Abstract
During replication, the physical state of a virus is controlled by assembly and disassembly processes, when particles are put together and dismantled by cellular cues, respectively. A fundamental question has been how a cell can assemble an infectious virus, and dismantle a virus entering an uninfected cell and thereby trigger a new round of infection. This apparent paradox might be explained by considering that infected and uninfected cells are functionally different, or that assembly and disassembly take place along different cellular pathways. A third possibility is that the physical properties of newly assembled viruses are different from the infection-ready viruses. Recent biophysical experiments measured the stiffness of single Influenza viruses and combined this with biochemical measurements and cell biological assays. Besides inducing the fusogenic state of hemagglutinin, low pH cues softened the virus and precluded aggregation of viral ribonucleoprotein particles with the matrix protein M1. The recent experiments suggest a two-step model for Influenza virus entry and uncoating involving low pH in early and late endosomes, respectively. I conclude with a short outlook into how combined biophysical and cell biological approaches might lead to the identification of new cellular cues controlling viral uncoating and infection.
Introduction
Influenza is a devastating human and animal disease, as indicated by the H1N1 pandemics in 1918 killing 50–100,000,000 people (1), or the H1N1 Swine Influenza pandemics in 2009 (2). The Influenza virus is difficult to combat, because high rates of mutation and shuffling of the genome segments between viruses give raise to new Influenza strains resistant against vaccines and chemical inhibitors.
Influenza viruses belong to the family of orthomyxoviridae, and are grouped into three genera—the Influenza A (IAV), Influenza B (IBV), and Influenza C (ICV) viruses. These viruses differ from each other in many respects. For example, IAV and IBV have eight viral RNA segments and ICV only seven, or the M2 channels from IAV and IBV have little homology or are lacking in ICV (3). IAVs comprise seasonal human Influenza viruses, and a range of subtypes in wild water birds, which are a major natural host for IAV. IAVs are highly transmissible, and cause severe disease in humans, with estimations in the range of several hundred-thousand deaths across the world each year (4).
IAV is an enveloped particle with two glycoproteins, the major hemagglutinin and minor neuraminidase, arranged mostly outside the lipid membrane (for a simplified schematic representation, see Fig. 1 A). The envelope also contains a small disulfide-bonded tetrameric protein M2, which has one trans-membrane domain and occurs in 4–16 tetramers per virion (5,6). M2 particularly conducts protons at acidic pH, but other ions, such as sodium and potassium, can also be transported by M2 (7–10). In acidic endosomes, this leads to proton influx into the lumen of the virus, and enhances infection, presumably by helping the capsid to complete uncoating, as suggested by the use of an antiviral agent, amantadine, that blocks the M2 channel by steric hindrance (11–13). Eight segments of the viral genome are located in the lumen of IAV, and they are in contact with a soluble viral protein, the matrix protein M1. Detergent solubilization and density gradient centrifugation studies suggested that M1 can be in a ribbonlike form or a coil-structure, suggesting that it may have multiple functions in the virion (14). Each of the viral ribonucleoprotein particles (RNPs) contains a single-stranded negative-sense RNA (a template for transcription) helically wrapped around many copies of the nucleoprotein, and one copy each of the polymerase protein complex (15).
Figure 1.
