Abstract
The ethanol extract of Pseudoalteromonas strain J010, isolated from the surface of the crustose coralline alga Neogoniolithon fosliei, yielded thirteen natural products. These included a new bromopyrrole, 4′-((3,4,5-tribromo-1H-pyrrol-2-yl)methyl)phenol (1) and five new korormicins G–K (2–6). Also isolated was the known inducer of coral larval metamorphosis, tetrabromopyrrole (TBP), five known korormicins (A–E, previously named 1, 1a–c and 3) and bromoalterochromide A (BAC-A). Structures of the new compounds were elucidated through interpretation of spectra obtained after extensive NMR and MS investigations and comparison with literature values. The antibacterial, antifungal and antiprotozoal potential of 1–6, TBP and BAC-A was assessed. Compounds 1–6 showed antibacterial activity while BAC-A exhibited antiprotozoal properties against Tetrahymena pyriformis. TBP was found to have broad-spectrum activity against all bacteria, the protozoan and the fungus Candida albicans.
Keywords: Pseudoalteromonas, korormicin, bromopyrrole, bromoalterochromide, antibacterial, antifungal, antiprotozoal
1. Introduction
In the marine environment chemical signals play critical ecological roles at many organizational levels [1]. Epibiotic bacteria (e.g., associated with animals, plants, algae) are often the source of these signals [2].
In a previous bioassay-guided study of coral larval settlement cues [3], the pigmented bacterium Pseudoalteromonas strain J010, isolated from the surface of the crustose coralline alga (CCA) Neogoniolithon fosliei, triggered larval metamorphosis in acroporid corals [3]. The causative bacterial metabolite for the transition of planula larvae into non-attached primary polyps was identified as tetrabromopyrrole (TBP) [3]. During the course of this investigation other potentially new bacterial metabolites were encountered, including polybrominated pyrrole derivatives and a family of korormicin derivatives. Antimicrobial metabolites belonging to these compound classes have been previously isolated from other Pseudoalteromonads or closely related species [4,5,6]. Pigmented bacteria affiliated with the genus Pseudoalteromonas (class Gammaproteobacteria) have gained significant attention during the past decade as producers of a wide range of bioactive compounds [7].
In this study chemical analysis of Pseudoalteromonas strain J010 was undertaken and the new metabolites identified to better understand the metabolic capacity and antibiotic potential of this epiphytic bacterium. This study was motivated by the general notion that chemically undefended algae may benefit from bioactive secondary metabolites produced by associated bacteria [8], for example against bacterial colonization and fouling [9,10].
Details of the isolation and structural elucidation of the new bromopyrrole, 4′-((3,4,5-tribromo-1H-pyrrol-2-yl)methyl)phenol (1) and five new korormicins G–K (2–6) along with the known coral larval metamorphosis cue, TBP [3,4], five known korormicins (A–E, previously named 1, 1a–c and 3) [5,11,12] and bromoalterochromide A (BAC-A) [13] are provided. Their antimicrobial, antifungal and antiprotozoal activities are also described.
2. Results and Discussion
2.1. Isolation and Structural Elucidation of Bacterial Metabolites
NMR data and a characteristic MS isotopic distribution (3:10:10:3) established the molecular formula of compound 1 to be C11H8ONBr3, requiring seven degrees of unsaturation. The IR spectrum indicated the presence of both hydroxy (3388 cm−1) and amine (3271, 2925 cm−1) groups. Multiplicity-edited adiabatic HSQC data (Table 1) revealed a disubstituted double bond (δC 129.9, CH, C-8; δH 7.05, d, 8.3, H-8; 115.8, CH, C-9; δH 6.81, d, 8.3, H-9) and an isolated methylene (δC 32.5, CH2, C-6; δH 3.87, s, H-6). Furthermore, each proton signal in the 1H NMR integrated for two protons establishing the presence of two identical disubstituted double bonds and hence symmetry within the molecule. This together with HMBC correlations from H-8 to C-10 and from H-9 to δC 128.7 (C-7) as well as a J8–9 coupling of 8.3 Hz established a 1,4-disubstituted phenyl ring, accounting for four degrees of unsaturation in 1. The position of the hydroxy moiety on the phenyl ring was determined by the downfield resonance at δC 155.1 (C-10) while a HMBC correlation from H8/12 to δC 32.5 (C-6) established the methylene side chain to be para substituted.
Table 1.
NMR data (600 MHz and 125 MHz in CDCl3, Supplementary Table S1 and Figures S1–S14) for 4′-((3,4,5-tribromo-1H-pyrrol-2-yl)methyl)phenol (1).
| Position | δC, mult. | δH (J Hz) | gCOSY | gHMBC a |
|---|---|---|---|---|
| NH | - | - | - | - |
| 2 | 131.1, qC | - | - | - |
| 3 | 97.8, qC | - | - | - |
| 4 | 100.0, qC | - | - | - |
| 5 | 100.0, qC | - | - | - |
| 6 | 32.5, CH2 | 3.87, s | - | 2, 3, 7, 8, 12 |
| 7 | 128.7, qC | - | - | - |
| 8 | 129.9, CH | 7.05, d (8.3) | 9 | 2 b, 6, 7, 9, 10, 11, 12 |
| 9 | 115.8, CH | 6.81, d (8.3) | 8 | 8, 10, 11, 12 |
| 10 | 155.1, qC | - | - | - |
| OH | - | - | - | - |
| 11 | 115.8, CH | 6.81, d (8.3) | 12 | 8, 10, 11, 12 |
| 12 | 129.9, CH | 7.05, d (8.3) | 11 | 6, 8, 9, 10, 11 |
a (nJCH = 7.5 Hz), b (nJCH = 12 Hz).
The downfield resonance of C-2 (δC 131.1), HMBC correlations from H-6 to δC 131.1 (C-2) and δC 97.8 (C-3), plus the requirement for three bromines, established a tetrasubstituted pyrrole ring accounting for all seven degrees of unsaturation. Comparison of the carbon shifts of 1 with those of the natural and synthetic analogues 2,3,4-tribromopyrrole [14], 2,3,5-tribromopyrrole [14], pentabromopseudilin [15] and iso-pentabromopseudilin [15], as well as the reported expontaneous decomposition of iso-pentabromopseudilin [15], indicated that the two subunits were likely linked at C-2. This was supported by a weak HMBC correlation from H-8 to C-2 (Table 1 and Supplementary Figure S10). The structure of 1, 4′-((3,4,5-tribromo-1H-pyrrol-2-yl)methyl)phenol, is as shown (Figure 1).
Figure 1.
