Abstract
Lipopolysaccharide (LPS) is a unique lipoglycan, with two major physiological roles: 1), as a major structural component of the outer membrane of Gram-negative bacteria and 2), as a highly potent mammalian toxin when released from cells into solution (endotoxin). LPS is an amphiphile that spontaneously inserts into the outer leaflet of lipid bilayers to bury its hydrophobic lipidic domain, leaving the hydrophilic polysaccharide chain exposed to the exterior polar solvent. Divalent cations have long been known to neutralize and stabilize LPS in the outer membrane, whereas LPS in the presence of monovalent cations forms highly mobile negatively-charged aggregates. Yet, much of our understanding of LPS and its interactions with the cell membrane does not take into account its amphiphilic biochemistry and charge polarization. Herein, we report fluorescence microscopy and atomic force microscopy analysis of the interaction between LPS and fluid-phase supported lipid bilayer assemblies (sLBAs), as model membranes. Depending on cation availability, LPS induces three remarkably different effects on simple sLBAs. Net-negative LPS-Na+ leads to the formation of 100-μm-long flexible lipid tubules from surface-associated lipid vesicles and the destabilization of the sLBA resulting in micron-size hole formation. Neutral LPS-Ca2+ gives rise to 100-μm-wide single- or multilamellar planar sheets of lipid and LPS formed from surface-associated lipid vesicles. Our findings have important implications about the physical interactions between LPS and lipids and demonstrate that sLBAs can be useful platforms to study the interactions of amphiphilic virulence factors with cell membranes. Additionally, our study supports the general phenomenon that lipids with highly charged or bulky headgroups can promote highly curved membrane architectures due to electrostatic and/or steric repulsions.
Introduction
Lipopolysaccharide (LPS) is of major medical importance, firstly, because it forms the outer surface of many pathogenic bacteria and, secondly, because LPS is a highly potent toxin when released from cells. The outer membrane of Gram-negative bacteria is a highly asymmetric complex lipid bilayer, comprised of an inner leaflet of various common phospholipids and the outer leaflet of the unique glycolipid, LPS (1–3). An Escherichia coli cell contains several million LPS molecules, covering 75% of the outer membrane surface, with the remaining area being occupied by proteins (1). LPS has several important functions for the bacteria including acting as a permeability barrier between the cell and the exterior, maintaining structural stability of the membrane, and functioning as a protective barrier against foreign particles (antimicrobial peptides, drugs, toxic heavy metals, salts, and enzymes) (4,5). LPS is critical to many pathogens’ ability to cause disease and is released from the outer membrane of the bacterium during infection. LPS, historically known as “endotoxin”, can cause overactivation of the immune system in toxic shock syndrome at μg/kg LPS/body mass ratios (6,7). Thus, LPS is an excellent target for diagnostics, vaccines, and treatment strategies against these pathogens (3,7).
The structure of LPS has been studied for many years (8). LPS is an amphiphilic molecule comprised of a hydrophobic domain named lipid A, covalently linked to a hydrophilic polysaccharide chain that extends away from the cell. The lipid A component contains six saturated fatty acid chains linked to a phosphate-substituted disaccharide (2,9). This is linked to the relatively conserved core-oligosaccharide and the variable O-polysaccharide of 0–50 oligosaccharide repeat units, dependent upon the particular bacterial species and strain. LPS preparations are heterogeneous, and a mixture of LPS structures are found in each preparation with partial modifications depending on growth conditions (3). Each LPS molecule has multiple negative charges from phosphate and acid groups in the lipid A and core-polysaccharide. In this study, we use LPS from E. coli serotype O111:B4 (structure shown in Fig. 1, see details in Raetz and Whitfield (3), Amor et al. (10), Kenne et al. (11), and Peterson et al. (12)). There are at least six negatively charged groups per LPS and a range of 1–18 O-polysaccharide repeat units. In the bacterial outer membrane, divalent cations such as Mg2+ and Ca2+ are essential to neutralize this negative charge, allowing cross-linking between LPS molecules, which maintains an effective barrier to drugs and other damaging molecules (2,13–16). Chelation of divalent cations leads to increased permeability to drugs, LPS release, and rapid disintegration of the outer membrane (14,17). In comparison to the common phospholipid DOPC (1,2-dioleoyl-sn-glycero-3-phosphocholine), LPS contains a much larger hydrophobic domain containing saturated fatty acids. In addition, LPS has a large hydrophilic, negatively-charged headgroup, whereas DOPC has a small zwitterionic headgroup (Fig. 1).
Figure 1.
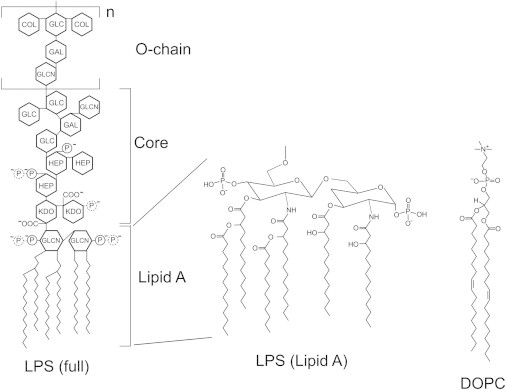
Comparison of the chemical structure of lipopolysaccharide from E. coli serotype O111 and the phospholipid DOPC (1,2-dioleoyl-sn-glycero-3-phosphocholine). Partial covalent modifications, which may result in additional phosphate groups, are shown (dotted lines) and are dependent on growth conditions and other factors. Number of repeat units in the O-chain, n, ranges from 1 to 18. COL, colitose (3,6-dideoxy-L-xylo-hexose); GAL, galactopyranose; GLC, glucopyranose; GLCN, 2-amino-2-deoxyglucopyranose; HEP, L-glycero-D-manno-heptopyranose; KDO, 3-deoxy-D-manno-oct-2-ulopyranosonic acid; P, phosphate.
LPS has previously been shown to form very different structures depending on its local ionic environment, observed in cell-free LPS extracts and reconstituted LPS-lipid membranes. LPS aggregates exposed to Na+ are not fully neutralized and have a net negative charge (16), resulting in formation of long tubular structures that were converted into bilayers by exposure to Ca2+ (18). Multiple studies have shown that divalent cations reduced the mobility of LPS aggregates and increased the rigidity of LPS bilayers, decreasing their permeability (18–24). Additionally, divalent cations have led to formation of highly ordered, stacked multilamellar LPS structures (20,21). The physiological activity of LPS also depends on its ion associations, and LPS-isolates rich in Na+ and K+ cations are significantly more active as endotoxins than those with Mg2+ or Ca2+ (25). The more rigid, multilamellar LPS formations induced by divalent cations were determined to be inactive as opposed to more freely mobile aggregates in monovalent cation-rich environments (26). These studies together indicate that the effect of cations on LPS structure and aggregate formation is an important factor that should be considered when studying the manifestation of endotoxic shock.
In studies where hybrid membranes were prepared by combining LPS with different lipids, LPS distribution and incorporation (27) and the membrane fluidity (28) were found to depend on lipid composition. The structure of LPS from different strains or species can be dramatically different. Studies have observed that LPS structural changes, self-association, and toxicity all depend on LPS subtype (8,22,29,30). In the human body, LPS interacts with intermediary factors of the human immune system including LPS binding protein, which has been shown to affect LPS interaction with membranes (31–33). This study focuses on the direct interaction of E. coli O111 LPS aggregates with fluid-phase DOPC membranes as a model for LPS-membrane interactions.
Although many studies have investigated the structures resulting from reconstitution of LPS-lipid membranes, the dynamic interaction of LPS with membrane architectures remains poorly characterized. In giant unilamellar vesicles (GUVs) formed from LPS and lipids, LPS was found to segregate into gel-like domains, showing that LPS lateral rearrangement and self-association of LPS molecules can occur within the lipid bilayer (29). Soluble LPS has been shown to insert into preformed lipid GUVs and cause shape changes and vesicle fission (34). Supported lipid bilayer assemblies (sLBAs) have been used for many years as models for biological phospholipid bilayers, as planar membrane systems with lateral lipid mobility (35–37). To our knowledge, there has not been direct visualization of the dynamic effects of free LPS aggregates on an sLBA, as a simple platform for evaluation of the interaction of amphiphilic toxins with membrane architectures. Herein, we evaluate the direct interaction of LPS with sLBAs using a combination of fluorescence microscopy and atomic force microscopy (AFM), powerful tools for investigating membrane organization (38–41). Our experiments were performed with concentrations of LPS ranging from 5 to 500 μg/mL, highly comparable to the lethal doses for various species ranging from 1 to 200 mg/kg body weight (6). Our findings could have significant ramifications on our understanding of the action of the important toxin LPS, and have general implications that should be considered for all amphiphilic pathogenic molecules.
