Abstract
Two new peptides, chujamides A (1) and B (2), were isolated from the marine sponge Suberites waedoensis, which was collected from Korean waters. Based upon the results of the combined spectroscopic analyses, the structures of these compounds were determined to be proline-riched and cyclic cystine bridged dodeca- and undecapeptides. The absolute configurations of all amino acid residues were determined to be l by advanced Marfey’s analysis. The new compounds exhibited weak cytotoxicities against A549 and K562 cell-lines, and compound 2 also demonstrated moderate inhibitory activity against Na+/K+-ATPase.
Keywords: Suberites waedoensis, cyclic cystine bridged peptides, chujamides, cytotoxicity, Na+/K+-ATPase
1. Introduction
Sponges are widely recognized to be one of the most prolific sources of natural marine products with diverse biogenetic origins. Although peptides account for a relatively minor proportion of sponge-derived metabolites, several of these peptides possess highly unique chemical structures and potent bioactivities [1]. Among the recently reported peptides, noticeable examples include the kapakahines [2,3] from Cribrochalina olemeda, the neopetrosiamides [4] from Neopetrosia sp., the koshikamides and mutremdamide A [5,6] from Theonella spp., the yaku’amides [7] from Ceratropsion sp., and the solomonamides [8] from Theonella swinhoei. These compounds possess unusual amino acid residues, uncommon linkages and/or significant bioactivities.
In our search for bioactive metabolites from sponges found in Korean waters, we recently reported the structural determination of gombamide A, a cyclic thiohexapeptide from Clathria gombawuiensis [9]. This highly modified peptide exhibited cytotoxicity against the A549 and K562 cell lines as well as inhibitory activity against Na+/K+-ATPase. In our continuing search for bioactive metabolites in sponges, we report here the isolation and structural determinations of chujamides A (1) and B (2), new cyclic cystine bridged peptides from the sponge Suberites waedoensis (Figure 1). These proline-rich dodeca- and undeca-peptides exhibited weak cytotoxicities, and compound 2 demonstrated moderate inhibitory activity against Na+/K+-ATPase.
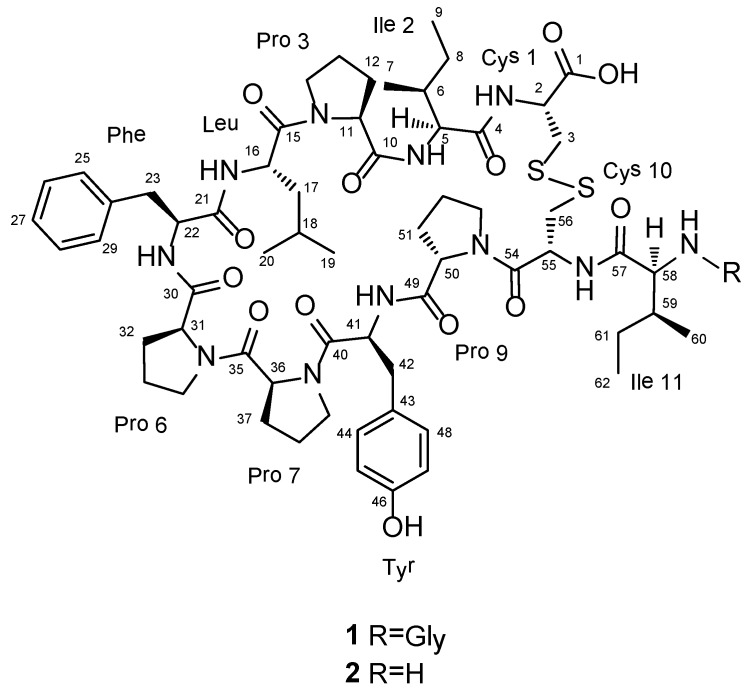
Figure 1.
Structures of compounds 1 and 2.