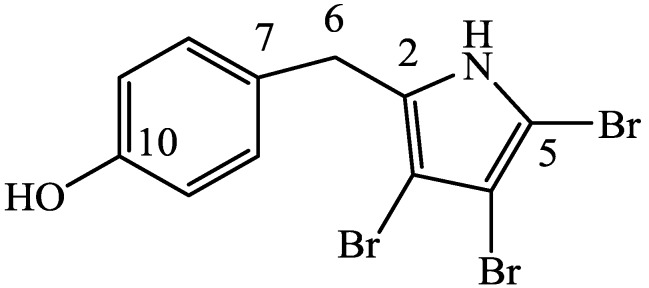
Structure of 4′-((3,4,5-tribromo-1H-pyrrol-2-yl)methyl)phenol (1).
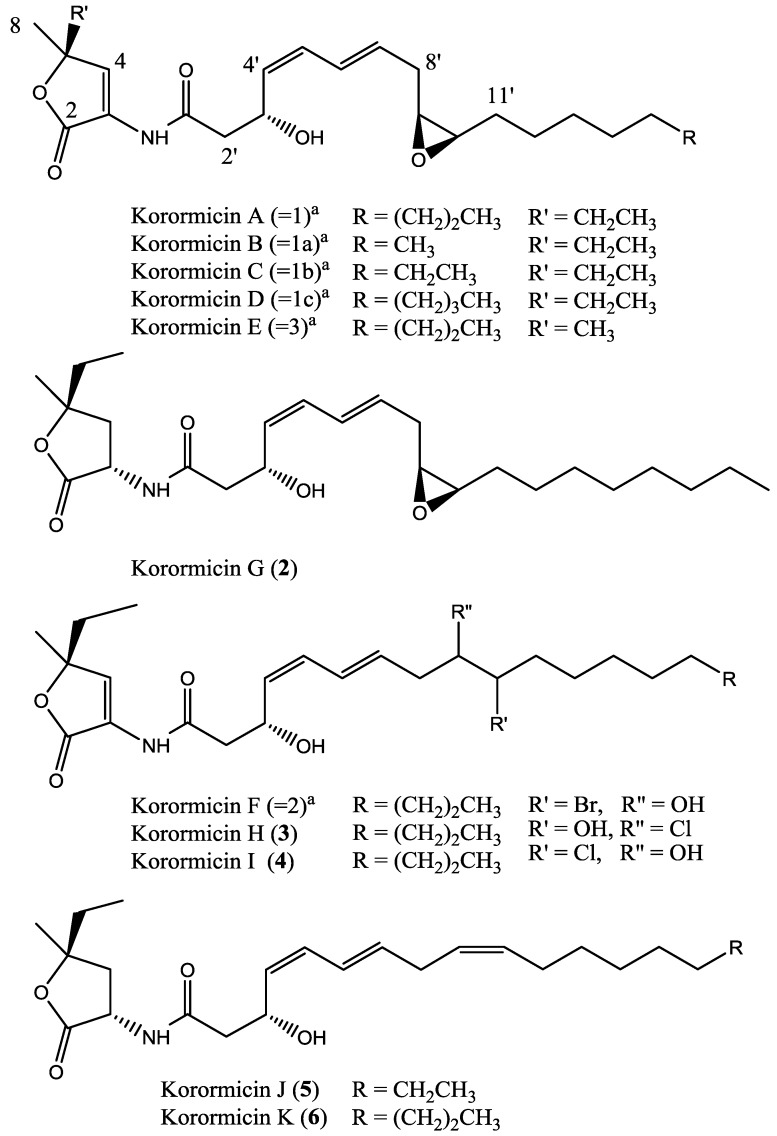
The molecular formula of compound 2 was established to be C25H41O5N from NMR and FTMS data. The presence of alcohol (3345 br cm−1), ester (1734 cm−1) and amide (1639 str, 1553 cm−1) functionality was determined by IR. In addition, NMR data (Table 2 and Table 3; Supplementary Figures S15–S18) for 2 was consistent with two conjugated, disubstituted double bonds (δC 129.3, CH, C-4′; 130.0, CH, C-5′; δH 5.39, dd, 11.3, 9.4, H-4′; δH 6.07, br t, 11.3, H-5′ and 126.4, CH, C-6′; 132.0, CH, C-7′; δH 6.47, br dd, 15.0, 11.3, H-6′; δH 5.82, br dt, 15.0, 6.8, H-7′) and two carbonyls (δC 174.7, C, C-2 and δC 172.0, C, C-1′), accounting for four of the six degrees of unsaturation; hence 2 is bicyclic. Also present (Table 2 and Table 3) was one quaternary carbon (δC 85.8, C, C-5), one C-N (δC 50.0, CH, C-3; δH 4.72, ddd, 11.5, 9.2, 5.6 Hz), three oxymethines (δC 64.7, CH; δH 5.01, br ddd, 9.4, 8.7, 3.2; δC 55.3, CH; δH 3.01, dt, 6.2, 4.2; δC 56.6, CH; δH 2.97, dt, 5.9, 4.2), eleven methylenes and three methyl groups.
Table 2.
13C NMR shifts of korormicins G–K (2–6).