Materials and Methods
Materials
All materials were used as received without further purification. Organic solvents were HPLC grade (Thermo Fisher Scientific, Waltham, MA). PBS (phosphate-buffered saline), HEPES, EDTA, NaCl, and CaCl2 were also purchased from Thermo Fisher Scientific. All aqueous buffers were prepared using 18 MΩ•cm H2O (Barnstead Nanopure filter; Thermo Fisher Scientific) and then passed through 0.22-μm filter membranes (Millipore, Billerica, MA). All lipids and fluorescence dyes were purchased in dry powdered form. DOPC (1,2-dioleoyl-sn-glycero-3-phosphocholine) was purchased from Avanti Polar Lipids (Alabaster, AL). Lipid-based dyes used for doping the lipid membranes were as follows: C5-BODIPY FL HPC (2-(4,4-difluoro-5,7-dimethyl-4-bora-3a,4a-diaza-s-indacene-3-pentanoyl)-1-hexadecanoyl-sn-glycero-3-phosphocholine) or Texas Red DHPE (Texas Red 1,2-dihexadecanoyl-sn-glycero-3-phosphoethanolamine, triethylammonium salt) (Molecular Probes, Eugene, OR). The standard LPS used in this study unless otherwise mentioned was from E. coli serotype O111:B4 (phenol extract); a FITC-conjugate of LPS from E. coli serotype O111:B4 was used for direct tracking (Sigma-Aldrich, St. Louis, MO).
Liposome and supported lipid bilayer formation
The standard DOPC liposome preparation contained 99.5% (mol/mol) DOPC and 0.5% C5-BODIPY-HPC or 0.5% Texas Red DHPE. Lipids and lipid dyes in chloroform were mixed in the desired molar ratios, dried overnight under vacuum, and rehydrated in buffer solution. The lipid suspension was subjected to three freeze-thaw cycles followed by probe sonication for 10 min in an ice bath to form small liposomes (41). Hydrophilic glass coverslips were used as substrates (cleaned with Piranha solution of 3:1 H2SO4/30% H2O2). Hydrophobic ultrathin adhesive imaging spacers (0.12-mm depth, 9-mm diameter) were attached to substrates to create small wells to confine a droplet of buffer (Electron Microscopy Sciences, Hatfield, PA), for an open sample setup to allow multiple buffer exchanges and top-down access for AFM. sLBAs were formed by deposition of liposomes onto the substrate. After 20-min incubation at room temperature, the sLBA was washed by exchanging the buffer solution 10 times to remove excess liposomes, although significant numbers of associated lipid vesicles evidently remain.
Treatment of surfaces with LPS
LPS was handled as per manufacturer’s guidelines, dissolved into buffer at 5 mg/mL, stored in silanized glass vials at 4°C and, before each usage, stocks were vortexed and bath-sonicated (15 min) at room temperature to homogenize immediately before sLBA treatment. sLBAs of DOPC were prepared and washed with buffer. The DOPC sLBA was then treated with LPS and analyzed with microscopy as described in the Results. For experiments testing LPS in the presence of monovalent cations, the buffer used was PBS (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 10 mM KH2PO4, pH 7.4), whereas for testing LPS in the presence of divalent cations, Ca2+ buffer was used (150 mM CaCl2 and 20 mM HEPES, pH 7.5). For experiments testing the effects of cation concentration, buffers containing 20 mM TRIS-HCl (pH 7.5) and 10–900 mM NaCl or 10–900 mM CaCl2 were used. Control experiments confirmed that buffering with phosphates, HEPES and TRIS, were equivalent for our studies (data not shown). Control experiments found that common small molecule contaminants had no observable effect on our sLBAs (RNA from baker’s yeast, bovine serum albumin, fetal bovine serum; data not shown) compared to the effects of LPS under the same conditions.
Microscopy of LPS-treated sLBAs
Lipid bilayers were imaged with laser scanning confocal fluorescence microscopy (LSCM), epifluorescence microscopy, total internal reflection fluorescence microscopy (TIRFM), and AFM. Samples were kept hydrated and never allowed to dry either during preparation or analysis.
LSCM used an FV-1000 inverted optical microscope (Olympus, Tokyo, Japan) equipped with multichannel photomultiplier detectors, operated in photon-counting mode (very low background noise, 0–2 counts), acquiring 512 × 512 pixel images unless otherwise stated. A 40× air objective (NA = 0.95) was used for the majority of experiments and a 60× water-immersion objective (NA = 1.20) was used for higher resolution confirmation. Excitation was provided by a multi-line Ar laser (488 nm, for BODIPY), a HeNe laser (543 nm, for Texas Red), or a diode laser (635 nm, for AlexaFluor 647). Appropriate high-performance band-pass emission filters were used (505–525 nm for BODIPY and FITC; 655–755 nm for Texas Red and AlexaFluor 647). Fluorescence recovery after photobleaching (FRAP) was performed using the manufacturer’s provided software.
Epifluorescence was performed using an IX-81 inverted optical microscope (Olympus) with a model No. C11440-22C charge-coupled device camera (Hamamatsu, Hamamatsu City, Japan). A 100× oil-immersion objective (NA = 1.40) was used. Excitation was provided by a 200W metal-halide lamp and a FITC filter set and appropriate neutral density (ND) filters were used. Images and movies were acquired at 1024 × 1024 pixels using the software.
TIRFM was performed using an IX-71 inverted optical microscope (Olympus) equipped with a model No. C7780-20C charge-coupled device camera (1344 × 1032 pixels; Hamamatsu). A 100× oil-immersion TIRFM objective (NA = 1.45) was used. Excitation was provided by a 488-nm Ar ion laser and a green filter set and appropriate ND filters were used.
AFM was performed using an MFP-3D-SA system (Asylum Research, Santa Barbara, CA), equipped with a closed loop XY scanner and an all-digital ARC2 Controller (Asylum Research). All imaging was performed under fluid using SNL probes (Bruker AFM Probes, Camarillo, CA) with a sharpened Si tip on a triangular SiN cantilever (k ∼ 0.12 N/m). High quality topographs were generally acquired at 512 × 512 pixels and 1 Hz scan speed. Images were processed using IGOR PRO-based software (WaveMetrics, Tigard, OR).
Results
sLBAs were formed on glass substrates by deposition of small unilamellar vesicles comprised of 99.5% DOPC and 0.5% C5-BODIPY FL HPC (41). AFM confirmed that the bilayers were continuous and defect-free over many microns with a few small protrusions (Fig. 2 A). LSCM showed a relatively homogenous fluorescence with small numbers of higher-intensity dots (Fig. 2 B, before LPS). The fluidity of DOPC sLBAs at room temperature was confirmed by FRAP (data not shown). The higher intensity fluorescence dots and protrusions in AFM data represent lipid vesicles that remain loosely associated with the surface even after rinsing with buffers. Lipid vesicles have an average size of ∼50 nm in solution (see Fig. S1 in the Supporting Material); the largest may be resolved as surface-associated vesicles in LSCM. Sequential images show that surface-associated vesicles are highly mobile and independent of the underlying sLBA (see Movie S1 in the Supporting Material).
Figure 2.
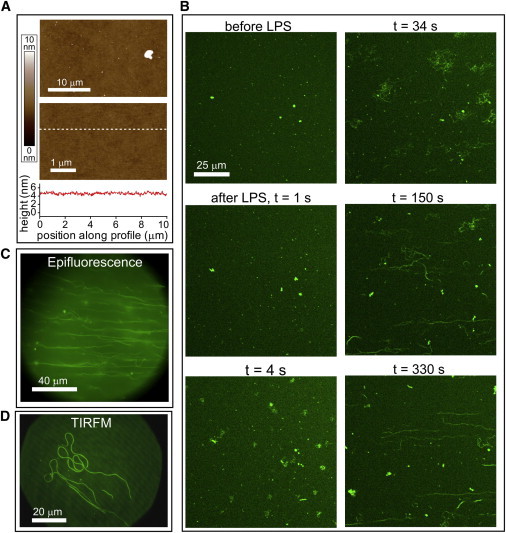
Lipid tubule formation induced by LPS in PBS. (A) AFM topographs showing a DOPC sLBA in PBS at low and high magnification. A height profile across the lower image (dashed white line) shows a relatively flat surface. (B) LSCM of the DOPC sLBA (doped with 0.5% green fluorescent lipids) in PBS before and after addition of 100 μg/mL LPS. Sequential images are shown at selected time periods after addition of the LPS. (C) Representative epifluorescence microscopy image of lipid tubules. (D) Representative TIRFM image showing long tubules. The background of green fluorescence suggested a homogenous lipid bilayer (note: 45° periodic noise is an optical artifact that should be ignored). To see this figure in color, go online.