2. Results and Discussion
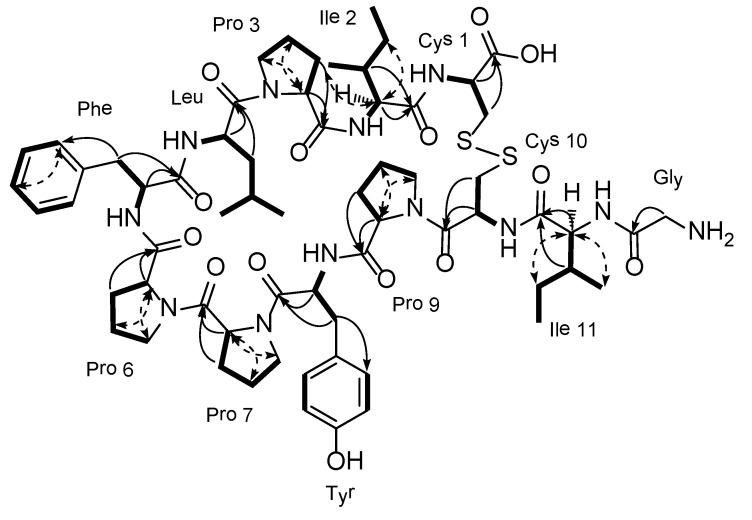
The molecular formula of chujamide A (1) was determined to be C64H92N12O14S2 by HRFABMS analysis. Evidence of the peptide nature of this compound was given by the presence of several carbonyl and methine carbons in the regions of δC 177-169 and 65-50, respectively, in the 13C NMR data. The structures of individual amino acid residues were determined by a combination of 1H COSY, TOCSY, HSQC, and HMBC experiments (See Supplementary Information, Figures S1–S12), which led to the identification of four prolines (Pro), two cysteines (Cys), two isoleucines (Ile), and one unit each of glycine (Gly), leucine (Leu), phenylalanine (Phe), and tyrosine (Tyr). Notably, due to poor resolution in DMSO-d6 and pyridine-d5, the 2-D NMR work was first performed in a MeOH-d4 solution to evaluate the protons attached to the carbon atoms. These data were supported by data concerning the amide protons obtained in MeOH-d3 solution. In this manner, all of the protons and carbon atoms, including the carbonyl carbon atoms, of each amino acid residue in 1 were assigned (Table 1 and Figure 2).
Table 1.
13C (150 MHz) and 1H (600 MHz) NMR assignments for chujamides A and B in MeOH-d4 and MeOH-d3 a.
| A. A. unit | Position | chujamide A | chujamide B | ||
|---|---|---|---|---|---|
| δC | δH (J in Hz) | δC | δH (J in Hz) | ||
| Cys 1 | 1 | 176.7, C | 176.9, C | ||
| 2 | 60.1, CH | 4.71, dd (11.4, 2.4) | 60.3, CH | 4.71, dd (11.4, 2.4) | |
| 3 | 43.0, CH2 | 3.45, m | 43.0, CH2 | 3.47, m | |
| 2.55, m | 2.58, m | ||||
| 2-NH a | 7.81, d (9.6) | 8.50, bs | |||
| Ile 2 | 4 | 172.4, C | 173.6, C | ||
| 5 | 59.1, CH | 4.82, d (3.6) | 59.2, CH | 4.82, m | |
| 6 | 37.7, CH | 2.34, m | 38.0, CH | 2.32, m | |
| 7 | 16.1, CH3 | 0.84, d (6.6) | 16.1, CH3 | 0.83, d (6.6) | |
| 8 | 25.2, CH2 | 1.54, m | 25.4, CH2 | 1.55, m | |
| 0.94, m | 0.95, m | ||||
| 9 | 12.2 CH3 | 0.90, t (6.6) | 12.2, CH3 | 0.90, t (6.6) | |
| 5-NH a | 7.52, d (10.2) | 7.67, bs | |||
| Pro 3 | 10 | 174.6, C | 173.4, C | ||
| 11 | 61.9, CH | 5.24, d (7.8) | 61.5, CH | 5.25, m | |
| 12 | 26.8, CH2 | 2.53, m | 27.0, CH2 | 2.48, m | |
| 1.72, m | 1.73, m | ||||
| 13 | 26.2, CH2 | 2.15, m | 26.3, CH2 | 2.13, m | |
| 1.97, m | 1.93, m | ||||
| 14 | 48.3, CH2 | 3.81, m | 48.3, CH2 | 3.80, m | |
| 3.63, dd (8.4, 8.4) | 3.64, m | ||||
| Leu | 15 | 176.4, C | 176.5, C | ||
| 16 | 51.7, CH | 4.50, dd (12.0, 1.8) | 51.6, CH | 4.51, dd (12.0, 1.8) | |
| 17 | 41.4, CH2 | 1.73, m | 41.6, CH2 | 1.72, m | |
| 1.40, m | 1.34, m | ||||
| 18 | 25.9, CH | 1.71, m | 26.0, CH | 1.70, m | |
| 19 | 20.5, CH3 | 0.97, d (6.6) | 20.5, CH3 | 0.97, d (6.6) | |
| 20 | 24.0, CH3 | 0.99, d (6.6) | 24.0, CH3 | 0.98, d (6.6) | |
| 16-NH a | 7.78, d (4.8) | 7.77, d (4.8) | |||