| No. | Korormicin | |||||
|---|---|---|---|---|---|---|
| G (2) | H (3) | I (4) | I (4) | J (5) | K (6) | |
| δC, mult. a | δC, mult. b | δC, mult. b | δC, mult. a | δC, mult. a | δC, mult. a | |
| 1 | - | - | - | - | - | - |
| 2 | 174.7, qC | 168.5, qC | 168.5, qC | 168.5, qC | nd c | 174.3, qC |
| 3 | 50.0, CH | 125.0, qC | 125.0, qC | 125.0, qC | 50.0, CH | 50.4, CH |
| 4 | 39.9, CH2 | 133.7, CH | 134.0, CH | 134.0, CH | 40.0, CH2 | 40.4, CH2 |
| 5 | 85.8, qC | 87.2, qC | 87.2, qC | 87.2, qC | nd c | 85.6, qC |
| 6 | 32.4, CH2 | 31.2, CH2 | 31.1, CH2 | 34.6, CH2 | 32.9, CH2 | 32.8, CH2 |
| 7 | 8.2, CH3 | 8.0, CH3 | 8.0, CH3 | 8.1, CH3 | 8.3, CH3 | 8.4, CH3 |
| 8 | 25.9, CH3 | 23.9, CH3 | 24.1, CH3 | 24.2, CH3 | 25.9, CH3 | 26.2, CH3 |
| NH | - | - | - | - | - | - |
| 1′ | 172.0, qC | 170.1, qC | 170.2, qC | 170.2, qC | nd c | 171.7, qC |
| 2′ | 42.3, CH2 | 43.9, CH2 | 44.0, CH2 | 43.1, CH2 | 42.7, CH2 | 42.7, CH2 |
| 3′ | 64.7, CH | 63.5, CH | 63.8, CH | 64.6, CH | 64.8, CH | 65.0, CH |
| OH | - | - | - | - | - | - |
| 4′ | 129.3 CH | 132.8 CH | 132.2 CH | 129.4, CH | 127.6, CH | 129.0 CH |
| 5′ | 130.0, CH | 128.2, CH | 128.5, CH | 130.7, CH | 130.6, CH | 131.0, CH |
| 6′ | 126.4, CH | 127.8, CH | 127.4, CH | 127.4, CH | 124.0, CH | 124.7, CH |
| 7′ | 132.0, CH | 131.9, CH | 132.5, CH | 132.7, CH | 136.6, CH | 136.4, CH |
| 8′ | 31.1, CH2 | 37.2, CH2 | 37.1, CH2 | 38.2, CH2 | 30.5, CH2 | 30.6, CH2 |
| 9′ | 55.3, CH | 67.0, CH | 72.1, CH | 73.1, CH | 128.3, CH | 126.1, CH |
| OH | - | - | - | - | - | - |
| 10′ | 56.6, CH | 71.4, CH | 67.7, CH | 68.0, CH | 129.2 CH | 131.4, CH |
| 11′ | 27.3, CH2 | 33.3, CH2 | 33.7, CH2 | 31.9, CH2 | 27.0, CH2 | 27.2, CH2 |
| 12′ | 29.1 d, CH2 | 25.4, CH2 | 26.3, CH2 | 26.0, CH2 | 25.4, CH2 | 29.1 d, CH2 |
| 13′ | 29.1 d, CH2 | 29.0 d, CH2 | 28.8 d, CH2 | 28.8 d, CH2 | 29.0 d, CH2 | 29.1 d, CH2 |
| 14′ | 29.1 d, CH2 | 29.0 d, CH2 | 28.6 d, CH2 | 28.58 d, CH2 | 28.6 d, CH2 | 29.1 d, CH2 |
| 15′ | 29.1 d, CH2 | 29.0, CH2 | 28.6 d, CH2 | 28.55 d, CH2 | 31.4, CH2 | 29.1 d, CH2 |
| 16′ | 31.6, CH2 | 31.3, CH2 | 31.2, CH2 | 31.7, CH2 | 22.0, CH2 | 31.6, CH2 |
| 17′ | 22.6, CH2 | 22.1, CH2 | 22.1, CH2 | 22.6, CH2 | 13.5, CH3 | 22.6, CH2 |
| 18′ | 13.9, CH3 | 13.9, CH3 | 13.9, CH3 | 13.9, CH3 | - | 14.1, CH3 |
a CDCl3; b DMSO-d6; c No HMBC data was recorded for the qC due to low yield of 5, these carbons remain unassigned; d interchangeable.
Table 3.
1H NMR shifts of korormicins G–K (2–6).
| No. | Korormicin | |||||
|---|---|---|---|---|---|---|
| G (2) | H (3) | I (4) | I (4) | J (5) | K (6) | |
| δH (J Hz) a | δH (J Hz) b | δH (J Hz) b | δH (J Hz) a | δH (J Hz) a | δH (J Hz) a | |
| 1 | - | - | - | - | - | - |
| 2 | - | - | - | - | - | - |
| 3 | 4.72, ddd (11.5, 9.2, 5.6) | - | - | - | 4.70, ddd (11.2, 9.2, 6.1) | 4.70, ddd (11.2, 9.2, 5.6) |
| 4 | 2.78, dd (12.8, 9.2) 1.90, dd (12.8, 11.5) | 7.39, s | 7.39, s | 7.37, s | 2.78, dd (12.8, 9.2) 1.90, dd (12.8, 11.2) | 2.78, dd (12.9, 9.2) 1.90, dd (12.9, 11.2) |
| 5 | - | - | - | - | - | - |
| 6 | 1.76, dq (14.7, 7.5) 1.68, dq (14.7, 7.5) | 1.76, q (7.5) | 1.76, q (7.4) | 1.82, q (7.4) | 1.77, dq (14.4, 7.5) 1.70, dq (14.4, 7.5) | 1.76, dq (14.7, 7.5) 1.69, dq (14.7, 7.5) |
| 7 | 1.0, t (7.5) | 0.76, t (7.5) | 0.76, t (7.4) | 0.90, t (7.4) | 1.00, t (7.5) | 1.00, t (7.5) |
| 8 | 1.47, s | 1.41, s | 1.41, s | 1.50, s | 1.49, s | 1.47, s |
| NH | 6.58, d (5.6) | 9.91, br s | 9.88, br s | 8.22, br s | 6.59, d (6.1) | 6.60, d (5.6) |
| 1′ | - | - | - | - | - | - |
| 2′ | 2.51, dd (15.4, 8.7) 2.45, dd (15.4, 3.2) | 2.60, dd (14.4, 8.0) 2.41, dd (14.4, 4.6) | 2.60, dd (14.4, 8.0) 2.41, dd (14.4, 5.2) | 2.63, dd (15.8, 8.7) 2.58, dd (15.8, 3.1) | 2.49, dd (15.5, 8.6) 2.45, dd (15.5, 3.2) | 2.49, dd (15.3, 8.5) 2.45, dd (15.3, 3.5) |
| 3′ | 5.01, br ddd (9.4, 8.7, 3.2) | 4.85, br dddd (9.2, 8.0, 4.6, 4.1) | 4.84, dddd (9.1, 8.0, 5.2, 4.7) | 5.06, ddt (9.4, 8.7, 3.1) | 5.01, m | 5.00, ddd (9.4, 8.5, 3.5) |
| OH | 5.16, br d (4.1) | 5.11, d (4.7) | ||||
| 4′ | 5.39, dd (11.3, 9.4) | 5.30, br dd (11.1, 9.2) | 5.28, dd (11.1, 9.1) | 5.41, dd (11.1, 9.4) | 5.35, dd (11.3, 9.4) | 5.33, br dd (11.3, 9.4) |
| 5′ | 6.07, br t (11.3) | 5.92, t (11.1) | 5.91, t (11.1) | 6.09, t (11.1) | 6.05, br t (11.3) | 6.05, br t (11.3) |
