Abstract
When engineering lipid membranes for applications, it is essential to characterize them to avoid artifacts introduced by manipulation and the experimental environment. Wide-angle X-ray scattering is a powerful structural characterization tool for well-ordered lipid systems. It reveals remarkable differences in rotational order parameters for samples prepared in different ways. New data and perspectives are presented here for multilamellar systems that support and extend the characterization work on unilamellar systems that is reported by Watkins et al. in this issue of ACS Nano.
Biomembranes are one of biology’s key nanomaterials (∼5 nm thick), and they are essential to all cellular life. Manipulation of membranes and of their constitutive core, lipid bilayers, is necessary to engineer systems toward applications.1−4 It is important to characterize how such manipulations and the resulting environment perturb the structure and to build systems that minimize unnatural perturbations.5−8 The study by Watkins et al.(9) described in this issue of ACS Nano has focused on a detailed characterization of single lipid bilayers supported on a solid substrate. Remarkable differences were found compared to lipid monolayers, for which the supported bilayers were constructed by the Langmuir–Blodgett/Langmuir–Schaefer (LB/LS) technique. Some data in the literature suggest that differences also occur for traditional systems that consist of multilamellar arrays of lipid bilayers,10−13 but there is some concern that those results were not well-documented.9
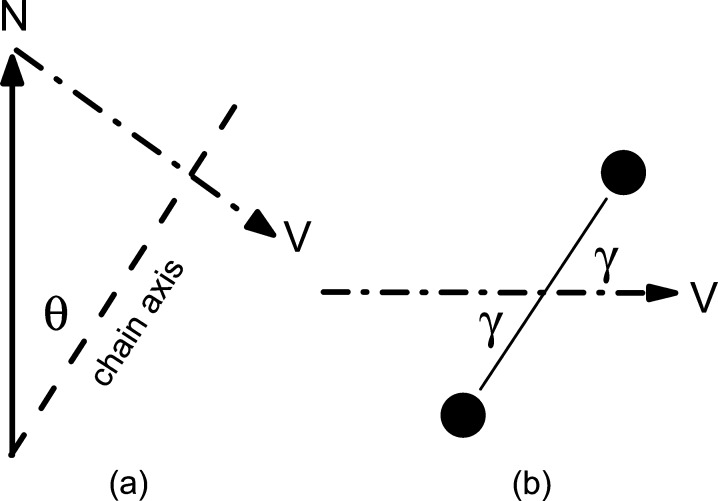
The primary experimental quantity exhibiting the differences found by Watkins et al.(9) is the wide-angle X-ray scattering (WAXS) chain packing reflections in the gel phase of dipalmitoylphosphatidylcholine (DPPC) lipid bilayers.10−13 Each lipid has a pair of hydrophobic saturated hydrocarbon chains 2 nm long capped on one end by a hydrophilic headgroup in contact with water, thereby forming a bilayer often thought of as two back-to-back monolayers. The chains tilt with an angle θ relative to the bilayer normal because the interfacial area occupied by the headgroup is greater than the area of the pair of chains.14 Remarkably, the chains in each monolayer tilt in the same azimuthal (ϕ) direction even though there are no covalent bonds between the monolayers.10,11,15 As the number of X-ray reflections is far smaller than occurs in polyethylene crystals or in the subgel phase of lipid bilayers,16 the classical interpretation of the gel phase WAXS data is that the chains are disordered with respect to rotations about their long axis, suggesting that they can be modeled as tilted cylinders packed together. However, this classical interpretation was refined to accommodate an apparent discrepancy that occurred in multilamellar arrays between the value of the tilt angle θ inferred from infrared (IR) spectroscopy and that inferred just from the positions of the WAXS peaks.17 Infrared is sensitive not only to the tilt angle but also to the rotational angle γ of the all-trans C–C···C–C plane about the chain long axis relative to the plane defined by the tilt direction and the bilayer normal (see Figure 1). A two-dimensional rotational order parameter, g = ⟨2 cos2 γ – 1⟩, is the appropriate statistically averaged quantity to describe this rotational disorder. Complete disorder has all γ equally probable and g = 0, whereas complete rotational order corresponds to |g| = 1 (g = −1 has the zigzag chain plane perpendicular to the tilt plane). A value g = −0.3 resolved the IR/WAXS difference for multilamellar arrays.17
Figure 1.