| Phe | 21 | 173.7, C | 173.7, C | ||
| 22 | 56.3, CH | 4.46, dd (12.0, 4.8) | 56.4, CH | 4.46, dd (12.0, 4.8) | |
| 23 | 33.8, CH2 | 3.41, m | 33.9, CH2 | 3.41, m | |
| 3.01, m | 3.00, m | ||||
| 24 | 140.2, C | 140.3, C | |||
| 25/29 | 131.1, CH | 7.27, dd (7.2, 1.8) | 131.2, CH | 7.27, dd (7.2, 1.8) | |
| 26/28 | 129.1, CH | 7.14, m | 129.1, CH | 7.14, m | |
| 27 | 127.3, CH | 7.14, m | 127.3, CH | 7.14, m | |
| 22-NH a | 8.79, d (7.2) | 8.80, d (7.2) | |||
| Pro 6 | 30 | 171.9, C | 171.9, C | ||
| 31 | 62.5, CH | 4.17, d (7.8) | 62.5, CH | 4.15, d (7.8) | |
| 32 | 30.3, CH2 | 2.55, m | 30.3, CH2 | 2.54, m | |
| 1.97, m | 1.99, m | ||||
| 33 | 23.1, CH2 | 1.97, m | 23.1,CH2 | 1.90, m | |
| 1.60, m | 1.62, m | ||||
| 34 | 47.8, CH2 | 3.50, m | 47.8, CH2 | 3.50, m | |
| 3.44, m | 3.44, m | ||||
| Pro 7 | 35 | 172.0, C | 172.1, C | ||
| 36 | 59.9, CH | 2.99, m | 59.9, CH | 2.99, m | |
| 37 | 29.4, CH2 | 2.00, m | 29.4, CH2 | 2.01, m | |
| 1.61, m | 1.62, m | ||||
| 38 | 26.0, CH2 | 2.00, m | 25.9, CH2 | 2.01, m | |
| 1.94, m | 1.93, m | ||||
| 39 | 48.8, CH2 | 3.61, m | 48.7, CH2 | 3.61, m | |
| 3.53, m | 3.53, m | ||||
| Tyr | 40 | 171.2, C | 171.3, C | ||
| 41 | 52.7, CH | 4.85, t (6.0) | 52.6, CH | 4.84, t (6.0) | |
| 42 | 36.9, CH2 | 3.00, m | 37.0, CH2 | 3.01, m | |
| 2.74, dd (14.4, 6.0) | 2.75, m | ||||
| 43 | 127.2, C | 127.4, C | |||
| 44/48 | 132.1, CH | 6.98, d (8.4) | 132.1, CH | 6.98, d (8.4) | |
| 45/47 | 116.1, CH | 6.71, d (8.4) | 116.3, CH | 6.70, d (8.4) | |
| 46 | 157.7, C | 157.7, C | |||
| 41-NH a | 7.05, d (7.2) | 7.11, m | |||
| Pro 9 | 49 | 172.5, C | 172.4, C | ||
| 50 | 62.2, CH | 4.68, d (8.4) | 62.2, CH | 4.69, d (8.4) | |
| 51 | 28.7, CH2 | 2.32, m | 28.6, CH2 | 2.32, m | |
| 2.00, m | 2.00, m | ||||
| 52 | 25.7, CH2 | 2.09, m | 25.7, CH2 | 2.09, m | |
| 1.80, m | 1.80, m | ||||
| 53 | 48.6, CH2 | 3.87, m | 48.6, CH2 | 3.87, m | |
| 3.73, m | 3.73, m | ||||
| Cys 10 | 54 | 172.7, C | 172.7, C | ||
| 55 | 51.2, CH | 4.77, dd (12.0, 2.4) | 51.5, CH | 4.76, ddd (12.0, 2.4, 2.4) | |
| 56 | 40.3, CH2 | 3.09, m | 40.5, CH2 | 3.08, m | |
| 2.77, dd (14.4, 3.0) | 2.73, m | ||||
| 55-NH a | 8.58, d (4.8) | 8.27, bs | |||
| Ile 11 | 57 | 174.3, C | 174.3, C | ||
| 58 | 59.7, CH | 4.01, d (10.2) | 60.0, CH | 3.95, m | |
| 59 | 37.2, CH | 1.72, m | 37.2, CH | 1.72, m | |
| 60 | 15.8, CH3 | 0.93, d (6.6) | 15.8, CH3 | 0.93, d (6.6) | |
| 61 | 26.7, CH2 | 1.55, m | 26.8, CH2 | 1.55, m | |
| 1.18, m | 1.17, m | ||||
| 62 | 10.7, CH3 | 0.86, t (6.6) | 10.8, CH3 | 0.86, t (6.6) | |
| 58-NH a | 8.73, d (6.6) | ||||
| Gly | 63 | 169.4, C | |||
| 64 | 41.8, CH2 | 3.57, m | |||
| 3.55, m | |||||
a Amide protons were observed in the spectra obtained in MeOH-d3 solution; A. A.: Amino Acid.
