fluorophores used: Atto 647 from ATTO-TEC (Siegen, Germany) and 7-diethylaminocoumarin-3-carbonyl (DAC) and nitrobenzoxadiazole (NBD) from Invitrogen. The activity of the purified labeled proteins was assessed by measuring their capacity to induce Bax-mediated pore formation using a well-established ANTS release assay (22).
Preparation of liposomes and SLBs
The reconstituted membranes used in this study had a lipid composition reflecting that of mitochondrial membranes: 48 mol % phosphatidylcholine (PC), 28 mol % phosphatidylethanolamine (PE), 10 mol % phosphatidylinositol (PI), 10 mol % phosphatidylserine (PS), and 4 mol % cardiolipin (CL). This mainly represents an average between the composition of the inner and outer mitochondrial membranes, as determined in yeast and several vertebrates (23,24). The exception is cardiolipin, which is found in large amounts (∼25%) in the inner membrane, but which we used here at the lower concentration (∼4%) at which it is usually found in the outer membrane. This mitochondria-like lipid composition has been shown to support Bid- and Bax-mediated pore formation in liposomes (7,22,25). All lipids were purchased from Avanti Polar Lipids (Alabaster, AL). Phosphatidylcholine and phosphatidylethanolamine were egg extracts (Avanti 840051 and 841118, both with 16:0/18:1 as the predominant acyl chain composition). Phosphatidylinositol was liver extract (Avanti 840042, predominant form 18:0/20:4). PS was synthesized as 18:1/18:1 PS (DOPS, Avanti 840035). CL was synthesized as 18:1/18:1/18:1/18:1 CL (TOCL, Avanti 710335). When needed, the membranes were fluorescently labeled by adding either 0.0025 mol % (for single-particle tracking) or 0.008 mol % (for membrane position determination or liposome detection) of lipophilic dye (DiO-C18 or DiD-C18, both from Invitrogen). Liposomes were formed as previously described (10,11). SLBs were formed on mica substrate by direct liposome fusion, following a protocol adapted from Chiantia et al. (26) and Oreopoulos and Yip (27). Mica-coated coverslips were mounted in an ∼400 μL FCS2 perfusion chamber (Bioptechs, Butler, PA). An assay buffer solution with 0.5 mg/mL liposome was injected into the chamber, and the samples were then incubated at 37°C for 1 h to encourage liposome fusion on the solid surface. Excess liposomes were then removed by washing the membrane with ∼5 mL assay buffer pumped through the chamber at a rate of ∼1.5 mL/min, after which the bilayer was allowed to recover at 37°C for 15 min. When required, a 1 mL solution of cBid diluted in assay buffer (with concentration 100 nM for ensemble binding measurements or 100–400 pM for single-particle measurements) was injected into the chamber at a rate of ∼1.5 mL/min, and incubated for 15 min at 37°C.
Steady-state fluorescence spectroscopy
The homooligomerization and membrane insertion of cBid were detected concurrently using FRET and environment-sensitive fluorescence, mainly as described before (11). To prevent protein adhesion, cuvettes were surfactant-coated with DOPC. Assay buffer containing up to 8 nM mitochondria-like liposomes (that is up to 0.5 mg/mL or 650 μM lipids) was then placed in the cuvette, followed by 20 nM cBid-DAC (donor) and finally 100 nM cBid-NBD (acceptor). Control reactions in the absence of acceptor, i.e., where cBid-NBD was replaced by unlabeled cBid, were carried out in parallel. The reaction was stirred and kept at 37°C. The NBD fluorescence (λex = 475 nm, λem = 530 nm) and the DAC fluorescence (λex = 380 nm, λem = 460 nm) were monitored simultaneously using a Quantamaster fluorometer (Photon Technology International, Birmingham, NJ). FRET efficiency was calculated from the donor fluorescence by accounting for background, progressive loss of protein to the walls of the cuvette, and environment sensitivity of DAC, as explained in detail in the Supporting Material.
Confocal microscopy
Instrument
All confocal measurements (fluorescence fluctuation and imaging) were performed on an Insight fluorescence fluctuation microscope (Evotec Technologies, Hamburg, Germany, now Perkin-Elmer, Waltham, USA), described in detail elsewhere (28). Fluorescence excitation was achieved using either a Sapphire 488-20 CDRH solid-state laser (Coherent, Santa Clara, CA), when detecting cBid-Alexa488 or DiO-labeled lipid membranes, or a Radius 635-25 solid-state laser (Coherent), when detecting cBid-Atto647 or DiD-labeled membranes.
Fluorescence fluctuation measurements
Unless otherwise stated, a ∼20 μW excitation power was used, and at least three repeats of 5–60 s each were collected for each condition. Analysis of the photon-counting histograms by FIDA was performed using the Acapella software (Evotec Technologies) and returned both the specific brightness, B, and the concentration of the different fluorescent species in solution (29). Analysis of the autocorrelation functions was done assuming diffusion in two or three dimensions (see Supporting Material for details), using either Acapella or Kaleidagraph (Synergy Software, Reading, PA), and returning the diffusion coefficient, D, of the different species present. In almost all cases, the analysis was done assuming the presence of a single fluorescent species. The exception was the analysis of the data collected for cBid-Alexa488 incubated with DiD-labeled liposomes. In that case, the photon-counting histograms and autocorrelation functions collected in the protein channel were analyzed assuming the coexistence of two species, corresponding to the bound and unbound fraction of the protein, as explained in more detail in Shamas-Din et al. (11) and Satsoura et al. (25).
Confocal image acquisition
To establish the membrane position at the beginning of each experiment and to quantify protein binding, images were acquired perpendicular to the plane of the membrane. Those were typically 100 × 100 pixel images with a pixel-to-pixel distance of d = 1 μm and a pixel dwell time of δ = 1 ms. These images were also used for membrane-binding quantification, as explained in the Supporting Material. For the imaging of single particles at the membrane, stacks of 10 images were acquired parallel to the plane of the membrane, starting below and finishing above the plane of the membrane, in increments of 1 μm. The imaging conditions used in this case (10 × 10 μm, d = 0.1 μm, δ = 1 ms, resulting in a line time of τL = 150 ms) offered the best compromise between obtaining sufficient signal to resolve single spots and reducing photobleaching.
Single-particle detection of mobile and immobile objects in confocal images
An ImageJ plugin was written for automated particle detection in stacks of images acquired parallel to the plane of the membrane, inspired both by the single-particle localization method described in Henriques et al. (30) and by the work on mobile-particle detection done previously by our group (20). The developed algorithm is described in detail in the Supporting Material. In brief, the brightest stack in the image was first duplicated. One copy was used for event detection and the other for event characterization. The position of the pixel with the highest intensity in the detection image was determined, and the corresponding region of the characterization image was submitted to a least-squared 2D-Gaussian fit to obtain the maximum fluorescence intenstiy, imax, noise level, iB, widths along and perpendicular to the scanning direction, wx and wy, respectively, and exact coordinates, x0 and y0, of this particular event. That event was then deleted from the detection image and the process was repeated until all events above the threshold value p = <iB> + B/2 had been detected, where B is the specific brightness of a protein monomer as measured in solution by FIDA, and <iB> is the running average value of the noise level in the image. Each fitted event with a normalized χ-squared >1.4 was tested for classification as a diffraction-limited spot (the expected morphology for immobile particles) or horizontal streak (the expected morphology for mobile particles), as explained in the Results section. The distribution of oligomer stoichiometry was obtained from the distribution of maximum fluorescence intensity by normalizing it by the monomer specific brightness and correcting for fluorophore labeling efficiency, as explained in detail in the Supporting Material.