| 6′ | 6.47, br dd (15.0, 11.3) | 6.45, br dd (14.8, 11.1) | 6.41, br dd (14.8, 11.1) | 6.45, br dd (14.9, 11.1) | 6.34, br dd (14.9, 11.3,1.5) | 6.34, ddd (14.7, 11.3, 0.8) |
| 7′ | 5.82, br dt (15.0, 6.8) | 5.71, br dt (14.8, 7.1) | 5.71, br dt (14.8, 7.5, 7.2) | 5.81, dt (14.9, 7.2) | 5.75, dt (14.9, 6.7) | 5.75, br dt (14.7, 6.9) |
| 8′ | 2.36, br dd (6.8, 6.2) | 2.62, m 2.45, br dd (15.7, 8.3) | 2.32, br ddd (14.2, 7.2, 6.8) 2.28, br ddd (14.2, 7.5, 6.5) | 2.44, br dd (7.2, 6.8) | 2.87, br ddd (7.5, 6.7, 1.5) | 2.87, ddd (7.3, 6.9, 1.0) |
| 9′ | 3.01, dt (6.2, 4.2) | 3.95, br ddd (8.3, 4.4, 3.1) | 3.60, dddd (6.8, 6.5, 6.1, 3.1) | 3.72, ddd (7.2, 6.8, 3.5) | 5.37, ddd (10.9, 7.5, 1.5) | 5.38, ddd (10.4, 7.3, 0.8) |
| 9′-OH | - | - | 5.01, br d (6.1) | - | - | - |
| 10′ | 2.97, dt (5.9, 4.2) | 3.56, m | 3.91, ddd (9.8, 3.7, 3.1) | 3.91, ddd (9.8, 3.9, 3.5) | 5.47, ddd (10.9, 7.3, 1.5) | 5.48, ddd (10.4, 7.3, 1.0) |
| 10′-OH | - | 4.95, br d (5.6) | - | - | - | - |
| 11′ | 1.55, m | 1.43, m | 1.76, ddt (14.4, 7.1, 3.7) 1.66, ddt (14.1, 4.6, 9.8) | 1.82, m 1.66, ddt (14.1, 4.6, 9.8) | 2.04, m | 2.05, dt (14.7, 7.3) |
| 12′ | 1.44, m | 1.26, m | 1.45, m 1.33, m | 1.43, m 1.33, m | 1.39, m | 1.36, m |
| 13′ | 1.28–1.41 | 1.20–1.36 | 1.20–1.36 | 1.20–1.36 | 1.20–1.35 | 1.28–1.41 |
| 14′ | 1.28–1.41 | 1.20–1.36 | 1.20–1.36 | 1.20–1.36 | 1.20–1.35 | 1.28–1.41 |
| 15′ | 1.28–1.41 | 1.20–1.36 | 1.20–1.36 | 1.20–1.36 | 1.24, m | 1.28–1.41 |
| 16′ | 1.30, m | 1.25, m | 1.24, m | 1.26, m | 1.32, m | 1.30, m |
| 17′ | 1.32, m | 1.27, m | 1.27, m | 1.28, m | 0.89, t (7.0) | 1.30, m |
| 18′ | 0.89, t (7.0) | 0.86, t (6.9) | 0.86, t (6.8) | 0.88, t (7.2) | - | 0.89, t (6.9) |
a CDCl3; b DMSO-d6.
The COSY spectrum of 2 revealed three spin systems. The first, from H-3 (δH 4.72, ddd, 11.5, 9.2, 5.6) to NH (δH 6.58, d, 5.6) and Ha-4 (δH 2.78, dd, 12.8, 9.2), and the second from H2-6 (δH 1.76, dq, 14.7, 7.5; 1.68, dq, 14.7, 7.5) to H3-7 (δH 1.0, t, 7.5), along with HMBC correlations from Ha-4 to C-2 and C-3, and from H3-8 to C-4, C-5 and C-6 confirmed the presence of a 3,5,5-trisubstituted γ-lactone ring. The third spin system extended from H2-2′ to H3-18′. Coupling constants of J4′–5′ 11.3 Hz and J6′–7′ 14.5 Hz confirmed the Z- and E-configurations for each double bond, respectively. Further HMBC correlations from H2-2′ to C-1′ and C-3′ located this side chain at the amide carbonyl C-1′ attached to the γ-lactone ring, reminiscent of korormicin A [5,12].
An epoxide was deduced at C-9′ and C-10′ based on the 13C chemical shifts (δC 55.3, CH; δC 56.6, CH), the COSY correlations from δH 3.01 (H-9′) to δH 2.97 (H-10′) and a C-O-C stretch at 1103 cm−1. As for korormicin A [5], the J9′–10′ vicinal coupling constant of 4.2 Hz confirmed the epoxide to be cis. The remaining hydroxy group was located at C-3′ (δC 64.7). Therefore, compound 2 (Figure 2) is the saturated γ-lactone analogue of korormicin A [5,12]. Yoshikawa et al. reported the isolation and structural elucidation of six korormicins (korormicin 1, 1a–c, 2 and 3) [5,11]. We propose that these korormicins be recognised for nomenclature purposes as korormicins A–F respectively, hence compound 2 is named korormicin G.
Figure 2.
Structures of korormicins G–K (2–6). a nomenclature used by Yoshikawa et al. [11].
The molecular formula of 3 was established as C25H40O5NCl based on 13C NMR (Table 2) and FTMS data, requiring six degrees of unsaturation. Analysis of the 1H and 13C NMR data (Table 2 and Table 3; Supplementary Figures S19–S23) of 3 indicated it was similar to that of 2, with two notable differences. Compound 3 contained a tri-substituted double bond (δC 125.0, C, C-3; 133.7, CH, C-4; δH 7.39, s, H-4) which, based on HMBC correlations from δH 7.39 to C-2 and C-5, confirmed it to have the same γ-lactone ring present in korormicin A [5]. In addition, the 13C chemical shifts of C-9′ (δC 67.0, CH) and C-10′ (δC 71.4, CH) along with the shift downfield of their respective protons (δH 3.95, br ddd, 8.3, 4.4, 3.1, H-9′; δH 3.56, m, H-10′) indicated the epoxide in 2 was no longer present in 3. Moreover, a COSY correlation from δH 4.95 (br d, 5.6) to H-10′ confirmed the presence of a hydroxy moiety at C-10′ and hence the location of the chlorine at C-9′. Comparison of the 1D NMR data of 3 with korormicin F [11] confirmed the planar structure of 3, korormicin H, is as shown in Figure 2.