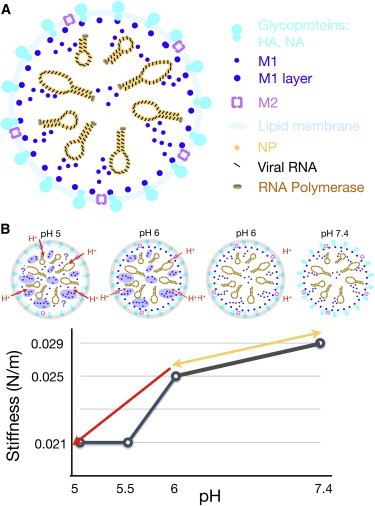
How pH impacts on Influenza virus stiffness. (A) Schematic representation of major structural features of the Influenza A virus, including the lipid envelope with the glycoproteins hemagglutinin (HA) and neuraminidase (NA), the M2 channel, the matrix protein M1, and the viral genome consisting of eight ribonucleoprotein particles made up of the nucleoprotein (NP) and negative-stranded RNA. (B) Two-step pH-dependent softening of IAV. (Upper part of panel) Changes in the virus are represented by four schematic drawings. (Lower part of panel) Simplified stiffness plot as a function of pH (taken from Li et al. (38)). The conversion from the neutral to the pH 6 form of IAV is reversible, and first leads to a softening of the glycoprotein layer (represented by shaded circles in the schematic figure). A small amount of protons will enter into the lumen of the virus through the M2 channel, although this process is thought to be inefficient, because the ion conductance of M2 is generally low at neutral or slightly acidic pH, and there are only a few M2 channels in a single virion. Regardless, protons or other endosomal cues prime the interior of the virus, as indicated by the M1 proteins (blue-shaded) in the viral lumen. This step might occur in early endosomes but it is unknown whether M1 aggregates. As the virus proceeds to more acidic late endosomes, more protons flux into the virus lumen through the acid-gated M2 channel, and, it is thought to lead to disturbance of the M1 layer underneath the viral envelope (38). To see this figure in color, go online.
The physical properties of viruses determine how viral genomes traffic in cells, how they penetrate membranes, or how they are uncoated from the viruses (16–19). Uncoating is required for activation of viral trafficking, transcription, and replication, and at the same time provides signals to the cell for triggering innate immune system responses (20–22). Viruses control the uncoating of their genome in many different ways, and each virus family has probably found a proprietary solution (recently reviewed in Yamauchi and Helenius (23)). Cellular cues include receptors, low pH, osmolytes, proteases, or physical forces by engaging cellular motor proteins (18,24–26).
The simplest forms of viruses, the so-called nonenveloped viruses, lack a lipid envelope. They uncoat their genome from the protein capsid by exposing or releasing proteins at strategic steps in entry, and thereby gain membrane penetration ability with, for example, members of human adenovirus or picornavirus families (27,28). The loss of proteins or the expansion of the capsid, triggered by low endosomal pH, can perforate a rhinovirus to enable genome exit (29,30). Alternatively, the loss of proteins from an adenovirus proceeds in a stepwise manner (31), starting with the shedding of the fibers at the cell surface and the exposure of the membrane-disrupting protein, which is triggered by acto-myosin-mediated motions of viral receptors (32). Further protein loss makes the genome accessible to small solutes independent of low endosomal pH, akin to the perforation of rhinoviruses triggered by low pH (33,34). It is important to note that the infection by adenovirus is independent of low endosomal pH, implying that cues other than pH prepare the virus for releasing the genome when the appropriate subcellular location has been reached, in this case the nucleus (for a review, see Wolfrum and Greber (35)).
Influenza Virus Entry—The Acid Cues
How do cellular cues control Influenza virus uncoating?
Enveloped viruses uncoat by fusing their limiting membrane with a host membrane, typically in an endosome, and thereby shed their outer layer. Virus-endosome fusion is in many instances triggered by low endosomal pH, for example for IAV (36). The endosome step is skipped in acid bypass assays, where the viral envelope is fused with the plasma membrane by lowering the extracellular pH together with adding an inhibitor to block endosomal acidification and thereby precluding the normal infection pathway, as initially shown for Semliki Forest virus (37). It has recently been shown that viral fusion with endosomal membranes is not sufficient, because acid bypass with Influenza A virus strain Panama/2007/99 (H3N2) at pH 5 restored only ∼5% of the normal infection (38). However, if the isolated virus was pretreated with pH 6, the amount of infection more than doubled in the acid bypass experiment. This boost was completely abrogated by amantadine, suggesting that the mechanism involved events in the lumen of the virus.