TIRF microscopy
SLBs either labeled with DiD or interacting with cBid-Atto647 were imaged at an excitation wavelength of 640 nm using a Nikon Eclipse Ti TIRF microscope equipped with a 100× oil immersion objective (NA 1.49). Series of 250 × 250 pixel images were recorded using an iXon3 897 EMCCD camera (Andor Technology, Belfast, United Kingdom) with a frame interval of 35 ms and a pixel size of 16 μm (corresponding to a pixel-to-pixel distance in the lens focal plane of 160 nm). Single-particle tracking was subsequently performed using a plugin written for ImageJ. The position of clearly identified objects was determined in successive frames using the same 2D Gaussian fitting procedure as for single-particle localization in confocal images. This procedure was applied directly on image series without any prior background subtraction, and it returned the position, dimension, intensity, and local background for the particle for each image. From this information, the average mean-squared displacement, <r2(t)>, was calculated for each particle and fitted with the function <r2(t)> = 4Dt to obtain the particle diffusion coefficient.
Results
tBid oligomerizes after inserting into the membrane
To confirm and extend the previous observation that tBid can homooligomerize in the presence of mitochondrial membranes (18), we used a reconstituted system composed of purified cBid labeled with either DAC (cBid-DAC, labeled at residue 126, i.e., on the tBid fragment of the protein) or NBD (cBid-NBD, also labeled at residue 126) and liposomes with mitochondria-like lipid composition. When 100 nM cBid-NBD was added to 20 nM cBid-DAC preincubated with liposomes, an interaction between the tBid fragments of the two types of protein occurred, as evidenced by the observed FRET between the donor fluorophore, DAC, and the acceptor fluorophore, NBD (Fig. 1 A). This interaction got noticeably stronger in the presence of liposomes. The FRET efficiency curves were fit using two exponential terms. The first term corresponded to the interaction between tBid fragments in solution, with a half-time fixed to the average half-time observed in the absence of lipids, 16 s. The second term corresponded to the interaction between membrane-bound tBid fragments, with a half-time on the order of 100–200 s, depending on lipid concentration. In addition, a third exponential term had to be added for samples at low lipid concentration, to account for protein loss to binding on sample surfaces. The tBid oligomerization process was compared to the insertion of the protein into the membrane, followed by the increase in fluorescence of the environment-sensitive dye NBD (Fig. 1 B). As we reported previously, the average hydrophobicity around residue 126 of cBid increases in the presence of lipid membranes with single-exponential kinetics, due to insertion of the protein into the membrane (11). Comparing the half-time for oligomerization of membrane tBid species (∼100–200 s) with that of insertion of different residues into the membrane (∼10–50 s), we see that tBid oligomerization proceeds or continues to proceed long after the protein is fully inserted into the membrane (Fig. 1 C). This is consistent with a model for the interaction of cleaved Bid with membranes, where after very rapid fragment separation and loose binding to the lipid bilayer, the protein undergoes a complex conformational change resulting in partial insertion into the membrane (as we recently proposed (11)), only then followed by oligomerization (Fig. 1 D, and see the Supporting Material). This model predicts that as lipid concentration increases, the fraction of tBid bound to liposomes increases, whereas the number of bound tBid per liposome decreases. Due to the interplay between these two opposing factors, the fraction of oligomerized protein at the membrane thus first increases and then decreases, as we indeed observed (Fig. 1 A).
Figure 1.
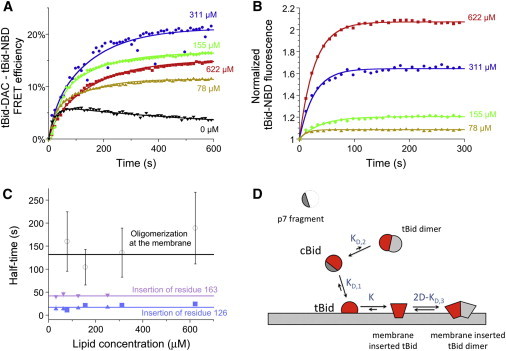
tBid forms oligomers in the membrane of mitochondria-like liposomes. (A and B) FRET efficiency calculated from the DAC fluorescence and corrected for environmental effects using the signal obtained in the absence of acceptor (A) and normalized NBD fluorescence (B) measured after addition of 100 nM acceptor cBid-NBD to 20 nM donor cBid-DAC premixed with liposomes. Each curve corresponds to the average of three repeats. Lines are single (B) or multiple (A) exponential fits of the data. (C) Comparison between the half-times associated with the insertion of Bid residue 126 (solid triangles and squares) or 163 (solid inverted triangles) in the membrane and those associated with tBid oligomerization (open circles), as obtained from the exponential fit of either NBD-fluorescence or DAC-NBD FRET data as shown in A and B (mean ± SD, n = 3). Data shown as squares and circles are from this study, whereas data shown as triangles and inverted triangles was taken from Shamas-Din et al. (11). (D) Simple model for the interaction of cBid with a lipid membrane. To see this figure in color, go online.
Mitochondria-like SLBs are a tenable model system for the study of the interaction of tBid with membranes
To further test the validity of the model for Bid interaction with membranes shown in Fig. 1 D, we turned to single-molecule experiments to resolve the different conformations and oligomeric states of tBid when bound to a lipid bilayer. We first tested the validity of using single SLBs with mitochondria-like composition as a model system to study the interaction of Bid with membranes. SLBs were formed on freshly cleaved mica substrates by liposome fusion. The membrane structure was probed by atomic force microscopy (see text and Fig. S1 in the Supporting Material). The membrane thickness was always found to correspond to that of a single lipid bilayer (∼4 nm), as expected when using the liposome fusion method (31,32). In addition, no observable lateral features were detected, consistent with the formation of a homogeneous fluid phase. The diffusion of a lipophilic dye placed in the membrane, DiD, was assessed by single-particle tracking, and found to be D = 2.2 μm2/s. Although this value is lower than that of dyes in free-standing lipid bilayers (33,34), it falls in the range measured for both dyes and fluorescently labeled lipids in SLBs in the fluid liquid-disordered phase, found to be between 1 and 6 μm2/s, depending on experimental conditions (33,35,36). This shows that the mitochondria-like SLBs retain fluid properties.