Figure 2.
Key correlations within the individual amino acid residues of the COSY (bold line), TOCSY (dashed arrow), and gHMBC (solid arrow) experiments for chujamide A.
Confirmation of the amino acid residues as well as the absolute configuration of each residue in compound 1 was accomplished by advanced Marfey’s analysis [10,11]. After the acid hydrolysis of 1, ESI-LC/MS analysis of the hydrolysate adducts with l-FDAA (1-fluoro-2-4-dinitrophenyl-5-l-alanine amide) and d-FDAA clearly confirmed the NMR-based amino acid residue assignments. Comparison of the ESI-LC/MS retention times of the l- and d-FDAA-derivatized hydrolysates allowed the assignment of the l configuration to all of the amino acid residues. In addition, the configurations of the β-carbons of the two l-Ile residues were both also assigned to be S by the co-injection of both l- and l-allo-Ile with the hydrolysates during ESI-LC/MS analysis.
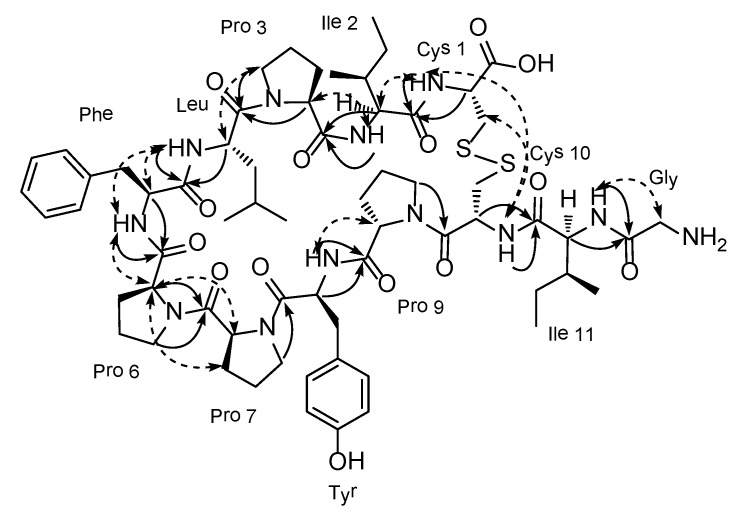
Given the identity of the amino acid residues, the structural construction of 1 was accomplished by a combination of HMBC and NOESY experiments (Figure 3). That is, the long-range correlations of the H-2 (δH 4.71) and 2-NH (δH 7.81) protons with the C-4 carbonyl carbon (δC 172.4) in the HMBC data suggested a peptide linkage between a Cys (Cys 1) and an Ile (Ile 2) that was supported by the NOESY cross peak at 2-NH/H-5. Similarly, the placement of a Pro (Pro 3) at the other end of this Ile was supported by the HMBC correlations at H-5/C-10 and 5-NH/C-10 as well as the NOESY cross peak at 5-NH/H-11. In this manner, the linear assembly of twelve amino acid residues connected to each other by peptide bonds was unambiguously identified as Cys 1-Ile 2-Pro 3-Leu-Phe-Pro 6-Pro 7-Tyr-Pro 9-Cys 10-Ile 11-Gly.
Figure 3.
Key correlations between the neighboring amino acid residues of chujamide A from gHMBC (solid arrow) and ROESY (dashed arrow) experiments.
Due to the lack of suitable carbons and protons within a two or three bond distance, the disulfide linkage between Cys-1 and Cys-10, inherent in the molecular formula as an additional degree of unsaturation, was not supported by the HMBC data but was indicated by the NOESY cross-peaks at 2-NH/55-NH and H-3/55-NH. The presence of a disulfide linkage between the cysteine residues was also supported by the chemical shifts of the Cβ carbons at δC 40.3 and 40.0, which were similar to those of other peptides possessing cysteines with a similar disulfide linkage [12]. For comparison, the shifts of free cysteine and cysteic acid were approximately δC 25.5 and 50, respectively [13,14]. Thus, chujamide A (1) was determined to be a novel cyclic cystine bridged dodecapeptide.
A related compound, chujamide B (2), was also isolated as an amorphous solid. Although its molecular formula was not directly determined from the high-resolution MS analysis, the MALDITOF-MS data, in conjunction with the 1H and 13C NMR data, established the molecular formula to be C62H89N11O13S2 [4]. The spectroscopic data of this compound were highly reminiscent of the data of 1, and the loss of signals for a carbonyl and a methylene were the most noticeable differences in both the 1H and 13C NMR data. These spectroscopic differences were readily accounted for by the loss of a terminal Gly residue, which was confirmed by combined 2-D NMR experiments in which the same proton-proton and proton-carbon correlations as those in 1 were obtained for the entire cyclic portion (See Supplementary Information, Figures S13–S24). Thus, chujamide B (2) was determined to be a cyclic cystine bridged undecapeptide.