Physical Properties of Viruses—Viral Mechanics in Entry
To address how chemicals affect viruses, researchers use atomic force microscopy (AFM) measurements, as has recently been done with IAV (38). AFM experimentation with viruses is typically conducted in two steps: Imaging occurs in so-called tapping mode with low indentation force to preserve the integrity of the virus. The second step occurs with higher needle force, and is recorded together with particle deformation. Force indentation profiles bear information about the mechanical properties of the viruses. In particular, because force correlates with indentation until the virus structure ruptures, this gives insight into the elastic behavior of the virus on the solid support, with single particle information. Depending on the thermal fluctuations of the sample and the diameter of the needle tip, this experiment can give information at high spatial resolution at the nanometer range. From such measurements, a spring constant (N/m) can be derived, which is meaningful, because it can reveal strengths and weaknesses of the particle and susceptibility to cellular cues (39,40).
For Influenza virus, a combination of AFM, cryo-EM, and cell biological experiments was recently used reporting spring constants of ∼0.03 N/m (38). This was surprisingly low, somewhat higher than liposomes reconstituted from extracted Influenza virus lipids. The stiffness of Influenza virus was nearly one order of magnitude lower than that previously measured for bacteriophages, herpes virus, adenovirus, hepatitis B virus, parvovirus, murine leukemia virus, or HIV (19,39,41–46). This low stiffness of Influenza virus may be due to flexible contacts of the helical RNPs with the viral envelope, in particular the matrix protein M1. Remarkably, considerable stiffness variation between particles was found, and measurements with ∼100 particles were performed to obtain average particle properties (38). The observed variations may be due to particle size and shape differences, flexibility of viral constituents, or lack of M1. The latter had been suggested by earlier cryo-EM work (47–49).
Interestingly, acidic pH softened the Influenza virus in two distinct steps (see Fig. 1 B and Li et al. (38)). The first step was reversible and occurred at pH 6, the pH of early endosomes. Under these conditions, the glycoproteins softened—a finding compatible with earlier notions that hemagglutinin is less compact at pH 6 compared to neutral pH (50). The second softening step was irreversible and occurred below pH 6, representing conditions in late endosomes or lysosomes triggering the conversion of hemagglutinin to the fusion active state. This irreversible step was, however, independent of the glycoproteins, because it also occurred with bald viruses, which lacked the glycoproteins as a consequence of protease treatment, and the stiffness of the envelope remained constant below pH 6. The irreversible pH step was dependent on amantadine, implicating softening events in the lumen of the virus. Cryo-EM data further suggested that low pH dissociated the M1 layer underneath the viral envelope. This was in agreement with previous cryo-electron tomography studies showing that a 5-min exposure of Influenza virus to pH 4.9 increased the proportion of virions lacking an M1 layer from 10 to 50% (51). One can speculate that a loosely organized protein layer is less stiff than a well-organized layer (52,53).
What does a late penetrating virus gain in early endosomes?
The combination of acid bypass assays, cryo-electron microscopy, and AFM measurements further showed that two pH steps softening IAV are important for infection (38). The priming step in early endosomes occurred at the slightly acidic pH of ∼6. This leads to reversible softening of the viral glycoproteins, apparently without affecting the M1 layer in the viral lumen. It is possible that M1-RNP interactions are weakened if the virus takes a bath at pH 6 (54–56). This priming appears to be important to preclude that M1-RNPs aggregate by immediate exposure to low pH prevalent in late endosomes (48). That M1-RNP interactions need to be dissolved for successful infection has been shown in earlier cell biological experimentations (11,12).
Outlook
Emerging questions from these experiments are the effect of protons and possibly other ions within the Influenza virus: Is it the disruption of the M1 layer and/or the dissociation of the M1 from the RNPs? The latter could occur distant from the M1 layer in the viral lumen. How does the structure of the genome respond to changes of the ionic environment?
Such questions can be answered by biophysical approaches using intact viruses in combination with cell biological assays taking into account that the luminal environment changes along the viral entry pathway (57). It will be important to define the in vitro uncoating conditions as close as possible to the conditions prevalent at the site of virus uncoating in cells. This requires consideration of multiple factors, such as the ionic milieu, pH, proteases, the reductive potential, and the mechanical forces.