Geometry of the hydrocarbon chains. (a) Normal N is perpendicular to the plane of the bilayer, and the long axis along the all-trans chain is tilted by θ. (b) Looking down the chain axis, the carbons in the zigzag chain are shown by dark circles that define a plane perpendicular to the paper that is rotated by γ relative to vector V that is in the chain tilt plane shown in (a).
Wide-angle X-ray scattering data contain information in addition to the peak positions in reciprocal space that suffice to obtain the chain tilt θ. The additional information is the relative intensity R of the WAXS in-plane reflections, usually called the (20) peak in the literature, and the out-of plane (11) peak. The classical model of tilted cyclinders gives R = 1. Even before the current paper of Watkins et al.,9 it has been known that R depends on the rotational order parameter g, but a simple model for the electron density of the chains suggested that R would only be reduced by 40% for complete rotational order compared to the value R = 1, so it was previously ignored when interpreting WAXS.15 In contrast, the new model of Watkins et al. suggests that R is more sensitive to g,9 so careful measurement of R opens a new window on the structure of lipid bilayer systems. For supported lipid bilayers, the Watkins paper reports quite small experimental values for R ranging downward from less than 0.4, giving values of g varying from −0.5 to −1.0 as the monolayer pressure at deposition was increased. In striking contrast, g was reported to be essentially zero for monolayers, corresponding to R = 1 and the classical cylindrical model. In view of these considerable differences between monolayers and supported bilayers, it is of interest to compare R and g to the more traditional multilamellar lipid bilayer systems. While the literature has some suggestive figures,10−13 values of R were not given quantitatively, so it is timely to view higher quality data that confirm R ≈ 1.
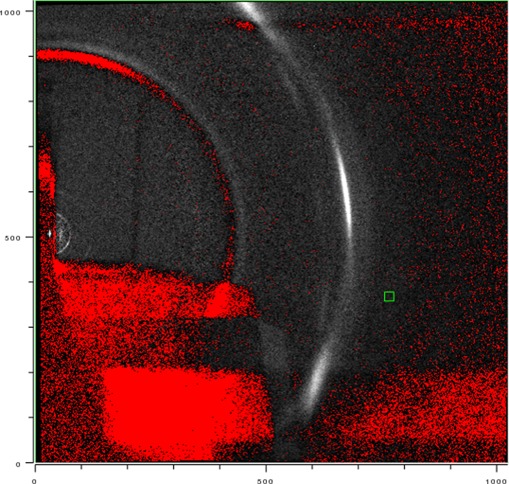
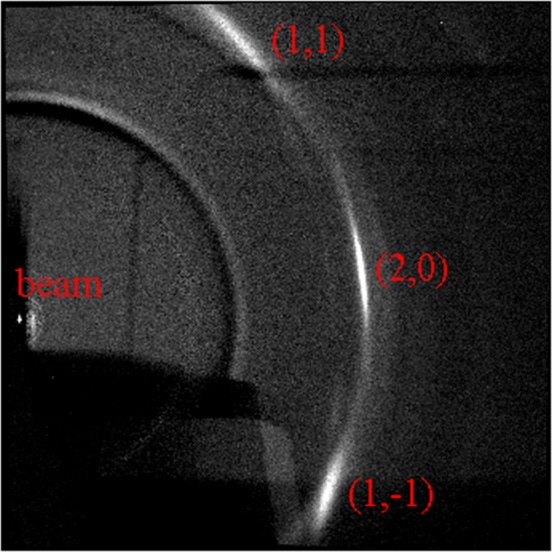
Figure 2 shows grazing incidence (GIXD) WAXS scattering from a multilamellar stack of ∼2000 DMPC lipid bilayers supported on a Si substrate. The lamellar repeat spacing D = 5.8 nm obtained from the lamellar orders on the left indicated that the stack was nearly fully hydrated from water vapor with an estimated RH greater than 98%. The intense peak in the upper right center is the out-of-plane (11) reflection. Directly below that is the first satellite on the (11) Bragg rod10 that is also seen in unoriented multilamellar vesicles.15 The (1,–1) reflection, though strongly attenuated by having to go through the thin (35 μm) substrate, can be seen in the lower right center. Only the tail of the (20) reflection is visible. The central part of the (20) reflection cannot be seen because, for a positive incident angle ω, those scattered X-rays must travel long distances through the substrate as well as the sample itself. When ω is decreased, more of the (20) reflection can be seen,13,18 but its intensity still appears somewhat weaker than the (11) reflection. Note that, historically, the (20) peak was often not observed and there was even concern that it did not exist.