Cyclic peptides with disulfide linkages have been rarely found in sponges, as the microcionamides from Clathria abietina [12], the neopetrosiamides from Neopetrosia sp. [4], asteropsin A from Asteropus sp. [15], and the recently reported gombamide A from Clathria gombawuiensis [9] are the only examples in the literature, which confirms the scarcity of these peptides. A literature survey also revealed that previous works on the peptides from sponges of the genus Suberites only yielded geodiamolides and serangamides [16], cyclic and linear lipotripeptides related to jasplakinolides [17] (=jaspamide [18]) whose frameworks differ significantly from those of the chujamides.
In our bioactivity measurements, chujamides A (1) and B (2) exhibited weak cytotoxicities toward the K562 and A549 cell-lines. The LC50 values were 37.0 and 10.1 µM for compound 1 and were 55.6 and 26.4 µM for compound 2, respectively (the LC50 values of doxorubicin were 1.5 and 1.3 µM, respectively). Chujamide B also moderately inhibited the action of Na+/K+-ATPase with an IC50 value of 17.2 µM (the IC50 value of ouabain was 6.1 µM). However, these compounds were inactive (MIC > 100 mM) against strains of Gram-positive and Gram-negative bacteria and pathogenic fungi [19].
In summary, two cyclic cystine bridged peptides rich in prolines, chujamides A (1) and B (2), were isolated from the Korean sponge Suberites waedoensis and were structurally elucidated by combining spectroscopic and Marfey’s analyses. These compounds exhibited weak cytotoxicities, and compound 2 demonstrated moderate inhibition against Na+/K+-ATPase.
3. Experimental Section
3.1. General Experimental Procedures
Optical rotations were measured on a JASCO P-1020 polarimeter using a 1-cm cell. UV spectra were recorded on a Hitachi U-3010 spectrophotometer, and IR spectra were recorded on a JASCO 300E FT-IR spectrometer. NMR spectra were recorded in MeOH-d4 and MeOH-d3 containing Me4Si as an internal standard on a Bruker Avance 600 spectrometer. Proton and carbon NMR spectra were measured at 600 and 150 MHz (1 and 2), respectively. Mass spectrometric data were obtained at the Korea Basic Science Institute (Daegu, Korea) and were acquired using a JEOL JMS 700 mass spectrometer with meta-nitrobenzyl alcohol (NBA) as the matrix for the FABMS. MALDI-TOF was provided by National Center for Inter-University Research Facilities (Seoul, Korea) and was acquired using a Voyager-DE™ STR Biospectrometry Workstation (Foster City, CA, USA) for compound 2. Low-resolution ESIMS data were recorded on an Agilent Technologies 6130 quadrupole mass spectrometer with an Agilent Technologies 1200 series HPLC (Agilent Technologies, Santa Clara, CA, USA). HPLC was performed on a SpectraSystem p2000 equipped with a SpectraSystem RI-150 (Thermo, Waltham, MA, USA) refractive index detector. All of the solvents were of spectroscopic grade or were distilled in glass prior to use.
3.2. Animal Materials
Specimens of Suberites waedoensis (voucher collection number 12CH-1) were collected by hand using scuba equipment off the shore of Chuja Island, Korea at a depth of 25 m during 8–11 October 2012. The sponge was cushion shaped, had a red color in life, and measured 12 × 10 cm with a thickness of 3 cm. The surface was lightly wrinkled but smooth, and the texture was elastic. The skeleton was small and had large tightly arranged tylostyles (200 − 400 × 5 μm and 600 − 900 × 10 − 16 μm). These morphological features agreed well with those reported in the literature [20]. A voucher specimen (registry no. spo 71) was deposited at the Natural History Museum, Hannam University, Korea, under the curatorship of C.J. Sim.
3.3. Extraction and Isolation
The freshly collected specimens were frozen immediately and kept at −25 °C until used for the chemical investigation. The freeze-dried sponge was sliced and repeatedly extracted with MeOH (3 × 3 L) and CH2Cl2 (3 × 3 L). The combined organic extract (354.5 g) was partitioned between H2O (210.8 g) and n-BuOH (140.7 g), and the latter was then repartitioned between n-hexane (120.1 g) and H2O–MeOH (15:85, 18.5 g). An aliquot (9.2 g) of the H2O–MeOH layer from the solvent partitioning was subjected to reversed-phase vacuum flash chromatography using a sequential mixture of H2O and MeOH (six fractions in the gradient, H2O–MeOH, from 50:50 to 0:100), acetone, and finally EtOAc as the eluents.