The implementation of physical measurements of viruses and cells opens new ways to analyze virus entry into host cells. For example, different levels of acidic pH in the endosomal pathway of Influenza virus exerted different effects on the virus, besides inducing the fusogenic state of hemagglutinin (38). In addition to low pH, other cellular cues from endosomes may also be important for Influenza virus infection, as suggested by the observation that a stepwise acid bypass (depicted in Fig. 1) achieved only ∼14% of the maximal infectivity. One way to hunt for such cues can be to use agents blocking vesicular trafficking (58,59). This would arrest the viruses in early endosomes, and expose them to a defined local environment. Washing in acidic pH together with other ions using, for example, ionophores (60), may then restore viral uncoating and infection to even higher levels than acid bypass from the plasma membrane.
Acknowledgments
The author thanks Dr. Maarit Suomalainen (University of Zurich, Switzerland) and Sarah Stauffer (ETH Zurich, Switzerland) for their valuable comments on the text, and acknowledges support from the Swiss National Science Foundation (under grant No. 31003A-141222/1).
References
- 1.Taubenberger J.K. The origin and virulence of the 1918 “Spanish” Influenza virus. Proc. Am. Philos. Soc. 2006;150:86–112. [PMC free article] [PubMed] [Google Scholar]
- 2.Trifonov V., Khiabanian H., Rabadan R. Geographic dependence, surveillance, and origins of the 2009 Influenza A (H1N1) virus. N. Engl. J. Med. 2009;361:115–119. doi: 10.1056/NEJMp0904572. [DOI] [PubMed] [Google Scholar]
- 3.Palese P., Shaw M.L. Orthomyxoviridae: the viruses and their replication. In: Knipe D.M., Howley P.M., editors. Fields’ Virology. Lippincott Williams & Wilkins; Philadelphia, PA: 2007. pp. 1647–1689. [Google Scholar]
- 4.Thompson W.W., Shay D.K., Fukuda K. Mortality associated with Influenza and respiratory syncytial virus in the United States. JAMA. 2003;289:179–186. doi: 10.1001/jama.289.2.179. [DOI] [PubMed] [Google Scholar]
- 5.Zebedee S.L., Lamb R.A. Influenza A virus M2 protein: monoclonal antibody restriction of virus growth and detection of M2 in virions. J. Virol. 1988;62:2762–2772. doi: 10.1128/jvi.62.8.2762-2772.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Holsinger L.J., Lamb R.A. Influenza virus M2 integral membrane protein is a homotetramer stabilized by formation of disulfide bonds. Virology. 1991;183:32–43. doi: 10.1016/0042-6822(91)90115-r. [DOI] [PubMed] [Google Scholar]
- 7.Pinto L.H., Holsinger L.J., Lamb R.A. Influenza virus M2 protein has ion channel activity. Cell. 1992;69:517–528. doi: 10.1016/0092-8674(92)90452-i. [DOI] [PubMed] [Google Scholar]
- 8.Tang Y., Zaitseva F., Pinto L.H. The gate of the Influenza virus M2 proton channel is formed by a single tryptophan residue. J. Biol. Chem. 2002;277:39880–39886. doi: 10.1074/jbc.M206582200. [DOI] [PubMed] [Google Scholar]
- 9.Leiding T., Wang J., Arsköld S.P. Proton and cation transport activity of the M2 proton channel from Influenza A virus. Proc. Natl. Acad. Sci. USA. 2010;107:15409–15414. doi: 10.1073/pnas.1009997107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shimbo K., Brassard D.L., Pinto L.H. Ion selectivity and activation of the M2 ion channel of Influenza virus. Biophys. J. 1996;70:1335–1346. doi: 10.1016/S0006-3495(96)79690-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Martin K., Helenius A. Nuclear transport of Influenza virus ribonucleoproteins: the viral matrix protein (M1) promotes export and inhibits import. Cell. 1991;67:117–130. doi: 10.1016/0092-8674(91)90576-k. [DOI] [PubMed] [Google Scholar]