A further validation of the SLB system was the ability of Bid to bind the membrane. This was tested by acquiring confocal images in the presence and absence of a DiO-labeled SLB incubated with solutions of cBid-Atto647 (Fig. 2 A). In the absence of an SLB, only slight nonspecific binding of cBid-Atto647 to the mica surface was observed. In contrast, in the presence of an SLB, the tBid fragment of cBid (observed by monitoring the fluorescence of the bound Atto647 dye) bound rapidly (within ∼2 min) and with high specificity to the SLB, similar to what has been reported for cBid binding to liposomes (10,11). The interaction between cBid and the membrane was further quantified by comparing the fluorescence coming from the protein at the membrane, <IM>, with that coming from the protein in solution, <IS>. Both were obtained from the average intensity profile (along the optical axis) of confocal images of the membrane (Fig. 2 B). As expected for a simple partition between the fluid phase and the membrane phase (see Supporting Material for details), we observed a linear relationship between <IM> and <IS> (Fig. 2 C). This means that there is no saturation of the binding in the considered concentration range. We find that <IM>/<IS> = 9.4, leading to an estimate for the value of the partition coefficient of PM/S = (7.0 ± 1.5) × 103. This corresponds to an apparent dissociation constant between cBid and the lipids in the SLB, KD = 160 ± 40 μM. This value is only slightly higher than that obtained for the binding of cBid to 100 nm liposomes with the same lipid composition, KD = 76 μM (11). Thus, tBid still efficiently binds the lipids in the SLB. Finally, incubations of fluorescently labeled Bax proteins with SLBs showed that Bax was significantly recruited to the membrane only when preincubated with cBid (data not shown), showing that tBid bound to mitochondria-like SLBs retains Bax activating capacity.
Figure 2.
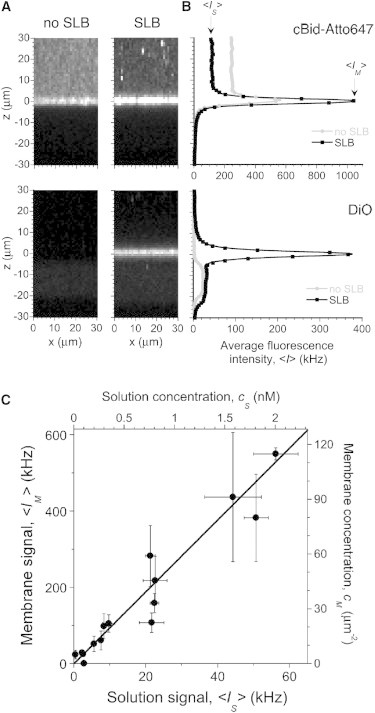
tBid binds specifically to mitochondria-like SLBs. (A) Representative confocal images in the absence (left) or presence (right) of a mitochondria-like SLB, incubated for 12 min with 25 nM cBid-Atto647. Images acquired at 635 nm excitation (upper) were used to localize cBid-Atto647, whereas those acquired at 488 nm excitation (lower) were used to confirm the formation of the DiO-labeled SLB. (B) Average intensity profiles along the optical axis of the images shown in A. (C) Average membrane signal, <IM>, as a function of average solution signal, <IS>, measured at 635 nm excitation, for SLBs incubated with different concentration of cBid-Atto647 (1–30 nM). Each point represents the mean ± SD of three different images acquired at different locations in the same sample. The continuous line is a linear fit to the data, with a slope of 9.4.
Stoichiometry of Bid membrane species
We next evaluated the stoichiometry of single tBid molecular assemblies bound to SLBs by acquiring and quantitatively analyzing confocal images. Solutions with a subnanomolar concentration (100–400 pM) of cBid-Atto647 were introduced into a perfusion chamber containing a mitochondria-like SLB. At these concentrations, the chance for coincidental detection of two different membrane-bound particles in the same pixel is very low. Stacks of confocal images were acquired with a spacing of 1 μm, such that one of the images in the stack could almost always be found with the plane of the membrane in focus (Fig. 3 A). Reliably, two types of fluorescent events could be observed. The first was diffraction-limited spots, such as the one indicated by a blue arrow in Fig. 3 A. Such a feature indicates the presence of fluorescent particles smaller than the instrument resolution (∼300 nm) and immobile on the timescale of the acquisition of the 10 lines necessary to image them, i.e., with D < w02/(40τL) ∼ 0.02 μm2/s. The second type was horizontal streaks, such as the one indicated by a red arrow in Fig. 3 A. This is the feature expected for particles that are immobile on the timescale of the pixel dwell time (D < w02/(4δ) ∼ 40 μm2/s) but mobile on the timescale of a line scan (D > w02/(4τL) ∼ 0.2 μm2/s), meaning that they are observed only for one or two lines before moving away.
Figure 3.
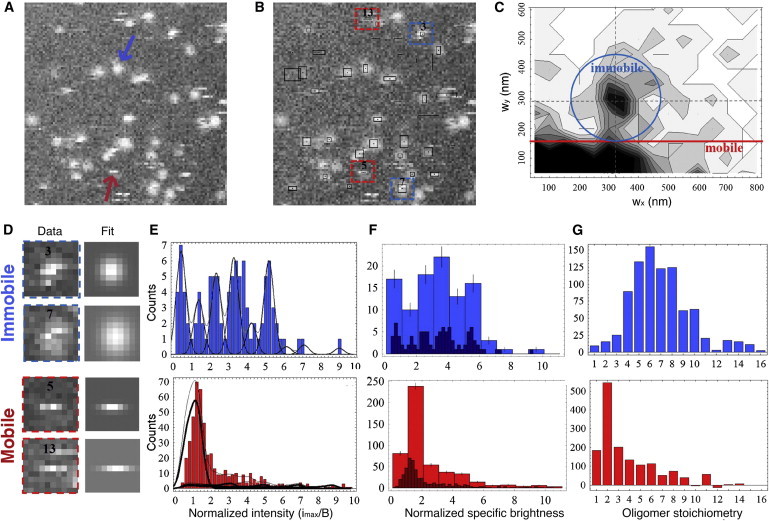
tBid bound to mitochondria-like SLBs forms both mobile low-order oligomers and immobile high-order oligomers. (A) Representative confocal image of a mitochondrial SLB incubated with 400 pM cBid-Atto647 (635 nm, 10 μm × 10 μm). (B) Same image as in A, annotated after detection of intensity maxima. The black boxes show the position and dimension of events as determined by the 2D-Gaussian fit. (C) Contour plot showing the distributions of 1/e2 dimensions (wx and wy) for a total of n = 723 events detected in the same sample. The regions in which diffraction-limited spots (corresponding to immobile particles) and horizontal streaks (corresponding to mobile particles) were found are shown. (D) Example of mobile and immobile particles (blue and red boxes in B) and corresponding 2D-Gaussian fit. (E) Distributions of normalized event intensity for immobile particles (upper) and mobile particles (lower). Thick continuous lines represent fits performed as explained in the text. Thin continuous lines represent the individual contributions of subpopulations of particles with different specific brightness. (F) Distributions of normalized specific brightness, constructed from the fits shown in E. (G) Distributions of oligomer stoichiometry (averaged for three different samples), as obtained after protein labeling efficiency correction, as explained in the text and in Figs. S2 and S3.