Figure 2.
Grazing incidence wide-angle X-ray scattering from an oriented stack of 1,2-dimyristoyl-sn-glycero-3-phosphocholine (DMPC) bilayers; white is most intense. The beam is located at the x near the center left, and lamellar repeat orders h = 2–7 and h = 9 are on the meridian above the beam. The bright (11) peak is located at (qr,qz) = (13.0 nm–1, 8.2 nm–1). The (1,–1) peak is strongly attenuated by the substrate. Most of the (20) peak, subsequently shown to be centered at (14.8 nm–1, 0), is even more strongly attenuated by the substrate and the sample itself. A satellite on the (1,1) Bragg rod occurs below the (1,1) peak. Temperature was 10 °C.
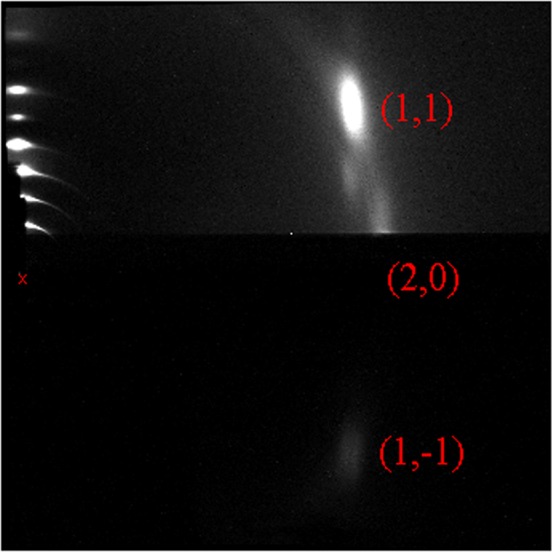
While the grazing incidence experimental geometry in Figure 2 might be taken to indicate R < 1, that is wrong. Figure 3 shows scattering from the same sample but tilted with respect to the beam by ω = 45°. As was described previously,11 this moves the reflections around on the area detector, but most importantly, the (20), (1,1), and (1,–1) reflections traverse nearly equal thickness of the thin substrate, so the differential attenuation is negligible.
Figure 3.
Transmission wide-angle X-ray scattering from the sample in Figure 1 with the beam incident at ω = 45°, so the three main labeled reflections are rotated in the laboratory frame according to formula11 that also gives the q values in the caption to Figure 2. The first satellites of the (1,1) and (1,–1) are visible, as are the overlapping second satellites15 centered on the equator (qr = 0), which follows a trajectory from the beam toward the (20) reflection. The weak ring closer to the beam than the WAXS reflections is from a Mylar window on the sample chamber and occurs because of imperfect subtraction of the background obtained from a substrate with no lipid. The intensity of the beam on the charge-coupled device was highly attenuated by an absorber downstream from the sample, and there are no lamellar reflections for large incidence angles ω.