Based on the results of the 1H NMR and the cytotoxicity analyses, the fraction that eluted with H2O-MeOH (30:70; 0.23 g) was selected for separation. This fraction was separated by semi-preparative reversed-phase HPLC (YMC ODS-A column, 10 × 250 mm, H2O–MeOH, 40:60) to yield, in order of their elution, compounds 2 and 1. The final purification of the individual compounds was then accomplished by reversed-phase HPLC (YMC-Pack CN column, 4.6 × 250 mm, H2O–MeOH, and 50:50 for 1 and YMC ODS-A column, 4.6 × 250 mm, and H2O–MeOH, 45:55 for 2). Compound 1 was also isolated from the flash chromatographic fraction eluted with H2O–MeOH (20:80; 0.29 g) using the same HPLC conditions. The purified metabolites were isolated in the following amounts: 27.0 mg for 1 and 4.0 mg for 2.
Chujamide A (1): white, amorphous solid, 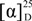 −15 (c 0.50, MeOH); UV (MeOH) λmax (log ε) 210 (4.40), 227 (4.14), 277 (3.16) nm; IR (ZnSe) vmax 3300, 2958, 1655 cm−1; 1H and 13C NMR data, see Table 1; HRFABMS m/z 1317.6371 [M + H]+ (calcd for C64H93N12O14S2, 1317.6367).
−15 (c 0.50, MeOH); UV (MeOH) λmax (log ε) 210 (4.40), 227 (4.14), 277 (3.16) nm; IR (ZnSe) vmax 3300, 2958, 1655 cm−1; 1H and 13C NMR data, see Table 1; HRFABMS m/z 1317.6371 [M + H]+ (calcd for C64H93N12O14S2, 1317.6367).
Chujamide B (2): white, amorphous solid, 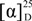 −52 (c 0.45, MeOH); UV (MeOH) λmax (log ε) 210 (4.41), 227 (4.10), 276 (3.16) nm; IR (ZnSe) vmax 3308, 2956, 1637 cm−1; 1H and 13C NMR data, see Table 1; MALDITOF-MS m/z 1260 [M]+ (calcd for C62H90N11O13S2, 1260).
−52 (c 0.45, MeOH); UV (MeOH) λmax (log ε) 210 (4.41), 227 (4.10), 276 (3.16) nm; IR (ZnSe) vmax 3308, 2956, 1637 cm−1; 1H and 13C NMR data, see Table 1; MALDITOF-MS m/z 1260 [M]+ (calcd for C62H90N11O13S2, 1260).
3.4. Advanced Marfey’s Analysis of Compound 1
Compound 1 (1.0 mg) was dissolved in 0.5 mL of 6 N HCl and heated at 110 °C for 15 h. This solution was evaporated, and traces of HCl were removed by repeatedly drying the compound under vacuum with distilled water. To the divided hydrolysate (0.5 mg), 100 μL of 1 N NaHCO3 and 50 μL of 1% l- or d-FDAA in acetone were added. The mixture was stirred at 70 °C for 1 h. After the reaction was quenched by the addition of 50 μL of 2 N HCl, the mixture was analyzed by ESI-LC/MS (YMC ODS-A column, 5 μm, 4.6 × 100 mm, H2O-MeCN gradient (80:20 to 30:70 in 40 min, v/v), 0.7 mL/min flow rate, UV detector, 360 nm) to assign the chirality of the amino acids. The retention times of the l- and d-FDAA-derivatized hydrolysates were 13.5 min and 14.4 min for l-Pro, 15.3 min and 16.5 min for l-Tyr, 16.9 min and 18.9 min for l-Cys, 20.6 min and 24.0 min for l-Ile, 21.3 min and 24.5 min for l-Leu, and 21.4 min and 23.9 min for l-Phe, respectively. Thus, all of the amino acids appeared to be l-amino acids.
3.5. Analysis of the Configuration of the l-Ile Residue in Compound 1
The absolute configuration of the β-carbon of l-Ile moiety was identified by an ESI-LC/MS experiment with the l-FDAA derivatives of standard l-allo-Ile and l-Ile. Under the given chromatographic conditions (YMC ODS-A column, 5 μm, 4.6 × 250 mm, H2O-MeCN gradient (80:20 to 30:70 in 40 min, v/v), 0.7 mL/min flow rate, UV detector, 360 nm), the l-FDAA derivatives of authentic l-allo-Ile and l-Ile had retention times of 38.349 and 37.845 min, respectively. In addition, co-injection with the l-FDAA derivative of the hydrolysate of the l-Ile moiety showed one single peak of l-Ile-l-FDAA at 37.845 min. Thus, the absolute configurations of both l-Ile moieties in compound 1 were determined to be S.
3.6. Biological Assays
The cytotoxicity assays were performed in accord with literature protocols [21,22]. The Na+/K+-ATPase inhibition assay was performed according to the previously described method [23].