To properly characterize both types of detected events, we developed a dedicated single-particle detection algorithm, as described in the Materials and Methods. Each detected event was fitted with a 2D-Gaussian function (Fig. 3 B), returning the peak fluorescence intensity, imax, and the 1/e2 radii of the particle along (wx) and perpendicular to (wy) the scanning direction. The distribution of events in the (wx,wy) plane (Fig. 3 C, corresponding to a total of 723 events detected in 15 images acquired for the same sample) renders obvious their separation into two distinct categories. tBid spots closely clustered around wx ≈ wy ≈ w0,PSF = 300 nm, between the expected value w0 = 0.4λ/NA = 235 nm, and the experimental value w0 = 380 nm obtained from FCS calibration measurements. We thus classified events as well-resolved diffraction-limited spots if they lay within w0,PSF/2 of the point (w0,PSF, w0,PSF) in the (wx,wy) plane. tBid streaks, on the other hand, clustered in the region defined by wx < 200 nm, as expected for mobile particles visible for only one or two lines in the confocal image. Thus, we classified events as well-resolved streaks if wy < w0,PSF/2. Examples of events corresponding to spots (immobile particles) and streaks (mobile particles), together with their Gaussian fit, are presented in Fig. 3 D (more particles are shown in Fig. S3).
The distributions of peak fluorescence intensity for mobile and immobile particles, obtained for the same set of 15 images as in Fig. 3 C, are shown in Fig. 3 E. The fluorescence intensity was normalized by the specific brightness of a cBid-Atto647 monomer in solution (as justified below). This quantity was measured by FIDA immediately after the images were recorded, using the same confocal microscope and the exact same settings, with a 10 nM solution of cBid-Atto647. It is immediately apparent that the normalized intensity distribution for the immobile particles consists of a series of well-defined peaks extending, for this sample, up to n ∼ 6, whereas that of the mobile particles mainly consists of a large peak around n ∼ 1.2 (Fig. 3 E). For the immobile particles, the data were fitted by a sum of equally spaced Gaussian distributions, as expected for a population of immobile oligomers with different stoichiometries. The spacing of the peaks, Δn = 0.96, was very close to 1. This confirms that the specific brightness value measured in solution by FIDA remained very similar at the membrane, and was not significantly modified by binding to the SLB or by the proximity of other fluorophores. On the other hand, the first peak (presumably corresponding to a monomer) did not fall at n1 = 1, but at n1 ∼ 0.6. This could be due to a systematic error in the measurement of imax, maybe due to an overestimate of the background fluorescence caused by the finite size of the regions of interest used for fitting of events. From this analysis, the normalized specific brightness distributions, PA(N), were generated (Fig. 3 F). The true stoichiometry distribution of tBid assemblies, PT(N), was then calculated from the specific brightness distribution by accounting for imperfect labeling efficiency (see the Supporting Material for details). The average stoichiometry distribution obtained for three separate experiments is shown in Fig. 3 G. It shows that the visible immobile spots have a broad distribution of stoichiometries centered around N ∼ 6.
The data obtained for mobile particles is harder to interpret, for two reasons, both associated with the fact that most mobile particles will have less-than-ideal trajectories passing away from the center of the confocal detection volume. A first consequence of this is that instead of a symmetric distribution of intensity values around the particle specific brightness, we expect an asymmetric distribution skewed toward values lower than the specific brightness (see Materials and Methods for details on the shape of the expected distribution). A second consequence is that mobile particles carrying a single fluorophore (e.g., monomers) might not always be detected due to a low signal/noise (S/N) ratio. The main contribution to noise in single-particle experiments comes from the photon shot noise of the signal (the photon noise of the background is negligible). The specific brightness of tBid-Atto647 was reproducibly found to be B ∼ 20 kHz in our experimental conditions, translating into an average signal S ∼ 20 photons for an immobile tBid monomer placed in the center of the detection volume (given a pixel dwell time of 1 ms). Thus, in our experiments, a signal/noise ratio of S/N ∼ S1/2 ∼ 4.5, just above the lowest acceptable value of S/N = 3, was associated with immobile monomeric tBid species. However, for mobile particles not passing through the center of the detection volume or not residing at its center for the full duration of the 1 ms pixel dwell time, the S/N is even lower. As a result, many of the mobile tBid monomers will escape detection. Since the FRET data presented in Fig. 1 suggest that the dissociation coefficient associated with dimer formation in the membrane is ∼100 μm−1 or more (the surface concentration of the protein in the FRET experiments), at the very low surface concentration used for our single-particle tracking experiments we expect monomeric species to dominate. Thus, many mobile particles escaped detection. We can still safely conclude that the stoichiometry distribution for mobile particles is dominated by low stoichiometry species, N = 1–3, and, of more importance, that mobile particles have low stoichiometry compared to immobile ones. Thus, tBid species in mitochondria-like SLBs mostly fall into two broad categories, mobile complexes with low stoichiometry, and immobile complexes with higher stoichiometry.
Mobility of Bid membrane species
To get an insight into their degree of insertion into the membrane, we next investigated the diffusion of mobile tBid species. We used two separate techniques—FCS and single-particle tracking in TIRF movies—to cover a large range of mobilities. TIRF movies were acquired for samples prepared in a manner similar to those used for the confocal experiments discussed in the previous section. These movies confirmed the presence of two different categories of tBid species, mobile and immobile. The detected particles were first identified using Gaussian filtering of the image, which was followed by thresholding and then tracked using the same 2D-Gaussian fit that was used for event detection in confocal images. Fig. 4 A shows examples of trajectories obtained for three different representative particles. Usually, mobile particles remained mobile throughout the 1.7 s tracking sequence (e.g., particle 1), whereas immobile particles remained immobile (e.g., particle 2). Occasionally, however, particles were observed to transition from mobile to immobile, sometimes several times, as was the case for particle 3. The motion of mobile particles was characterized through mean-squared displacement analysis. The observed mobile Bid species displayed a mainly diffusive behavior, characterized by a roughly linear mean-squared displacement (Fig. 4 B), allowing the measurement of a diffusion coefficient for each detected mobile particle. The distribution of diffusion coefficients obtained for the visible mobile Bid species is shown in Fig. 4 C and compared to the distribution of diffusion coefficients measured separately for the lipophilic dye DiD inserted into mitochondria-like SLBs. The average diffusion coefficient of the tBid membrane species followed by single-particle tracking, D = (0.8 ± 0.5) μm2/s (mean ± SD, n = 123), was about a third of that measured for the lipophilic dye, D = (2.2 ± 2.7) μm2/s (n = 100), suggesting that these proteins were partially inserted into the membrane.
Figure 4.