- 12.Bui M., Whittaker G., Helenius A. Effect of M1 protein and low pH on nuclear transport of Influenza virus ribonucleoproteins. J. Virol. 1996;70:8391–8401. doi: 10.1128/jvi.70.12.8391-8401.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Takeda M., Pekosz A., Lamb R.A. Influenza a virus M2 ion channel activity is essential for efficient replication in tissue culture. J. Virol. 2002;76:1391–1399. doi: 10.1128/JVI.76.3.1391-1399.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ruigrok R.W., Barge A., Whittaker G.R. Membrane interaction of Influenza virus M1 protein. Virology. 2000;267:289–298. doi: 10.1006/viro.1999.0134. [DOI] [PubMed] [Google Scholar]
- 15.Portela A., Digard P. The Influenza virus nucleoprotein: a multifunctional RNA-binding protein pivotal to virus replication. J. Gen. Virol. 2002;83:723–734. doi: 10.1099/0022-1317-83-4-723. [DOI] [PubMed] [Google Scholar]
- 16.Helenius A. Unpacking the incoming Influenza virus. Cell. 1992;69:577–578. doi: 10.1016/0092-8674(92)90219-3. [DOI] [PubMed] [Google Scholar]
- 17.Greber U.F., Singh I., Helenius A. Mechanisms of virus uncoating. Trends Microbiol. 1994;2:52–56. doi: 10.1016/0966-842x(94)90126-0. [DOI] [PubMed] [Google Scholar]
- 18.Suomalainen M., Greber U.F. Uncoating of non-enveloped viruses. Curr. Opin. Virol. 2013;3:27–33. doi: 10.1016/j.coviro.2012.12.004. [DOI] [PubMed] [Google Scholar]
- 19.Pang H.B., Hevroni L., Rousso I. Virion stiffness regulates immature HIV-1 entry. Retrovirology. 2013;10:4. doi: 10.1186/1742-4690-10-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Greber U.F. Viral trafficking violations in axons: the herpesvirus case. Proc. Natl. Acad. Sci. USA. 2005;102:5639–5640. doi: 10.1073/pnas.0501696102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mercer J., Greber U.F. Virus interactions with endocytic pathways in macrophages and dendritic cells. Trends Microbiol. 2013;21:380–388. doi: 10.1016/j.tim.2013.06.001. [DOI] [PubMed] [Google Scholar]
- 22.Hendrickx R., Stichling N., Greber U.F. Innate immunity to adenovirus. Hum. Gene Ther. 2014;25:265–284. doi: 10.1089/hum.2014.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yamauchi Y., Helenius A. Virus entry at a glance. J. Cell Sci. 2013;126:1289–1295. doi: 10.1242/jcs.119685. [DOI] [PubMed] [Google Scholar]
- 24.Greber U.F. Virus assembly and disassembly: the adenovirus cysteine protease as a trigger factor. Rev. Med. Virol. 1998;8:213–222. doi: 10.1002/(sici)1099-1654(1998100)8:4<213::aid-rmv225>3.0.co;2-w. [DOI] [PubMed] [Google Scholar]
- 25.Cataläo M.J., Gil F., Pimentel M. Diversity in bacterial lysis systems: bacteriophages show the way. FEMS Microbiol. Rev. 2012;37:554–571. doi: 10.1111/1574-6976.12006. [DOI] [PubMed] [Google Scholar]
- 26.Burckhardt C.J., Greber U.F. Redox rescues virus from ER trap. Nat. Cell Biol. 2008;10:9–11. doi: 10.1038/ncb0108-9. [DOI] [PubMed] [Google Scholar]
- 27.Hogle J.M. Poliovirus cell entry: common structural themes in viral cell entry pathways. Annu. Rev. Microbiol. 2002;56:677–702. doi: 10.1146/annurev.micro.56.012302.160757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wiethoff C.M., Wodrich H., Nemerow G.R. Adenovirus protein VI mediates membrane disruption following capsid disassembly. J. Virol. 2005;79:1992–2000. doi: 10.1128/JVI.79.4.1992-2000.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Garriga D., Pickl-Herk A., Verdaguer N. Insights into minor group rhinovirus uncoating: the x-ray structure of the HRV2 empty capsid. PLoS Pathog. 