Lipid tubule formation is induced by LPS in PBS
To test the effect of soluble LPS on our model lipid bilayer system, experiments were first performed in PBS, containing monovalent cations (Na+). LSCM fluorescence images (time-lapse series) obtained after addition of 100 μg/mL LPS in PBS to an sLBA are shown in Fig. 2 B (and see Movie S2). The surface-associated vesicles that originally exist were observed to split into multiple vesicles, leading to an increase in mobility and the formation of fluorescent strandlike structures. Initially disordered webs of strands stretch out over a few minutes into elongated strands that we term lipid tubules, analogous to the tubules characterized by other studies of membrane-perturbing molecules (42,43). The lipid tubules retain a point of association to the membrane surface and often extend up to 100 μm in length and span many microns above the surface, as shown by epifluorescence microscopy (Fig. 2 C). Movies of sequential epifluorescence images confirmed the high degree of mobility of lipid tubules (see Movie S3). The lipid tubules stretch away from the boundary of the substrate, to orient in a radial organization during the course of the experiment, suggesting that fluid flow plays a role in tubule extension (see Fig. S2).
AFM imaging of LPS-treated sLBAs revealed a flat surface (data not shown) and no lipid tubules were observed, as may be expected because of the relatively poor stability and high mobility of these formations. Membrane-inserted LPS was not detected by AFM, which could be due to the instability of the protruding polysaccharide chain that can be pushed aside by the AFM probe (44) or due to the transient nature of LPS insertion into lipid bilayers, previously observed in GUVs (34). TIRFM, which has higher signal/noise and restricted penetration depth of excitation, was used in an attempt to observe any subtle height variations in the sLBA, such as bubbles, ripples, or other perturbations that may not have been observed with other optical techniques. TIRFM images (Fig. 2 D) did not show any evidence of disruptions to the underlying lipid bilayer, but both lipid tubules and surface-associated vesicles were prominent. The lack of any observable changes to the underlying lipid bilayer by both AFM and TIRFM indicate that it is either intact or that any perturbations cannot be observed by these strategies.
Hole formation in the sLBA after LPS treatment in PBS and washing
After treatment of the sLBA with LPS in PBS, the DOPC bilayer was washed to remove any residual unassociated LPS. This resulted in the unexpected formation of voids lacking fluorescence of ∼1–5 μm in width (Fig. 3 A). Intensity profiles from single photon counting (SPC) LSCM data show that there is essentially zero fluorescence at the center of these voids (Fig. 3 B). The fluorescence intensity of membrane areas other than voids was roughly similar before and after LPS treatment, suggesting a continuous membrane surface. It should be noted that these voids were stable, with no detectable change in shape or size over the course of 80 min at the ∼250-nm resolution of our microscope (see Fig. S3). To test whether the voids of fluorescence were actually physical holes in the lipid bilayer, AFM was performed on an LPS-treated sample in PBS. The DOPC bilayer appeared as a relatively smooth film with many holes of ∼5-nm depth via AFM, consistent with expected height of an sLBA (Fig. 3 E). The lateral size of holes varied from <1 μm up to 5 μm in width. High-resolution topographs (Fig. 3 F) demonstrated that holes are essentially empty with only small amounts of debris visible, which correlated with the SPC fluorescence data. TIRFM (Fig. 3 C) images were likewise consistent with LSCM and AFM. FRAP showed that lipids had lateral mobility within the remaining membrane, with holes unperturbed (Fig. 3 D). These observations support the notion of static holes within an otherwise continuous fluid lipid bilayer.
Figure 3.
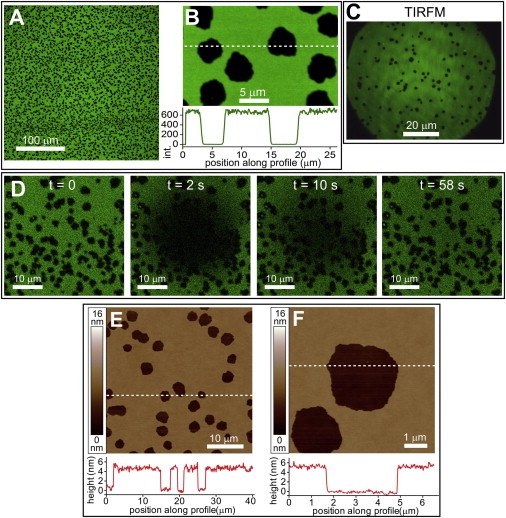
Holes in sLBAs after LPS treatment and washing. (A) LSCM fluorescence images showing DOPC sLBAs after treatment with 100 μg/mL LPS in PBS followed by washing the surface (PBS, 10 changes). (B) Higher-magnification LSCM image. A profile of the fluorescence intensity (below) shows the SPC counts along a line drawn across the image (white dashed line). (C) A representative TIRFM image of a similar sample. (D) FRAP experiment from the sample in panel A. A circular region was photobleached and then sequential images acquired to show the lateral diffusion of fluorescent lipids. (E) AFM topograph showing accurate width and depth of holes induced by LPS, similar sample to panel A. Height profiles (below, red lines) show the height data (across white dashed lines) in the image, chosen to show the depth of holes in the lipid bilayer. (F) Higher magnification topograph from the field of holes in panel E. To see this figure in color, go online.
Hole formation was found to depend on both LPS concentration and incubation time. To test concentration effects, sLBAs were treated with different concentrations of LPS for a standard 20 min and then rinsed. Above 20 μg/mL, which is higher than the critical micelle concentration (CMC) of LPS (10–14 μg/mL) (45,46), hole formation increased with LPS concentration (Fig. 4, A–C). Holes were not initially detected using LPS at concentrations ≤20 μg/mL. To test incubation time effects, sLBAs were treated with a standard LPS concentration and then rinsed with buffer after different time periods (Fig. 4, D–F). At sub-CMC concentration (5 μg/mL), the number and size of holes increased with LPS incubation time from 20 to 180 min as the numbers of surface-associated vesicles decreased (related to their increased conversion to tubules and removal after rinsing). Thus, hole formation can occur above or below the CMC of LPS; however, more time is necessary for hole formation at sub-CMC concentrations.
Figure 4.
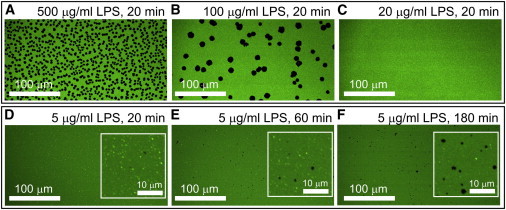
LPS concentration and time dependence on hole formation. LSCM fluorescence images of a DOPC sLBA treated for 20 min with LPS at varying concentrations of LPS in PBS: (A) 500 μg/mL, (B) 100 μg/mL, or (C) 20 μg/mL, and then washed with PBS, showing decreasing numbers of holes with LPS concentration. Parallel samples were treated with 5 μg/mL LPS in PBS for (D) 20 min, (E) 60 min, or (F) 180 min, and then washed and imaged immediately. More holes are observed with increased incubation time. These images were acquired at higher pixel density (4096 × 4096) to resolve small holes. (D–F, inset) Digitally magnified areas of these images showing small holes more clearly. To see this figure in color, go online.
Although AFM showed that holes in the DOPC bilayer were mostly empty, we could not rule out whether small amounts of LPS were present but undetected by AFM. To directly detect the presence of LPS, an FITC-labeled LPS was used with a DOPC sLBA doped with Texas Red DHPE lipid dye (see Fig. S4). A fluid lipid bilayer was observed with low background in the FITC channel. Addition of FITC-LPS (100–500 μg/mL) resulted in a significant FITC signal increase. Unfortunately, we were unable to discriminate FITC-LPS in solution (or in lipid tubules) from that interacting with the surface, due to the overwhelming signal. After surface rinsing holes were observed, but there was no enhanced FITC fluorescence in holes relative to background counts. This suggests that little or no LPS remained associated with the lipid bilayer or in solution after rinsing.
LPS in Ca2+ buffer induces lamellar sheet formation instead of lipid tubules
The action of LPS on lipid membranes under different cation conditions was tested using LPS in Ca2+ buffer (divalent cations, see Materials and Methods). Time-lapse LSCM fluorescence images were acquired during treatment of DOPC sLBAs with 100 μg/mL LPS in Ca2+ buffer (Fig. 5 A, and see Movie S4). Within seconds, small patches of fluorescence were observed that slowly grew in size over the course of minutes. SPC fluorescence intensity profiles from LSCM data show that these LPS-induced membranes had fluorescence intensity of roughly double that of the normal lipid bilayers (Fig. 5 B), suggesting a second lipid-containing bilayer stacked on top of the original sLBA. Overlapping sheets with fluorescence intensity at multiples of the original lipid bilayer were observed (Fig. 5 C), indicating that LPS can induce formation of multiple lipid-LPS membranes stacked on top of each other, in the presence of Ca2+. FRAP experiments revealed slightly reduced lateral lipid mobility in the newly-formed lamellar sheets and underlying sLBA compared to a DOPC sLBA before LPS-Ca2+ treatment (see Fig. S5, A–C). Recovery of fluorescence indicative of multilayers after almost complete photobleaching of stacked membranes suggests that lipids can exchange from the underlying sLBA into the lipid-LPS stacks (see Fig. S5, D–E). Multilamellar membranes were not observed by AFM; instead, a smooth surface expected to represent the original lipid bilayer was found (data not shown), implying that these membranes were too unstable for AFM imaging. LSCM fluorescence images after washing the surface with fresh Ca2+ buffer showed the apparent removal of LPS-induced lamellar membranes (Fig. 5 D), confirming their lack of stability.