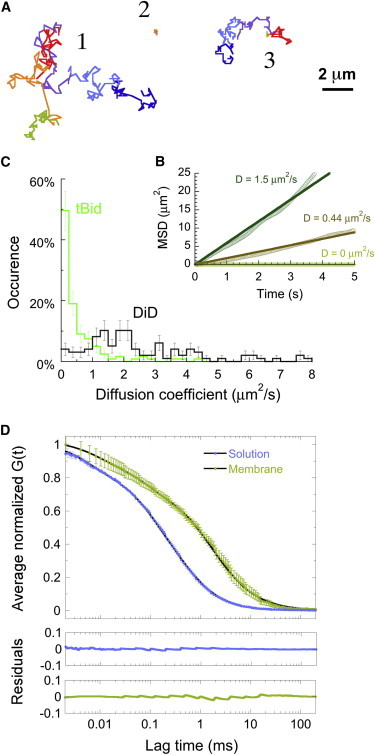
Mobility of membrane-bound tBid as observed by single-particle tracking and FCS. (A) Representative trajectories for three different kinds of particles (mobile (particle 1), immobile (particle 2), alternatively mobile, and immobile (particle 3)) tracked in TIRF movies of cBid associated with an SLB. All trajectories are 10.2 s long, and each color represents a 1.7 s time interval. (B) Mean-squared displacement (MSD) calculated for the three trajectories shown in A. Lines are linear fits of the data. (C) Histogram of diffusion coefficients measured at the membrane by single-particle tracking and MSD analysis for tBid (green, n = 122) and DiD (black, n = 98). (D) Average autocorrelation function obtained for cBid in solution (blue symbols, mean ± SD, n = 15) and when associated with an SLB (green symbols, n = 24). Lines correspond to fits of the data assuming one fluorescent species, yielding D = 118 μm2/s for the protein in solution and D = 22 μm2/s for the protein at the membrane. Residuals are shown in the lower panels.
Because TIRF movies were acquired here with a frame time of t = 35 ms, particles with a diffusion coefficient of D > ∼L2/(4t) (where L = 5 pxl = 0.8 μm is the maximum displacement allowed for a particle between frames to be considered the same particle), i.e., particles with D > ∼5 μm2/s, were not detected in the TIRF experiments. We therefore also performed FCS experiments. The average autocorrelation functions measured for cBid in solution and tBid membrane species are shown in Fig. 4 D. The cBid monomer diffuses in solution with D = (118 ± 6) μm2/s, corresponding to a hydrodynamic radius of 3.9 nm. Compared to the solution species, the membrane species have a noticeably reduced diffusion coefficient, D = (22 ± 2) μm2/s. However, remarkably, this value is still one order of magnitude higher than what is detected by TIRF, emphasizing that both methods detect distinct tBid membrane species, with very different mobility. In comparison, when measuring the diffusion coefficient of DiD by fluorescence fluctuation methods (FCS or image correlation spectroscopy, data not shown), we obtained values that were either the same or only slightly higher than that obtained from single-particle tracking, showing that the discrepancy observed for the protein is meaningful. We note that this fast-diffusing membrane-bound tBid species was not detected in a previous line-scanning FCS study of tBid interacting with giant unilamellar liposomes (19). The reason is that the temporal resolution of line-scanning FCS (just as that of TIRF) is not high enough to resolve fast membrane species. The fact that the diffusion coefficient of this fast-diffusing species is larger than that of the lipids suggests that these proteins were only superficially bound to the membrane.
Discussion
Early models for the activity of Bcl-2 family proteins considered the system as a simple rheostat regulated by the balance between the total concentrations of pro- and antiapoptotic proteins (37). However, it is now clear that a very complex network of interactions controls the cellular response to the life-or-death question at the heart of apoptosis (2,38). This is manifest in the number of possible binding partners, conformations, and stoichiometries for each Bcl-2 family protein. Characterizing these different species represents an experimental challenge, made harder by the fact that most are membrane species. Here, we met this challenge by employing a range of complementary fluorescence techniques. A new type of analysis in confocal images allowed the observation of membrane-bound tBid molecular complexes with distinct specific brightness and mobility. Techniques with complementary time resolution (such as FCS and TIRF) allowed characterizing the mobility of these species. Based on our results, we propose a model for the sequence of interactions between cBid and mitochondria-like membranes, illustrated in Fig. 5. By including oligomer formation, it refines and extends a model we had recently proposed based on ensemble fluorescence measurements that had already demonstrated the coexistence of different membrane tBid species (11). The tenets of this refined model are that cBid first loosely associates with the membrane as a monomer from solution, then partially inserts into the membrane before forming dimers, some of which later oligomerize into larger structures.
Figure 5.
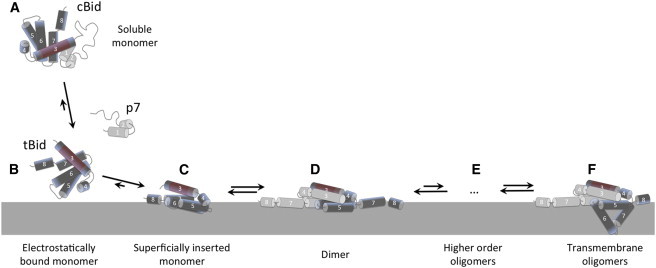
Proposed sequence for the interaction of cBid with lipid bilayers. (A) In solution, the two fragments of cBid remain noncovalently bound. The depicted tertiary structure was drawn according to the known soluble cBid structure (16). The red region on helix 3 represents the BH3 region. (B) Upon approaching the membrane, the fragments separate and tBid becomes electrostatically trapped to the lipid bilayer. (C) A progressive conformational change leads to superficial insertion in the lipid bilayer, as described in Shamas-Din et al. (11). A structure with shallow insertion of all helices as pictured here is supported by the NMR data from Wang and Tjandra (45). (D) Two inserted tBid monomers associate into a dimer. For illustration purposes, the tBid dimer structure was depicted with a symmetric BH3-in-the-groove structure inspired by that proposed for Bax dimers in Czabotar et al. (58). (E) Further oligomerization takes place leading to the formation of higher order oligomers. (F) Large oligomers can adopt a transmembrane configuration. To see this figure in color, go online.
One important, to our knowledge novel, finding of our study is the existence of a loosely membrane-bound tBid species. In the absence of a lipid membrane, the two fragments of cBid remain associated via noncovalent hydrophobic interactions (11) and cBid adopts a globular α-helical conformation similar to that of full-length Bid (39). Accordingly, we find that soluble cBid has a diffusion coefficient (D = 118 μm2/s) corresponding to an object with a hydrodynamic radius r = 3.9 nm, just slightly larger than what has been reported for tBid (D = 143 μm2/s, r = 3.2 nm (19)). However, as soon as a membrane is present, the two fragments of cBid separate (11). Electrostatics must play a major role in fragment separation, since the negatively charged mitochondrial membrane must attract and retain the positively charged tBid fragment while driving back the negatively charged p7 fragment (16,40). The fast-diffusing species that we detected by FCS then almost certainly corresponds to tBid proteins loosely electrostatically bound to the membrane (Fig. 5 B). The diffusion coefficient of this species (D = 22 μm2/s) is too low to correspond to a solution species, but too high to correspond to a membrane-inserted one (even partially inserted proteins cannot diffuse faster than the lipids in the bilayer (41,42)). Once inserted into the membrane (Fig. 4 C), the mobility of the protein is significantly reduced, with D ∼ 0.8 μm2/s, i.e., significantly less than that of the surrounding lipids. It must therefore involve the (maybe partial) insertion of several helices into the membrane (41,42). Likely candidates are helices 4, 6, 7, and 8, which all have been implicated in membrane binding (11,40,43,44). Alternatively, as suggested by a recent NMR study, all of tBid helices might in fact be shallowly inserted into the membrane in that conformation (45).