As for 3, the molecular formula of 4 was determined to be C25H40O5NCl. The 13C NMR data (Table 2; Supplementary Figures S24–S32) of 4 closely resembled that of 3, except the 13C chemical shifts of C-9′ and C-10′ were reversed. COSY, HSQC and HMBC correlations, and comparison with literature values of korormicin F [11], confirmed the position of the chlorine at C-10′ and the planar structure of 4, korormicin I, to be as shown in Figure 2. The configuration at C-9′ and C-10′ remains unresolved for both 3 and 4.
Compound 5 was determined, by NMR and FTMS to have the molecular formula C24H39O4N. The NMR data (Table 2 and Table 3; Supplementary Figures S33–S35) of 5 showed strong similarity to 2, with signals diagnostic of the amide substituted γ-lactone ring (δC 50.0, CH; δH 4.70, ddd, 11.2, 9.2, 6.1; δC 40.0, CH2; δH 2.78, dd, 12.8, 9.2, and 1.90, dd, 12.8, 11.2; δH 6.59, NH, d, 6.1). Downfield resonances indicating the presence of an isolated double bond (δC 128.3, CH; δH 5.37, dd, 10.9, 7.5, 1.5; δC 129.2, CH δH 5.47, ddd, 10.9, 7.3, 1.5; cis geometry) were observed in place of those for the epoxide in 2. FTMS and 13C NMR data also confirmed that 5 had one less methlyene unit than 2, hence the planar structure of 5, korormicin J, is as shown (Figure 2). Several attempts to re-isolate 5 were futile; as a result the quaternary carbons remain unassigned (Table 2).
The final compound isolated in this study, compound 6, was established as having the molecular formula C25H41O4N by FTMS. The NMR data (Table 2 and Table 3; Supplementary Figures S36–S39) for 6 were similar to that of 5, the only difference being the addition of a methylene unit in the alkyl side chain. The configuration of the three double bonds in both compounds 5 and 6 was determined from their 1H NMR coupling constants, J4′,5′ (11.3 and 11.3), J6′,7′ (14.9 and 14.7), and J9′,10′ (10.9 and 10.4), to be Z, E and Z, respectively. The planar structure of 6, korormicin K, is as shown in Figure 2.
The biosynthetic pathway of the korormicins has not yet been elucidated, however, this compound class shares structural similarity with acyl-homoserine lactones (AHL), which are also produced by Pseudoalteromonas spp. and whose biosynthesis has been described [16]. Given this, it is plausible that there would be some homology between the two biosynthetic pathways giving rise to compound classes which share the same configuration, especially at C-3 in the γ-lactone ring. Based on this assumption it is likely that compound 2 shares the same configurations at positions C-5, C-3′, C-9′ and C-10′ (5S*, 3′R*, 9′S *, 10′R*) with korormicin A. Likewise, positions C-5 and C-3′ for compounds 5 and 6 were assumed to be 5S* and 3′R*. For compounds 2, 5 and 6 key nOe correlations from H-3 to H-6 and H-7 confirmed that in each case H-3 and the ethyl group were both on the same face of the molecule, hence 2, 5 and 6 have the relative configuration 3S*, 5S*. The configurations at C-9′ and C-10′ remain unassigned for compounds 3 and 4.
2.2. Antibacterial, Antifungal and Antiprotozoal Activities of Bacterial-Derived Metabolites
The activities of TBP, BAC-A and compounds 1–4, 6 against twelve bacterial strains and one fungal strain are presented in Table 4. Due to the low yield, compound 5 was not tested.
Table 4.
Antibacterial, antifungal and antiprotozoal activities of bromoalterochromide A (BAC-A), tetrabromopyrrole (TBP), 4′-((3,4,5-tribromo-1H-pyrrol-2-yl)methyl)phenol (1) and korormicins G–I and K (1–4, 6); bacterial and fungal inhibition was determined with 40 µg compound/diffusion disc. Growth media: LB10—LB10 agar, MA—Marine Agar, NSS—Nine Salts Solution.
| Strains (Growth media; temperature °C) | Bromopyrroles | Korormicins G–I and K | |||||
|---|---|---|---|---|---|---|---|
| BAC-A | TBP | 1 | 2 | 3 | 4 | 6 | |
| Pseudoalteromonas haloplanctis (MA; 28) | n | y | y | y | y | y | y |
| Pseudoalteromonas piscicida (MA; 28) | n | y | y | y | y | y | y |
| Pseudoalteromonas undina (MA; 28) | n | y | y | y | y | y | y |
| Pseudoalteromonas strain. J010 (MA; 28) | n | y | n | n | n | n | n |
| Pseudomonas aeruginosa (LB10; 37) | n | y | y | y | y | y | y |
| Vibrio campbellii (MA; 28) | n | y | y | y | y | y | y |
| Vibrio vulnificus (LB10; 37) | n | y | y | y | y | y | y |
| Vibrio coralliilyticus (MA; 28) | n | y | y | y | y | y | y |
| Vibrio harveyi (MA; 28) | n | y | y | y | y | y | y |
| Shewanella aquimarina (MA; 28) | n | y | y | y | y | y | y |
| Staphylococcus aureus (LB10; 37) | n | y | y | n | n | n | n |
| Candida albicans (MA; 28) | n | y | n | n | n | n | n |
| Tetrahymena pyriformis (NSS; ambient) | y | y | n | n | n | n | n |
TBP exhibited broad-spectrum activity against all bacterial strains tested and was also the only compound to display antifungal activity. Similarly, the bromopyrrole 1 showed broad-spectrum antibacterial activity; however, it did not inhibit fungal growth nor was it active against the source Pseudoalteromonas strain J010. BAC-A, on the other hand, did not show any antibacterial or antifungal activity. TBP and 1 were also the only compounds to exhibit activity against the gram-positive bacterium, Staphylococcus aureus. The antibacterial activity of the new korormicins in this study is in accordance with previous reports [5,11,17]. The korormicins 2–4 and 6 did not, however, inhibit the growth of either the gram-positive bacterium or the fungus, and similar to 1, did not inhibit the CCA-derived Pseudoalteromonas strain J010.
These in vitro results imply an allelopathic role for Pseudoalteromonas strain J010-derived metabolites particularly against the pathogenic Vibrios indicating a putative role(s) in protecting the algal host. However, it remains to be shown if these metabolites evoke similar effects in vivo.
The antiprotozoal activity of TBP and BAC-A was 2.6 µM and 59.2 µM, respectively. At these concentrations all flagellates formed cysts within 5 min of incubation, with complete cell lysis within 30 min. These results raise the possibility that TBP- and BAC-A-producing bacteria may consequently be defended against protozoan grazing. None of the new compounds 1–4 and 6 exhibited antiprotozoal activity.