Figure 5.
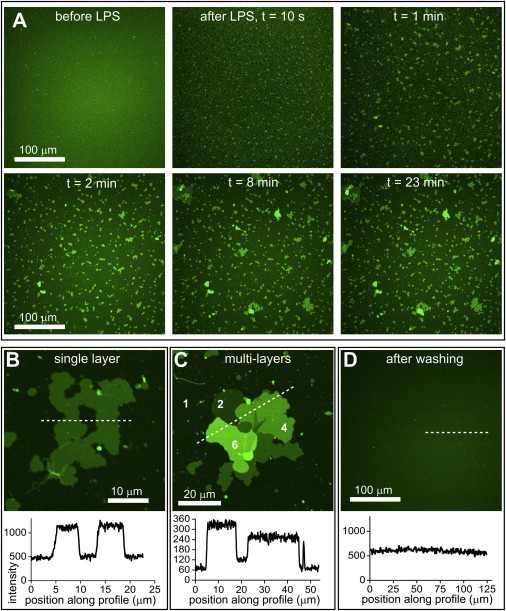
LPS in Ca2+ buffer causes formation and growth of multilamellar stacks. (A) LSCM of the DOPC sLBA in Ca2+ buffer before and after addition of 100 μg/mL LPS in Ca2+ buffer. Sequential images are shown from selected time points after addition of the LPS. Fluorescent patches are observed in images immediately after LPS addition (10 s) and continue to grow in size over the following minutes (1–23 min). (B) Image at high magnification showing a patch of contiguous fluorescence of approximately double the intensity of the DOPC bilayer. (C) Image at a high magnification showing a fluorescent patch with multiple distinct step-changes in the intensity. Numbers (2), (4), and (6) indicate expected stacked bilayers with multiples of intensity of a single bilayer (1). (D) Fluorescence image after washing the LPS-Ca2+-treated surface with 10 changes of Ca2+ buffer. (Note: images in panel C acquired with lower exposure settings than in panels B and D, hence, lower fluorescence intensity.) To see this figure in color, go online.
Formation of lipid-LPS-Ca2+ membranes was dependent on the concentration of LPS. Patches formed using 5 μg/mL LPS in Ca2+ buffer were smaller, fewer in number and less stable, appearing to disintegrate over time (see Fig. S6), relative to the larger, more stable patches formed at higher concentrations. This LPS concentration dependence confirms the direct relationship between LPS (Ca2+) and sheet formation.
Concentration of cations also modified the effect of LPS, as observed when sLBAs were treated with 100 μg/mL LPS in buffers at a range of NaCl or CaCl2 concentrations (see Fig. S7). Concentrations from 10 to 300 mM NaCl all produced lipid tubules, whereas at and above 450 mM NaCl both lipid tubules and potential lamellar sheets were observed (see Fig. S7 A). Holes were always observed with NaCl buffers after rinsing the surface. In contrast, all concentrations of CaCl2 from 10 to 900 mM produced lipid sheets, which were removed without hole formation by rinsing the surface (see Fig. S7 B). Lipid sheet size increased with CaCl2 concentration, and the largest sheets were >100 μm in width (see Fig. S7 C).
Discussion
Considerations of the model membrane system
sLBAs are model membranes that are relatively stable, robust, and relevant to biological systems. Previous studies have demonstrated the ability of a single component sLBA to form highly curved structures, including curved lipid caps and vesicle budding (41). In this study, we report that LPS, a biological toxin of significant concern, can cause three remarkably different deformations by insertion into the controlled environment of a model lipid membrane. LPS insertion into membranes, as observed in previous studies (34), may be driven by LPS in aggregates in a polar solvent that transfer into a more thermodynamically favorable environment as the large hydrophobic lipid A tail of LPS becomes buried in the hydrophobic core of lipid bilayer. LPS insertion occurs into both surface-associated vesicles and the solid-supported LBA. Once inserted, the effect of LPS varies due to differences in membrane curvature, interactions with the support, and the local ionic environment.
The first two effects (lipid tubules and holes in the sLBA) were generated by exposure of DOPC sLBAs to LPS in the presence of (<450 mM) monovalent cations (LPS-Na+). The third effect (lamellar membranes) was observed by exposing the same DOPC system to LPS in the presence of divalent cations (LPS-Ca2+) or very high (≥450 mM) monovalent cation concentration. It is clear that ionic strength and valency influence both the structure of LPS, and its interaction with lipidic architectures. Hundreds of publications have investigated the nature of different membrane deformations induced by biological or synthetic nanoparticles, including theoretical simulations (47,48). We focus on how LPS could induce such changes, discussing examples of similar membrane rearrangements.
Membrane effects induced by LPS-Na+
Treatment of sLBAs with LPS-Na+ induced the formation of highly mobile, fluorescent lipid-based tubules (Fig. 2; and see Movie S2 and Movie S3). Tubules are known to form when there is a high degree of local membrane curvature and an abundant supply of lipids (42,43). sLBAs can have multiple surface-associated liposomes, observed as brighter spots in fluorescence images and protrusions in AFM topography, even after washing the surface. We established the presence of surface-associated lipid vesicles on our sLBAs in Fig. 2 A (and see Fig. S1 and Movie S1). We conclude that the surface-associated lipid vesicles and any remaining solution-based vesicles are the source of lipids in formation of tubules, along with LPS, based on the following evidence:
-
1.
In some images, we can observe apparent tubule nucleation from surface-associated liposomes (Fig. 2 B; and see Movie S2).
-
2.
If multiple cycles of LPS treatment are performed, fewer tubules are formed each time, as surface-associated vesicles are used up (see Fig. S8 A).
We can estimate the amount of lipids required for an average tubule by calculating the outer surface area of a tubule (modeling as a cylinder) and equating this to the surface area of multiple liposomes (modeling as spheres). A tubule of 10 μm in length and 25 nm in diameter (as previously reported for LPS tubes (18)) would require ∼100 vesicles of 50 nm diameter. This could represent, for example, 50 DOPC lipid vesicles, and 50 LPS aggregates, a reasonable quantity considering that LSCM may resolve only the larger surface-associated vesicles. LPS appears to promote fusion of multiple lipid-LPS vesicles, leading to the observed growth of tubular structures. We postulate that the destabilizing inserting/excising effect of LPS (34) and its natural propensity to form tubules on its own (18) could cause nanoscale membrane defects that increase the exposure of lipids to the external environment, promoting further lipid-lipid associations and vesicle fusion, however, further studies are needed to confirm this. Fluid flow due to convection currents within an open droplet, as described by studies of the “coffee-ring effect” (49–51), may then direct the stretching out of these tubules observed in LSCM (see Fig. S2). In contrast, tubules observed by TIRFM in which a closed-box sample enclosure was employed, appeared to rest onto the sLBA surface (Fig. 2 D), possibly due to a reduction in fluid flow.
Upon rinsing the surface of LPS-Na+-treated sLBA with fresh buffer, holes devoid of lipids were observed in a concentration- and time-dependent manner (Figs. 3 and 4). In addition to the removal of lipid tubules, these observations indicate that LPS accumulates in the sLBA over time. Apart from holes, the remaining lipid appeared to be in the form of a normal sLBA with high lateral mobility of lipids and a flat, continuous membrane. This evidence, along with the fact that LPS is not found to be associated with the sLBA after hole formation (see Fig. S4), suggests that LPS clusters into large domains that are then dislodged from the surface during the rinsing process. If LPS induces membrane curvature in surface-associated liposomes resulting in tubules, it is logical that LPS could insert and disrupt the sLBA, although the LPS-treated sLBA appears flat by microscopy. The likely explanation is that LPS causes minor curvature in sLBAs below the detection limit of our instruments, and that we only observe the result of this destabilization when holes are formed after rinsing the surface. Subtle, LPS-induced membrane curvature could lead to separation of the lipid bilayer and the solid support by only a few nanometers, which would not be resolved by optical measurements. LPS insertion into lipid bilayers may be highly transient, as previously reported in Alam and Yamazaki (34) and Stoica et al. (44), which would hinder detection of membrane deformations by AFM. The interaction of the sLBA and the solid support could also limit curvature and cause a strained system as compared to the relatively unrestricted surface-associated liposomes. Based on these arguments, we conclude that LPS induces subtle membrane curvature in sLBAs, which leads to localized delamination sufficient to form holes upon washing the surface.