Intensity profiles through the orders in Figure 3 are shown in Figure 4. The in-plane (20) peak is narrow with greater maximum intensity than the out-of-plane (11) and (1,–1) peaks, so the integrated intensity is about the same for all peaks, consistent with the conventional view that R = 1 for multilamellar systems. (The (11) peak width is instrumentally resolved here. The (20) width was even narrower when the instrumental resolution was greater,15 but instrumental resolution does not affect R.) However, for a different exposure of the same sample, R was about 0.8, more consistent with the g = −0.3 obtained from using IR and WAXS.17 Nevertheless, as Watkins et al. suppose,9 there is a considerable difference compared to the much smaller value of R they obtain for supported single bilayers.
Figure 4.
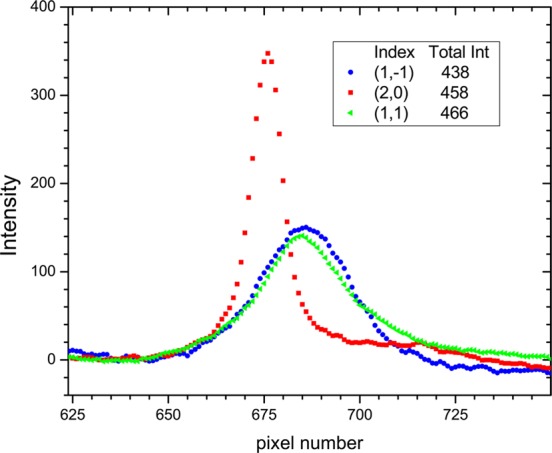
Integrated intensity versus pixel number in the direction perpendicular to the elongated reflections on the charge-coupled device in Figure 3.
As supported single lipid bilayers are used for many purposes, it is important that the method of forming them and subsequent distortions be documented. It could be argued that the most relevant lipid bilayer system to be studied is the fluid phase that does not have a rotational order parameter. However, obtaining fluid phase structure is relatively difficult and differences even more so. Another advantage of the gel phase for this kind of study is that it is relatively rigid compared to fluid phases and would be expected to be more robust under the stresses of deposition and the effect of a substrate. Detection of even subtle changes in the gel phase, like the rotational order parameter, would indicate stresses that would then likely deform a fluid phase even more, albeit undetectably so, but not necessarily without consequences for studies employing supported bilayers. Concerns about the substrate have, indeed, led to devising many ways to support single bilayers.1−8,12
The result for supported single bilayers that R ∼ 0.3 from the study of Watkins et al.(9) together with the result that R = 1 for multilamellar stacks makes it clear that there is a substantial difference. As the multilamellar stacks have an ample 2 nm water cushion between adjacent bilayers, it is unlikely that stacking affects the properties of the individual bilayers; this is supported by the absence of three-dimensional WAXS reflections, indicating no correlation of the in-plane structure between adjacent bilayers.10 Differences with the results from supported bilayers would then be attributed either to interactions with the substrate or to the deposition method. Because Watkins et al. obtain the same small R using a different deposition method, the focus shifts to the substrate. While multilamellar stacks are also deposited on substrates and the closest bilayer would likely have similar properties to the single supported bilayer, WAXS overwhelmingly comes from the other ∼2000 bilayers that do not directly interact with the substrate.
A concern for the results of Watkins et al.(9) might have been that they did not perform transmission WAXS, so their R value from GIXD might have been small for the same reason that it has been artifactually small for multilamellar samples. One difference is that they use higher energy X-rays to make water more transparent, but that is similar to having air above multilayer stacks. Their substrate was quartz, which is 20 times as absorptive as water at their X-ray energy, so that is similar to previous studies where the substrate is also much more absorptive than air. Crucially, they performed an experiment that allays this concern; namely, they found that monolayers on water and on quartz both have R ∼ 1. A likely explanation for R smaller than 1 for GIXD of oriented stacks is that the (20) scattering is attenuated from those bilayers deep within the 10 μm multilamellar stack because they have to go farther through the upper bilayers than the (11) scattering. This explanation could be tested by examining multilayer stacks with higher energy X-rays, which have much longer attenuation lengths in lipids.