4. Conclusions
Two new peptides, chujamides A (1) and B (2), were isolated from the marine sponge Suberites waedoensis, from Korea. These compounds were structurally classified to a group of the proline-riched and cyclic cystine bridged dodeca- and undecapeptides. These compounds exhibited weak cytotoxicity and moderate inhibition against Na+/K+-ATPase (2).
Acknowledgments
We are grateful to the Basic Science Research Institute in Daegu, Korea for providing the mass data. This study was supported by National Research Foundation of Korea (NRF) grants funded by the Korean government (Ministry of Science, ICT and Future Planning) (2010-0020429 and 2009-0083533).
Supplementary Files
Supplementary Information (PDF, 2809 KB)
Author Contributions
J. Shin and K.-B. Oh designed and supervised whole experimental procedures and wrote the manuscript. J. Song and J.-e. Jeon isolated compounds and got a spectroscopic data. T.H. Won contributed to analyzing data and chemical analysis. D.-C. Oh evaluated whole processes of structure determination. C.J. Sim taxonomically identified the sponge specimen.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Blunt J.W., Copp B.R., Keyzers R.A., Munro M.H.G., Prinsep M.R. Marine natural products. Nat. Prod. Rep. 2013;30:237–323. doi: 10.1039/c2np20112g. [DOI] [PubMed] [Google Scholar]
- 2.Yeung B.K.S., Nakao Y., Kinnel R.B., Carney J.R., Yoshida W.Y., Scheuer P.J., Kelly-Borges M. The kapakahines, cyclic peptides from the marine sponge Cribrochalina olemda. J. Org. Chem. 1996;61:7168–7173. doi: 10.1021/jo960725e. [DOI] [PubMed] [Google Scholar]
- 3.Nakao Y., Kuo J., Yoshida W.Y., Kelly M., Scheuer P.J. More kapakahines from the marine sponge Cribrochalina olemda. Org. Lett. 2003;5:1387–1390. doi: 10.1021/ol026830u. [DOI] [PubMed] [Google Scholar]
- 4.Williams D.E., Austin P., Diaz-Marrero A.R., van Soest R., Matainaho T., Roskelley C.D., Roberge M., Andersen R.J. Neopetrosiamides, peptides from the marine sponge Neopetrosia sp. that inhibit ameoboid invasion by human tumor cells. Org. Lett. 2005;7:4173–4176. doi: 10.1021/ol051524c. [DOI] [PubMed] [Google Scholar]
- 5.Araki T., Matsunaga S., Nakao Y., Furihata K., West L., Faulkner D.J., Fusetani N. Koshikamide B, a cytotoxic peptide lactone from a marine sponge Theonella sp. J. Org. Chem. 2008;73:7889–7894. doi: 10.1021/jo801032n. [DOI] [PubMed] [Google Scholar]
- 6.Plaza A., Bifulco G., Masullo M., Lloyd J.R., Keffer J.L., Colin P.L., Hooper J.N.A., Bell L.J., Bewley C.A. Mutremdamide A and koshikamides C-H, peptide inhibitors of HIV-1 entry from different Theonella species. J. Org. Chem. 2010;75:4344–4355. doi: 10.1021/jo100076g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yeoka R., Ise Y., Ohtsuka S., Okada S., Yamori T., Matsunaga S. Yaku’amide A and B, cytotoxic linear peptides rich in dehydroamino acids from the marine sponge Ceratopsion sp. J. Am. Chem. Soc. 2010;132:17692–17694. doi: 10.1021/ja109275z. [DOI] [PubMed] [Google Scholar]
- 8.Festa C., de Marino S., Sepe V., D’Auria M.V., Bifulco G., Débitus C., Bucci M., Vellecco V., Zampella A. Solomonamides A and B, new anti-inflammatory peptides from Theonella swinhoei. Org. Lett. 2011;13:1532–1535. doi: 10.1021/ol200221n. [DOI] [PubMed] [Google Scholar]
- 9.Woo J.-K., Jeon J.-E., Kim C.-K., Sim C.J., Oh D.-C., Oh K.-B., Shin J. Gombamide A, a cyclic thiopeptide from the sponge Clathra gombawuiensis. J. Nat. Prod. 2013;76:1380–1383. doi: 10.1021/np4003367. [DOI] [PubMed] [Google Scholar]
- 10.Fujii K., Ikai Y., Mayumi T., Oka H., Suzuki M., Harada K. A nonempirical method using LC/MS for determination of the absolute configuration of constituent amino acids in a peptide: Elucidation of limitations of Marfey’s method and of its separation mechanism. Anal. Chem. 1997;69:3346–3352. doi: 10.1021/ac9701795. [DOI] [Google Scholar]