We expect that tubules and holes are different manifestations of similar effects of LPS-Na+ acting on different starting materials, either surface-associated liposomes or an sLBA. Both require:
-
1.
LPS insertion into the outer leaflet of lipid membranes,
-
2.
LPS self-association or clustering, and
-
3.
LPS induction of membrane curvature.
We will briefly describe other studies in which one or more of these effects are observed, supporting our conclusions. Spontaneous insertion of LPS from solution into the lipid membranes has been previously observed in GUVs, whereupon it appeared to induce subtle, localized curvature resulting in shape changes from spherical to pear-shaped or pearls-on-a-string vesicles (34). Other studies have shown that short-length LPS can self-associate to form gel-like domains (29). Although we are using full-length LPS, for which these authors did not observe phase segregation, self-association of LPS in our sLBA system may be transient or on smaller scales. LPS has been observed to form highly curved structures depending on its ionic environment. Exposure of native LPS to Na+ resulted in formation of long, tubular LPS-based structures of 9–18 nm diameter, as observed by electron microscopy (18). These could be related to the membrane curvature and lipid tubules formed in our experiments with LPS in the Na+-containing buffer PBS. Each LPS molecule contains at least six negatively-charged groups, phosphates and carboxylates (12) (Fig. 1), which typically cannot be fully neutralized by monovalent cations due to electrostatic repulsion of individual ions, resulting in LPS with a net-negative charge (16). The authors concluded that incomplete charge neutralization leads to electrostatic repulsion between LPS chains, causing a high degree of curvature and formation of LPS tubes. We extend these previous findings to show that not only does LPS-Na+ form tubules in isolation but it can also induce curvature in preformed sLBAs. In our sLBA system, attractive hydrophobic self-associations of saturated fatty acids in the lipid A domains of LPS, in contrast to unsaturated DOPC lipids, and size/shape mismatch may drive phase segregation of LPS away from DOPC lipids, outweighing the electrostatic repulsion that would otherwise drive negatively-charged LPS apart into a maximally separated configuration. Then, the negative charge-charge repulsion of the membrane extrinsic hydrophilic domains of nearby LPS would induce membrane curvature.
We may compare our findings of membrane curvature induced by proteins in both natural and artificial systems. In nature, lipid tubules and vesicle budding is found in multiple specific situations, e.g., endocytosis, exocytosis, phagocytosis, and endoplasmic reticulum- and cytoskeleton- associated protein trafficking (52–54). In each case, specific proteins are targeted to a local area of the membrane leading to either the budding of vesicles or the protrusion of long tubules from the membrane. Common mechanisms involve insertion of amphiphilic protein helices and wedge-shaped proteins into the cellular lipid membranes (55–59), which could be related to LPS insertion. Lipid tubules have been induced in vitro from lipid GUVs by promoting protein crowding at a localized point on the vesicle surface; tubule growth was observed to occur in real time from the point of protein binding (42,43). Relevant to our study is the finding that any generic protein was sufficient to induce tubule formation so long as it leads to the buildup of a sufficient high density of protein packing. This lead to the conclusion that steric repulsion between any bulky particles packed at the surface has the potential to cause severe membrane bending. Crowding of LPS could have an analogous effect due to the bulky polysaccharide chain; however, it seems that electrostatic, rather than steric repulsive effects, dominate in the case of LPS.
Perforation of lipid membranes by LPS was suggested by previous indirect evidence of electrical resistance of lipid membranes stretched across an aperture where treatment with LPS (110–720 μg/mL) led to a decreased resistance and eventual collapse (60). Holes and other lesions can be formed in lipid bilayers by various small molecules, including pore-forming protein toxins (61), highly charged synthetic nanoparticles (62–66), and polycationic polymers (67–71). Hole formation in each case is dependent upon the specific interactions between the membrane and the disruptive molecule of interest. It appears that LPS belongs to this list of membrane-disruptive molecules.
Membrane effects induced by LPS-Ca2+
LPS is known to change its structure, aggregation state, and mobility depending on the availability and concentration of monovalent and divalent cations (18–24). In our experiments, we conclude that LPS-Ca2+ inserts into surface-adsorbed liposomes and causes these LPS-lipid assemblies to fuse, resulting in very different structures in comparison to LPS-Na+ of similar concentrations. Whereas LPS-Na+ induces membrane curvature, LPS-Ca2+ induces planar membrane formation (Fig. 5). We find that surface-associated and solution-based vesicles are the source of lipids along with LPS for planar sheet formation, analogous to lipid tubule formation, with evidence provided by multiple cycles of LPS (Ca2+) treatment (see Fig. S8 B). Furthermore, increasing Ca2+ concentration (10–900 mM) results in increasing the extent of multilamellar formation (see Fig. S7 B). In contrast, concentrations of ≥450 mM Na+ are required for LPS to induce even small lamellar sheets, which occur in addition to tubules and holes (see Fig. S7 A), suggesting that LPS induces planar membranes only at very high Na+, and even then, not to the extent as those observed with Ca2+. Although we were unable to verify the topography of these structures by AFM due to their seemingly unstable nature, we are able to infer their multilamellar membrane-like nature from fluorescence data. The slight decreased lateral lipid mobility in planar lamellar membranes compared to a normal DOPC sLBA (see Fig. S5) suggests that LPS-Ca2+ within the lipid bilayers impedes lipid diffusion or increases the rigidity of the membrane. Although the precise structure of stacked membranes is uncertain, we did find they associated closely enough to the underlying sLBA for lipid exchange to occur. Previous spectroscopy and electron microscopy studies found that divalent cations reduce the molecular mobility of LPS within aggregates and cause LPS to reorganize into stacked multibilayers (18,20,21). Our findings are congruent with these studies, and suggest that not only does LPS change its own organization due to Ca2+, it also induces lipids to rearrange with it.
It is instructive to look at the natural environment of LPS in the outer membrane of bacteria. LPS is found in the outer leaflet of a lipid bilayer forming a selectively permeable barrier between the cell and the exterior (4,5). The outer membrane is relatively flat, when compared with small lipid vesicles, with a gentle curvature over many hundreds of nanometers to micrometers. Studies have shown that the outer membrane is enriched in divalent cations relative to the cytoplasmic membrane (16,18) and that divalent cations are essential for outer membrane stability (2,17). The prevailing view from these studies is that Mg2+ or Ca2+ neutralizes the negative charge of LPS where Na+ cannot at physiological concentrations. This neutralization allows the self-association of LPS where otherwise it would be electrostatically unfavorable, and LPS-LPS bridging and linkages to transmembrane proteins stabilize the membrane (72). Our findings agree with this consensus and further demonstrate that LPS can spontaneously insert into lipid membranes and induce self-assembly into an outer-membrane-like structure in the presence of Ca2+ at low concentration (10 mM), whereas much higher Na+ (450 mM) is required for similar effects. Divalent ions are known to be significantly more effective at screening electrostatic interactions at lower concentrations when compared to similar solutions of simple monovalent ions (73), and our results confirm that this applies to the neutralization of LPS, visualizing the biological importance of divalent cations.
Understanding LPS-sLBA interaction
We have demonstrated three very different rearrangements of a simple, single component lipid system, all caused by one membrane-inserting amphiphile in different local environments. A schematic of LPS-induced rearrangements of lipid membranes dependent on monovalent or divalent cations is shown in Fig. 6. The thermodynamically favorable insertion of LPS into the lipid membranes and clustering leads to a high density of LPS, at which point the net charge of the extrinsic polysaccharide portion of LPS appears to determine whether this causes curved or planar structures. The net-negative LPS-Na+ leads to electrostatic charge repulsion between adjacent LPS and induces membrane curvature (Fig. 6 A). Surface-associated lipid vesicles merge and the high membrane curvature drives formation and elongation of lipid tubules (Fig. 6, (1)). In supported lipid bilayers, LPS-induced curvature disrupts the lipid bilayer’s interaction with the solid support, causing unstable delaminated regions that can be excised from the remainder of the lipid bilayer by washing the surface, leading to hole formation (Fig. 6 (2)). In contrast, even low concentrations of Ca2+ promote planar self-associations of LPS (Fig. 6 B). Fusion of surface-associated lipid vesicles in this case leads to growth of planar lamellar sheets of lipid and LPS on top of the sLBA surface (Fig. 6, (3)). To form multilayers, we postulate that mobile lipid-LPS particles deposit on top of the first lipid-LPS planar sheet, leading to growth of a second layer and potentially further layers, held together by interlayer interactions between LPS. The ability of LPS to switch from curvature-inducing to planar-sheet formation simply from changing its net charge by adjusting the buffering cations indicates that LPS-induced membrane curvature is due to electrostatic repulsion rather than steric repulsive effects (42,43).
Figure 6.