While it would be attractive to suppose that the substrate alone causes the difference in R, this is refuted by the finding of Watkins et al. that monolayers also have R = 1 when deposited on a quartz substrate.9 A difference between monolayers and bilayers, both on the substrate, is the interactions between the monolayers in a bilayer that manifests itself in parallel alignment of the chains in both monolayers.9−11Watkins et al. therefore suggest that obtaining a small R and rotational ordering requires both the substrate interaction and the interactions between the two monolayers in a bilayer. They also carefully document the interesting and important variation of structural parameters in monolayers and supported lipid bilayers as the surface pressure in the LB/LS deposition method is varied. Their Figure 6 shows that the chain tilt θ of the supported bilayer is larger than the monolayer from which it was formed. That would be expected to make the area per molecule larger for the bilayer, but the opposite occurs because the original packing density reflected in the inverse chain area Achain is considerably smaller for the monolayer. This is additional evidence supporting the perspective that bilayers are not simply two back-to-back monolayers and that, instead of using one system to elucidate the other, efforts should focus on the differences between them.19 These new data nicely do so.9
Methods for the Data in the Included Figures
Samples were prepared on thin (35 μm) Si wafers as previously described.13 Experiments were performed at the G1 station at CHESS. The X-ray energy was 10.5 keV at which the absorption length of lipid is 2.6 mm. The W/B4C multilayer monochromator had 1.2% energy dispersion. The beam size was 0.2 mm × 0.2 mm, and measured divergences were less than 0.0001 radians. Reflections recorded on the 1024 × 1024 CCD detector (Finger Lakes Instrumentation, Lima, NY) were broadened by the X-ray energy dispersion and divergence; additionally, they were geometrically broadened in Figure 2 by the beam width and the 25 mm length of the sample along the beam and in Figure 3 by the beam width and height. The pixel size of the detector was 71.13 μm, and the distance from sample to detector was 175 mm. At these distances, there were scattering rings from the Mylar windows of the hydration chamber.13 Highly accurate background for Figure 2 was obtained by rotating the sample to negative angle to block scattering from the sample.13 For the background for Figure 3, the sample had to be replaced by a different bare thin Si substrate, resulting in incomplete subtraction of the Mylar ring.
Acknowledgments
The new data were taken for a project supported by NIH Grant R01GM44976 at the Cornell High Energy Synchrotron Source (CHESS) supported by the National Science Foundation (NSF) and the NIH/NIGMS under NSF Award DMR-0936384.