2012;8:e1002473. doi: 10.1371/journal.ppat.1002473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Harutyunyan S., Kumar M., Blaas D. Viral uncoating is directional: exit of the genomic RNA in a common cold virus starts with the poly-(A) tail at the 3′-end. PLoS Pathog. 2013;9:e1003270. doi: 10.1371/journal.ppat.1003270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Greber U.F., Willetts M., Helenius A. Stepwise dismantling of adenovirus 2 during entry into cells. Cell. 1993;75:477–486. doi: 10.1016/0092-8674(93)90382-z. [DOI] [PubMed] [Google Scholar]
- 32.Burckhardt C.J., Suomalainen M., Greber U.F. Drifting motions of the adenovirus receptor CAR and immobile integrins initiate virus uncoating and membrane lytic protein exposure. Cell Host Microbe. 2011;10:105–117. doi: 10.1016/j.chom.2011.07.006. [DOI] [PubMed] [Google Scholar]
- 33.Suomalainen M., Luisoni S., Greber U.F. A direct and versatile assay measuring membrane penetration of adenovirus in single cells. J. Virol. 2013;87:12367–12379. doi: 10.1128/JVI.01833-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang I.H., Suomalainen M., Greber U.F. Tracking viral genomes in host cells at single-molecule resolution. Cell Host Microbe. 2013;14:468–480. doi: 10.1016/j.chom.2013.09.004. [DOI] [PubMed] [Google Scholar]
- 35.Wolfrum N., Greber U.F. Adenovirus signaling in entry. Cell. Microbiol. 2013;15:53–62. doi: 10.1111/cmi.12053. [DOI] [PubMed] [Google Scholar]
- 36.White J., Kartenbeck J., Helenius A. Membrane fusion activity of Influenza virus. EMBO J. 1982;1:217–222. doi: 10.1002/j.1460-2075.1982.tb01150.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Helenius A., Kartenbeck J., Fries E. On the entry of Semliki Forest virus into BHK-21 cells. J. Cell Biol. 1980;84:404–420. doi: 10.1083/jcb.84.2.404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li S., Sieben C., Schaap I.A.T. pH-controlled two-step uncoating of Influenza virus. Biophys. J. 2014;106:1447–1456. doi: 10.1016/j.bpj.2014.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Snijder J., Reddy V.S., Wuite G.J. Integrin and defensin modulate the mechanical properties of adenovirus. J. Virol. 2013;87:2756–2766. doi: 10.1128/JVI.02516-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ortega-Esteban A., Perez-Berna A.J., de Pablo P.J. Monitoring dynamics of human adenovirus disassembly induced by mechanical fatigue. Sci. Rep. 2013;3:1434. doi: 10.1038/srep01434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Evilevitch A., Roos W.H., Wuite G.J. Effects of salts on internal DNA pressure and mechanical properties of phage capsids. J. Mol. Biol. 2011;405:18–23. doi: 10.1016/j.jmb.2010.10.039. [DOI] [PubMed] [Google Scholar]
- 42.Carrasco C., Carreira A., de Pablo P.J. DNA-mediated anisotropic mechanical reinforcement of a virus. Proc. Natl. Acad. Sci. USA. 2006;103:13706–13711. doi: 10.1073/pnas.0601881103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kol N., Shi Y., Rousso I. A stiffness switch in human immunodeficiency virus. Biophys. J. 2007;92:1777–1783. doi: 10.1529/biophysj.106.093914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kol N., Gladnikoff M., Rousso I. Mechanical properties of murine leukemia virus particles: effect of maturation. Biophys. J. 2006;91:767–774. doi: 10.1529/biophysj.105.079657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Roos W.H., Gibbons M.M., Wuite G.J. Squeezing protein shells: how continuum elastic models, molecular dynamics simulations, and experiments coalesce at the nanoscale. Biophys. J. 2010;99:1175–1181. doi: 10.1016/j.bpj.2010.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liashkovich I., Hafezi W., Shahin V. Exceptional mechanical and structural stability of HSV-1 unveiled with fluid atomic force microscopy. J. Cell Sci. 2008;121:2287–2292. doi: 10.1242/jcs.032284. [DOI] [PubMed] [Google Scholar]