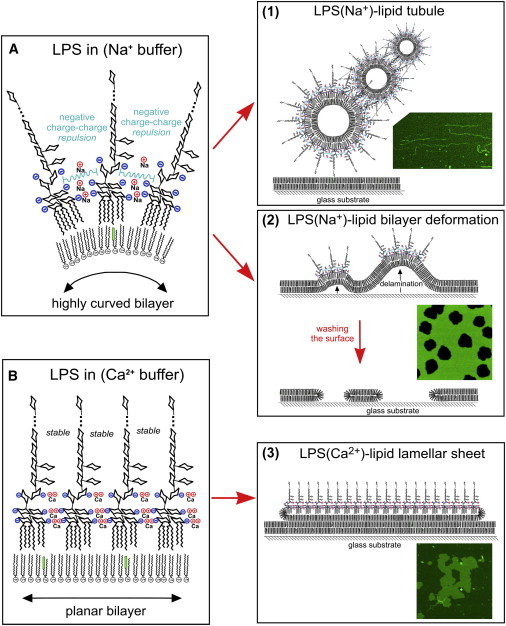
Schematic of the mechanism of LPS-induced lipid bilayer deformation. See text for description. LPS is represented by a simplified molecular structure showing the hydrophobic domain with six fatty acid tails linked to core sugar units and the extended O-chain. Sugar units are represented by their cyclic rings. Note that, for clarity, side groups are not displayed and the polysaccharide chain is greatly shortened, represented by (…). Normal phospholipids are represented by their two fatty acid tails linked to a headgroup (green boxes represent BODIPY dye). Negatively-charged groups of LPS are represented by their charge symbols (blue). Cations that associate with LPS are represented by their elemental symbol and single or double charge (in red). Electrostatic repulsion is shown (Cyan lines). To see this figure in color, go online.
Previous studies with another amphiphilic pathogenic molecule, lipoarabinomannan, detected insertion into lipid membranes without any disruption (74). Lipoarabinomannan and LPS are both amphiphilic virulence factors that each interact with similar factors during infection of a mammalian host (TLR2, TLR4, HDL) (3). Structurally, they share similarities, both with hydrophobic fatty acid tails and a membrane extrinsic domain, but they evidently interact very differently with sLBAs. Whereas lipoarabinomannan inserts passively without destabilizing the membrane (74), LPS causes major disruptions, as shown by this study. Thus, each membrane-inserting amphiphile may cause a very different effect, depending on its unique physical properties.
Finally, we consider three examples of other complex lipids that have been reported to induce membrane curvature:
-
1.
Studies on poly(ethylene glycol)-derivatized-lipids (PEG-lipids) at low concentration within fluid lipid bilayers in hydrogels (75,76) are informative because of similarities between LPS and PEG-lipids (negatively-charged headgroup, bulky hydrophilic domain). PEG-lipids were found to phase-segregate into domains that stabilize regions of high curvature based on steric and electrostatic repulsions (76).
-
2.
Gangliosides, lipids with a single negative charge and a bulky aromatic headgroup, have also been reported to induce formation of tubules and pearls in DOPC GUVs (77).
-
3.
Multivalent cationic lipids (MVLs) have been shown to cause formation of narrow tubules and pearling instabilities in DOPC lipid vesicles (78–80). In these studies, the authors hypothesized that increased membrane tension due to electrostatic repulsion between the highly-positively charged lipid headgroups leads to phase segregation whereby curved regions become enriched in and stabilized by MVLs. In common with our findings on LPS, screening of MVL charges by increased salt concentrations lead to a transition from tubular to multilamellar stacked membranes (79).
It is remarkable that both cationic (MVLs) and anionic lipids (gangliosides, PEG-lipids, LPS) can induce similar effects, bolstering suggestions of a general phenomenon (78) that lipids with highly-charged (or very bulky) headgroups have the potential to cause dramatic membrane curvature and reorganizations, dependent on repulsive effects: electrostatic, steric, or a combination of both.
Conclusions
We have demonstrated that LPS, a biologically important molecule, causes dynamic rearrangements of DOPC lipid bilayers dependent upon cation availability, indicating potential driving forces behind physiological effects. It was not the purpose of this study to investigate physiological effects of LPS, which are influenced by a myriad of immunological and other factors in the human body; however, it is our hope that the work presented will facilitate the future design of experimental systems to investigate the role of this complex toxin on the host cells. The continuum of effects observed suggests an ability to tune the membrane deformation by adjusting conditions and components. Further variations may exist if one used different lipid mixtures (e.g., charged or gel-phase), LPS from different bacteria, or alternative cations. Our study supports the general notion that highly curved membrane architectures can be generated by clustering of membrane amphiphiles that have an effectively conical shape, due to charged or bulky headgroups, causing electrostatic and/or steric repulsions.
Acknowledgments
The authors thank Dr. Nathan F. Bouxsein (Sandia National Laboratory) and Dr. Aaron Collins (Los Alamos National Laboratory) for helpful discussions.
This work was performed, in part, at the Center for Integrated Nanotechnologies, an Office of Science User Facility operated for the United States Department of Energy’s Office of Science. Los Alamos National Laboratory, an affirmative action equal opportunity employer, is operated by Los Alamos National Security, LLC, for the National Nuclear Security Administration of the United States Department of Energy under contract No. DE-AC52-06NA25396. Work by P.G.A. and K.L.S. was supported by Photosynthetic Antenna Research Center, an Energy Frontier Research Center funded by the United States Department of Energy, Office of Science, Basic Energy Sciences under Award No. DE-SC0001035. H.M. and L.L. were supported by Agriculture and Food Research Initiative Competitive Grant No. 2012-68003-30155 from the United States Department of Agriculture’s National Institute of Food and Agriculture.
Supporting Material
References
- 1.Nikaido H., Vaara M. Molecular basis of bacterial outer membrane permeability. Microbiol. Rev. 1985;49:1–32. doi: 10.1128/mr.49.1.1-32.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Raetz C.R.H. Bacterial endotoxins: extraordinary lipids that activate eucaryotic signal transduction. J. Bacteriol. 1993;175:5745–5753. doi: 10.1128/jb.175.18.5745-5753.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Raetz C.R.H., Whitfield C. Lipopolysaccharide endotoxins. Annu. Rev. Biochem. 2002;71:635–700. doi: 10.1146/annurev.biochem.71.110601.135414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nikaido H. Molecular basis of bacterial outer membrane permeability revisited. Microbiol. Mol. Biol. Rev. 2003;67:593–656. doi: 10.1128/MMBR.67.4.593-656.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Delcour A.H. Outer membrane permeability and antibiotic resistance. Biochim. Biophys. Acta. 2009;1794:808–816. doi: 10.1016/j.bbapap.2008.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berczi I., Bertók L., Bereznai T. Comparative studies on the toxicity of Escherichia coli lipopolysaccharide endotoxin in various animal species. Can. J. Microbiol. 1966;12:1070–1071. doi: 10.1139/m66-143. [DOI] [PubMed] [Google Scholar]
- 7.Cohen J. The immunopathogenesis of sepsis. Nature. 2002;420:885–891. doi: 10.1038/nature01326. [DOI] [PubMed] [Google Scholar]
- 8.Erridge C., Bennett-Guerrero E., Poxton I.R. Structure and function of lipopolysaccharides. Microbes Infect. 2002;4:837–851. doi: 10.1016/s1286-4579(02)01604-0. [DOI] [PubMed] [Google Scholar]
- 9.Rietschel E.T., Kirikae T., Brade H. Bacterial endotoxin: molecular relationships of structure to activity and function. FASEB J. 1994;8:217–225. doi: 10.1096/fasebj.8.2.8119492. [DOI] [PubMed] [Google Scholar]
- 10.Amor K., Heinrichs D.E., Whitfield C. Distribution of core oligosaccharide types in lipopolysaccharides from Escherichia coli. Infect. Immun. 2000;68:1116–1124. doi: 10.1128/iai.68.3.1116-1124.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kenne L., Lindberg B., Griffith D.W. Structural studies of the O-antigens from Salmonella greenside and Salmonella adelaide. Carbohydr. Res. 1983;111:289–296. doi: 10.1016/0008-6215(83)88313-x. [DOI] [PubMed] [Google Scholar]