3. Experimental Section
3.1. General Experimental Procedures
General experimental details and instrumentation have been previously reported [18]. Reversed-phase C18 material (Sepra C18-e, 50 µM) and HPLC columns were purchased from Phenomenex (Sydney, Australia). Solvents used were HPLC grade (Mallinckrodt, Sydney, Australia). All other chemicals were sourced from Sigma-Aldrich (Sydney, Australia). A Perkin Elmer Spectrum 100 FTIR spectrophotometer was used to record all IR spectra. 1H and 13C NMR spectra were recorded in either CDCl3 or DMSO-d6 (Cambridge Isotopes Laboratories Inc., Novachem, Melbourne, Australia) using 3 mm Bruker MATCH NMR tubes on a Bruker Avance 600 MHz NMR spectrometer with cryoprobe. Low-resolution MS data of the fractions were measured by direct infusion on a Bruker Daltonics Esquire 3000 ion-trap mass spectrometer (MS). Accurate mass for each compound was measured with a Bruker BioApex 47e FT-ICR mass spectrometer.
3.2. Isolation of Bacterial Metabolites
Petri dishes (n = 300, 2% marine agar) were inoculated with a stock culture of Pseudoalteromonas strain J010 and incubated for 48 h at 28 °C. Bacterial colonies were harvested with a sterile spatula, pooled and the bacterial biomass (25 g) extracted with ethanol (3 × 50 mL). The pooled ethanol extract was dried in vacuo, dissolved in 20 mL of methanol (MeOH), and purified on a pre-equilibrated C18 flash chromatography column (5 × 100 cm). The column was flushed with two bed volumes of MeOH and the eluant dried in vacuo then re-dissolved in 2 mL MeOH. The sample was separated on a Luna Phenyl-Hexyl column (250 × 21.2 mm, 5 µM) under gradient elution from 70:30 MeOH:H2O to 100% MeOH over 30 min, followed by 6 min with 100% MeOH with a flow rate of 8 mL/min. Fractions collected according to the peak profile at λ 220 nm. Low resolution MS data of the fractions was also measured by direct infusion. All individual compounds were further purified by C18 analytical HPLC (250 × 4.6 mm, 5µ Luna 2); 1 mL/min with gradient elution using H2O and acetonitrile. The chromatography yielded (in order of elution) BAC-A ([13]; rt 10.9 min, 5.2 mg, 0.02%); 4′-((3,4,5-tribromo-1H-pyrrol-2-yl)methyl)phenol (1; rt 19.25 min, 18.3 mg, 0.06%), korormicin B (previously 1a [11]; rt 20 min, 24.6 mg, 0.08%), TBP ([3,4]; rt 20.8 min, 26.2 mg, 0.10%), korormicin G (2; rt 21.3 min, 14.7 mg, 0.05%), korormicin E (previously 1c [11]; rt 21.6 min, 7.5 mg, 0.03%), korormicin H (3; rt 21.6 min, 7.9 mg, 0.03%), korormicin I (4; rt 21.7 min, 31.2 mg, 0.12%), korormicin A (previously 1 [5]; rt 24.6 min, 121.3 mg, 0.48%), korormicin C (previously 1b [11]; rt 23.8 min, 15.0 mg, 0.06%), korormicin J (5, rt 24.60 min, 1.0 mg, 0.004%), korormicin D (previously 3 [11]; rt 24.8 min, 26.4 mg, 0. 11%) and korormicin K (6; rt 25.38 min, 12.8 mg, 0.05%). The percentage yield of all compounds isolated was based on the weight of bacterial biomass. The known compounds had identical physical and spectroscopic properties to those previously published [3,4,5,11,13].
3.2.1. 4′-((3,4,5-Tribromo-1H-pyrrol-2-yl)methyl)phenol (1)
Colourless oil. UV (PDA) λmax nm: 229, 278; IR νmax cm−1: 3388 br, 3271, 2925, 1655, 1024, 992, 825; 1H (600 MHz, CDCl3 and DMSO-d6) and 13C (125 MHz, CDCl3 and DMSO-d6) NMR data Table 1 and Supplementary Figures S1–S13; (−)-ESI-FTMS m/z 405.8100 (calcd for C11H7ONBr3− monoisotopic 405.8083).
3.2.2. (3S*, 5S*, 3′R*, 4′Z, 6′E, 9′S*, 10′R*) Korormicin G (2)
Colourless oil. [α]21D not determined UV (PDA) λmax nm: 232; IR νmax cm−1: 3345, 2914, 1639 str, 1620, 1553, 1388, 1237, 1103, 1044; 1H (600 MHz, CDCl3) Table 3 and 13C (125 MHz, CDCl3) NMR data Table 2 and Supplementary Figures S14–S17; (+)-ESI-FTMS m/z 458.2863 (calcd for C25H41O5NNa+ 458.2877).
3.2.3. (3S*, 3′R*, 4′Z, 6′E) Korormicin H (3)
Colourless oil. [α]21D not determined UV (PDA) λmax nm: 230; 1H (600 MHz, DMSO-d6) Table 3 and 13C (125 MHz, DMSO-d6) NMR data Table 2 and Supplementary Figures S18–S22; (+)-ESI-FTMS m/z 492.2497 (calcd for C25H40O5NClNa+ 492.2487).
3.2.4. (3S*, 3′R*, 4′Z, 6′E) Korormicin I (4)
Colourless oil. [α]21D not determined UV (PDA) λmax nm: 232; 1H (600 MHz, DMSO-d6) Table 3 and 13C (125 MHz, DMSO-d6) NMR data Table 2 and Supplementary Figures S23–S31; (+)-ESI-FTMS m/z 492.2499 (calcd for C25H40O5NClNa+ 492.2487).
3.2.5. (3S*, 5S*, 3′R*, 4′Z, 6′E, 9′Z*) Korormicin J (5)
Colourless oil. [α]21D not determined UV (PDA) λmax nm: 233; 1H (600 MHz, CDCl3) Table 3 and 13C (125 MHz, CDCl3) NMR data Table 2 and Supplementary Figure S32–S34; (+)-ESI-FTMS m/z 428.2768 (calcd for C24H39O4NNa+ 428.2771).
3.2.6. (3S*, 5S*, 3′R*, 4′Z, 6′E, 9′Z*) Korormicin K (6)
Colourless oil. [α]21D 17° (CH3OH; c 0.6); UV (PDA) λmax nm: 232; 1H (600 MHz, CDCl3) Table 3 and 13C (125 MHz, CDCl3) NMR data Table 2 and Supplementary Figures S35–S38; (+)-ESI-FTMS m/z 442.2919 (calcd for C25H41O4NNa+ 442.2928).