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
References
- Branton D.; Deamer D. W.; Marziali A.; Bayley H.; Benner S. A.; Butler T.; Di Ventra M.; Garaj S.; Hibbs A.; Huang X. H.; et al. The Potential and Challenges of Nanopore Sequencing. Nat. Biotechnol. 2008, 26, 1146–1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta S.; Dura J. A.; Freites J. A.; Tobias D. J.; Blasie J. K. Structural Characterization of the Voltage-Sensor Domain and Voltage-Gated K+-Channel Proteins Vectorially Oriented within a Single Bilayer Membrane at the Solid/Vapor and Solid/Liquid Interfaces via Neutron Interferometry. Langmuir 2012, 28, 10504–10520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh W. T.; Hsu C. J.; Capraro B. R.; Wu T. T.; Chen C. M.; Yang S.; Baumgart T. Curvature Sorting of Peripheral Proteins on Solid-Supported Wavy Membranes. Langmuir 2012, 28, 12838–12843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung M.; Koo B. J.; Boxer S. G. Formation and Analysis of Topographical Domains between Lipid Membranes Tethered by DNA Hybrids of Different Lengths. Faraday Discuss. 2013, 161, 333–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes L. D.; Boxer S. G. DNA-Based Patterning of Tethered Membrane Patches. Langmuir 2013, 29, 12220–12227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jablin M. S.; Dubey M.; Zhernenkov M.; Toomey R.; Majewski J. Influence of Lipid Membrane Rigidity on Properties of Supporting Polymer. Biophys. J. 2011, 101, 128–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budvytyte R.; Valincius G.; Niaura G.; Voiciuk V.; Mickevicius M.; Chapman H.; Goh H. Z.; Shekhar P.; Heinrich F.; Shenoy S.; et al. Structure and Properties of Tethered Bilayer Lipid Membranes with Unsaturated Anchor Molecules. Langmuir 2013, 29, 8645–8656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fragneto G.; Charitat T.; Daillant J. Floating Lipid Bilayers: Models for Physics and Biology. Eur. Biophys. J. 2012, 41, 863–874. [DOI] [PubMed] [Google Scholar]
- Watkins E. B.; Miller C. E.; Liao W.-P.; Kuhl T. L.. Equilibrium or Quenched: Fundamental Differences between Lipid Monolayers, Supported Bilayers, and Membranes. ACS Nano 2014, DOI: 10.1021/nn4052953. [DOI] [PubMed] [Google Scholar]
- Smith G. S.; Sirota E. B.; Safinya C. R.; Plano R. J.; Clark N. A. X-ray Structural Studies of Freely Suspended Ordered Hydrated DMPC Multimembrane Films. J. Chem. Phys. 1990, 92, 4519–4529. [Google Scholar]
- Tristram-Nagle S.; Zhang R.; Suter R. M.; Worthington C. R.; Sun W. J.; Nagle J. F. Measurement of Chain Tilt Angle in Fully Hydrated Bilayers of Gel Phase Lecithins. Biophys. J. 1993, 64, 1097–1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziblat R.; Leiserowitz L.; Addadi L. Crystalline Lipid Domains: Characterization by X-ray Diffraction and Their Relation to Biology. Angew. Chem., Int. Ed. 2011, 50, 3620–3629. [DOI] [PubMed] [Google Scholar]
- Mills T. T.; Toombes G. E. S.; Tristram-Nagle S.; Smilgies D. M.; Feigenson G. W.; Nagle J. F. Order Parameters and Areas in Fluid-Phase Oriented Lipid Membranes Using Wide Angle X-ray Scattering. Biophys. J. 2008, 95, 669–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagle J. F. Theory of the Main Lipid Bilayer Phase-Transition. Annu. Rev. Phys. Chem. 1980, 31, 157–195. [Google Scholar]
- Sun W. J.; Suter R. M.; Knewtson M. A.; Worthington C. R.; Tristram-Nagle S.; Zhang R.; Nagle J. F. Order and Disorder in Fully Hydrated Unoriented Bilayers of Gel Phase Dipalmitoylphosphatidylcholine. Phys. Rev. E 1994, 49, 4665–4676. [DOI] [PubMed] [Google Scholar]
- Blaurock A. E.; Mcintosh T. J. Structure of the Crystalline Bilayer in the Subgel Phase of Dipalmitoylphosphatidylglycerol. Biochemistry 1986, 25, 299–305. [DOI] [PubMed] [Google Scholar]
- Nagle J. F. Evidence for Partial Rotational Order in Gel Phase DPPC. Biophys. J. 1993, 64, 1110–1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tristram-Nagle S.; Liu Y. F.; Legleiter J.; Nagle J. F. Structure of Gel Phase DMPC Determined by X-ray Diffraction. Biophys. J. 2002, 83, 3324–3335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagle J. F.; Tristram-Nagle S. Structure of Lipid Bilayers. Biochim. Biophys. Acta, Rev. Biomembr. 2000, 1469, 159–195. [DOI] [PMC free article] [PubMed] [Google Scholar]