The concept of electrostatic scanning was put forward when advancing the notion that a reactant adsorbed on a surface would take less time to find its target than if it were diffusing in the bulk, since its diffusion would be confined to two dimensions (46). However, for this strategy to be efficient (for an organelle the size of mitochondria) the diffusion coefficient of the protein on the surface should be at least a tenth of its diffusion coefficient in the bulk (47). The loosely bound tBid species uncovered by our FCS experiments (D = 22 μm2/s) fulfills this condition, but the partially inserted one (D ∼ 0.8 μm2/s) does not, suggesting that in its loosely membrane-bound form tBid can more efficiently search for binding partners (other proteins, e.g., Mtch2 or Bax, or lipid domains). Why has this loosely bound state proven so elusive? Our experiments show that single-particle techniques are too slow to capture fast-diffusing membrane species. In general, diffusion experiments with a characteristic length scale d and a characteristic timescale T will only detect and characterize well particles with D < d2/T. For microscopy, d ∼ 0.5 μm is the pixel size and T is either the frame rate (TIRF, T ∼ 1–30 ms) or the line time (confocal microscopy and line-scanning FCS, T ∼ 1–100 ms). Thus, particles with D > 5 μm2/s will easily be overlooked when using these techniques. Signs of their presence in single-particle experiments may include a larger-than-expected fluorescent background and step-size distributions deviating from the expected χ distribution (48).
A second believed novel finding reported here is that tBid readily forms dimers in membranes. We previously reported that fragment separation is closely followed by a complex and progressive conformational change of tBid, leading to superficial insertion of helices 6 and 7, and to the coexistence of two main membrane-inserted conformations distinguished by a change in environment for the N- and C-terminal regions (11). Based on the results presented here, we now propose that these two membrane-inserted conformations represent a monomeric state and a dimeric state of the protein (Fig. 5, C and D). Indeed, helix 1 at the N-terminal of tBid (helix 3 of Bid) contains the BH3 region that mediates the interaction of tBid with other family members (49). The detection of tBid-tBid FRET localized at the mitochondria in apoptotic cells demonstrates that tBid homooligomers form in vivo (18). Although it had initially been suggested that the dominant homooligomer species was trimers (18), the same group later showed that the ∼45 kDa complex identified as a tBid homotrimer was in fact a complex between tBid and the 33 kDa mitochondrial protein Mtch2 (50). Thus, it seems reasonable that membrane tBid would preferentially form dimers in our reconstituted system, even though in cells, numerous interactions (e.g., with Mtch2, Bax, Bak, and Bcl-XL) may compete with the formation of tBid homooligomers. It is important to note that the forced homooligomerization of tBid has been shown to promote apoptosis (18). What then is the function of the tBid oligomers? A first possibility is that tBid oligomerization changes its affinity for some binding partners, e.g. Bax. A second possibility is that the tBid dimer acts as a sink state, which, if favored, e.g., by the correct lipid composition, could enhance the binding of tBid to specific membranes. The tBid dimer would then have a function similar to that of the tBid-Mtch2 heterodimer, whose formation renders the interaction with the membrane quasi-irreversible (11). A third possibility is that the tBid dimer simply represents the default stable state of the protein in the membrane while waiting to interact with other proteins for which it may have a higher affinity, e.g., Bax or Bcl-XL.
Assemblies of tBid molecules beyond the dimers were also clearly detected, and were often found to be immobile. None of the helices of tBid is thought to completely span the membrane bilayer (43,45). However, the presence of immobile tBid complexes, and of tBid complexes alternating between a mobile and an immobile state, could be explained by a (transiently or permanently) transmembrane helix interacting with the mica substrate. Another possibility is that immobilization occurs because of the interaction between the substrate and the lipids. We note that whereas the FRET signal we observe in the presence of membrane proves that the interaction of the two tBid molecules in a homodimer is a close physical interaction, the interaction between two dimers could instead be an indirect interaction mediated, for example, by lipid domains (i.e., it could represent colocalization into diffraction-limited lipid domains). In any case, the observation of tBid assemblies larger than dimers is interesting, because so far, among Bcl-2 family proteins, only Bax and Bak are believed to associate in large complexes.
Classically, Bid has been characterized as a proapopotic BH3 protein and a direct activator of Bax and Bak pore formation, meaning that it physically interacts with Bax and Bak, even if only transiently (5,7,8). However, Bid also presents similarities with multidomain Bcl-2 family proteins, and our work reinforces this idea in several ways. First, it shows that, like Bax and Bcl-XL, tBid is able to oligomerize in mitochondria-like membranes. Second, the loosely bound tBid conformation reported here is reminiscent of the loosely associated Bax species characterized in previous works (22,25,51). Third, we showed that the sequence of interaction of tBid with the membrane is similar to that reported for Bax (10,52): Like Bax, tBid first binds the membrane as a monomer, then inserts into the membrane, and only subsequently dimerizes. Obviously, the functionalities of Bid and Bax are ultimately different. In contrast to Bax, tBid does not cause membrane thinning (25), and although it can cause ion release from liposomes and form ion channels in planar membranes (53), it cannot by itself form pores large enough for cytochrome c release. However, the steps shown in Fig. 5 could constitute a universal sequence for other Bcl-2 family proteins, such as Bcl-XL or Bad, that switch from a soluble to a membrane-bound state.
Acknowledgments
This work was supported by grants FRN86657 to C.F. and FRN12517 to D.W.A. and B.L. from the Canadian Institutes of Health Research (CIHR).
Footnotes
David W. Andrews’s present address is Sunnybrook Research Institute, Toronto, ON, M4N 3M5, Canada.