- 12.Peterson A.A., Haug A., McGroarty E.J. Physical properties of short- and long-O-antigen-containing fractions of lipopolysaccharide from Escherichia coli 0111:B4. J. Bacteriol. 1986;165:116–122. doi: 10.1128/jb.165.1.116-122.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shroll R.M., Straatsma T.P. Molecular structure of the outer bacterial membrane of Pseudomonas aeruginosa via classical simulation. Biopolymers. 2002;65:395–407. doi: 10.1002/bip.10279. [DOI] [PubMed] [Google Scholar]
- 14.Hancock R.E.W. The bacterial outer membrane as a drug barrier. Trends Microbiol. 1997;5:37–42. doi: 10.1016/S0966-842X(97)81773-8. [DOI] [PubMed] [Google Scholar]
- 15.Kotra L.P., Golemi D., Mobashery S. Dynamics of the lipopolysaccharide assembly on the surface of Escherichia coli. J. Am. Chem. Soc. 1999;121:8707–8711. [Google Scholar]
- 16.Coughlin R.T., Tonsager S., McGroarty E.J. Quantitation of metal cations bound to membranes and extracted lipopolysaccharide of Escherichia coli. Biochemistry. 1983;22:2002–2007. doi: 10.1021/bi00277a041. [DOI] [PubMed] [Google Scholar]
- 17.Amro N.A., Kotra L.P., Liu G.-y. High-resolution atomic force microscopy studies of the Escherichia coli outer membrane: structural basis for permeability. Langmuir. 2000;16:2789–2796. [Google Scholar]
- 18.Coughlin R.T., Haug A., McGroarty E.J. Physical properties of defined lipopolysaccharide salts. Biochemistry. 1983;22:2007–2013. doi: 10.1021/bi00277a042. [DOI] [PubMed] [Google Scholar]
- 19.Brandenburg K., Koch M.H.J., Seydel U. Phase diagram of lipid A from Salmonella minnesota and Escherichia coli rough mutant lipopolysaccharide. J. Struct. Biol. 1990;105:11–21. doi: 10.1016/1047-8477(90)90093-r. [DOI] [PubMed] [Google Scholar]
- 20.Naumann D., Schultz C., Labischinski H. New insights into the phase behavior of a complex anionic amphiphile: architecture and dynamics of bacterial deep rough lipopolysaccharide membranes as seen by FTIR, x-ray, and molecular modeling techniques. J. Mol. Struct. 1989;214:213–246. [Google Scholar]
- 21.van Alphen L., Verkleij A., Lugtenberg B. 31P Nuclear magnetic resonance and freeze-fracture electron microscopy studies on Escherichia coli. II. Lipopolysaccharide and lipopolysaccharide-phospholipid complexes. Biochim. Biophys. Acta. Biomembr. 1980;597:502–517. doi: 10.1016/0005-2736(80)90223-0. [DOI] [PubMed] [Google Scholar]
- 22.Snyder D.S., McIntosh T.J. The lipopolysaccharide barrier: correlation of antibiotic susceptibility with antibiotic permeability and fluorescent probe binding kinetics. Biochemistry. 2000;39:11777–11787. doi: 10.1021/bi000810n. [DOI] [PubMed] [Google Scholar]
- 23.Kučerka N., Papp-Szabo E., Katsaras J. Effect of cations on the structure of bilayers formed by lipopolysaccharides isolated from Pseudomonas aeruginosa PAO1. J. Phys. Chem. B. 2008;112:8057–8062. doi: 10.1021/jp8027963. [DOI] [PubMed] [Google Scholar]
- 24.Jeworrek C., Evers F., Winter R. Effects of specific versus nonspecific ionic interactions on the structure and lateral organization of lipopolysaccharides. Biophys. J. 2011;100:2169–2177. doi: 10.1016/j.bpj.2011.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Garidel P., Rappolt M., Brandenburg K. Divalent cations affect chain mobility and aggregate structure of lipopolysaccharide from Salmonella minnesota reflected in a decrease of its biological activity. Biochim. Biophys. Acta. Biomembr. 2005;1715:122–131. doi: 10.1016/j.bbamem.2005.07.013. [DOI] [PubMed] [Google Scholar]
- 26.Schromm A.B., Brandenburg K., Seydel U. Biological activities of lipopolysaccharides are determined by the shape of their lipid A portion. Eur. J. Biochem. 2000;267:2008–2013. doi: 10.1046/j.1432-1327.2000.01204.x. [DOI] [PubMed] [Google Scholar]
- 27.Nomura K., Inaba T., Kusumoto S. Interaction of lipopolysaccharide and phospholipid in mixed membranes: solid-state 31P-NMR spectroscopic and microscopic investigations. Biophys. J. 2008;95:1226–1238. doi: 10.1529/biophysj.108.131706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cañadas O., Keough K.M.W., Casals C. Bacterial lipopolysaccharide promotes destabilization of lung surfactant-like films. Biophys. J. 2011;100:108–116. doi: 10.1016/j.bpj.2010.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kubiak J., Brewer J., Bagatolli L.A. Lipid lateral organization on giant unilamellar vesicles containing lipopolysaccharides. Biophys. J. 2011;100:978–986. doi: 10.1016/j.bpj.2011.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tong J., McIntosh T.J. Structure of supported bilayers composed of lipopolysaccharides and bacterial phospholipids: raft formation and implications for bacterial resistance. Biophys. J. 2004;86:3759–3771. doi: 10.1529/biophysj.103.037507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roes S., Mumm F., Gutsmann T. Localization of the lipopolysaccharide-binding protein in phospholipid membranes by atomic force microscopy. J. Biol. Chem. 2006;281:2757–2763. doi: 10.1074/jbc.M507634200. [DOI] [PubMed] [Google Scholar]
- 32.Schromm A.B., Brandenburg K., Seydel U. The charge of endotoxin molecules influences their conformation and IL-6-inducing capacity. J. Immunol. 1998;161:5464–5471. [PubMed] [Google Scholar]
- 33.Gutsmann T., Müller M., Seydel U. Dual role of lipopolysaccharide (LPS)-binding protein in neutralization of LPS and enhancement of LPS-induced activation of mononuclear cells. Infect. Immun. 2001;69:6942–6950. doi: 10.1128/IAI.69.11.6942-6950.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Alam J.M., Yamazaki M. Spontaneous insertion of lipopolysaccharide into lipid membranes from aqueous solution. Chem. Phys. Lipids. 2011;164:166–174. doi: 10.1016/j.chemphyslip.2010.12.008. [DOI] [PubMed] [Google Scholar]
- 35.Castellana E.T., Cremer P.S. Solid supported lipid bilayers: from biophysical studies to sensor design. Surf. Sci. Rep. 2006;61:429–444. doi: 10.1016/j.surfrep.2006.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Johnson S.J., Bayerl T.M., Sackmann E. Structure of an adsorbed dimyristoylphosphatidylcholine bilayer measured with specular reflection of neutrons. Biophys. J. 1991;59:289–294. doi: 10.1016/S0006-3495(91)82222-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tamm L.K., McConnell H.M. Supported phospholipid bilayers. Biophys. J. 1985;47:105–113. doi: 10.1016/S0006-3495(85)83882-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Adams P.G., Hunter C.N. Adaptation of intracytoplasmic membranes to altered light intensity in Rhodobacter sphaeroides. Biochim. Biophys. Acta. Bioenerg. 2012;1817:1616–1627. doi: 10.1016/j.bbabio.2012.05.013. [DOI] [PubMed] [Google Scholar]
- 39.Adams P.G., Mothersole D.J., Hunter C.N. Monomeric RC-LH1 core complexes retard LH2 assembly and intracytoplasmic membrane formation in PufX-minus mutants of Rhodobacter sphaeroides. Biochim. Biophys. Acta. Bioenerg. 2011;1807:1044–1055. doi: 10.1016/j.bbabio.2011.05.019. [DOI] [PubMed] [Google Scholar]
- 40.Montaño G.A., Adams P.G., Goodwin P.M. Scanning probe microscopy of nanocomposite membranes and dynamic organization. Adv. Funct. Mater. 2013;23:2576–2591. [Google Scholar]
- 41.Goertz M.P., Goyal N., Bunker B.C. Lipid bilayer reorganization under extreme pH conditions. Langmuir. 2011;27:5481–5491. doi: 10.1021/la2001305. [DOI] [PubMed] [Google Scholar]
- 42.Stachowiak J.C., Schmid E.M., Hayden C.C. Membrane bending by protein-protein crowding. Nat. Cell Biol. 2012;14:944–949. doi: 10.1038/ncb2561. [DOI] [PubMed] [Google Scholar]
- 43.Stachowiak J.C., Hayden C.C., Sasaki D.Y. Steric confinement of proteins on lipid membranes can drive curvature and tubulation. Proc. Natl. Acad. Sci. USA. 2010;107:7781–7786. doi: 10.1073/pnas.0913306107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stoica O., Tuanyok A., Beveridge T.J. Elasticity of membrane vesicles isolated from Pseudomonas aeruginosa. Langmuir. 2003;19:10916–10924. [Google Scholar]