3.3. Antibacterial, Antifungal and Antiprotozoal Bioassays
The disc diffusion assay was performed according to protocols described in Bauer et al. [19]. Briefly, fractions were applied without drying while pure compounds 1, BAC-A and TBP were dissolved in MeOH and 2–6 in CHCl3 at 200 μg/mL. Aliquots (20 μL) of fractions and compounds were evaporated on filter paper discs (d = 6 mm, Bio-Rad, Sydney, Australia) and placed on petri dishes containing Marine Agar or LB agar depending on the target strain with solvent as the negative control. Each petri dish was inoculated with one of the following bacterial strains: the Gram positive Staphylococcus aureus, or the Gram negative bacterial strains: Pseudoalteromonas strain J010, Pseudoalteromonas haloplanctis, Pseudoalteromonas piscicida, Pseudoalteromonas undina, Pseudomonas aeruginosa, Shewanella aquimarina, Vibrio campbellii, V. coralliilyticus, V. harveyi and V. vulnificus. Fractions and compounds were also tested against the fungus Candida albicans. Petri dishes were incubated for 24 h (at the temperature given in Table 4) after which clearance zones >10 mm in diameter (2 mm zone around the disc) were deemed as being inhibited by the target compound.
An axenic culture of the ciliate Tetrahymena pyriformis was used to assess antiprotozoal effects according to Matz et al. [20]. Briefly, samples were dried under vacuum, suspended in EtOH and 1 to 200 μg/mL transferred into 24-well plates. Once dry, Nine Salts Solution medium [21] with 103 flagellates per mL was added to each well (500 µL final volume per well) and total flagellate and active cell numbers monitored using an inverted light microscope.
4. Conclusions
Chemical screening of the epiphytic Pseudoalteromonas strain J010, isolated from the surface of the CCA N. fosliei, yielded six new compounds: the unprecedented bromopyrrole, 4′-((3,4,5-tribromo-1H-pyrrol-2-yl)methyl)phenol (1) and the five korormicins G–K (2–6), together with the seven known compounds: korormicins A–E [5,11], BAC-A [4] and TBP [3,4]. The metabolites identified in this study were shown to elicit antagonistic effects against bacteria, fungi, and protozoa, similar to previous reports [22,23,24,25]. Given the korormicins share a structural scaffold with the AHLs it is also possible that these compounds, like the long-chain (C13–16 and C18) AHLs, may play a role in quorum sensing [26]. Bromopyrroles have also previously been shown to have feeding deterrent [27,28] and antineoplastic properties [29].
Based on its metabolic capacity and antibiotic potential, it is tempting to speculate that the epiphytic bacterium Pseudoalteromonas strain J010 is highly defended against both eukaryotic and prokaryotic microbes in its ecological niche. This may have further implications for the macroscopic host. To test these hypotheses, both the bacterial abundance and the concentrations of bioactive metabolites need to be measured and shown to occur above active thresholds in vivo.
Acknowledgments
Funding: T. Harder was partially supported by a Research Fellowship awarded by the German Research Foundation (HA 3496/5-1). Funding was also provided by the Australian Institute of Marine Science’s Futures Project, Appropriation Fund 2233.
Supplementary Files
Supplementary Information (PDF, 3009 KB)
Author Contributions
T. Harder lead the research team and supervised J. Tebben (PhD). J. Tebben and T. Harder designed the experiment. J. Tebben and P. Thomas-Hall cultured the bacterium. J. Tebben, D. Tapiolas and P. Thomas-Hall isolated the metabolites. D. Tapiolas, C. Motti and J. Tebben conducted chemical analyses. J. Tebben, C. Motti and D. Tapiolas analyzed the data. C. Motti and D. Tapiolas determined the structures. J. Tebben evaluated the bioactivity. C. Motti, J. Tebben and T. Harder wrote the manuscript. C. Motti prepared tables, figures and Supplementary Information.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Hay M.E. Marine chemical ecology: Chemical signals and cues structure marine populations, communities, and ecosystems. Annu. Rev. Mar. Sci. 2009;1:193–212. doi: 10.1146/annurev.marine.010908.163708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hadfield M.G. Biofilms and marine invertebrate larvae: What bacteria produce that larvae use to choose settlement sites. Annu. Rev. Mar. Sci. 2011;3:453–470. doi: 10.1146/annurev-marine-120709-142753. [DOI] [PubMed] [Google Scholar]
- 3.Tebben J., Tapiolas D.M., Motti C.A., Abrego D., Negri A.P., Blackall L.L., Steinberg P.D., Harder T. Induction of larval metamorphosis of the coral Acropora millepora by tetrabromopyrrole isolated from a Pseudoalteromonas bacterium. PLoS One. 2011;6:e19082. doi: 10.1371/journal.pone.0019082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Andersen R.J., Wolfe M.S., Faulkner D.J. Autotoxic antibiotic production by a marine Chromobacterium. Mar. Biol. 1974;27:281–285. doi: 10.1007/BF00394363. [DOI] [Google Scholar]
- 5.Yoshikawa K., Takadera T., Adachi K., Nishijima M., Sano H. Korormicin, a novel antibiotic specifically active against marine Gram-negative bacteria, produced by a marine bacterium. J. Antibiot. 1997;50:949–953. doi: 10.7164/antibiotics.50.949. [DOI] [PubMed] [Google Scholar]
- 6.Fehér D., Barlow R., Mcatee J., Hemscheidt T.K. Highly brominated antimicrobial metabolites from a marine Pseudoalteromonas sp. J. Nat. Prod. 2010;73:1963–1966. doi: 10.1021/np100506z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bowman J.P. Bioactive compound synthetic capacity and ecological significance of marine bacterial genus Pseudoalteromonas. Mar. Drugs. 2007;5:220–241. doi: 10.3390/md504220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Egan S., Harder T., Burke C., Steinberg P., Kjelleberg S., Thomas T. The seaweed holobiont: Understanding bacteria-seaweed interactions. FEMS Microbiol. Rev. 2013;37:462–476. doi: 10.1111/1574-6976.12011. [DOI] [PubMed] [Google Scholar]