Supporting Material
References
- 1.Antonsson B., Martinou J.C. The Bcl-2 protein family. Exp. Cell Res. 2000;256:50–57. doi: 10.1006/excr.2000.4839. [DOI] [PubMed] [Google Scholar]
- 2.Leber B., Lin J., Andrews D.W. Embedded together: the life and death consequences of interaction of the Bcl-2 family with membranes. Apoptosis. 2007;12:897–911. doi: 10.1007/s10495-007-0746-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Certo M., Del Gaizo Moore V., Letai A. Mitochondria primed by death signals determine cellular addiction to antiapoptotic BCL-2 family members. Cancer Cell. 2006;9:351–365. doi: 10.1016/j.ccr.2006.03.027. [DOI] [PubMed] [Google Scholar]
- 4.Chipuk J.E., Fisher J.C., Green D.R. Mechanism of apoptosis induction by inhibition of the anti-apoptotic BCL-2 proteins. Proc. Natl. Acad. Sci. USA. 2008;105:20327–20332. doi: 10.1073/pnas.0808036105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eskes R., Desagher S., Martinou J.C. Bid induces the oligomerization and insertion of Bax into the outer mitochondrial membrane. Mol. Cell. Biol. 2000;20:929–935. doi: 10.1128/mcb.20.3.929-935.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Letai A., Bassik M.C., Korsmeyer S.J. Distinct BH3 domains either sensitize or activate mitochondrial apoptosis, serving as prototype cancer therapeutics. Cancer Cell. 2002;2:183–192. doi: 10.1016/s1535-6108(02)00127-7. [DOI] [PubMed] [Google Scholar]
- 7.Kuwana T., Mackey M.R., Newmeyer D.D. Bid, Bax, and lipids cooperate to form supramolecular openings in the outer mitochondrial membrane. Cell. 2002;111:331–342. doi: 10.1016/s0092-8674(02)01036-x. [DOI] [PubMed] [Google Scholar]
- 8.Terrones O., Antonsson B., Basañez G. Lipidic pore formation by the concerted action of proapoptotic BAX and tBID. J. Biol. Chem. 2004;279:30081–30091. doi: 10.1074/jbc.M313420200. [DOI] [PubMed] [Google Scholar]
- 9.Li H., Zhu H., Yuan J. Cleavage of BID by caspase 8 mediates the mitochondrial damage in the Fas pathway of apoptosis. Cell. 1998;94:491–501. doi: 10.1016/s0092-8674(00)81590-1. [DOI] [PubMed] [Google Scholar]
- 10.Lovell J.F., Billen L.P., Andrews D.W. Membrane binding by tBid initiates an ordered series of events culminating in membrane permeabilization by Bax. Cell. 2008;135:1074–1084. doi: 10.1016/j.cell.2008.11.010. [DOI] [PubMed] [Google Scholar]
- 11.Shamas-Din A., Bindner S., Fradin C. tBid undergoes multiple conformational changes at the membrane required for Bax activation. J. Biol. Chem. 2013;288:22111–22127. doi: 10.1074/jbc.M113.482109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tait S.W., Green D.R. Mitochondria and cell death: outer membrane permeabilization and beyond. Nat. Rev. Mol. Cell Biol. 2010;11:621–632. doi: 10.1038/nrm2952. [DOI] [PubMed] [Google Scholar]
- 13.Billen L.P., Kokoski C.L., Andrews D.W. Bcl-XL inhibits membrane permeabilization by competing with Bax. PLoS Biol. 2008;6:e147. doi: 10.1371/journal.pbio.0060147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Billen L.P., Shamas-Din A., Andrews D.W. Bid: a Bax-like BH3 protein. Oncogene. 2008;27(Suppl 1):S93–S104. doi: 10.1038/onc.2009.47. [DOI] [PubMed] [Google Scholar]
- 15.Petros A.M., Olejniczak E.T., Fesik S.W. Structural biology of the Bcl-2 family of proteins. Biochim. Biophys. Acta. 2004;1644:83–94. doi: 10.1016/j.bbamcr.2003.08.012. [DOI] [PubMed] [Google Scholar]
- 16.McDonnell J.M., Fushman D., Cowburn D. Solution structure of the proapoptotic molecule BID: a structural basis for apoptotic agonists and antagonists. Cell. 1999;96:625–634. doi: 10.1016/s0092-8674(00)80573-5. [DOI] [PubMed] [Google Scholar]
- 17.Shamas-Din A., Brahmbhatt H., Andrews D.W. BH3-only proteins: orchestrators of apoptosis. Biochim. Biophys. Acta. 2011;1813:508–520. doi: 10.1016/j.bbamcr.2010.11.024. [DOI] [PubMed] [Google Scholar]
- 18.Grinberg M., Sarig R., Gross A. tBID homooligomerizes in the mitochondrial membrane to induce apoptosis. J. Biol. Chem. 2002;277:12237–12245. doi: 10.1074/jbc.M104893200. [DOI] [PubMed] [Google Scholar]
- 19.García-Sáez A.J., Ries J., Schwille P. Membrane promotes tBID interaction with BCL(XL) Nat. Struct. Mol. Biol. 2009;16:1178–1185. doi: 10.1038/nsmb.1671. [DOI] [PubMed] [Google Scholar]
- 20.Friaa O., Furukawa M., Shamas-Din A., Leber B., Andrews D.W., Fradin C. Optimizing the acquisition and analysis of confocal images for quantitative single-mobile-particle detection. Chemphyschem. 2013;14:2476–2490. doi: 10.1002/cphc.201201047. [DOI] [PubMed] [Google Scholar]
- 21.Dlugosz P.J., Billen L.P., Andrews D.W. Bcl-2 changes conformation to inhibit Bax oligomerization. EMBO J. 2006;25:2287–2296. doi: 10.1038/sj.emboj.7601126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yethon J.A., Epand R.F., Andrews D.W. Interaction with a membrane surface triggers a reversible conformational change in Bax normally associated with induction of apoptosis. J. Biol. Chem. 2003;278:48935–48941. doi: 10.1074/jbc.M306289200. [DOI] [PubMed] [Google Scholar]
- 23.Horvath S.E., Daum G. Lipids of mitochondria. Prog. Lipid Res. 2013;52:590–614. doi: 10.1016/j.plipres.2013.07.002. [DOI] [PubMed] [Google Scholar]
- 24.van Meer G., Voelker D.R., Feigenson G.W. Membrane lipids: where they are and how they behave. Nat. Rev. Mol. Cell Biol. 2008;9:112–124. doi: 10.1038/nrm2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Satsoura D., Kučerka N., Fradin C. Interaction of the full-length Bax protein with biomimetic mitochondrial liposomes: a small-angle neutron scattering and fluorescence study. Biochim. Biophys. Acta. 2012;1818:384–401. doi: 10.1016/j.bbamem.2011.10.007. [DOI] [PubMed] [Google Scholar]
- 26.Chiantia S., Kahya N., Schwille P. Dehydration damage of domain-exhibiting supported bilayers: an AFM study on the protective effects of disaccharides and other stabilizing substances. Langmuir. 2005;21:6317–6323. doi: 10.1021/la050115m. [DOI] [PubMed] [Google Scholar]
- 27.Oreopoulos J., Yip C.M. Combined scanning probe and total internal reflection fluorescence microscopy. Methods. 2008;46:2–10. doi: 10.1016/j.ymeth.2008.05.011. [DOI] [PubMed] [Google Scholar]
- 28.Satsoura D., Leber B., Fradin C. Circumvention of fluorophore photobleaching in fluorescence fluctuation experiments: a beam scanning approach. ChemPhysChem. 2007;8:834–848. doi: 10.1002/cphc.200600589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kask P., Palo K., Gall K. Fluorescence-intensity distribution analysis and its application in biomolecular detection technology. Proc. Natl. Acad. Sci. USA. 1999;96:13756–13761. doi: 10.1073/pnas.96.24.13756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Henriques R., Lelek M., Mhlanga M.M. QuickPALM: 3D real-time photoactivation nanoscopy image processing in ImageJ. Nat. Methods. 2010;7:339–340. doi: 10.1038/nmeth0510-339. [DOI] [PubMed] [Google Scholar]