- 45.Bergstrand A., Svanberg C., Nydén M. Aggregation behavior and size of lipopolysaccharide from Escherichia coli O55:B5. Colloids Surf. B Biointerfaces. 2006;53:9–14. doi: 10.1016/j.colsurfb.2006.06.007. [DOI] [PubMed] [Google Scholar]
- 46.Santos N.C., Silva A.C., Saldanha C. Evaluation of lipopolysaccharide aggregation by light scattering spectroscopy. ChemBioChem. 2003;4:96–100. doi: 10.1002/cbic.200390020. [DOI] [PubMed] [Google Scholar]
- 47.van Lehn R.C., Alexander-Katz A. Penetration of lipid bilayers by nanoparticles with environmentally-responsive surfaces: simulations and theory. Soft Matter. 2011;7:11392–11404. [Google Scholar]
- 48.Makarucha A.J., Todorova N., Yarovsky I. Nanomaterials in biological environment: a review of computer modeling studies. Eur. Biophys. J. 2011;40:103–115. doi: 10.1007/s00249-010-0651-6. [DOI] [PubMed] [Google Scholar]
- 49.Weon B.M., Je J.H. Capillary force repels coffee-ring effect. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 2010;82:015305. doi: 10.1103/PhysRevE.82.015305. [DOI] [PubMed] [Google Scholar]
- 50.Fang X., Li B., Rafailovich M.H. Drying of DNA droplets. Langmuir. 2006;22:6308–6312. doi: 10.1021/la060479u. [DOI] [PubMed] [Google Scholar]
- 51.Deegan R.D., Bakajin O., Witten T.A. Capillary flow as the cause of ring stains from dried liquid drops. Nature. 1997;389:827–829. [Google Scholar]
- 52.Aderem A., Underhill D.M. Mechanisms of phagocytosis in macrophages. Annu. Rev. Immunol. 1999;17:593–623. doi: 10.1146/annurev.immunol.17.1.593. [DOI] [PubMed] [Google Scholar]
- 53.Conner S.D., Schmid S.L. Regulated portals of entry into the cell. Nature. 2003;422:37–44. doi: 10.1038/nature01451. [DOI] [PubMed] [Google Scholar]
- 54.Brownhill K., Wood L., Allan V. Molecular motors and the Golgi complex: staying put and moving through. Semin. Cell Dev. Biol. 2009;20:784–792. doi: 10.1016/j.semcdb.2009.03.019. [DOI] [PubMed] [Google Scholar]
- 55.Antonny B. Membrane deformation by protein coats. Curr. Opin. Cell Biol. 2006;18:386–394. doi: 10.1016/j.ceb.2006.06.003. [DOI] [PubMed] [Google Scholar]
- 56.Farsad K., De Camilli P. Mechanisms of membrane deformation. Curr. Opin. Cell Biol. 2003;15:372–381. doi: 10.1016/s0955-0674(03)00073-5. [DOI] [PubMed] [Google Scholar]
- 57.Campelo F., McMahon H.T., Kozlov M.M. The hydrophobic insertion mechanism of membrane curvature generation by proteins. Biophys. J. 2008;95:2325–2339. doi: 10.1529/biophysj.108.133173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Doherty G.J., McMahon H.T. Mechanisms of endocytosis. Annu. Rev. Biochem. 2009;78:857–902. doi: 10.1146/annurev.biochem.78.081307.110540. [DOI] [PubMed] [Google Scholar]
- 59.Shibata Y., Voeltz G.K., Rapoport T.A. Rough sheets and smooth tubules. Cell. 2006;126:435–439. doi: 10.1016/j.cell.2006.07.019. [DOI] [PubMed] [Google Scholar]
- 60.Schuster B.G., Palmer R.F., Aronson R.S. The effect of endotoxin on thin lipid bilayer membranes. J. Membr. Biol. 1970;3:67–72. doi: 10.1007/BF01868007. [DOI] [PubMed] [Google Scholar]
- 61.Czajkowsky D.M., Iwamoto H., Shao Z. The vacuolating toxin from Helicobacter pylori forms hexameric pores in lipid bilayers at low pH. Proc. Natl. Acad. Sci. USA. 1999;96:2001–2006. doi: 10.1073/pnas.96.5.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Goodman C.M., McCusker C.D., Rotello V.M. Toxicity of gold nanoparticles functionalized with cationic and anionic side chains. Bioconjug. Chem. 2004;15:897–900. doi: 10.1021/bc049951i. [DOI] [PubMed] [Google Scholar]
- 63.Xiao X., Montaño G.A., Brozik S.M. Lipid bilayer templated gold nanoparticles nanoring formation using zirconium ion coordination chemistry. Langmuir. 2011;27:9484–9489. doi: 10.1021/la2014754. [DOI] [PubMed] [Google Scholar]
- 64.Xiao X., Montaño G.A., Brozik S.M. Surface charge dependent nanoparticle disruption and deposition of lipid bilayer assemblies. Langmuir. 2012;28:17396–17403. doi: 10.1021/la303300b. [DOI] [PubMed] [Google Scholar]
- 65.Arvizo R.R., Miranda O.R., Mukherjee P. Effect of nanoparticle surface charge at the plasma membrane and beyond. Nano Lett. 2010;10:2543–2548. doi: 10.1021/nl101140t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chen J., Hessler J.A., Orr B.G. Cationic nanoparticles induce nanoscale disruption in living cell plasma membranes. J. Phys. Chem. B. 2009;113:11179–11185. doi: 10.1021/jp9033936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hong S., Bielinska A.U., Banaszak Holl M.M. Interaction of poly(amidoamine) dendrimers with supported lipid bilayers and cells: hole formation and the relation to transport. Bioconjug. Chem. 2004;15:774–782. doi: 10.1021/bc049962b. [DOI] [PubMed] [Google Scholar]
- 68.Hong S., Leroueil P.R., Banaszak Holl M.M. Interaction of polycationic polymers with supported lipid bilayers and cells: nanoscale hole formation and enhanced membrane permeability. Bioconjug. Chem. 2006;17:728–734. doi: 10.1021/bc060077y. [DOI] [PubMed] [Google Scholar]
- 69.Leroueil P.R., Berry S.A., Holl M.M.B. Wide varieties of cationic nanoparticles induce defects in supported lipid bilayers. Nano Lett. 2008;8:420–424. doi: 10.1021/nl0722929. [DOI] [PubMed] [Google Scholar]
- 70.Leroueil P.R., Hong S., Banaszak Holl M.M. Nanoparticle interaction with biological membranes: does nanotechnology present a Janus face? Acc. Chem. Res. 2007;40:335–342. doi: 10.1021/ar600012y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mecke A., Majoros I.J., Orr B.G. Lipid bilayer disruption by polycationic polymers: the roles of size and chemical functional group. Langmuir. 2005;21:10348–10354. doi: 10.1021/la050629l. [DOI] [PubMed] [Google Scholar]
- 72.van Alphen L., Verkleij A., Lugtenberg B. Architecture of the outer membrane of Escherichia coli. III. Protein-lipopolysaccharide complexes in intramembranous particles. J. Bacteriol. 1978;134:1089–1098. doi: 10.1128/jb.134.3.1089-1098.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Israelachvili J. Academic Press; New York: 1991. Intermolecular and Surface Forces. [Google Scholar]
- 74.Mukundan H., Price D.N., Swanson B.I. Understanding the interaction of lipoarabinomannan with membrane mimetic architectures. Tuberculosis (Edinb.) 2012;92:38–47. doi: 10.1016/j.tube.2011.09.006. [DOI] [PubMed] [Google Scholar]
- 75.Needham D., McIntosh T.J., Lasic D.D. Repulsive interactions and mechanical stability of polymer-grafted lipid membranes. Biochim. Biophys. Acta. Biomembr. 1992;1108:40–48. doi: 10.1016/0005-2736(92)90112-y. [DOI] [PubMed] [Google Scholar]
- 76.Warriner H.E., Idziak S.H.J., Safinya C.R. Lamellar biogels: fluid-membrane-based hydrogels containing polymer lipids. Science. 1996;271:969–973. doi: 10.1126/science.271.5251.969. [DOI] [PubMed] [Google Scholar]
- 77.Akiyoshi K., Itaya A., Yoshikawa K. Induction of neuron-like tubes and liposome networks by cooperative effect of gangliosides and phospholipids. FEBS Lett. 2003;534:33–38. doi: 10.1016/s0014-5793(02)03743-2. [DOI] [PubMed] [Google Scholar]
- 78.Zidovska A., Ewert K.K., Safinya C.R. Block liposome and nanotube formation is a general phenomenon of two-component membranes containing multivalent lipids. Soft Matter. 2011;7:8363–8369. doi: 10.1039/C1SM05481C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zidovska A., Ewert K.K., Safinya C.R. The effect of salt and pH on block liposomes studied by cryogenic transmission electron microscopy. Biochim. Biophys. Acta. Biomembr. 2009;1788:1869–1876. doi: 10.1016/j.bbamem.2009.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zidovska A., Ewert K.K., Safinya C.R. Block liposomes from curvature-stabilizing lipids: connected nanotubes, -rods, or -spheres. Langmuir. 2009;25:2979–2985. doi: 10.1021/la8022375. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