- 9.Egan S., Thomas T., Kjelleberg S. Unlocking the diversity and biotechnological potential of marine surface associated microbial communities. Curr. Opin. Microbiol. 2008;11:219–225. doi: 10.1016/j.mib.2008.04.001. [DOI] [PubMed] [Google Scholar]
- 10.Engel S., Jensen P.R., Fenical W. Chemical ecology of marine microbial defense. J. Chem. Ecol. 2002;28:1971–1985. doi: 10.1023/A:1020793726898. [DOI] [PubMed] [Google Scholar]
- 11.Yoshikawa K., Adachi K., Nishida F., Mochida K. Planar structure and antibacterial activity of korormicin derivatives isolated from Pseudoalteromonas sp. F-420. J. Antibiot. 2003;56:866–870. doi: 10.7164/antibiotics.56.866. [DOI] [PubMed] [Google Scholar]
- 12.Kobayashi Y., Yoshida S., Nakayama Y. Total synthesis of korormicin. Eur. J. Org. Chem. 2001;2001:1873–1881. doi: 10.1002/1099-0690(200105)2001:10<1873::AID-EJOC1873>3.0.CO;2-O. [DOI] [Google Scholar]
- 13.Speitling M., Smetanina O.F., Kuznetsova T.A., Laatsch H. Bromoalterochromides A and A′, unprecedented chromopeptides from a marine Pseudoalteromonas maricaloris strain KMM 636T. J. Antibiot. 2007;60:36–42. doi: 10.1038/ja.2007.5. [DOI] [PubMed] [Google Scholar]
- 14.John E.A., Pollet P., Gelbaum L., Kubanek J. Regioselective syntheses of 2,3,4-tribromopyrrole and 2,3,5-tribromopyrrole. J. Nat. Prod. 2004;67:1929–1931. doi: 10.1021/np0498399. [DOI] [PubMed] [Google Scholar]
- 15.Schwalm C.S., Castro I.B.D.D., Ferrari J., Oliveira F.L.D., Aparicio R., Correia C.R.D. Synthesis of pentabromopseudilin and other arylpyrrole derivatives via Heck arylations. Tetrahedron Lett. 2012;53:1660–1663. doi: 10.1016/j.tetlet.2012.01.086. [DOI] [Google Scholar]
- 16.Watson W.T., Minogue T.D., von Bodman S.B., Val D.L., Churchill M.E.A. Structural basis and specificity of acyl-homoserine lactone signal production in bacterial quorum sensing. Mol. Cell. 2004;9:685–694. doi: 10.1016/s1097-2765(02)00480-x. [DOI] [PubMed] [Google Scholar]
- 17.Hayashi M., Shibata N., Nakayama Y., Yoshikawa K., Unemotoa T. Korormicin insensitivity in Vibrio alginolyticus is correlated with a single point mutation of Gly-140 in the NqrB subunit of the Na+-translocating NADH-quinone reductase. Arch. Biochem. Biophys. 2002;401:173–177. doi: 10.1016/S0003-9861(02)00007-3. [DOI] [PubMed] [Google Scholar]
- 18.Tapiolas D.M., Bowden B.F., Abou-Mansour E., Willis R.H., Doyle J.R., Muirhead A.N., Liptrot C., Llewellyn L.E., Wolff C.W.W., Wright A.D., et al. Eusynstyelamides A, B, and C, nNOS inhibitors, from the ascidian Eusynstyela latericius. J. Nat. Prod. 2009;72:1115–1120. doi: 10.1021/np900099j. [DOI] [PubMed] [Google Scholar]
- 19.Bauer A.W., Kirby W.M.M., Sherris J.C., Turck M. Antibiotic sensitivity testing by a standardized single disk method. Am. J. Clin. Pathol. 1966;45:493–496. [PubMed] [Google Scholar]
- 20.Matz C., Webb J.S., Schupp P.J., Phang S.Y., Penesyan A., Egan S., Steinberg P., Kjelleberg S. Marine biofilm bacteria evade eukaryotic predation by targeted chemical defense. PLoS One. 2008;3:e2744. doi: 10.1371/journal.pone.0002744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vaatanen P. The Walter and Andree De Nottbeck Foundation Scientific Report. Volume 1. University of Helsinki; Hanko, Finland: 1976. Microbiological studies in coastal waters of the Northern Baltic sea, I. Distribution and abundance of bacteria and yeasts in the Tvärminne area; pp. 1–58. Preliminary report for 1973-74, ISBN 951-95341-0-5. [Google Scholar]
- 22.Penesyan A., Marshall-Jones Z., Holmström C., Kjelleberg S., Egan S. Antimicrobial activity observed among cultured marine epiphytic bacteria reflects their potential as a source of new drugs. FEMS Microbiol. Ecol. 2009;69:113–124. doi: 10.1111/j.1574-6941.2009.00688.x. [DOI] [PubMed] [Google Scholar]
- 23.Dobretsov S., Dahms H.-U., Qian P.-Y. Inhibition of biofouling by marine microorganisms and their metabolites. Biofouling. 2006;22:43–54. doi: 10.1080/08927010500504784. [DOI] [PubMed] [Google Scholar]
- 24.Wiese J., Thiel V., Nagel K., Staufenberger T., Imhoff J. Diversity of antibiotic-active bacteria associated with the brown alga Laminaria saccharina from the Baltic Sea. Mar. Biotechnol. 2009;11:287–300. doi: 10.1007/s10126-008-9143-4. [DOI] [PubMed] [Google Scholar]
- 25.Egan S., Thomas T., Holmstrom C., Kjelleberg S. Phylogenetic relationship and antifouling activity of bacterial epiphytes from the marine alga Ulva lactuca. Environ. Microbiol. 2000;2:343–347. doi: 10.1046/j.1462-2920.2000.00107.x. [DOI] [PubMed] [Google Scholar]
- 26.Wagner-Döbler I., Thiel V., Eberl L., Allgaier M., Bodor A., Meyer S., Ebner S., Hennig A., Pukall R., Schulz S. Discovery of complex mixtures of novel long-chain quorum sensing signals in free-living and host-associated marine Alphaproteobacteria. ChemBioChem. 2005;6:2195–2206. doi: 10.1002/cbic.200500189. [DOI] [PubMed] [Google Scholar]
- 27.Haber M., Carbone M., Mollo E., Gavagnin M., Ilan M. Chemical defense against predators and bacterial fouling in the Mediterranean sponges Axinella polypoides and A. verrucosa. Mar. Ecol. Prog. Ser. 2011;422:113–122. doi: 10.3354/meps08921. [DOI] [Google Scholar]
- 28.Scala F., Fattorusso E., Menna M., Taglialatela-Scafati O., Tierney M., Kaiser M., Tasdemir D. Bromopyrrole alkaloids as lead compounds against protozoan parasites. Mar. Drugs. 2010;8:2162–2174. doi: 10.3390/md8072162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xiong S., Pang H.-D. In vitro and in vivo antineoplastic activity of a novel bromopyrrole and its potential mechanism of action. Br. J. Pharmacol. 2010;159:909–918. doi: 10.1111/j.1476-5381.2009.00573.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information (PDF, 3009 KB)