- 31.Reviakine I., Brisson A. Formation of supported phospholipid bilayers from unilamellar vesicles investigated by atomic force microscopy. Langmuir. 2000;16:1806–1815. [Google Scholar]
- 32.Richter R.P., Brisson A.R. Following the formation of supported lipid bilayers on mica: a study combining AFM, QCM-D, and ellipsometry. Biophys. J. 2005;88:3422–3433. doi: 10.1529/biophysj.104.053728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sonnleitner A., Schütz G.J., Schmidt T. Free Brownian motion of individual lipid molecules in biomembranes. Biophys. J. 1999;77:2638–2642. doi: 10.1016/S0006-3495(99)77097-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Przybylo M., Sýkora J., Hof M. Lipid diffusion in giant unilamellar vesicles is more than 2 times faster than in supported phospholipid bilayers under identical conditions. Langmuir. 2006;22:9096–9099. doi: 10.1021/la061934p. [DOI] [PubMed] [Google Scholar]
- 35.Chiantia S., Ries J., Kahya N., Schwille P. Combined AFM and two-focus SFCS study of raft-exhibiting model membranes. Chemphyschem. 2006;7:2409–2418. doi: 10.1002/cphc.200600464. [DOI] [PubMed] [Google Scholar]
- 36.Guo L., Har J.Y., Sankaran J., Hong Y., Kannan B., Wohland T. Molecular diffusion measurement in lipid bilayers over wide concentration ranges: a comparative study. Chemphyschem. 2008;9:721–728. doi: 10.1002/cphc.200700611. [DOI] [PubMed] [Google Scholar]
- 37.Korsmeyer S.J., Shutter J.R., Oltvai Z.N. Bcl-2/Bax: a rheostat that regulates an anti-oxidant pathway and cell death. Semin. Cancer Biol. 1993;4:327–332. [PubMed] [Google Scholar]
- 38.Chipuk J.E., Green D.R. How do BCL-2 proteins induce mitochondrial outer membrane permeabilization? Trends Cell Biol. 2008;18:157–164. doi: 10.1016/j.tcb.2008.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chou J.J., Li H., Wagner G. Solution structure of BID, an intracellular amplifier of apoptotic signaling. Cell. 1999;96:615–624. doi: 10.1016/s0092-8674(00)80572-3. [DOI] [PubMed] [Google Scholar]
- 40.Veresov V.G., Davidovskii A.I. Monte Carlo simulations of tBid association with the mitochondrial outer membrane. Eur. Biophys. J. 2007;37:19–33. doi: 10.1007/s00249-007-0149-z. [DOI] [PubMed] [Google Scholar]
- 41.Gambin Y., Lopez-Esparza R., Urbach W. Lateral mobility of proteins in liquid membranes revisited. Proc. Natl. Acad. Sci. USA. 2006;103:2098–2102. doi: 10.1073/pnas.0511026103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gambin Y., Reffay M., Urbach W. Variation of the lateral mobility of transmembrane peptides with hydrophobic mismatch. J. Phys. Chem. B. 2010;114:3559–3566. doi: 10.1021/jp911354y. [DOI] [PubMed] [Google Scholar]
- 43.Oh K.J., Barbuto S., Korsmeyer S.J. Conformational changes in BID, a pro-apoptotic BCL-2 family member, upon membrane binding. A site-directed spin labeling study. J. Biol. Chem. 2005;280:753–767. doi: 10.1074/jbc.M405428200. [DOI] [PubMed] [Google Scholar]
- 44.Lutter M., Fang M., Wang X. Cardiolipin provides specificity for targeting of tBid to mitochondria. Nat. Cell Biol. 2000;2:754–761. doi: 10.1038/35036395. [DOI] [PubMed] [Google Scholar]
- 45.Wang Y., Tjandra N. Structural insights of tBid, the caspase-8-activated Bid, and its BH3 domain. J. Biol. Chem. 2013;288:35840–35851. doi: 10.1074/jbc.M113.503680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Adam G., Delbruck M. Reduction of dimensionality in biological diffusion processes. In: Rich A., Davidson N., editors. Structural Chemistry and Molecular Biology. W. H. Freeman; New York: 1968. pp. 198–215. [Google Scholar]
- 47.McCloskey M.A., Poo M.M. Rates of membrane-associated reactions: reduction of dimensionality revisited. J. Cell Biol. 1986;102:88–96. doi: 10.1083/jcb.102.1.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rozovsky S., Forstner M.B., Groves J.T. Single molecule kinetics of ENTH binding to lipid membranes. J. Phys. Chem. B. 2012;116:5122–5131. doi: 10.1021/jp210045r. [DOI] [PubMed] [Google Scholar]
- 49.Wang K., Yin X.M., Korsmeyer S.J. BID: a novel BH3 domain-only death agonist. Genes Dev. 1996;10:2859–2869. doi: 10.1101/gad.10.22.2859. [DOI] [PubMed] [Google Scholar]
- 50.Grinberg M., Schwarz M., Gross A. Mitochondrial carrier homolog 2 is a target of tBID in cells signaled to die by tumor necrosis factor α. Mol. Cell. Biol. 2005;25:4579–4590. doi: 10.1128/MCB.25.11.4579-4590.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Antonsson B., Montessuit S., Martinou J.C. Bax is present as a high molecular weight oligomer/complex in the mitochondrial membrane of apoptotic cells. J. Biol. Chem. 2001;276:11615–11623. doi: 10.1074/jbc.M010810200. [DOI] [PubMed] [Google Scholar]
- 52.Annis M.G., Soucie E.L., Andrews D.W. Bax forms multispanning monomers that oligomerize to permeabilize membranes during apoptosis. EMBO J. 2005;24:2096–2103. doi: 10.1038/sj.emboj.7600675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schendel S.L., Azimov R., Reed J.C. Ion channel activity of the BH3 only Bcl-2 family member, BID. J. Biol. Chem. 1999;274:21932–21936. doi: 10.1074/jbc.274.31.21932. [DOI] [PubMed] [Google Scholar]
- 54.Petrásek Z., Schwille P. Precise measurement of diffusion coefficients using scanning fluorescence correlation spectroscopy. Biophys. J. 2008;94:1437–1448. doi: 10.1529/biophysj.107.108811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Loman, A., C. B. Muller, …, J. Enderlein. 2008. Absolute and precise measurements of the diffusion of small fluorescent dye molecules across the visible spectrum. Poster. Int. Workshop Single-Mol. Spectrosc. Ultrasens. Anal. Life Sci.,14th, Berlin.
- 56.Abu-Arish A., Kalab P., Fradin C. Spatial distribution and mobility of the Ran GTPase in live interphase cells. Biophys. J. 2009;97:2164–2178. doi: 10.1016/j.bpj.2009.07.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kasai R.S., Suzuki K.G., Kusumi A. Full characterization of GPCR monomer-dimer dynamic equilibrium by single molecule imaging. J. Cell Biol. 2011;192:463–480. doi: 10.1083/jcb.201009128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Czabotar P.E., Westphal D., Colman P.M. Bax crystal structures reveal how BH3 domains activate Bax and nucleate its oligomerization to induce apoptosis. Cell. 2013;152:519–531. doi: 10.1016/j.cell.2012.12.031. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