- 47.Calder L.J., Wasilewski S., Rosenthal P.B. Structural organization of a filamentous Influenza A virus. Proc. Natl. Acad. Sci. USA. 2010;107:10685–10690. doi: 10.1073/pnas.1002123107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fontana J., Cardone G., Steven A.C. Structural changes in Influenza virus at low pH characterized by cryo-electron tomography. J. Virol. 2012;86:2919–2929. doi: 10.1128/JVI.06698-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Harris A., Cardone G., Steven A.C. Influenza virus pleiomorphy characterized by cryoelectron tomography. Proc. Natl. Acad. Sci. USA. 2006;103:19123–19127. doi: 10.1073/pnas.0607614103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Remeta D.P., Krumbiegel M., Blumenthal R. Acid-induced changes in thermal stability and fusion activity of Influenza hemagglutinin. Biochemistry. 2002;41:2044–2054. doi: 10.1021/bi015614a. [DOI] [PubMed] [Google Scholar]
- 51.Fontana J., Steven A.C. At low pH, Influenza virus matrix protein M1 undergoes a conformational change prior to dissociating from the membrane. J. Virol. 2013;87:5621–5628. doi: 10.1128/JVI.00276-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schaap I.A., Eghiaian F., Veigel C. Effect of envelope proteins on the mechanical properties of Influenza virus. J. Biol. Chem. 2012;287:41078–41088. doi: 10.1074/jbc.M112.412726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mateu M.G. Mechanical properties of viruses analyzed by atomic force microscopy: a virological perspective. Virus Res. 2012;168:1–22. doi: 10.1016/j.virusres.2012.06.008. [DOI] [PubMed] [Google Scholar]
- 54.Baudin F., Petit I., Ruigrok R.W. In vitro dissection of the membrane and RNP binding activities of Influenza virus M1 protein. Virology. 2001;281:102–108. doi: 10.1006/viro.2000.0804. [DOI] [PubMed] [Google Scholar]
- 55.Ye Z., Liu T., Levandowski R.A. Association of Influenza virus matrix protein with ribonucleoproteins. J. Virol. 1999;73:7467–7473. doi: 10.1128/jvi.73.9.7467-7473.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Noda T., Sugita Y., Kawaoka Y. Three-dimensional analysis of ribonucleoprotein complexes in Influenza A virus. Nat. Commun. 2012;3:639. doi: 10.1038/ncomms1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Scott C.C., Gruenberg J. Ion flux and the function of endosomes and lysosomes: pH is just the start: the flux of ions across endosomal membranes influences endosome function not only through regulation of the luminal pH. BioEssays. 2011;33:103–110. doi: 10.1002/bies.201000108. [DOI] [PubMed] [Google Scholar]
- 58.Sieczkarski S.B., Brown H.A., Whittaker G.R. Role of protein kinase C βII in Influenza virus entry via late endosomes. J. Virol. 2003;77:460–469. doi: 10.1128/JVI.77.1.460-469.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gillespie E.J., Ho C.L., Bradley K.A. Selective inhibitor of endosomal trafficking pathways exploited by multiple toxins and viruses. Proc. Natl. Acad. Sci. USA. 2013;110:E4904–E4912. doi: 10.1073/pnas.1302334110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jurgeit A., McDowell R., Greber U.F. Niclosamide is a proton carrier and targets acidic endosomes with broad antiviral effects. PLoS Pathog. 2012;8:e1002976. doi: 10.1371/journal.ppat.1002976. [DOI] [PMC free article] [PubMed] [Google Scholar]