- 11.Fujii K., Ikai Y., Oka H., Suzuki M., Harada K. A nonempirical method using LC/MS for determination of the absolute configuration of constituent amino acids in a peptide: Combination of Marfey’s method with mass spectrometry and its practical application. Anal. Chem. 1997;69:5146–5151. doi: 10.1021/ac970289b. [DOI] [Google Scholar]
- 12.Davis R.A., Mangalindan G.C., Bojo Z.P., Antemano R.R., Rodriguez N.O., Concepcion G.P., Samson S.C., Guzman D., Cruz L.J., Tasdemir D., et al. Microcionamides A and B, bioactive peptides from the Philippine sponge Clathria (Thalysias) abietina. J. Org. Chem. 2004;69:4170–4176. doi: 10.1021/jo040129h. [DOI] [PubMed] [Google Scholar]
- 13.Katritzky A.R., Tala S.R., Abo-Dya N.E., Ibrahim T.S., El-Feky S.A., Gyanda K., Pandya K.M. Chemical ligation of S-scylated cysteine peptides to form native peptides via 5-, 11-, and 14-membered cyclic transition states. J. Org. Chem. 2011;76:85–96. doi: 10.1021/jo1015757. [DOI] [PubMed] [Google Scholar]
- 14.Laird D.W., LaBarbera D.V., Feng X., Bugni T.S., Harper M.K., Ireland C.M. Halogenated cyclic peptides isolated from the sponge Corticium sp. J. Nat. Prod. 2007;70:741–746. doi: 10.1021/np060489v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li H., Bowling J.J., Fronczek F.R., Hong J., Jabba S.V., Murray T.F., Ha N.C., Hamman M.T., Jung J.H. Asteropsin A: An unusual cystine-crosslinked peptide from porifera enhances neuronal Ca2+ influx. Biochim. Biophys. Acta. 2013;1830:2591–2599. doi: 10.1016/j.bbagen.2012.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tanaka C., Tanaka J., Bolland R., Marriott G., Higa T. Seragamides A-F, new actin-targeting depsipeptides from the sponge Suberites japonicus Thiele. Tetrahedron. 2006;62:3536–3542. doi: 10.1016/j.tet.2006.01.099. [DOI] [Google Scholar]
- 17.Crews P., Manes L.V., Boehler M. Jasplakinolide, a cyclodepsipeptide from the marine sponge, Jaspis sp. Tetrahedron Lett. 1986;27:2797–2800. doi: 10.1016/S0040-4039(00)84645-6. [DOI] [Google Scholar]
- 18.Zabriskie T.M., Klocke J.A., Ireland C.M., Marcus A.H., Molinski T.F., Faulkner D.J., Xu C., Clardy J.C. Jaspamide, a modified peptide from a Jaspis Sponge, with insecticidal and antifungal activity. J. Am. Chem. Soc. 1986;108:3123–3124. doi: 10.1021/ja00271a062. [DOI] [Google Scholar]
- 19.Oh K.-B., Lee J.H., Chung S.-C., Shin J., Shin H.J., Kim H.-K., Lee. H.-S. Antimicrobial activities of the bromophenols from the red alga Odonthalia corymbifera and some synthetic derivatives. Bioorg. Med. Chem. Lett. 2008;18:104–108. doi: 10.1016/j.bmcl.2007.11.003. [DOI] [PubMed] [Google Scholar]
- 20.Shim E.J., Sim C.J. Two new species of genus Suberites (Hadromerida: Suberitidae) from Korea. Korean J. Syst. Zool. 2008;24:215–218. doi: 10.5635/KJSZ.2008.24.2.215. [DOI] [Google Scholar]
- 21.Mosmann T. Rapid colometric assay for cellular growth and survival: Application to proliferation and cytotoxicity assays. J. Immunol. Methods. 1983;65:55–63. doi: 10.1016/0022-1759(83)90303-4. [DOI] [PubMed] [Google Scholar]
- 22.Ulukaya E., Ozdikicioglu F., Oral A.Y., Demirci M. The MTT assay yields a relatively lower result of growth inhibition than the ATP assay depending on the chemotherapeutic drugs tested. Toxicol. In Vitro. 2008;22:232–239. doi: 10.1016/j.tiv.2007.08.006. [DOI] [PubMed] [Google Scholar]
- 23.Johansson M., Karlsson L., Wennergren M., Jansson T., Powell T.L. Activity and protein expression of Na+/K+ ATPase are reduced in microvillous syncytiotrophoblast plasma membranes isolated from pregnancies complicated by intrauterine growth restriction. J. Clin. Endocrinol. MeTab. 2003;88:2831–2837. doi: 10.1210/jc.2002-021926. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information (PDF, 2809 KB)